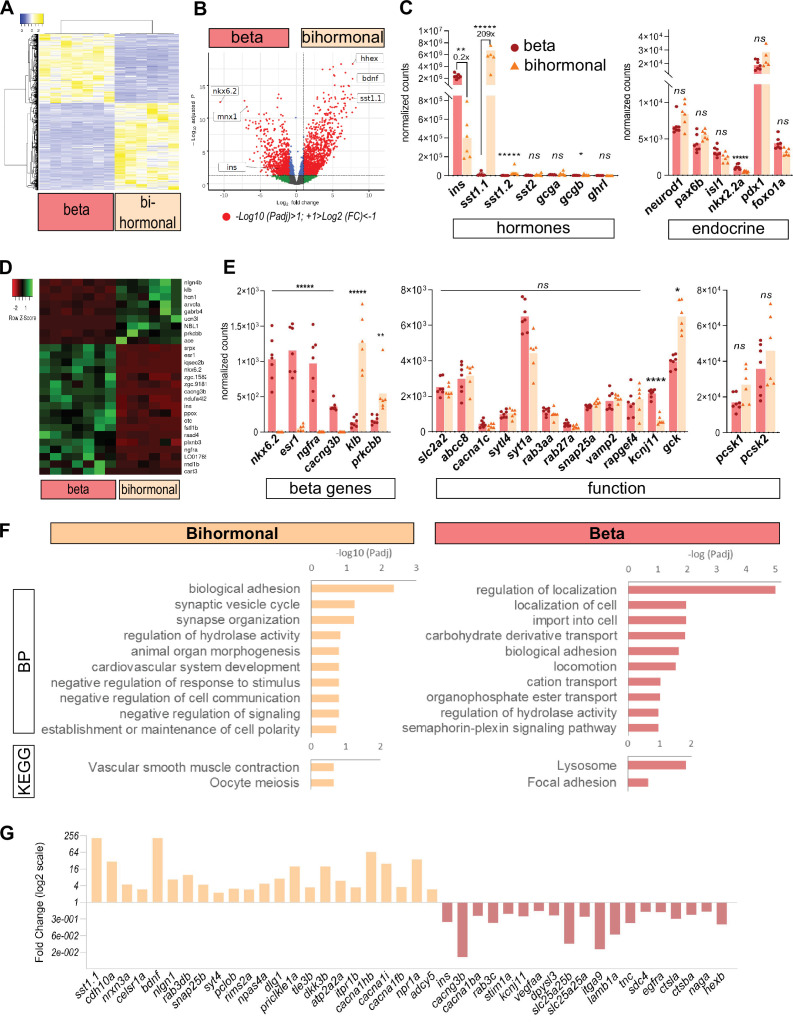
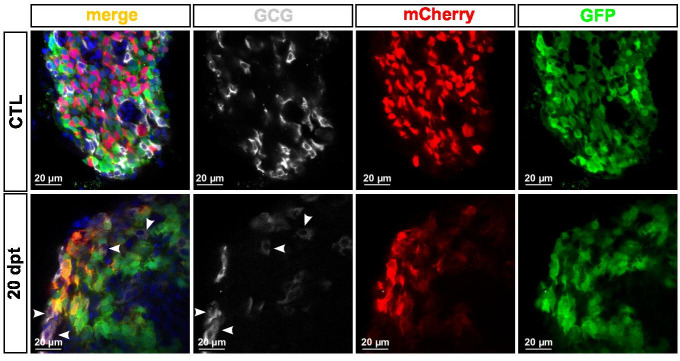
Figure 3. Transcriptomic comparison of bihormonal cells and β-cells.
(A) Heatmap representation of the transcriptomes of 20 dpt bihormonal (six replicates) and β-cells (seven replicates) (significant DE genes). (B) Volcano plot showing the distribution of genes in β-cells without ablation and bihormonal cells. The x-axis represents the log2 of fold change (FC) and the y-axis the log10 of adjusted P value (Padj) provided by DESeq. The red dots highlight the significantly DE genes (Padj <0.05). A full list of significant DE genes is provided in Figure 3—source data 1. (C) Expression values (mean normalized reads) as provided by DESeq of the main hormones and endocrine genes in β-cell and bihormonal cell transcriptomes. sst1.1 and ins are the two highest expressed hormones. Padj are calculated by DESeq. ns: no significant DE between the two conditions, 0.05< P* < 0.005, 0.005< P** < 0.0005, P***** < 0.000005. (D) Heatmap plot showing the direction and amplitude of changes in expression of the β-cell markers between normal β-cells and bihormonal cells (significant DEG only). The 62 β-cell markers are provided in Figure 3—source data 2. (E) Expression values (mean normalized reads) as provided by DESeq of selected β-cell markers and genes important for β-cell function in β-cells and bihormonal cells. Padj are calculated by DESeq. ns: no significant DE between the two conditions, 0.05<* < 0.005, 0.005<** < 0.0005, 0.00005<**** < 0.000005, ***** < 0.000005. (F) Enriched Gene Ontology (GO) terms. Top 10 or Padj (FDR) < 0.25 Biological Processes (BP) and KEGG pathways are shown. The plots represent the enrichment ratio of Biological Processes and KEGG pathways identified with WebGestalt (Liao et al., 2019) using the genes over- and underexpressed in bihormonal cells compared to β-cells obtained with a twofold differential expression and Padj <0.05. All overrepresented Biological Processes and Pathways (< FDR 0.25) are listed in Figure 3—source data 3 (bihormonal cells) and Figure 3—source data 4 (β-cells). (G) Over- and underexpression of selected significantly DE genes from the BP and KEGG pathways identified in β-cells and bihormonal cells (Fold Change, log2 scale).