Abstract
Mycobacterium triplex, a recently described slowly growing nontuberculous mycobacterium, was isolated from a Finnish patient with pulmonary mycobacteriosis. The disease was successfully treated with antimycobacterial drugs. The strain isolated, which was similar to the type strain but differed slightly from the species description, was regarded as a variant of M. triplex sensu stricto. According to present knowledge this variant of the species has never been isolated before.
Recent advances in diagnostic methodology for mycobacteria other than Mycobacterium tuberculosis (MOTT), and new possibilities introduced for drug therapy of the diseases they induce, have renewed interest in mycobacterial research. The recognition that most patients with AIDS are at high risk of complications and/or terminal illness caused by MOTT has further stimulated research efforts. The principles of diagnosis and therapy of diseases caused by MOTT have also recently been updated by the American Thoracic Society (1). These guidelines include recommendations for specific drug regimens which recognize the major impact of the newer chemotherapeutic agents.
Improved diagnostic means allow enhanced detection and more-accurate species identification of mycobacteria isolated from clinical specimens, including the classification of species not recognized earlier. In 1996, Floyd et al. (5) characterized a new species of slow-growing MOTT, which they designated Mycobacterium triplex. The primary characterization of this novel species was based on conventional testing, but the conclusive genetic evidence relied on 16S rRNA analyses. Although the species is uncommon, it was found to be geographically widespread in the United States (5). There is one very recent report of M. triplex in Europe (4).
We isolated M. triplex from a Finnish patient suffering from pulmonary mycobacteriosis. M. triplex was the only mycobacterium isolated from the specimens. As illustrated in this paper, the strain found in this patient differed slightly from the earlier description of the species. To our knowledge, this is the first report of a pulmonary disease associated with M. triplex.
Case report.
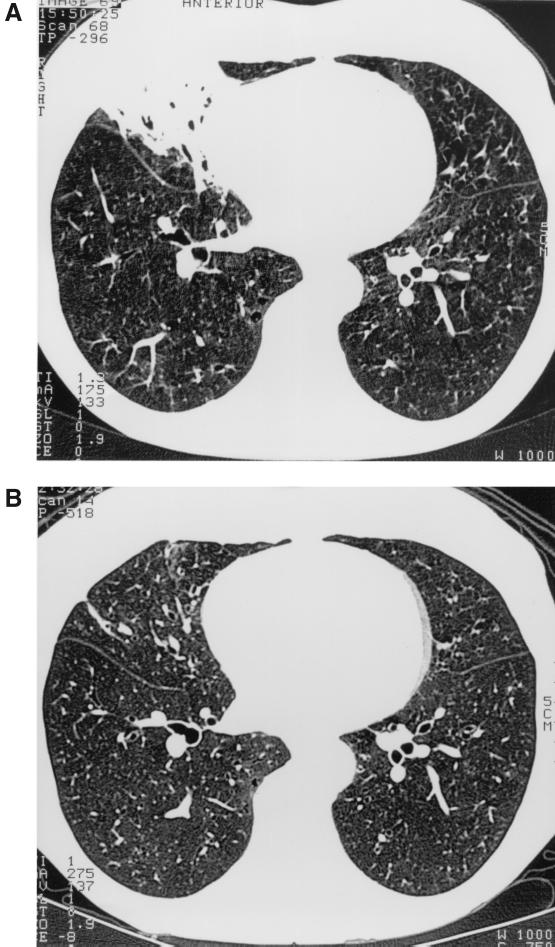
A 67-year-old, human immunodeficiency virus-negative dentist presented in hospital with hemoptysis in 1995. On chest radiographs, thin fibrotic changes were detected in both lower fields, but no cavitary changes were detected. High-resolution computed tomography (HRCT), performed in December 1995, revealed multifocal bronchiectases in both lungs and multiple small nodules in the middle and lower lobes of the right lung and the lower lobe of the left lung, indicating mycobacteriosis (1). Bronchoscopy showed nonspecific inflammatory mucosal changes on the right side. Smears of sputum and bronchoalveolar lavage specimens were negative for acid-fast bacilli, but cultures of sputum samples taken on three successive days grew a slow-growing mycobacterium. It was identified using conventional tests, gas-liquid chromatographic (GLC) analysis of cellular fatty acid and alcohol composition, and 16S rRNA gene sequencing as described earlier in detail (6, 7, 11). The isolates met the identification criteria for M. triplex (5). Since this was the only organism growing in the three cultures, we believe that the isolation of M. triplex represents infection rather than colonization. Moreover, the patient has bronchiectases throughout both lungs, which would predispose her to this kind of lung infection. Treatment was initiated using a standard antituberculous regimen for 2 months, to which the lesions responded only partially. Treatment was continued with rifampin, ciprofloxacin, ethambutol, and clarithromycin for a total of 10 months, with the exception of a 3-week period in March and April 1996 due to a temporary rise in liver enzymes. This regimen proved to be well tolerated. The bacteriological and clinical responses were good. An HRCT scan in October 1996, after 10 months of treatment, showed a good radiological response (Fig. 1). The patient was regarded as cured at follow-up, after taking antituberculosis medication for 18 months.
FIG. 1.
HRCT scans before (A) and after (B) treatment.
The growth and biochemical characteristics of the patient's isolates were most closely compatible with M. triplex (Table 1) (5). By in vitro testing, performed using the proportion method in Löwenstein-Jensen medium, the strain was susceptible to rifampin, streptomycin, and clarithromycin and moderately sensitive to ciprofloxacin, but it had decreased susceptibility to ethambutol, isoniazid, and pyrazinamide. The isolates failed to hybridize with the commercially available genetic probe for Mycobacterium avium complex (Accuprobe; Gen-Probe Inc., San Diego, Calif.). Two of the patient's isolates were analyzed by GLC for cellular fatty acid methyl esters and alcohols, prepared by acid methanolysis, and analyzed and identified as described earlier in detail (10). These two isolates had an identical GLC profile, which closely resembled that of the M. triplex type strain (ATCC 70071). However, repeated analyses of the fatty acid composition verified minor but constantly detectable differences from the M. triplex type strain, including a higher relative amount of tetracosanoate (24:0) and a low relative amount of tuberculostearic acid (10-Me-18:0) (Table 1). The fatty acid profile of the M. triplex type strain was found to be indistinguishable from that of M. simiae (3), a slow-growing photochromogenic species. Pigment production easily separates these two species. They both have GLC profile is grossly similar to that of M. tuberculosis. As shown in Table 1, the high relative content of tetradecanoate (14:0) in both the type strain of M. triplex and the patient isolates (7%) differentiated them from M. tuberculosis (<2%). Others have verified the same difference between M. tuberculosis and M. simiae (3). Overall, it was difficult to identify the species of the isolated strains using conventional techniques.
TABLE 1.
Biochemical tests and markers in the GLC profile of fatty acids and mycolic acid cleavage products found useful in differentiation of M. triplex and the isolate described
| Characteristic | Resulta
|
||
|---|---|---|---|
| Patient isolates | M. triplex | M. tuberculosis | |
| Biochemical characteristics | |||
| Growth at | |||
| 25°C | + | V | − |
| 37°C | + | + | + |
| 45°C | − | V | − |
| Pigment production | − | − | − |
| Niacin | − | − | + |
| Arylsulfatase | |||
| 3 days | − | − | − |
| 14 days | (Weak +) | V | − |
| Semiquantitative catalase | + | + | − |
| Nitrate reductase | + | + | + |
| Urease | + | + | + |
| Tween 80 hydrolysis | − | − | − |
| M. avium complex DNA probe | − | − | − |
| Marker peaks in GLC fatty acid profileb | |||
| 14:0 | 7.2 | 7.8 | 1.6 |
| 10-Me-18:0 | 7.2 | 12.6 | 19.4 |
| 24:0 | 8.9 | 5.9 | 3.2 |
| 26:0 | 8.7 | 13.6 | 9.1 |
Biochemical tests were performed by the procedure of Floyd et al. (5). +, positive; −, negative; V, variable.
14:0, tetradecanoate; 18:0, octadecanoate; 10-Me-18:0, tuberculostearate; 24:0, tetracosanoate; 26:0, hexacosanoate. Results are mean area percentages for the patient's isolates (eight analyses), for the M. triplex type strain (three analyses), and for 20 clinical M. tuberculosis strains.
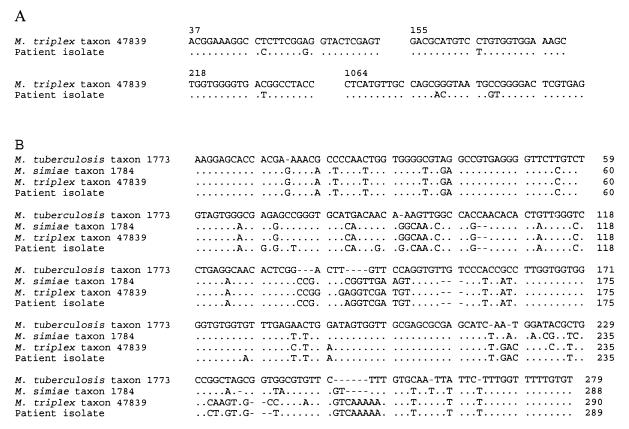
Sequencing of the 16S rRNA gene is widely used for species identification of mycobacteria. The complete 16S rRNA gene was amplified, and both strands of the amplified DNA were sequenced as described elsewhere (6). A BLAST sequence similarity search (EBI, Hinxton, United Kingdom) of the 16S sequence obtained showed a 99.5% similarity to M. triplex (Fig. 2A). The internal transcribed spacer (ITS) sequences have been shown to be useful phylogenetic markers for species differentiation and identification (8, 9). The ITS region of the patient isolate was amplified and sequenced as described previously (10). Comparison of the sequences showed 8 differences in the 16S rRNA gene and 14 differences in the ITS region between the patient isolate and the M. triplex type strain (Fig. 2A and B, respectively) (5, 8).
FIG. 2.
Comparison of ribosomal gene sequences. Dots indicate nucleotide identity, and dashes indicate deletions. (A) Differences in the 16S rRNA gene between the isolate (AJ276890) and M. triplex (U57362). Numbering is according to the reference sequence. (B) Alignment of ITS region sequences of selected species including the patient isolate. Reference sequence accession numbers are as follows: M. triplex, Y14189; M. simiae, AB026694; M. tuberculosis, AB026698.
The result of 16S rRNA sequencing, with 99.5% sequence similarity with M. triplex, indicated that the isolate was very closely related to M. triplex. We found the sequencing of the ITS region necessary because of the eight differences in the 16S rRNA gene. The result of the ITS sequencing confirmed the other results obtained in this study, indicating that this organism was an M. triplex-like mycobacterium. According to currently available data, the patient's strain is regarded as a variant of M. triplex. Further analyses are needed to evaluate the relevance of the differences detected and to answer the open question of whether the strain should be regarded as a subspecies of M. triplex or as another as yet unclassified but closely related species.
Several recent reports describe infections caused by unusual Mycobacterium species (2, 12). The development of molecular biological methods has allowed more-accurate species identification and the study of species diversity. The clinical case reported here indicated that the M. triplex variant described had to be regarded as a potential human pathogen. The results also demonstrated that the regimen often used to treat pulmonary mycobacteriosis caused by M. avium is also sufficient for the treatment of M. triplex infections.
Nucleotide sequence accession number.
The 16S and ITS sequences determined in this study have been deposited in the EMBL nucleotide sequence databank under accession number AJ276890.
Acknowledgments
S.S. was supported by a grant from the Oskar Öflund Foundation.
REFERENCES
- 1.American Thoracic Society. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am J Respir Crit Care Med. 1997;156:S1–S25. doi: 10.1164/ajrccm.156.2.atsstatement. [DOI] [PubMed] [Google Scholar]
- 2.Bajolet O, Beguinot I, Brasme L, Jaussaud R, Ingrand D, Vincent V. Isolation of an unusual Mycobacterium species from an AIDS patient with acute lymphadenitis. J Clin Microbiol. 2000;38:2018–2020. doi: 10.1128/jcm.38.5.2018-2020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou S, Chedore P, Kasatiya S. Use of gas chromatographic fatty acid and mycolic acid cleavage product determination to differentiate among Mycobacterium genavense, Mycobacterium fortuitum, Mycobacterium simiae, and Mycobacterium tuberculosis. J Clin Microbiol. 1998;36:577–579. doi: 10.1128/jcm.36.2.577-579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cingolani A, Sanguinetti M, Antinori A, Larocca L M, Ardito F, Posteraro B, Federico G, Fadda G, Ortona L. Brief report: disseminated mycobacteriosis caused by drug-resistant Mycobacterium triplex in a human immunodeficiency virus-infected patient during highly active antiretroviral therapy. Clin Infect Dis. 2000;31:177–179. doi: 10.1086/313903. [DOI] [PubMed] [Google Scholar]
- 5.Floyd M M, Guthertz L S, Silcox V A, Duffey P S, Jang Y, Desmond E P, Crawford J T, Butler W R. Characterization of an SAV organism and proposal of Mycobacterium triplex sp. nov. J Clin Microbiol. 1996;34:2963–2967. doi: 10.1128/jcm.34.12.2963-2967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koukila-Kähkölä P, Springer B, Böttger E C, Paulin L, Jantzen E, Katila M-L. Mycobacterium branderi sp. nov., a new potential human pathogen. Int J Syst Bacteriol. 1995;45:549–553. doi: 10.1099/00207713-45-3-549. [DOI] [PubMed] [Google Scholar]
- 7.Koukila-Kähkölä P, Paulin L, Brander E, Jantzen E, Eho-Remes M, Katila M-L. Characterisation of a new isolate of Mycobacterium shimoidei from Finland. J Med Microbiol. 2000;49:937–940. doi: 10.1099/0022-1317-49-10-937. [DOI] [PubMed] [Google Scholar]
- 8.Roth A, Fischer M, Hamid M E, Michalke S, Ludwig W, Mauch H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S–23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth A, Reischl U, Streubel A, Naumann L, Kroppenstedt R M, Habicht M, Fischer M, Mauch H. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S–23S rRNA gene spacer and restriction endonucleases. J Clin Microbiol. 2000;38:1094–1104. doi: 10.1128/jcm.38.3.1094-1104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torkko P, Suutari M, Suomalainen S, Paulin L, Katila M-L. Separation among the species of Mycobacterium terrae complex by lipid analysis: comparison with biochemical tests and 16S rRNA sequencing. J Clin Microbiol. 1998;36:499–505. doi: 10.1128/jcm.36.2.499-505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torkko P, Suomalainen S, Iivanainen E, Suutari M, Tortoli E, Paulin L, Katila M-L. Mycobacterium xenopi and related organisms isolated from stream waters in Finland and description of Mycobacterium botniense sp. nov. Int J Syst Evol Microbiol. 2000;50:283–289. doi: 10.1099/00207713-50-1-283. [DOI] [PubMed] [Google Scholar]
- 12.Tortoli E, Kischner P, Bartoloni A, Burrini C, Mantella A, Scagnelli M, Scarparo C, Simonetti M T, Böttger E C. Cervical lymphadenitis due to an unusual mycobacterium. Eur J Clin Microbiol Infect Dis. 1997;16:308–311. doi: 10.1007/BF01695636. [DOI] [PubMed] [Google Scholar]