Abstract
A real-time PCR assay was developed to detect and quantify Chlamydophila felis infection of cats. The assay uses a molecular beacon to specifically identify the major outer membrane protein gene, is highly reproducible, and is able to detect fewer than 10 genomic copies.
The family Chlamydiaceae has recently been revised on the basis of recent phylogenetic analyses of the 16S and 23S rRNA genes and genetic and phenotypic data (1). In this revision, the feline strain of Chlamydia psittaci is given specific status in a new genus, Chlamydophila. Chlamydophila felis infection is common in cats and is a major cause of conjunctivitis (2, 8, 9). It can also cause fever, lethargy, lameness, reduction in weight gain, and upper respiratory symptoms (5, 7). It can be difficult to diagnose from conjunctival swabs by isolation due to the low number of organisms present and the presence of tear antibodies in chronic infection. It has been shown that conventional PCR is more sensitive than isolation in detecting this infection (3, 6). Some major problems with conventional PCR are amplicon contamination, resulting in false positives, and the inability to accurately quantify the amount of starting template in the reaction mixture. We have developed a real-time quantitative PCR assay using a molecular beacon for C. felis that can detect fewer than 10 genomic equivalents and has a linear dilution curve of 9 log10.
A conjunctival swab was used to obtain cells from a cat known to be infected with C. felis. Genomic DNA was extracted from the swab using a DNeasy tissue kit (Qiagen, Crawley, United Kingdom) according to the manufacturer's instructions and was used as a template to generate an amplicon using primers Chl for and Chl rev designed to target the major outer membrane protein gene (Table 1) (4). The PCR mixture consisted of 25 μl of Qiagen 2X master mix (Qiagen), 0.2 μM Chl for, 0.2 μM Chl rev (both synthesized by Life Technologies Ltd., Paisley, Scotland), 1 μl of genomic DNA, and water to 50 μl. The PCR was carried out in a PTC 200 thermal cycler (MJ Research Inc., Waltham, Mass.) for 30 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The 129-bp amplicon was purified using the QIAquick PCR purification kit (Qiagen), electrophoresed on a 1% agarose gel containing 0.1 μg of ethidium bromide per ml, and visualized on a UVP GDS-8000 image capture system (Ultra Violet Products, Cambridge, United Kingdom). By comparison to the molecular weight markers, the concentration of the amplicon was estimated at 20 ng/μl. A standard curve was constructed using serial 10-fold dilutions (from 10−2 to 10−12) of the purified amplicon and used to quantify the results.
TABLE 1.
PCR primers and probe used in the C. felis real-time PCR assay
| Primer or probe | Primer or probe sequence | Region of MOMP gene (nt)a |
|---|---|---|
| Chl for | 5′-ATGCTTGTTCCATACATTGGGG-3′ | 965–986 |
| Chl rev | 5′-TCCTAAAAGAGTTGGGTTCCAGG-3′ | 1071–1093 |
| Chl molecular beaconb | 5′-FAM-CGGCGACACTATCCGCATTG CTCAACCGCCG-DABCYL-3′ | 1016–1038 |
From sequence with GenBank accession no. X61096. MOMP, major outer membrane protein.
The molecular beacon was labeled with a fluorescein residue at the 5′ end and a 4-(4′-dimethylaminophenylazo)benzoic acid (DABCYL) quencher at the 3′ end. The region of stem-loop formation is underlined.
Real-time PCR was performed using an iCycler (Bio-Rad Laboratories Ltd., Hemel Hempstead, United Kingdom) with an optical upgrade system. The PCR mixture consisted of 10 μl of Platinum Q PCR SuperMix-UDG (Life Technologies Ltd.), 0.2 μM Chl for, 0.2 μM Chl rev, 5 μl of template DNA, 120 nM C. felis molecular beacon (Cruachem Ltd., Glasgow, Scotland) (Table 1), and water to 20 μl. After an initial incubation at 50°C for 3 min to allow uracil DNA glycosylase (UDG) to digest any amplicon carryover and at 94°C for 2 min to inactivate the UDG, 45 cycles of 94°C for 10 s and 50°C for 30 s were carried out. Fluorescence was detected at 525 nm at each annealing step (50°C). All reactions were run in triplicate.
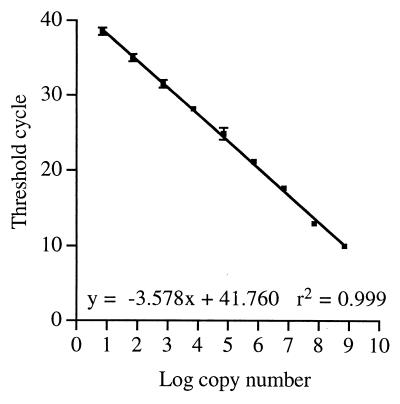
Figure 1 shows a plot of threshold cycle versus log10 copy number for the PCR standard in triplicate. It can be seen that the dilution is linear over a 9-log10 range with a correlation coefficient of 0.999. The estimated starting copy number was 7 × 109 molecules per PCR, and it was possible to detect as few as 7 molecules of template in two of the three reactions performed at this dilution. None of the reactions at 0.7 molecule per reaction gave a positive result.
FIG. 1.
Standard calibration curve for the C. felis real-time PCR assay. A C. felis PCR amplicon of known concentration was diluted from 7 × 109 molecules per 5 μl to 7 molecules per 5 μl, and 5 μl was used in the PCR assay. The threshold cycle was measured and plotted against the log10 of the starting copy number. Each point represents the average ± standard deviation for three PCRs.
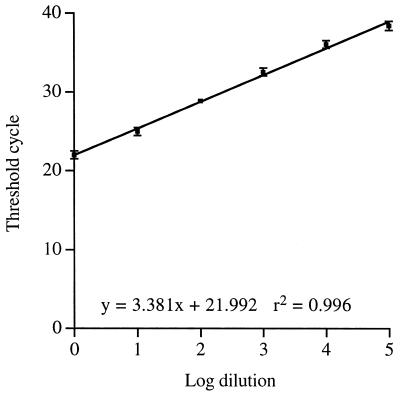
Figure 2 shows a dilution curve for a sample isolated from a cat known to have a C. felis infection. Genomic DNA was diluted 10-fold and used as a template in the PCR. It can be seen that, by using a clinical sample of genomic DNA, a dilution curve that is linear over a range of 5 log10 can be produced. Using the standard curve, it is possible to accurately quantify the number of genomic copies present in the starting material. This equates to fewer than 10 genomic equivalents in the 105 dilution. At a 106 dilution no signal was seen for any of the three PCRs. However, it must be noted that incomplete lysis of the sample or the presence of inhibitors would reduce the number of genomic copies available for PCR or reduce the efficiency of amplification. Hence, there may be more organisms in the original sample than are calculated from the standard curve.
FIG. 2.
Dilution curve of a clinical sample of C. felis DNA. Genomic DNA was isolated from a conjunctival swab taken from a cat. This was serially diluted 10-fold, and 5 μl was used in the PCR assay. The threshold cycle was measured and plotted against the log10 of the dilution. Each point represents the average ± standard deviation for three PCRs.
These results demonstrate for the first time the use of real-time PCR and molecular beacons to detect C. felis DNA isolated from a conjunctival swab. The assay is very sensitive, highly reproducible, and can be accomplished in less than 2 h. This assay is about 10 to 100 times more sensitive than our current nested PCR assay for C. felis and has equal specificity (results not shown). This increase in sensitivity is due to the highly sensitive fluorescence detection system used in real-time PCR machines compared to ethidium bromide-stained gels. From a diagnostic point of view, the main advantages of real-time PCR are that the closed tube system results in a decrease in false positives and that the assay is very easy to use. The inclusion of UDG and dUTP further reduces the chance of false positives occurring. The use of a molecular beacon ensures that only the desired target is detected and gives very low background fluorescence. The accurate quantitative nature of the assay lends itself to the determination of the number of organisms in the swab sample. This can be very useful in determining whether antibiotic treatment has been effective. However, for routine use the assay could be used for qualitative analysis of the presence or absence of C. felis.
The use of real-time PCR to diagnose infection and disease will allow results to be obtained more quickly than is currently possible with conventional PCR or isolation. Real-time PCR is also more sensitive than conventional PCR and allows accurate quantification if desired. The use of multiplex real-time PCR with molecular beacons can allow several infectious organisms to be identified in the same sample at the same time, yielding a quicker turnaround time and saving resources.
REFERENCES
- 1.Everett K D, Bush R M, Andersen A A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49(Part 2):415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 2.Johnson F W. Isolation of Chlamydia psittaci from nasal and conjunctival exudate of a domestic cat. Vet Rec. 1984;114:342–344. doi: 10.1136/vr.114.14.342. [DOI] [PubMed] [Google Scholar]
- 3.McDonald M, Willett B J, Jarrett O, Addie D D. A comparison of DNA amplification, isolation and serology for the detection of Chlamydia psittaci infection in cats. Vet Rec. 1998;143:97–101. doi: 10.1136/vr.143.4.97. [DOI] [PubMed] [Google Scholar]
- 4.Storey C, Lusher M, Yates P, Richmond S. Evidence for Chlamydia pneumoniae of non-human origin. J Gen Microbiol. 1993;139:2621–2626. doi: 10.1099/00221287-139-11-2621. [DOI] [PubMed] [Google Scholar]
- 5.Sykes J E, Anderson G A, Studdert V P, Browning G F. Prevalence of feline Chlamydia psittaci and feline herpesvirus 1 in cats with upper respiratory tract disease. J Vet Intern Med. 1999;13:153–162. doi: 10.1892/0891-6640(1999)013<0153:pofpaf>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Sykes J E, Studdert V P, Browning G F. Comparison of the polymerase chain reaction and culture for the detection of feline Chlamydia psittaci in untreated and doxycycline-treated experimentally infected cats. J Vet Intern Med. 1999;13:146–152. doi: 10.1892/0891-6640(1999)013<0146:cotpcr>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 7.TerWee J, Sabara M, Kokjohn K, Sandbulte J, Frenchick P, Dreier K J. Characterization of the systemic disease and ocular signs induced by experimental infection with Chlamydia psittaci in cats. Vet Microbiol. 1998;59:259–281. doi: 10.1016/s0378-1135(97)00185-5. [DOI] [PubMed] [Google Scholar]
- 8.Wills J, Gruffydd-Jones T J, Richmond S, Paul I D. Isolation of Chlamydia psittaci from cases of conjunctivitis in a colony of cats. Vet Rec. 1984;114:344–346. doi: 10.1136/vr.114.14.344. [DOI] [PubMed] [Google Scholar]
- 9.Wills J M, Howard P, Gruffydd-Jones T J, Wathes C M. Prevalence of Chlamydia psittaci in different cat populations in Britain. J Small Anim Pract. 1988;29:327–339. [Google Scholar]