Abstract
PURPOSE
Soft tissue and bone sarcomas are rare malignancies that exhibit significant pathologic and molecular heterogeneity. Deregulation of the CDKN2A-CCND-CDK4/6-retinoblastoma 1 (Rb) pathway is frequently observed in about 25% of unselected sarcomas and is pathognomonic for specific sarcoma subtypes. This genomic specificity has fueled the clinical evaluation of selective CDK4/6 inhibitors in sarcomas. Here, we highlight successes, opportunities, and future challenges for using CDK4/6 inhibitors to treat sarcoma.
MATERIALS AND METHODS
This review summarizes the current evidence for the use of CDK4/6 inhibitors in sarcoma while identifying molecular rationale and predictive biomarkers that provide the foundation for targeting the CDK4/6 pathway in sarcoma. A systematic review was performed of articles indexed in the PubMed database and the National Institutes of Health Clinical Trials Registry (ClinicalTrials.gov). For each sarcoma subtype, we discuss the preclinical rationale, case reports, and available clinical trials data.
RESULTS
Despite promising clinical outcomes in a subset of sarcomas, resistance to CDK4/6 inhibitors results in highly heterogeneous clinical outcomes. Current clinical data support the use of CDK4/6 inhibitors in subsets of sarcoma primarily driven by CDK4/6 deregulation. When dysregulation of the Rb pathway is a secondary driver of sarcoma, combination therapy with CDK4/6 inhibition may be an option. Developing strategies to identify responders and the mechanisms that drive resistance is important to maximize the clinical utility of these drugs in patients with sarcoma. Potential biomarkers that indicate CDK4/6 inhibitor sensitivity in sarcoma include CDK4, CCND, CCNE, RB1, E2F1, and CDKN2A.
CONCLUSION
CDK4/6 inhibitors represent a major breakthrough for targeted cancer treatment. CDK4/6 inhibitor use in sarcoma has led to limited, but significant, early clinical success. Targeted future clinical research will be key to unlocking the potential of CDK4/6 inhibition in sarcoma.
INTRODUCTION
Sarcomas are rare malignant tumors that originate from the connective tissues of bone or soft tissues including fat, muscle, blood vessels, or nerves. Sarcomas make up < 1% of all adult malignancies and approximately 20% of pediatric cancers.1 The rarity and heterogeneity of these tumors have led to challenges in both diagnosis and treatment development.2 Even with multimodal therapy including surgery, radiation, and chemotherapy in the advanced setting, sarcomas are incurable malignancies associated with dismal prognosis.1
CONTEXT
Key Objective
Sarcomas exhibit significant molecular heterogeneity including dysregulation of the CDKN2A-CCND-CDK4/6-retinoblastoma 1 (Rb) pathway. The use of CDK4/6 inhibitors in sarcomas has led to mixed results, indicating the need to identify biomarkers of sensitivity and resistance across sarcomas.
Knowledge Generated
While highlighting the promising clinical outcomes in subsets of sarcomas, we present the current knowledge of intrinsic and/or acquired resistance to CDK4/6 inhibitors identified across sarcomas. Current data support the use of CDK4/6 inhibitors in sarcomas primarily driven by CDK4/6 signaling. In sarcomas where Rb pathway dysregulation is a secondary driver, combinations of CDK4/6 inhibition therapy with additional therapies may be effective.
Relevance
The use of CDK4/6 inhibitors across sarcomas remains limited; however, subtypes of the disease are sensitive to this therapeutic strategy as either monotherapy or combination therapy.
Despite the molecular and cellular heterogeneity of sarcoma, with more than 100 subtypes exhibiting unique and defining genomic alterations, dysregulation of the cyclin D (CCND)-cyclin-dependent kinase 4/6 (CDK4/6)-retinoblastoma 1 (Rb) pathway is common across several sarcoma subtypes (Fig 1). These recurrent genetic aberrations result in the deregulation of activator E2 promoter binding factors 1-3 (E2F1-3). These potent transcription factors directly bind to and induce the transcription of genes required for the G1/S, G2/M cell cycle transition, apoptosis, and metabolism. The activity of E2F1-3 is tightly controlled by the Rb protein, which acts as a key cell cycle checkpoint regulator and tumor suppressor. In sarcomas, RB1 deletion and/or mutation, changes to kinase regulators of Rb stability (CDK4/6 and CCND1-3), or deletion/silencing of the CDK inhibitor family (p16[CDKN2A]/p15[CDKN2B]) all result in diminished Rb-mediated repression of E2F1-3 activity and deregulated cellular growth.
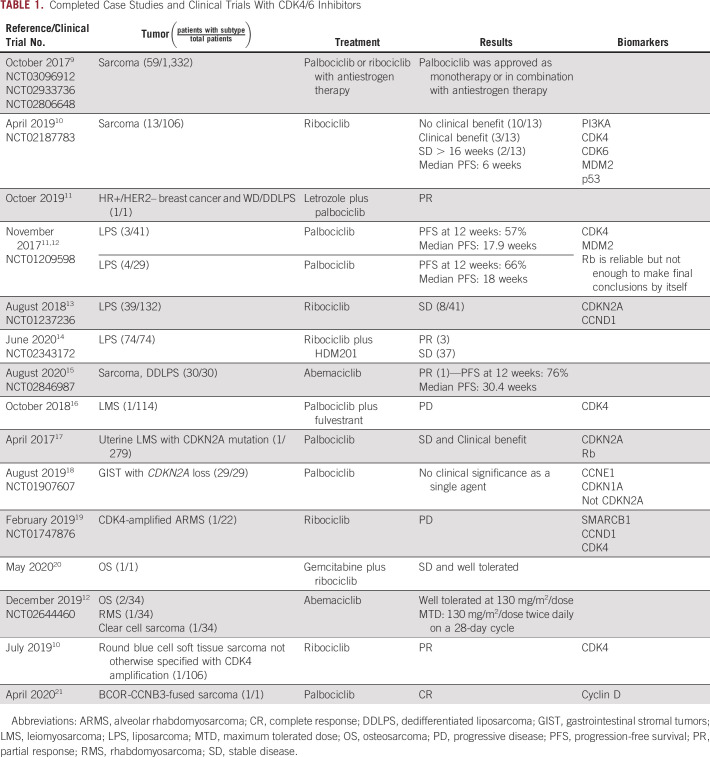
FIG 1.
Common derangements in CCND-CDK4/6-RB pathway in sarcoma. The CDK4-cyclin D complex can phosphorylate the Rb-E2F complex to promote cyclin E translation for G1/S progression. CDK4/6 inhibitors can prevent this phosphorylation process. Common genes that are overexpressed (red), underexpressed/loss (teal), or normal (orange) in the majority of sarcoma subtypes are depicted.
DNA sequencing of nearly 10,000 sarcomas identified widespread alterations in RB1 (14.6%) and CDK4 (12%).3 A similar analysis of soft tissue sarcomas in The Cancer Genomic Atlas (TCGA) also found that alterations in the CDKN2A-CCND-CDK4-Rb pathway are highly prevalent. Common alterations in the TCGA data included RB1 deletion (16.1%), CDKN2A/CDKN2B deletion (12.9%), CCND3 amplification (4.4%), and CDK4 amplification (18.5%).4 An additional study explored the genetic alterations in leiomyosarcoma and found frequent genomic alterations including TP53 mutation (34%), TP53 deletion (8%), CDKN2A deletion (21%), RB1 deletion (11%), MDM2 amplification (8%), and PIK3CA mutation (6%). In this study, CDKN2A deletion was correlated with poor overall survival.5 Collectively, these genomic data highlight that the CDKN2A-CCND-CDK4/6-Rb pathway is commonly altered in more than 25% of sarcoma and represents a key oncogenic driver in these tumors. On the basis of these results, targeting this pathway represents a promising therapeutic strategy for treating sarcoma.
Currently, the US Food and Drug Administration (FDA) has approved the clinical use of several CDK4/6 inhibitors that prevent G1/S cell cycle progression by blocking CDK4 and/or CDK6 from binding to their regulatory partner, CCND.6 The dependence of sarcomas on activation of the CDKN2A-CCND-CDK4/6-Rb pathway has opened opportunities for the use of CDK4/6 inhibitors in the clinic.7,8 This review outlines the current state of CDK4/6 inhibition as a treatment option for soft tissue and bone sarcomas. In addition, we focus on new clinical and preclinical research on the use of predictive biomarkers for CDK4/6-directed therapy in sarcoma.
CLINICAL USE OF CDK4/6 INHIBITORS IN SARCOMA
Given the diversity of sarcoma types, this review focuses primarily on the six common sarcoma subtypes: liposarcoma (LPS), leiomyosarcoma (LMS), gastrointestinal stromal tumors (GIST), osteosarcoma (OS), rhabdomyosarcoma (RMS), and Ewing's sarcoma (ES). The methods for reviewing the literature are included in Appendix 1. In each section, we discuss the preclinical rationale, available clinical trials and case reports specific to the sarcoma subtype, and potential biomarkers for therapeutic efficacy (Table 1). Using TCGA data and PubMed searches, subtypes with a characteristic genetic alteration in the CDKN2A-CDK4/6-CCND-Rb pathway including, but not limited to, CDK4, CDK6, CCND1, RB1, E2F, CDKN2A, CDKN1A, CCNE1, and MDM2 were identified and related studies were analyzed for significant biomarkers (Fig 2). A brief overview regarding CDK4/6 inhibitor development is included in Appendix 2.3,54-64
TABLE 1.
Completed Case Studies and Clinical Trials With CDK4/6 Inhibitors
FIG 2.
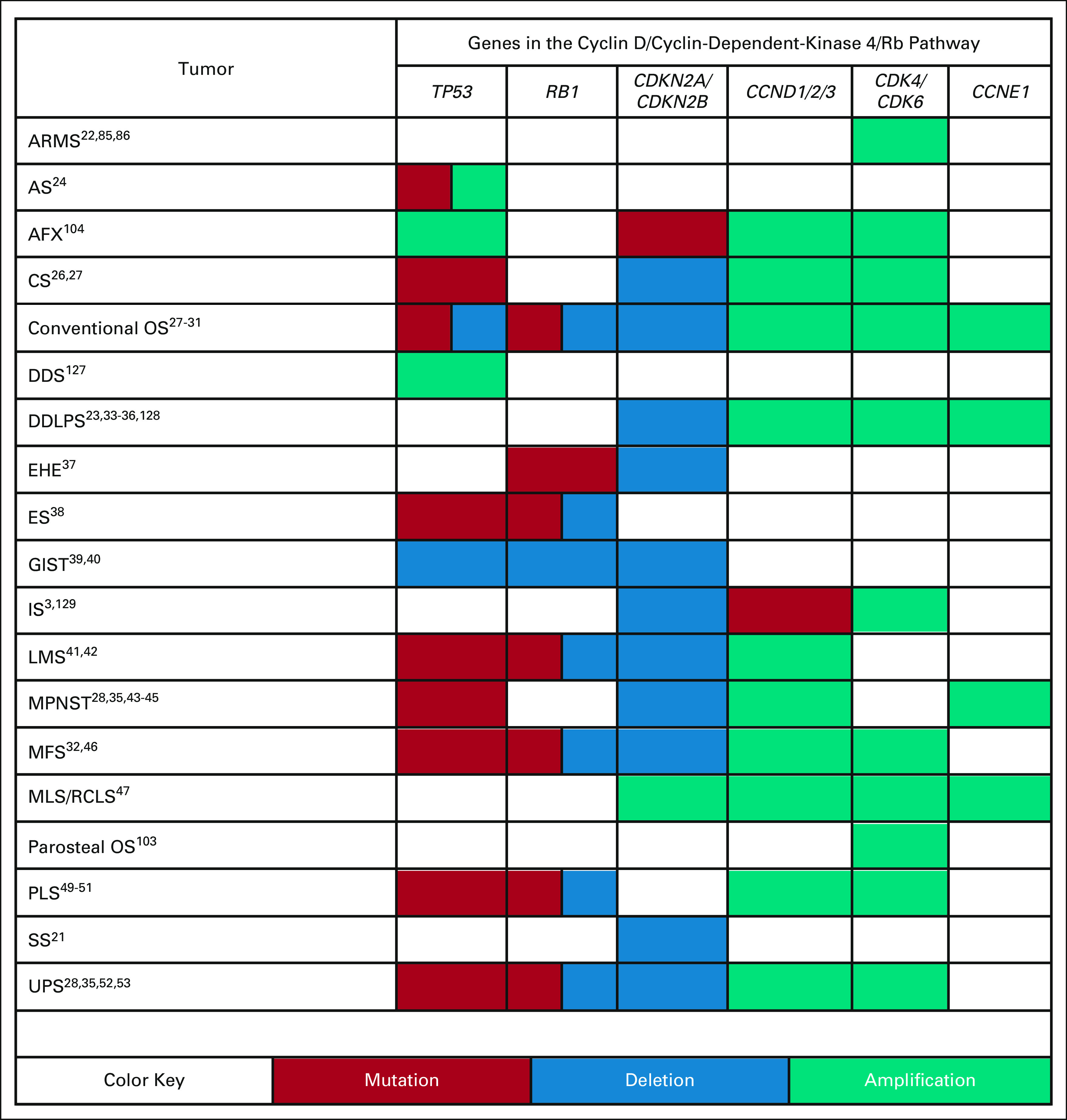
Common genetic alterations in the CDKN2A-CCND-CDK4/6-Rb pathway in various sarcoma subtypes. Dysregulation of the CDKN2A-CCND-CDK4/6-Rb pathway is common across sarcomas. Despite the common pathway dysregulation in sarcomas, molecular variations exist between subtypes. Subtypes with common mutations (red), deletions (blue), and amplifications (teal) in TP53, RB1, CDKN2A/CDKN2B, CCND1/2/3, CDK4/CDK6, and CCNE1 are indicated by the colored table cells. References are indicated by the numbers in each cell. ARMS, alveolar rhabdomyosarcoma; AS, angiosarcoma; AFX, atypical fibroxanthoma; CS, chondrosarcoma; DDS, dedifferentiated chondrosarcoma; DDLPS, dedifferentiated liposarcoma; EHE, epithelioid hemangioendothelioma; ES, Ewing's sarcoma; GIST, gastrointestinal sarcoma tumors; IS, intimal sarcoma; LMS, leiomyosarcoma; MPNST, malignant peripheral nerve sheath tumors; MFS, myxofibrosarcoma; MLS/RCLS, myxoid/round cell liposarcoma; OS, osteosarcoma; PLS, pleomorphic liposarcoma; SS, synovial sarcoma; UPS, undifferentiated pleomorphic sarcoma.
LPS
LPS, primarily arising from fat tissue in the thigh or retroperitoneum, comprise approximately 13% of all sarcomas.65,66 Subclasses of LPS, including well-differentiated/dedifferentiated liposarcomas (WDLPS/DDLPS), myxoid/round cell liposarcoma (MRCLS), and pleomorphic liposarcoma (PLS), have demonstrated consistent dysregulation of the Rb pathway. DDLPS frequently present with amplifications of the 12q region of chromosome 12, resulting in MDM2 and CDK4 overexpression.33,34 In addition, amplification and/or activation events of CDK4 are essential for the growth and survival of WDLPS/DDLPS.67 MRCLS harbors a FUS-DDIT3 chimeric gene, which causes abnormal expression of G1 checkpoint regulators including CCND, CCNE, CDK4, and CDK2.47 Widespread genetic alterations in PLS frequently present with the loss of TP53 and RB1 tumor suppressor genes as well as overexpression of CCND1, CDK4, MDM2, MYB, and GL11.49
To date, LPS represents the largest subgroup of sarcomas to be treated with CDK4/6 inhibitors and these trials have resulted in clinical success. In a pair of phase II clinical trials, two different doses of palbociclib were compared in WDLPS and DDLPS.68,69 Patients treated with a 200-mg daily dose for days 1-14 of a 21-day cycle showed a median progression-free survival (PFS) of 18 weeks and 66% of patients reached a PFS of at least 12 weeks or longer on this regimen.68 The second trial examined the effects of a 125-mg dose for days 1-21 of a 28-day cycle where the median PFS was also 17.9 weeks and 57% of patients reached a PFS of 12 weeks or longer.69 As a single agent in CDK4 amplified, Rb-intact LPSs, palbociclib demonstrated significant clinical activity and a favorable adverse event profile. On the basis of these results, palbociclib is currently listed as category 2A evidence in the soft tissue sarcoma National Comprehensive Cancer Network (NCCN) guidelines but is not currently approved by the FDA or EMA for this indication.70
In a phase I clinical trial to determine the maximum tolerated dose of ribociclib in advanced solid tumors, patients with LPS made up 30% (39 of 132) of the study population with six patients with LPS demonstrating stable disease (SD) at 6 months on treatment. Three of the four patients who remained on therapy for the longest durations harbored CCND1 amplification without CDKN2A/CDKN2B codeletion. Comparatively, seven (29%) of the 24 patients who received treatment for < 8 weeks had CDKN2A/CDKN2B codeletion.13
In a different tissue agnostic phase II trial of ribociclib, three of 13 (23.1%) enrolled patients with sarcoma exhibited clinical benefit, as defined by a response of stabilized disease or better at 16 weeks of therapy. Of these 13 enrolled patients with sarcoma, only four were considered DDLPS with none of these patients achieving clinical benefit by the study's definition. Across tumor types, cancers with a single hit in the cyclin D-CDK4/6 complex tended to see the greatest benefit from ribociclib.10 Interim analysis was conducted of a phase Ib study of the MDM2 inhibitor, HDM201, in combination with ribociclib in 74 patients with WDLPS/DDLPS with multiple dosing regimens. Partial response was achieved in three patients (4%), whereas 36 patients achieved stabilized disease (49%). Median PFS ranged from 2.1 to 4.8 months across three dosing regimens.14 In a key interim analysis of a phase II study of abemaciclib in patients with DDLPS, the observed PFS at 12 weeks was 76% (95% CI, 57 to 90), whereas the median PFS was 30.4 weeks (95% CI, 28.9 to not evaluable).15
The use of CDK4/6 inhibitors shows promise as an active therapy regimen in patients with WDLPS/DDLPS. Although phase I and II trials with CDK4/6 inhibitors include both DDLPS and WDLPS, varied representation of patients with DDLPS/WDLPS makes interpretation of each subtype difficult to determine. Overall, the genomic amplification of CDK4 and possibly CCND1 are likely biomarkers for CDK4/6 inhibitor sensitivity in this disease. MDM2, which is commonly upregulated in LPS and results in altered Rb function, is a biomarker that has been investigated but shows varied biomarker potential.67,71 The utility of CDKN2A levels as a biomarker for CDK4/6 inhibitor sensitivity in WDLPS/DDLP has also yet to be fully defined.72
LMS
LMS are smooth muscle sarcomas primarily arising from cells of the abdomen, uterus, and gastrointestinal tract, and less commonly from the vasculature or cutaneous structures.73 LMS commonly have genomic alterations of TP53, RB1, chromatin remodeling, and homologous recombination DNA repair pathways.74,75 21.4% of LMS exhibit RB1-deactivating events, and subpopulations of LMS demonstrate genomic alterations in CDK4, CDK6, CDKN2A, CDKN2B, CCND1, and CCND3.16,17,28,76 Uterine myxoid LMS also frequently harbor TP53 and/or CDKN2A genomic alterations. In cultured CDKN2A-deleted LMS cell lines, palbociclib exerts a strong growth-inhibitory effect and, conversely, high levels of p16 in combination with Rb loss correlate with palbociclib resistance.76,77
To date, there are no formal clinical studies with published results for CDK4/6 inhibitors in LMS. Retrospective analyses and case reports have documented mixed success. In particular, one patient with CDKN2A/CDKN2B loss, estrogen receptor–positive, LMS was treated with a combination of fulvestrant and palbociclib without evidence of clinical benefit.16 A second report described a patient with uterine LMS harboring loss of CDKN2A and heterozygous mutation in NF2 treated with palbociclib resulting in 8 months of disease stabilization.17 Although the Rb pathway is frequently dysregulated by genomic variants at RB1, in Rb-intact disease with dysregulating upstream variants, CDK4/6 inhibition may be a therapeutic option. A phase II trial of ribociclib and everolimus, an mTOR inhibitor, in DDLPS and Rb-positive LMS is currently underway and will provide insight into the benefits of CDK4/6 inhibitors in Rb-intact diseases.78 Given the predilection for alterations in RB1 in LMS, de novo or secondary resistance to CDK4/6 inhibitors may complicate, or limit the efficacy, of single-agent treatment.71,79
GIST
GIST are often misdiagnosed as LMS as they develop in the intestinal tract. However, GIST present with genomic features distinct from LMS, as 85% of patients with GIST have oncogenic changes within either KIT or PDGFRA.39,80 Less frequent GIST driver alterations include NF1, BRAF, or SDH. Interestingly, concomitant secondary alterations in CDKN2A, RB1, MDM2, and CCND1 amplifications are common. Recent work has shown that loss or deregulation of the Rb-CDK4 pathway is linked to increased risk of metastasis in patients with GIST.40,81
Genomic analysis of these tumors supports this hypothesis. Recent whole-exome sequencing on 29 high-grade metastatic KIT-mutant GIST found that CDKN2A, RB1, or TP53 mutation or loss was associated with poor patient prognosis.39 However, to date, targeting GIST with CDK4/6 inhibitors has not provided clinical utility. A phase II study examining palbociclib in CDKN2A-deleted advanced GIST demonstrated no significant clinical activity as a single agent with 19 of 22 (86.4%) patients experiencing progressive disease at 4 months on therapy. Therefore, the clinical and biological relevance of CDK4/6 inhibitors as a single-agent in GIST is unclear. Combination therapy and biomarkers to better understand the pathways fueling tumor growth are required, and the changes within the CDK4/CCND pathway remain under investigation.18
OS
OS is the most common bone cancer in children and is usually found around the knee or long bones of the extremities where immature bone is produced by mesenchymal cells.82 OS is prone to aberrations of cell cycle control regulators including RB1, TP53, CDKN2A, PTEN, CDK4, MDM2, MYC, TWIST1, CCND3, and CCNE1.28 Parosteal OS, not to be confused with periosteal OS that arises from the inner layer of the periosteum, originates from the outer fibrous layer of the periosteum, and has frequent alterations in 12q13-15 of chromosome 12 that include SAS, CDK4, and MDM2 genes. In particular, SAS and CDK4 genes were found to be amplified commonly in grade II and dedifferentiated tumors.83 One important consideration for the use of CDK4/6 inhibitor use in OS is that CDK4 amplification may be related to parosteal tumor grade, which should be considered with designing these trials.84
Because of its rarity, prospective trials with CDK4-directed therapy in OS are not available. The limited data we have are anecdotal. A patient with chemotherapy-resistant, metastatic, CDK6-amplified OS experienced SD for 10 cycles of treatment with ribociclib and gemcitabine. Treatment was ultimately stopped because of toxicity and not progression. Similar to CDK4, CDK6 may serve as a biomarker for CDK4/6 inhibitor success and these findings set the foundation for further testing.20
RMS
RMS develops from rhabdomyoblasts in soft tissues, especially skeletal muscle tissue, bladder, or uterus.85 RMS, specifically alveolar rhabdomyosarcoma (ARMS), is prone to gene amplifications of 12q13-q14 and 2p24, leading to an increase in the expression of MYCN, CDK4, CDK6, CDC25A, and SKP2. RMS comprises two major subtypes: fusion-positive and fusion-negative. Although most fusion-positive RMS tumors are characterized by the PAX3-FOXO1 gene fusion, a smaller subset of cases have a PAX7-FOXO1 fusion. Fusion-positive RMS tumors have equal rates of amplification of 2p24, but amplifications of 12q13-q14 are specific to PAX3-FOXO1 fusion-positive RMS.86 In ARMS, 80% of cases have PAX3-FOXO1 and PAX7-FOXO1 gene fusions. Fusion-negative RMS tumors have lower Rb protein levels than fusion-positive RMS.87-89
Downregulation of p21 has also been linked with tumorigenesis of transcription factor, FoxF1- or FoxF2-, elevated RMS.90 In contrast to other sarcoma subtypes, in vitro experimental data in RMS cultured samples show little correlation between CDK4 overexpression and CDK4/6 inhibitor sensitivity. Additional data also suggest that the RMS cells lines with CDK4 amplification have reduced sensitivity to ribociclib. These findings suggest that CDK4 may represent a biomarker for resistance to CDK4/6 inhibitors in RMS.87 In a phase I trial of ribociclib in pediatric solid tumors, a patient with CDK4-amplified RMS progressed after 5 months of therapy.19 It appears that CDK4 amplification may be related to CDK4/6 inhibitor sensitivity for patients with RMS; however, further research is required to confirm these findings.
ES and Ewing Family of Tumors
ES represents a large family of mesenchymal tumors with varied molecular fusions. Classical ES is a small round cell tumor that develops in the bones and/or soft tissue around the bones. ES’ characteristic fusion gene, EWS-FLI1, is an oncogenic transcription factor that upregulates MYC and CDK4 and downregulates CDK inhibitors, CDKN1A and CDKN1C.91 Secondary genomic alterations in ES include STAG2 (15%-17%), TP53 (6%-9%), CDKN2A (11%-22%), RB1, and CCND1.91-95 In EWSR1-PATZ1–positive sarcomas, secondary alterations in CDKN2A are highly prevalent (71%).92 The clinical evaluation of CDK4/6 inhibitors in the treatment of ES has yet to be described.
In addition, the BCOR-CCNB3 fusion-positive sarcoma, another Ewing's family variant, upregulates CCND1, SATB2, TLE1, and BCL2.96 Comprehensive genomic profiling in BCOR-fusion uterine sarcoma revealed CDK4 and MDM2 coamplifications or homozygous deletion of CDKN2A. These genetic changes closely mirror the genomic profile of DDLPS and, on the basis of the successes of CDK4/6 inhibitors in DDLPS, a therapeutic opportunity to use CDK4/6 inhibitors as a single agent or in combination with MDM2 inhibitors to treat BCOR-CCNB3 fusion-positive sarcomas exists.97 A report of a patient with a BCOR-CCNB3 fusion sarcoma and germline CDKN2B missense variant pediatric sarcoma demonstrated sustained complete response following treatment with palbociclib. Despite harboring a CDKN2B variant, CDKN2B was normally expressed, although RNA analysis suggested overactivation of the Rb pathway.21 It is unclear how CDK4/6 inhibition might provide benefit in the Ewing family of tumors.
Other Sarcomas
Synovial sarcomas (SS) frequently upregulate EGFR, MDM2, CDK2, and CDK4, and downregulate CCND1 to fuel tumor development.98 SS commonly harbor CDKN2A deletion (74%) and an array of additional mutations in Rb pathway genes: CCND1, CDK4, CDK6, and RB1.99 It has also been reported that heart sarcomas also have frequent CDK4 (38%) and CCND3 alterations (14%)3; this heart analysis did not delineate a more specific diagnosis, indicating more research is necessary to identify enriched disease subtypes harboring these CDK4 and CCND3 alterations. Similarly, intimal sarcoma also have amplified CDK4, mutated CCND, and deleted CDKN2A.129 Epithelioid hemangioendothelioma is a rare sarcoma subtype commonly characterized by a WWTR1-CAMTA1 fusion and altered CDKN2A/CDKN2B, RB1, APC, and FANCA genes.37 An estimated 96% of high-grade chondrosarcoma have affected RB1 pathways via TP53, RB1, or CDKN2A loss, CDK4 or CCND1 overexpression, or inactivated CDK inhibitors such as p16INK4a.26,100 On the basis of the continued theme of CDKN2A-CCND-CDK4-Rb pathway mutations and altered G1/S checkpoint regulations, these tumors may represent excellent candidates for CDK4/6 inhibitor treatment. However, similar to LMS, sarcoma subtypes with loss of Rb or further downstream proteins may limit the efficacy of CDK4/6 inhibitors as a single-agent treatment.
Sarcoma histology-agnostic studies of CDK4/6 inhibition have yielded a mixed response. A phase II study in CDK4/6 pathway activated soft tissue sarcomas had a median PFS of 6 weeks when treated with 600 mg doses of ribociclib for days 1-21 of a 28-day cycle. Out of 13 patients with sarcoma on this trial, 10 showed no clinical benefit. Of the three who did have clinical benefits, two had SD over 16 weeks on CDK4/6 inhibitor treatment. These patients presented with CDKN2A/CDKN2B loss and CCND1 variants plus CDK4 and MDM2 amplifications, respectively. The patient with partial response had a CDK4 amplification. Notably, the 10 patients with progressive disease also frequently harbored CDKN2A/CDKN2B loss and one had CCND3 amplification. Additionally, a patient with poorly differentiated, round blue cell sarcoma not otherwise specified with CDK4 amplification had a 100% reduction after ribociclib treatment. More work is required in this space; however, these findings do further support CDK4 as a potential biomarker for CDK4 inhibitor utility in treating sarcoma.10
FUTURE DIRECTIONS
Dysregulation of the CDKN2A-CDK4-CCND-Rb pathway in patients with sarcoma represents a promising opportunity for therapeutic treatments with CDK4/6 inhibitors. Although sensitivity to CDK4/6 inhibitors varies across sarcoma subtypes, biomarkers for CDK4/6 inhibitor sensitivity may serve as a driving force in diagnosing and treating sarcoma subtypes. Since CDK4/6 inhibitor sensitivity in many sarcoma subtypes remain untested or have yet to be reported, further in vitro, in vivo, and clinical testing of these agents is necessary for advancing our understanding of these compounds in sarcoma. To expand biomarker development, clinical trials with non-LPS subtypes treated with CDK4/6 inhibitors would help to draw connections between their shared genomic aberrations. Overall, CDK4/6 inhibitors have demonstrated modest clinical success when used as monotherapy; however, data from other cancer types suggest that combination strategies for CDK4/6 inhibitors may offer improved PFS and patient outcome.
In many sarcomas defined by characteristic genomic events, often an oncogenic fusion or activating kinase mutation, variants in the Rb pathway represent a second hit. These alterations are highly prevalent, but not exclusively seen, across the disease. This suggests that Rb pathway alterations are not necessary for the development of the initial disease but are enriched through the evolution of the tumor. These variants are potentially reasonable targets for CDK4/6 inhibitors either as single agents or in combination.
Notably, subtypes with Rb loss or mutation may have limited sensitivity to CDK4/6 inhibitors. In Rb-positive cell lines, combination with palbociclib and doxorubicin or Wee1 inhibitor, AZD1775, was synergistic; however, Rb knockdown cell lines displayed resistance to palbociclib treatment.101 As CDKN2A/CDKN2B loss is a common secondary alteration in sarcomas, potentially combining CDK4/6 inhibition with other agents to target primary and secondary drivers of oncogenesis is an attractive therapy. CDK4/6 inhibition in combination with hormone-directed therapies, DNA-damaging chemotherapy, antibodies against programmed cell death, Wee1, MEK, or mTOR inhibitors have been shown to have promising preliminary outcomes.102
A significant number of sarcoma types, at least in part, driven by CDK4 amplifications include DDLPS, undifferentiated pleomorphic sarcoma, LMS, ARMS, ES, SS, and parosteal sarcoma.21,25,28,39,48,76,103-105 Ongoing clinical trials including the phase II multicenter trial of palbociclib in advanced sarcomas with CDK4 overexpression, phase II study in bone and soft tissue sarcoma with CDK pathway alterations treated with abemaciclib, and the phase III study on molecular profiling of soft tissue sarcomas will provide a wealth of data for biomarker development and CDK4/6 inhibitor clinical utility.106-108
Further research examining the effect that CDK4, CCND, CCNE, Rb, E2F, and p16 proteins have on CDK4/6 inhibitor treatments in sarcoma is necessary to make any concrete decisions. While researchers continue to look for biomarkers that can predict sensitivity to CDK4/6 inhibitors in sarcomas, data from other tumor types may help guide these decisions. Recently published transcriptome profiling studies from CDK4/6 inhibitor–resistant and –sensitive breast cancer cells lines identified an Rb-loss signature RBsig that can discriminate between CDK4/6 inhibitor–resistant and –sensitive lines.109 Currently, no single biomarker can accurately predict CDK4/6 inhibitor sensitivity; however, gene signatures may provide insight into potential genetic profiles, specifically in sarcomas.
Precision Medicine: NCI-MATCH and TAPUR Studies
Pairing a patient's genomic profile with available treatments has recently been available from the National Cancer Institute's Molecular Analysis for Therapy Choice (NCI-MATCH) and ASCO's Targeted Agent and Profiling Utilization Registry (TAPUR).110 A TAPUR study of 29 patients with advanced non–small-cell lung cancer with CDKN2A loss or mutation and no RB mutations demonstrated antitumor effects from CDK4/6 inhibition. In a previous study including pancreatic adenocarcinoma and cholangiocarcinoma with CDKN2A loss or mutation, there was a lack of clinical activity from CDK4/6 inhibition, indicating that CDKN2A may not be a universal biomarker and should be tested in individual sarcoma subtypes.111,112 More information on cell cycle biomarkers for sarcoma subtypes will be collected from an ongoing phase II Pediatric MATCH trial of palbociclib in Rb-intact solid tumors with active mutations in cell cycle genes.113 A subgroup of this trial that focused on CCND1, 2, or 3 amplification had prolonged SD in 13% of patients but CCND1 or 3 amplification was not a predictor of palbociclib sensitivity.114
Combination Therapies and the Role of Treatment Sequencing
Several sarcoma subtypes have dual characteristic molecular alterations that may make them attractive targets for combination therapies. For example, LMS, angiosarcoma, and OS tend to exhibit activation of the mTOR pathway either through mTOR overexpression or direct loss of PTEN. In these sarcoma subtypes, mTOR or PI3K inhibitors may be logical synergistic partners alongside CDK4/6 inhibitors.102,115 In CDK4/6 inhibitor–resistant ES, which overexpresses IGF1R, a combination of CDK4/6 and IGF1R inhibitors was synergistic in vitro and in mouse models.116
Although PI3K or MEK inhibitors may be predicted to have synergy with combined CDK4/6 inhibition, other cotargets including Wee1 require further preclinical testing. Combination therapies with the CDK4 inhibitor, palbociclib, and MDM2 inhibitors have also been found to have both synergistic and antagonistic results in sarcomas. Reduced tumor growth and increased progression-free survival were evident in DDLPS when treated with palbociclib and MDM2 inhibitor, RG7388, but antagonistic effects were evident in myxofibrosarcoma and LMS cell lines.128 Similarly, in MDM2-amplified sarcomas, the MDM2-p53 binding inhibitor, nutlin, was antagonistic with palbociclib in preclinical sarcoma models.118
It should be noted that CDK4/6 inhibitors are frequently found to be antagonistic of drugs that require cells to enter the mitotic phase of the cell cycle to exert their effect. Nevertheless, the sequencing and timing of drug delivery can combat potential antagonism. Successful synergy was seen in combinations of CDK4/6 inhibitors and taxanes or microtubule stabilizers when taxanes were administered after CDK4/6 inhibitors.119 Another preclinical study suggested that Wee1 inhibitor, AZD1775, should only be treated after palbociclib treatment and a recovery period for cells to traverse through S phase.101 Although chemotherapies that target faster growing cells may be less likely to work when CDK4 is inhibited and slows cell growth, a study in ES and other nonsarcoma models had success when CDK4/6 inhibitors were used in combination with chemotherapy. Other in vitro and in vivo models in nonsarcoma tumors showed synergy between CDK4/6 inhibitors and chemotherapies using concurrent and sequential dosing schedules. CDK4/6 inhibitors’ ability to deregulate DNA repair, metabolism, and cell plasticity, and reduce thymidylate synthase, topoisomerase 1, and topoisomerase 2 alpha expression limit the dose required for chemotherapy efficacy and may enhance chemotherapy-induced apoptosis.115,116,120-126 Despite the various successful combination therapies and the fact that preclinical reports that described antagonistic relationships only recorded short-term effects, the potential for antagonism should not be discounted.119
Rational drug combination strategies in sarcoma will require a molecular understanding of both the interactions between genomic drivers of disease as well as the interactions between the drug combinations themselves.
In conclusion, CDK4/6 inhibitors represent a major breakthrough for targeted cancer treatment. CDK4/6 inhibitor use in sarcoma has led to limited, but significant, early clinical success. These therapies, as single agents, represent relatively well-tolerated therapies with the flexibility of oral administration, both of which are uncommon in these diseases. Current clinical data support the use of CDK4/6 inhibitors in subsets of sarcoma, which are primarily driven by CDK4/6 deregulation such as DDLPS and WDLPS. Alteration in the Rb-CDK4/6 pathway also serves a role as secondary drivers of sarcoma oncogenicity. Thus, combination therapy with CDK4/6 inhibition to target dual genomic derangements is attractive. Combination therapies with mTOR or PI3K inhibitors may provide promising response in subtypes with mTOR overexpression or PTEN loss such as LMS, angiosarcoma, and OS. However, further research should be conducted to determine the synergy of MDM2 inhibitors with CDK4 inhibitors. Overall, targeted future clinical research will be key to unlocking the potential of CDK4/6 inhibition in sarcoma.
APPENDIX 1. LITERATURE REVIEW
A systematic review was performed of articles indexed in the PubMed database and the National Institutes of Health Clinical Trials Registry (ClinicalTrials.gov) between January 1, 1996, and October 1, 2021. Search terms included sarcoma, CDK4/6 inhibitor, palbociclib, and biomarkers. Only articles published in English and articles with results were included. Gene expression of a sarcoma data set from 255 patients was obtained from The Cancer Genome Atlas.
APPENDIX 2. CDK4/6 INHIBITORS
CCND-dependent kinase activities of CDK4/6 are essential for progression through G1. CDK4/6-CCND phosphorylation of Rb and subsequent activation of E2F family of transcription factors then enhance transcription of CCNE. The CDK2/CCNE complex then starts a phosphorylation cascade that results in hyperphosphorylation and the degradation of Rb, which then allows transitioning through the G1/S checkpoint. The G1/S checkpoint represents the most important checkpoint for ensuring genome and cellular fidelity.3 Progression through this checkpoint requires adequate cell size, nutrients, and growth factors, in addition to a low threshold of DNA damage in the cell.54
CDK4/6 inhibitors are small molecules (approximately 500 Da) that were first approved in 2015. These agents were identified for their ability to selectively target and block CDK4/6-CCND activation compared with older, pan-CDK inhibitors.54-57 CDK4/6 inhibitors directly compete with CCND for binding to the ATP cleft of CDK4 and CDK6.58 By blocking the formation of the CDK4/6-CCND, the Rb tumor suppressor remains unphosphorylated and tightly bound to E2F1-3. This directly inhibits cell cycle progression by preventing E2F-mediated transcription of genes required for G1/S progression. Dysregulation of the Rb pathway and its role in genome integrity, cellular programming, and proliferation in sarcoma are key topics for continuing research.59
Current US Food and Drug Administration (FDA)-approved CDK4/6 inhibitors include palbociclib, ribociclib, and abemaciclib with an additional 15 new agents under development.60 Palbociclib (IBRANCE) was the first CDK4/6 inhibitor FDA-approved in 2015 for the treatment of hormone receptor (HR)-positive, human epidermal growth factor receptor 2 (HER2)-negative advanced or metastatic breast cancer.55 Ribociclib (KISQALI) and abemaciclib (VERZENIO) were both approved in 2017 for the treatment of HR-positive, HER2-negative advanced or metastatic breast cancer. Although palbociclib and ribociclib demonstrate significant selectivity for CDK4/6, abemaciclib has additional affinity for CDK9.56-61 Despite an increase in off-target activity associated with abemaciclib, preferential binding to CDK4 over CDK6 results in a lower prevalence of severe neutropenia with abemaciclib compared with either palbociclib or ribociclib.54-64
John L. Hays
Consulting or Advisory Role: AstraZeneca, Merck, Tesaro, Clovis Oncology, Deciphera, Ipsen
Travel, Accommodations, Expenses: Tesaro, Merck, AstraZeneca
James L. Chen
Consulting or Advisory Role: Syapse, Tempus
Speakers' Bureau: Foundation Medicine
Research Funding: Eisai
Patents, Royalties, Other Intellectual Property: MatchTX
No other potential conflicts of interest were reported.
Footnotes
J.Y.H. and N.D.S. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Jocelyn Y. Hsu, Nathan D. Seligson, John L. Hays, James L. Chen
Collection and assembly of data: Jocelyn Y. Hsu, Nathan D. Seligson, John L. Hays, James L. Chen
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
John L. Hays
Consulting or Advisory Role: AstraZeneca, Merck, Tesaro, Clovis Oncology, Deciphera, Ipsen
Travel, Accommodations, Expenses: Tesaro, Merck, AstraZeneca
James L. Chen
Consulting or Advisory Role: Syapse, Tempus
Speakers' Bureau: Foundation Medicine
Research Funding: Eisai
Patents, Royalties, Other Intellectual Property: MatchTX
No other potential conflicts of interest were reported.
REFERENCES
- 1.Bleloch JS, Ballim RD, Kimani S, et al. Managing sarcoma: Where have we come from and where are we going? Ther Adv Med Oncol 9637–6592017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katz D, Palmerini E, Pollack SM.More than 50 subtypes of soft tissue sarcoma: Paving the path for histology-driven treatments Am Soc Clin Oncol Ed Book 38925–9382018 [DOI] [PubMed] [Google Scholar]
- 3.Jardim DL, Millis SZ, Ross JS, et al. Cyclin pathway genomic alterations across 190,247 solid tumors: Leveraging large-scale data to inform therapeutic directions Oncologist 26e78–e892021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abeshouse A, Adebamowo C, Adebamowo SN, et al. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas Cell 171950–965.e282017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hensley ML, Chavan SS, Solit DB, et al. Genomic landscape of uterine sarcomas defined through prospective clinical sequencing Clin Cancer Res 263881–38882020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu Y, Xu J, Sun T.Cyclin-dependent kinases 4/6 inhibitors in breast cancer: Current status, resistance, and combination strategies J Cancer 105504–55172019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groisberg R, Hong DS, Holla V, et al. Clinical genomic profiling to identify actionable alterations for investigational therapies in patients with diverse sarcomas Oncotarget 839254–392672017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hay MA, Severson EA, Miller VA, et al. Identifying opportunities and challenges for patients with sarcoma as a result of comprehensive genomic profiling of sarcoma specimens JCO Precis Oncol 4176–1822020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty KT, Lorusso PM, Demichele A, et al. Phase I, dose-escalation trial of the oral cyclin-dependent kinase 4/6 inhibitor PD 0332991, administered using a 21-day schedule in patients with advanced cancer Clin Cancer Res 18568–5762012 [DOI] [PubMed] [Google Scholar]
- 10.Peguero J, Sohal DPS, O’Neil BH, et al. Tissue/site-agnostic study of ribociclib for tumors with cyclin D–CDK4/6 pathway genomic alterations: A phase II, open-label, single-arm basket study JCO Precis Oncol 31–102019 [DOI] [PubMed] [Google Scholar]
- 11.Loretan L, Moskovszky LE, Kurrer M, et al. Efficacy of a CDK4/6 inhibitor in a patient with breast cancer and liposarcoma: A case report and review of the literature Breast Care (Basel) 14325–3282019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cash T, Aguilera D, Macy ME, et al. Phase 1 study of abemaciclib in children with recurrent and refractory solid tumors including malignant brain tumors [abstract. Mol Cancer Ther. 2019;18 (suppl 12; abstr C002). Proceedings of the AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics, October 26-30, 2019, Boston, MA, Philadelphia, PA: AACR. [Google Scholar]
- 13.Infante JR, Cassier PA, Gerecitano JF, et al. A phase I study of the cyclin-dependent kinase 4/6 inhibitor ribociclib (LEE011) in patients with advanced solid tumors and lymphomas Clin Cancer Res 225696–57052016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Razak AA, Bauer S, Blay J-Y, et al. Results of a dose- and regimen-finding Phase Ib study of HDM201 in combination with ribociclib in patients with locally advanced or metastatic liposarcoma. Cancer Res. 2018;78 suppl; abstr CT009. [Google Scholar]
- 15. Dickson MA, Koff A, D'Angelo SP, et al. Phase 2 study of the CDK4 inhibitor abemaciclib in dedifferentiated liposarcoma. J Clin Oncol. 2019;37 suppl; abstr 11004. [Google Scholar]
- 16.Boddu S, Walko CM, Bienasz S, et al. Clinical utility of genomic profiling in the treatment of advanced sarcomas: A single-center experience JCO Precis Oncol 21–82018 [DOI] [PubMed] [Google Scholar]
- 17.Elvin JA, Gay LM, Ort R, et al. Clinical benefit in response to palbociclib treatment in refractory uterine leiomyosarcomas with a common CDKN2A alteration Oncologist 22416–4212017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toulmonde M, Blay JY, Bouche O, et al. Activity and safety of palbociclib in patients with advanced gastrointestinal stromal tumors refractory to imatinib and sunitinib: A biomarker-driven phase II study Clin Cancer Res 254611–46152019 [DOI] [PubMed] [Google Scholar]
- 19. Geoerger B, Bourdeaut F, Dubois SG, et al. Phase I study of Lee011 (Cdk4/6 inhibitor) in patients with malignant rhabdoid tumors, neuroblastoma, and cyclin D–Cdk4/6 pathway-activated tumors. Ann Oncol. 2014;25:iv151. [Google Scholar]
- 20.Lazow MA, Johnson SL, Johnson ND, et al. Genome-driven therapy for chemotherapy-resistant metastatic CDK6-amplified osteosarcoma JCO Precis Oncol 4498–5042020 [DOI] [PubMed] [Google Scholar]
- 21.Tramontana TF, Marshall MS, Helvie AE, et al. Sustained complete response to palbociclib in a refractory pediatric sarcoma with BCOR-CCNB3 fusion and germline CDKN2B variant JCO Precis Oncol 4466–4712020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan J, Simon R, Bittner M, et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays Cancer Res 585009–50131998 [PubMed] [Google Scholar]
- 23.Italiano A, Bianchini L, Gjernes E, et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas Clin Cancer Res 155696–57032009 [DOI] [PubMed] [Google Scholar]
- 24.Naka N, Tomita Y, Nakanishi H, et al. Mutations of p53 tumor-suppressor gene in angiosarcoma Int J Cancer 71952–9551997 [DOI] [PubMed] [Google Scholar]
- 25.Pappo AS, Dirksen U.Rhabdomyosarcoma, Ewing sarcoma, and other round cell sarcomas J Clin Oncol 36168–1792017 [DOI] [PubMed] [Google Scholar]
- 26.Schrage YM, Lam S, Jochemsen AG, et al. Central chondrosarcoma progression is associated with pRb pathway alterations: CDK4 down-regulation and p16 overexpression inhibit cell growth in vitro J Cell Mol Med 132843–28522009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speetjens FM, de Jong Y, Gelderblom H, et al. Molecular oncogenesis of chondrosarcoma: Impact for targeted treatment Curr Opin Oncol 28314–3222016 [DOI] [PubMed] [Google Scholar]
- 28. Kohlmeyer JL, Gordon DJ, Tanas MR, et al. CDKs in sarcoma: Mediators of disease and emerging therapeutic targets. Int J Mol Sci. 2020;21:3018. doi: 10.3390/ijms21083018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kansara M, Teng MW, Smyth MJ, et al. Translational biology of osteosarcoma Nat Rev Cancer 14722–7352014 [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Shen JK, Yu Z, et al. Expression and therapeutic implications of cyclin-dependent kinase 4 (CDK4) in osteosarcoma Biochim Biophys Acta Mol Basis Dis 18641573–15822018 [DOI] [PubMed] [Google Scholar]
- 32. Ogura K, Hosoda F, Arai Y, et al. Integrated genetic and epigenetic analysis of myxofibrosarcoma. Nat Commun. 2018;9:2765. doi: 10.1038/s41467-018-03891-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bill KLJ, Seligson ND, Hays JL, et al. Degree of MDM2 amplification affects clinical outcomes in dedifferentiated liposarcoma Oncologist 24989–9962019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Vita A, Mercatali L, Recine F, et al. Current classification, treatment options, and new perspectives in the management of adipocytic sarcomas Onco Targets Ther 96233–62462016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bui NQ, Przybyl J, Trabucco SE, et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res. 2019;9:12. doi: 10.1186/s13569-019-0122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuppens T, Moisse M, Depreeuw J, et al. Integrated genome analysis of uterine leiomyosarcoma to identify novel driver genes and targetable pathways Int J Cancer 1421230–12432018 [DOI] [PubMed] [Google Scholar]
- 37. Seligson ND, Awasthi A, Millis SZ, et al. Common secondary genomic variants associated with advanced epithelioid hemangioendothelioma. JAMA Netw Open. 2019;2:e1912416. doi: 10.1001/jamanetworkopen.2019.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toomey EC, Schiffman JD, Lessnick SL.Recent advances in the molecular pathogenesis of Ewing's sarcoma Oncogene 294504–45162010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heinrich MC, Patterson J, Beadling C, et al. Genomic aberrations in cell cycle genes predict progression of KIT-mutant gastrointestinal stromal tumors (GISTs) Clin Sarcoma Res. 2019;9:3. doi: 10.1186/s13569-019-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Du X, Lazar AJ, et al. Genetic aberrations of gastrointestinal stromal tumors Cancer 1131532–15432008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agaram NP, Zhang L, LeLoarer F, et al. Targeted exome sequencing profiles genetic alterations in leiomyosarcoma Genes Chromosomes Cancer 55124–1302016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dei Tos AP, Maestro R, Doglioni C, et al. Tumor suppressor genes and related molecules in leiomyosarcoma Am J Pathol 1481037–10451996 [PMC free article] [PubMed] [Google Scholar]
- 43. Brohl AS, Kahen E, Yoder SJ, et al. The genomic landscape of malignant peripheral nerve sheath tumors: Diverse drivers of Ras pathway activation. Sci Rep. 2017;7:14992. doi: 10.1038/s41598-017-15183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kourea HP, Cordon-Cardo C, Dudas M, et al. Expression of p27(kip) and other cell cycle regulators in malignant peripheral nerve sheath tumors and neurofibromas: The emerging role of p27(kip) in malignant transformation of neurofibromas Am J Pathol 1551885–18911999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krawczyk MA, Karpinsky G, Izycka-Swieszewska E, et al. Immunohistochemical assessment of cyclin D1 and p53 is associated with survival in childhood malignant peripheral nerve sheath tumor Cancer Biomark 24351–3612019 [DOI] [PubMed] [Google Scholar]
- 46.Wang T, Goodman MA, McGough RL, et al. Immunohistochemical analysis of expressions of RB1, CDK4, HSP90, cPLA2G4A, and CHMP2B is helpful in distinction between myxofibrosarcoma and myxoid liposarcoma Int J Surg Pathol 22589–5992014 [DOI] [PubMed] [Google Scholar]
- 47.Olofsson A, Willén H, Göransson M, et al. Abnormal expression of cell cycle regulators in FUS-CHOP carrying liposarcomas Int J Oncol 251349–13552004 [PubMed] [Google Scholar]
- 48.Weidema ME, Versleijen-Jonkers YMH, Flucke UE, et al. Targeting angiosarcomas of the soft tissues: A challenging effort in a heterogeneous and rare disease Crit Rev Oncol Hematol 138120–1312019 [DOI] [PubMed] [Google Scholar]
- 49.Fritz B, Schubert F, Wrobel G, et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma Cancer Res 622993–29982002 [PubMed] [Google Scholar]
- 50.Gardner JM, Dandekar M, Thomas D, et al. Cutaneous and subcutaneous pleomorphic liposarcoma: A clinicopathologic study of 29 cases with evaluation of MDM2 gene amplification in 26 Am J Surg Pathol 361047–10512012 [DOI] [PubMed] [Google Scholar]
- 51.Hofvander J, Jo VY, Ghanei I, et al. Comprehensive genetic analysis of a paediatric pleomorphic myxoid liposarcoma reveals near-haploidization and loss of the RB1 gene Histopathology 69141–1472016 [DOI] [PubMed] [Google Scholar]
- 52.Pérot G, Chibon F, Montero A, et al. Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics Am J Pathol 1772080–20902010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid AH, Tsai MM, Venzon DJ, et al. MDM2 amplification, P53 mutation, and accumulation of the P53 gene product in malignant fibrous histiocytoma Diagn Mol Pathol 565–731996 [DOI] [PubMed] [Google Scholar]
- 54.Marra A, Curigliano G.Are all cyclin-dependent kinases 4/6 inhibitors created equal? NPJ Breast Cancer 51–92019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibrance (palbociclib) package insert. New York, NY: Pfizer; 2019. [Google Scholar]
- 56.Kisqali (ribociclib) package insert. East Hanover, NJ: Novartis Pharmaceuticals; 2019. [Google Scholar]
- 57.Verzenio (abemaciclib) package insert. Indianapolis, IN: Eli Lilly; 2019. [Google Scholar]
- 58.Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy Nat Rev Drug Discov 14130–1462015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spring L, Bardia A, Modi S.Targeting the cyclin D-cyclin-dependent kinase (CDK) 4/6-retinoblastoma pathway with selective CDK 4/6 inhibitors in hormone receptor-positive breast cancer: Rationale, current status, and future directions Discov Med 2165–742016 [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan K, Wang X, Dong H, et al. Selective inhibition of CDK4/6: A safe and effective strategy for developing anticancer drugs Acta Pharmaceutica Sinica B 1130–542020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.https://oncpracticemanagement.com/videos/187-what-differentiates-abemaciclib-ribociclib-and-palbociclib Rugo HS: What Differentiates Abemaciclib, Ribociclib, and Palbociclib? [Google Scholar]
- 62.Kim S, Tiedt R, Loo A, et al. The potent and selective cyclin-dependent kinases 4 and 6 inhibitor ribociclib (LEE011) is a versatile combination partner in preclinical cancer models Oncotarget 935226–352402018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie N, Qin T, Ren W, et al. Efficacy and safety of cyclin-dependent kinases 4 and 6 inhibitors in HR+/HER2- advanced breast cancer Cancer Manag Res 124241–42502020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickler MN, Tolaney SM, Rugo HS, et al. MONARCH 1, A phase II study of abemaciclib, a CDK4 and CDK6 inhibitor, as a single agent, in patients with refractory HR+/HER2− metastatic breast cancer Clin Cancer Res 235218–52242017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crago AM, Dickson MA.Liposarcoma: Multimodality management and future targeted therapies Surg Oncol Clin N Am 25761–7732016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang L, Chen S, Luo P, et al. Liposarcoma: Advances in cellular and molecular genetics alterations and corresponding clinical treatment J Cancer 11100–1072020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang YX, Sicinska E, Czaplinski JT, et al. Antiproliferative effects of CDK4/6 inhibition in CDK4-amplified human liposarcoma in vitro and in vivo Mol Cancer Ther 132184–21932014 [DOI] [PubMed] [Google Scholar]
- 68.Dickson MA, Schwartz GK, Keohan ML, et al. Progression-free survival among patients with well-differentiated or dedifferentiated liposarcoma treated with CDK4 inhibitor palbociclib: A phase 2 clinical trial JAMA Oncol 2937–9402016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dickson MA, Tap WD, Keohan ML, et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma J Clin Oncol 312024–20282013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.von Mehren M, et al. Soft tissue sarcoma, version 2.2018, NCCN Clinical Practice Guidelines in OncologyJNCCN 16536–5632018 [DOI] [PubMed] [Google Scholar]
- 71.Knudsen ES, Witkiewicz AK.The strange case of CDK4/6 inhibitors: Mechanisms, resistance, and combination strategies Trends Cancer 339–552017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez M, Muñoz-Galván S, Jiménez-García MP, et al. Efficacy of CDK4 inhibition against sarcomas depends on their levels of CDK4 and p16ink4 mRNA Oncotarget 640557–405742015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gladdy RA, Qin LX, Moraco N, et al. Predictors of survival and recurrence in primary leiomyosarcoma Ann Surg Oncol 201851–18572013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo X, Jo VY, Mills AM, et al. Clinically relevant molecular subtypes in leiomyosarcoma Clin Cancer Res 213501–35112015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seligson ND, Kautto EA, Passen EN, et al. BRCA1/2 functional loss defines a targetable subset in leiomyosarcoma Oncologist 24973–9792019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Böhm MJ, Marienfeld R, Jäger D, et al. Analysis of the CDK4/6 cell cycle pathway in leiomyosarcomas as a potential target for inhibition by palbociclib. Sarcoma. 2019;2019:3914232. doi: 10.1155/2019/3914232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaefer IM, Hornick JL, Sholl LM, et al. Abnormal p53 and p16 staining patterns distinguish uterine leiomyosarcoma from inflammatory myofibroblastic tumour Histopathology 701138–11462017 [DOI] [PubMed] [Google Scholar]
- 78. Movva S, von Mehren M, Handorf EA, et al. SAR-096: A phase II trial of ribociclib in combination with everolimus in advanced dedifferentiated liposarcoma (DDL), and leiomyosarcoma (LMS) J Clin Oncol. 2020;38 doi: 10.1158/1078-0432.CCR-23-2469. suppl; abstr 11544. [DOI] [PubMed] [Google Scholar]
- 79.Dickson MA.Molecular pathways: CDK4 inhibitors for cancer therapy Clin Cancer Res 203379–33832014 [DOI] [PubMed] [Google Scholar]
- 80.Koumarianou A, Economopoulou P, Katsaounis P, et al. Gastrointestinal stromal tumors (GIST): A prospective analysis and an update on biomarkers and current treatment concepts Biomark Cancer 71–72015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmieder M, Wolf S, Danner B, et al. p16 expression differentiates high-risk gastrointestinal stromal tumor and predicts poor outcome Neoplasia 101154–11622008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J, Yang Z, Li Y, et al. Cell apoptosis, autophagy and necroptosis in osteosarcoma treatment Oncotarget 744763–447782016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nouri H, Ben Maitigue M, Abid L, et al. Surface osteosarcoma: Clinical features and therapeutic implications J Bone Oncol 4115–1232015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen PC, Yen CC, Hung GY, et al. Gene amplification and tumor grading in parosteal osteosarcoma J Chin Med Assoc 82889–8942019 [DOI] [PubMed] [Google Scholar]
- 85.Parham DM, Barr FG.Classification of rhabdomyosarcoma and its molecular basis Adv Anat Pathol 20387–3972013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barr FG, Duan F, Smith LM, et al. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: A report from the Children's Oncology Group Genes Chromosomes Cancer 48661–6722009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arnold MA, Barr FG.Molecular diagnostics in the management of rhabdomyosarcoma Expert Rev Mol Diagn 17189–1942017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olanich ME, Sun W, Hewitt SM, et al. CDK4 amplification reduces sensitivity to CDK4/6 inhibition in fusion-positive rhabdomyosarcoma Clin Cancer Res 214947–49592015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nguyen TH, Barr FG. Therapeutic approaches targeting PAX3-FOXO1 and its regulatory and transcriptional pathways in rhabdomyosarcoma. Molecules. 2018;23:2798. doi: 10.3390/molecules23112798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Milewski D, Pradhan A, Wang X, et al. FoxF1 and FoxF2 transcription factors synergistically promote Rhabdomyosarcoma carcinogenesis by repressing transcription of p21Cip1 CDK inhibitor Oncogene 36850–8622017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dauphinot L, De Oliveira C, Melot T, et al. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression Oncogene 203258–32652001 [DOI] [PubMed] [Google Scholar]
- 92.Bridge JA, Sumegi J, Druta M, et al. Clinical, pathological, and genomic features of EWSR1-PATZ1 fusion sarcoma Mod Pathol 321593–16042019 [DOI] [PubMed] [Google Scholar]
- 93. Cidre-Aranaz F, Alonso J. EWS/FLI1 target genes and therapeutic opportunities in Ewing sarcoma. Front Oncol. 2015;5:162. doi: 10.3389/fonc.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lerman DM, Monument MJ, McIlvaine E, et al. Tumoral TP53 and/or CDKN2A alterations are not reliable prognostic biomarkers in patients with localized Ewing sarcoma: A report from the Children's Oncology Group Pediatr Blood Cancer 62759–7652015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Grünewald TGP, Cidre-Aranaz F, Surdez D, et al. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5. doi: 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 96.Kao YC, Owosho AA, Sung YS, et al. BCOR-CCNB3 fusion positive sarcomas: A clinicopathologic and molecular analysis of 36 cases with comparison to morphologic spectrum and clinical behavior of other round cell sarcomas Am J Surg Pathol 42604–6152018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin DI, Hemmerich A, Edgerly C, et al. Genomic profiling of BCOR-rearranged uterine sarcomas reveals novel gene fusion partners, frequent CDK4 amplification and CDKN2A loss Gynecol Oncol 157357–3662020 [DOI] [PubMed] [Google Scholar]
- 98. Schettini F, De Santo I, Rea CG, et al. CDK 4/6 inhibitors as single agent in advanced solid tumors. Front Oncol. 2018;8:608. doi: 10.3389/fonc.2018.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Subramaniam MM, Noguera R, Piqueras M, et al. p16INK4A (CDKN2A) gene deletion is a frequent genetic event in synovial sarcomas Am J Clin Pathol 126866–8742006 [DOI] [PubMed] [Google Scholar]
- 100. Ouyang Z, Wang S, Zeng M, et al. Therapeutic effect of palbociclib in chondrosarcoma: Implication of cyclin-dependent kinase 4 as a potential target. Cell Commun Signal. 2019;17:17. doi: 10.1186/s12964-019-0327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Francis AM, Alexander A, Liu Y, et al. CDK4/6 inhibitors sensitize Rb-positive sarcoma cells to Wee1 kinase inhibition through reversible cell-cycle arrest Mol Cancer Ther 161751–17642017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cortés J, Im SA, Holgado E, et al. The next era of treatment for hormone receptor-positive, HER2-negative advanced breast cancer: Triplet combination-based endocrine therapies Cancer Treat Rev 6153–602017 [DOI] [PubMed] [Google Scholar]
- 103.Gamberi G, Ragazzini P, Benassi MS, et al. Analysis of 12q13-15 genes in parosteal osteosarcoma Clin Orthop Relat Res 195–2042000 [DOI] [PubMed] [Google Scholar]
- 104.Helbig D, Ihle MA, Pütz K, et al. Oncogene and therapeutic target analyses in atypical fibroxanthomas and pleomorphic dermal sarcomas Oncotarget 721763–217742016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ragazzini P, Gamberi G, Pazzaglia L, et al. Amplification of CDK4, MDM2, SAS and GLI genes in leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma Histol Histopathol 19401–4112004 [DOI] [PubMed] [Google Scholar]
- 106.Grupo Espanol de Investigacion en Sarcomas . Phase II Multicenter Trial of Palbociclib in Second Line of Advanced Sarcomas With CDK4 Overexpression. 2020. https://clinicaltrials.gov/ct2/show/NCT03242382 [Google Scholar]
- 107.Charlson J. Abemaciclib for Treatment of Advanced Bone and Soft Tissue Sarcoma Identified as Having CDK Pathway Alteration. 2020. https://clinicaltrials.gov/ct2/show/NCT04040205 [Google Scholar]
- 108.Institut National de la Santé Et de la Recherche Médicale, France . Molecular Profiling of Advanced Soft-Tissue Sarcomas. A Phase III Study. 2020. https://clinicaltrials.gov/ct2/show/NCT03784014 [Google Scholar]
- 109.Malorni L, Piazza S, Ciani Y, et al. A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer Oncotarget 768012–680222016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Andrews A. ASCO and NCI launch largest precision medicine trials using real-world evidence. Am Health Drug Benefits. 2015;8:37. [PMC free article] [PubMed] [Google Scholar]
- 111.Al Baghdadi T, Halabi S, Garrett-Mayer E, et al. Palbociclib in patients with pancreatic and biliary cancer with CDKN2A alterations: Results from the targeted agent and profiling utilization registry study JCO Precis Oncol 31–82019 [DOI] [PubMed] [Google Scholar]
- 112.Ahn ER, Mangat PK, Garrett-Mayer E, et al. Palbociclib in patients with non–small-cell lung cancer with CDKN2A alterations: Results from the targeted agent and profiling utilization registry study JCO Precis Oncol 4757–7662020 [DOI] [PubMed] [Google Scholar]
- 113.National Cancer Institute (NCI) Molecular Analysis for Therapy Choice (MATCH) 2021. https://clinicaltrials.gov/ct2/show/NCT02465060 [Google Scholar]
- 114.Clark AS, Hong F, Finn RS, et al. Molecular analysis for therapy choice (NCI-MATCH, EAY131) arm Z1B: Phase II trial of palbociclib for CCND1, 2 or 3 amplified tumors. American Association for Cancer Research annual meeting, Atlanta, GA, March 29–April 3, 2019 (abstr LB-010)
- 115.Dillon LM, Miller TW.Therapeutic targeting of cancers with loss of PTEN function Curr Drug Targets 1565–792014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guenther LM, Dharia NV, Ross L, et al. A combination CDK4/6 and IGF1R inhibitor strategy for Ewing sarcoma Clin Cancer Res 251343–13572019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Laroche-Clary A, Chaire V, Algeo MP, et al. Combined targeting of MDM2 and CDK4 is synergistic in dedifferentiated liposarcomas. J Hematol Oncol. 2017;10:123. doi: 10.1186/s13045-017-0482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sriraman A, Dickmanns A, Najafova Z, et al. CDK4 inhibition diminishes p53 activation by MDM2 antagonists. Cell Death Dis. 2018;9:918. doi: 10.1038/s41419-018-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fassl A, Sicinski P.Chemotherapy and CDK4/6 inhibition in cancer treatment: Timing is everything Cancer Cell 37265–2672020 [DOI] [PubMed] [Google Scholar]
- 120.Dean JL, McClendon AK, Knudsen ES.Modification of the DNA damage response by therapeutic CDK4/6 inhibition J Biol Chem 28729075–290872012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hamilton G, Klameth L, Rath B, et al. Synergism of cyclin-dependent kinase inhibitors with camptothecin derivatives in small cell lung cancer cell lines Molecules 192077–20882014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gao Y, Shen J, Choy E, et al. Inhibition of CDK4 sensitizes multidrug resistant ovarian cancer cells to paclitaxel by increasing apoptosiss Cell Oncol (Dordr) 40209–2182017 [DOI] [PubMed] [Google Scholar]
- 123.Roberts PJ, Kumarasamy V, Witkiewicz AK, et al. Chemotherapy and CDK4/6 inhibitors: Unexpected bedfellows Mol Cancer Ther 191575–15882020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang X, Di Liberto M, Jayabalan D, et al. Prolonged early G(1) arrest by selective CDK4/CDK6 inhibition sensitizes myeloma cells to cytotoxic killing through cell cycle-coupled loss of IRF4 Blood 1201095–11062012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang XH, Cheng Y, Shin JY, et al. A CDK4/6 inhibitor enhances cytotoxicity of paclitaxel in lung adenocarcinoma cells harboring mutant KRAS as well as wild-type KRAS Cancer Biol Ther 14597–6052013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Min A, Kim JE, Kim YJ, et al. Cyclin E overexpression confers resistance to the CDK4/6 specific inhibitor palbociclib in gastric cancer cells Cancer Lett 430123–1322018 [DOI] [PubMed] [Google Scholar]
- 127.Bovée JV, Cleton-Jansen AM, Rosenberg C, et al. Molecular genetic characterization of both components of a dedifferentiated chondrosarcoma, with implications for its histogenesis J Pathol 189454–4621999 [DOI] [PubMed] [Google Scholar]
- 128.Singer S, Socci ND, Ambrosini G, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma Cancer Res 676626–66362007 [DOI] [PubMed] [Google Scholar]
- 129. Roszik J, et al. Unique aberrations in intimal sarcoma identified by Next-Generation Sequencing as potential therapy targets. Cancers (Basel) 2019;11:1283. doi: 10.3390/cancers11091283. [DOI] [PMC free article] [PubMed] [Google Scholar]