Abstract
Breakthrough SARS-CoV-2 infections in fully vaccinated individuals are considered a consequence of waning immunity. Serum antibodies represent the most measurable outcome of vaccine-induced B cell memory. When antibodies decline, memory B cells are expected to persist and perform their function, preventing clinical disease. We investigated whether BNT162b2 mRNA vaccine induces durable and functional B cell memory in vivo against SARS-CoV-2 3, 6, and 9 months after the second dose in a cohort of health care workers (HCWs). While we observed physiological decline of SARS-CoV-2-specific antibodies, memory B cells persist and increase until 9 months after immunization. HCWs with breakthrough infections had no signs of waning immunity. In 3–4 days, memory B cells responded to SARS-CoV-2 infection by producing high levels of specific antibodies in the serum and anti-Spike IgA in the saliva. Antibodies to the viral nucleoprotein were produced with the slow kinetics typical of the response to a novel antigen.
Keywords: SARS-CoV-2, breakthrough infections, memory B cells, COVID-19, waning immunity, salivary IgA, mucosal immunity, mRNA vaccine
Graphical abstract
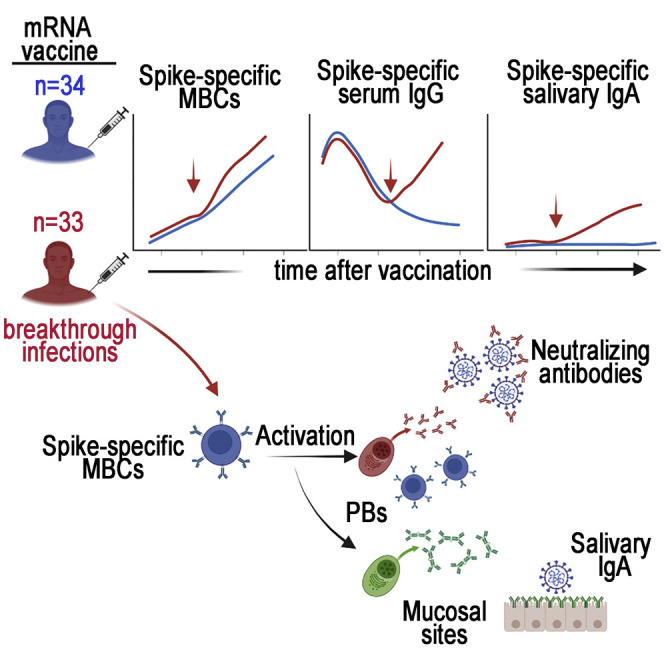
Terreri, Piano Mortari, and colleagues show that memory B cells persist and increase months after SARS-CoV-2 vaccination even if specific antibodies physiologically decline. Health care workers with breakthrough infections had no signs of waning immunity, as memory B cells produce high levels of specific antibodies in the serum and saliva.
Introduction
Breakthrough infections in individuals fully immunized against SARS-CoV-2 with the BNT162b2 mRNA vaccine continue to increase worldwide (Barda et al., 2021; Bergwerk et al., 2021). In most individuals with a positive nasopharyngeal swab (NPS), SARS-CoV-2 infection is clinically a- or pauci-symptomatic, demonstrating that vaccination protects against severe disease but is less effective in preventing contagion (Bergwerk et al., 2021). The epidemiological situation is complicated by the rapid surge and prevalence of the highly infective SARS-CoV-2 Delta variant (Monto, 2021) at a time when the increased vaccination rates have led to the removal of most restrictions, including the extensive use of masks and social distancing.
The occurrence of breakthrough infections has been correlated with the progressive decline of specific and neutralizing antibodies in the serum months after vaccination (Levin et al., 2021; Goldberg et al., 2021), leading to the conclusion that immunity induced by vaccination is waning. Consequently, the administration of a third vaccine dose has been recommended, starting from high-risk groups.
Antibodies in the serum are a reliable biological readout of vaccine efficacy because they demonstrate the successful response of the immune system, but they do not represent our main or only protection against infection (Goel et al., 2021a). Memory B and T cells, produced in response to immunization, are indispensable for protection because they migrate to the site of viral entry (Allie et al., 2019; Palm and Henry, 2019). Here, where the immune defenses are mostly needed, memory B cells (MBCs) secrete high amounts of neutralizing antibodies and memory T cells kill infected cells, thus preventing viral multiplication and spread. Whereas antibodies physiologically decline after every vaccination, B and T memory cells persist and perform their function, resulting in the prevention of clinical disease (Pollard and Bijker, 2021). In unvaccinated patients infected by SARS-CoV-2, innate immunity and T cell responses play a major defensive role, and B cell function is of minor importance (Carsetti et al., 2020). In contrast, B cells are most relevant for prevention of infection after vaccination, thanks to the production of neutralizing antibodies able to block the binding between the viral Spike protein and its ACE-2 receptor (Wang et al., 2021b; Goel et al., 2021b).
Our study addressed two equally important aims. We first investigated whether the BNT162b2 mRNA vaccine induces durable B cell memory by measuring the frequency of specific MBCs 3, 6, and 9 months after a two-dose vaccine cycle. The second aim was to prove the performance of vaccine-induced MBCs. We measured the humoral and B cellular response of fully vaccinated health care workers (HCWs) who had SARS-CoV-2 breakthrough infections 3 and 6 months after the second dose. We also investigated the mucosal response in vaccinated health care workers (HCWs) with a positive NPS. All the results were compared to those obtained in vaccinated HCWs never infected by SARS-CoV-2.
Results
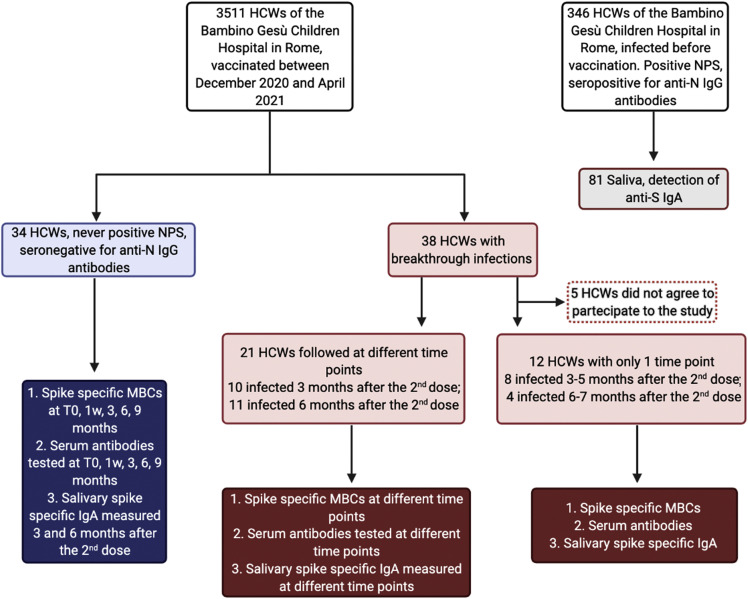
Demographic and clinical characteristic of study participants are reported in Table 1 , and Figure 1 describes the experimental plan.
Table 1.
Demographic and clinical characteristics of vaccinated HCWs without and with breakthrough infections
| Age (years) | Control Group∗ n = 34 46.3 (12.15) | Breakthrough Infections∗, ∗∗ n = 33 39.9 (11.3) |
|---|---|---|
| Sex | ||
| Female | 26 (77%) | 23 (70%) |
| Male | 8 (23%) | 10 (30%) |
| Hospitalization status | ||
| Never hospitalized | 34 (100%) | 33 (100%) |
| Hospitalized | n/a | 0 (0%) |
| Unknown if hospitalized | n/a | 0 (0%) |
| Sample Collection Date | December 2020-September 2021 | April 2021-September 2021 |
| SARS-CoV-2 PCR Positivity | ||
| Positive | 0 (0%) | 33 (100%) |
| Negative | 34 (100%) | 0 (0%) |
| Peak Disease Severity (Female [F], Male [M]) | ||
| Asymptomatic | n/a | 24 (73%) (16 [F], 8 [M]) |
| Mild (Non-hospitalized) | n/a | 9 (27%) (7 [F], 2 [M]) |
| Moderate (Hospitalized) | n/a | 0 (0%) |
| Severe (Hospitalized) | n/a | 0 (0%) |
| Unknown | n/a | 0 (0%) |
| SARS-CoV-2 clade∗∗∗ | ||
| Alpha | n/a | 4 (12%) |
| Gamma | n/a | 3 (14%) |
| Delta | n/a | 20 (61%) |
analyzed before vaccination and 1 week, 3, 6, and 9 months after the second dose
for 21, multiple samples were analyzed; from 12 cases, only one sample was available
information available for 27 patients with a nasopharyngeal viral load sufficient for genome sequencing
Figure 1.
Flowchart showing the experimental plan
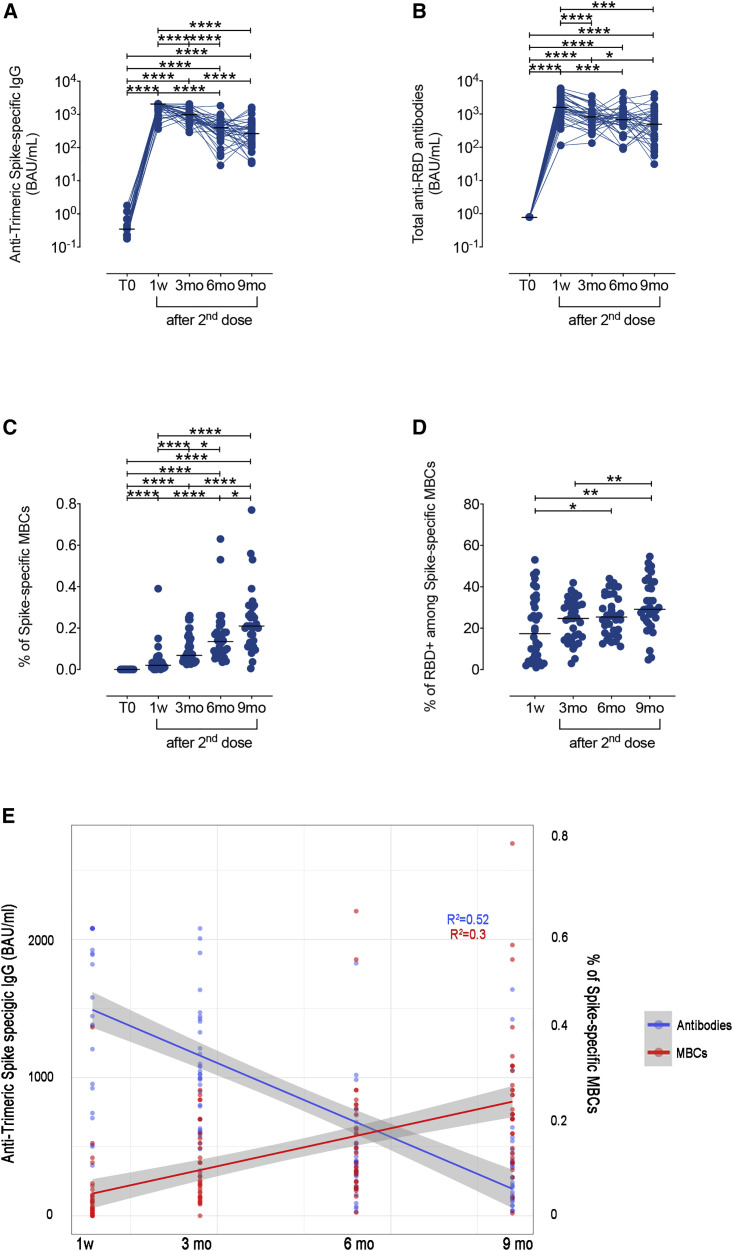
We first evaluated the duration of B cell memory after a complete vaccine cycle (two doses, 21 days apart), in fully vaccinated HCWs (n = 34) who never had a positive NPS. The concentration of serum anti-Trimeric Spike IgG and total anti-RBD antibodies at different time points is shown in Figure 2 . As reported before (Piano Mortari et al., 2021), serum anti-Trimeric Spike and total anti-RBD antibodies reached the highest levels 7 days after the second dose (Figures 2A and 2B; 1w). Anti-Trimeric Spike IgG progressively and significantly declined 3 and 6 months after vaccination (Figures 2A and S1A). Between 6 and 9 months, the reduction of anti-Trimeric Spike-specific IgG serum concentrations (Figures 2A and S1A) was not significant. Anti-RBD antibodies declined with similar kinetics between 1 week and 3 months after the second dose. Afterward, the progressive loss of anti-RBD antibodies was significant only between 3 and 9 months (Figures 2B and S1B).
Figure 2.
SARS-CoV-2 antibody responses and specific memory B cells in vaccinated HCWs
(A) Anti-Trimeric Spike-specific IgG, (B) total anti-RBD antibody levels, (C) percentage of Spike-specific MBCs, and (D) frequency of RBD-specific cells, identified inside the total Spike-positive MBCs, measured at different time points in vaccinated HCWs: before vaccine administration (T0), 7 days (1w), 3 (3mo), 6 (6mo), and 9 (9mo) months after the second dose (n = 34). (E) Simple linear regression models for Spike-specific MBCs and time (red line), and for anti-Trimeric Spike-specific IgG and time (blue line). Antibody titer is shown on a log10 scale. Medians are indicated, and statistical significance was determined using Wilcoxon matched pairs signed rank test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
The frequency of Spike-specific MBCs in the peripheral blood was also measured. MBCs were identified as CD19+CD24+CD27+, as shown in the gating strategy in Figure S2. Spike-specific MBCs that were absent before vaccination (T0) and significantly increased 7 days after the second dose (1w) (Piano Mortari et al., 2021) continued to significantly expand at each time point. At 3 months, the median value of Spike-specific MBCs was 0.07% (IQR 0.04–0.15) and 0.14% at 6 months (IQR 0.08–0.19) (Figure 2C). Finally, 9 months after vaccination, they further increased and were 0.21% (IQR 0.13–0.3) of all MBCs. The progressive increase of Spike-specific B cells was also detected in the total B cell population (Figure S3). At each time point after vaccination, about 20%–25% of the Spike-specific MBCs were directed to RBD (Figure 2D).
The changes in time of the frequency of Spike-specific MBCs and anti-Trimeric Spike IgG are represented in Figure 2E by the simple linear regression model for Spike-specific MBCs and time (red line) and for anti-Trimeric Spike-specific IgG and time (blue line). For Spike-specific MBCs, this model represents 30% of the variance present in the data, and time has a significant (p < 0.001) positive effect on the percentage of circulating Spike-specific MBCs. For Trimeric Spike-specific IgG, the model represents 52% of the variance present in the data, and time has a significant (p < 0.001) negative effect on the amount of antibodies.
Thus, in contrast to the decline of serum antibodies, MBCs specific for recombinant Spike and RBD persist and continue to increase for at least 9 months after the second vaccine dose.
We then asked the question of whether breakthrough infections were associated with waning immunity. The 33 HCWs with SARS-CoV-2 breakthrough infections who agreed to participate to the study were identified by the SARS-CoV-2 NPS performed either because of the presence of at least one symptom (8/33) or because of a reported contact (25/33) with an infected individual (mostly a family member); all of them were a- or pauci-symptomatic and none were hospitalized. Eighteen SARS-CoV-2 breakthrough infections occurred around 3 months after the second dose, and 15 were diagnosed after 6 months. Breakthrough infections were caused by the Alpha (12%), Gamma (9%), or Delta (61%) variants. In 6 cases (18%), the identification of the variant was impossible due to the low viral load.
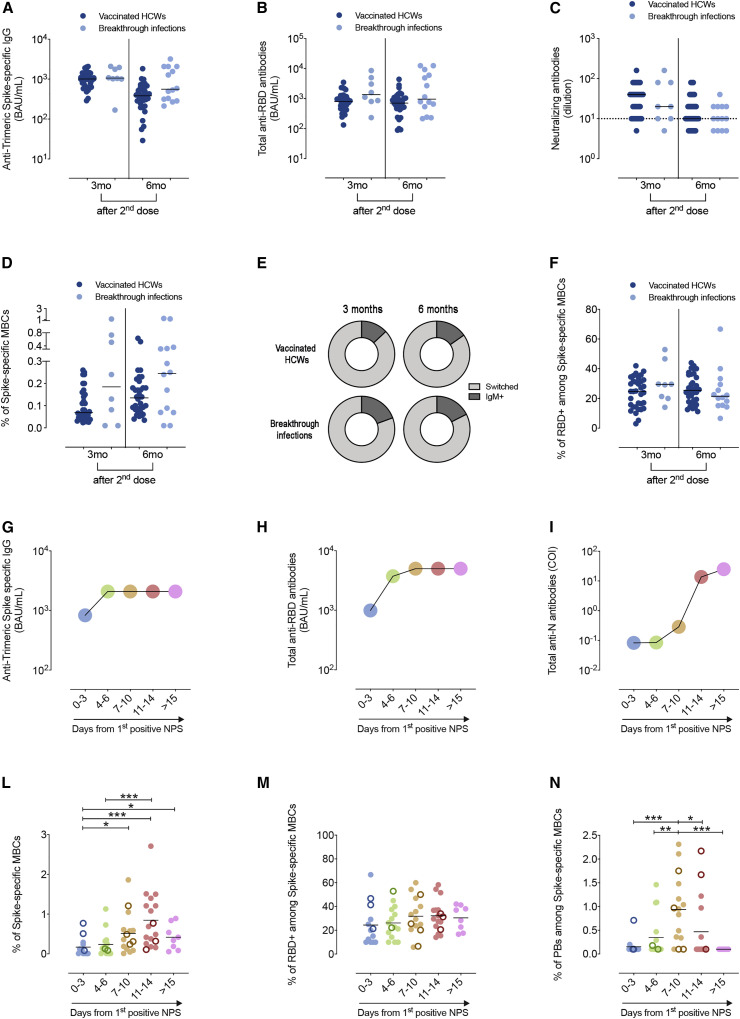
Figure 3 shows the levels of specific antibodies and MBCs in HCWs with breakthrough infections on the day of the first positive NPS compared to fully vaccinated HCWs matched for sex, age, and interval from vaccination. There was no difference in the level of antibodies specific for Trimeric Spike and RBD and in the titer of neutralizing antibodies (Figures 3A–3C). Spike-specific MBCs were not reduced in HCWs who had breakthrough infections (Figure 3D). Among Spike-specific MBCs, we determined the percentage of IgM+ and switched cells 3 and 6 months in HCWs with or without breakthrough infections after the first positive NPS (Figure 3E). There was no significant difference between the two groups analyzed. The fraction of Spike-specific MBCs able to bind the viral RBD was also similar in fully vaccinated HCWs with or without breakthrough infection (Figure 3F).
Figure 3.
Levels of specific antibodies and frequency of Spike-specific memory B cells in vaccinated HCWs who never had a positive NPS and in HCWs with breakthrough infections
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.chom.2022.01.003.
(A) Anti-Trimeric Spike-specific IgG, (B) total anti-RBD antibodies, and (C) neutralizing antibodies. The values measured in SARS-CoV-2-negative HCWs 3 and 6 months after vaccination (n = 34) are compared to those of HCWs with breakthrough infections that occurred either 3 (3mo) (n = 8) or 6 (6mo) (n = 14) months after the second dose. (D) Plot depicts the percentage of Spike-specific MBCs in vaccinees with and without breakthrough infections. (E) Pie charts show the distribution of switched and IgM+ Spike-specific MBCs in fully vaccinated HCWs without and with breakthrough infections 3 and 6 months after vaccination. In (F), the percentage of RBD+ cells among Spike-specific MBCs in the two groups of HCWs is shown. Medians of the antibody values at different time points showing the kinetics of anti-Trimeric Spike-specific IgG (G), anti-RBD Ig (H), and anti-N Ig (I) are shown. (L) Percentage of Spike-specific MBCs, (M) RBD-specific MBCs, identified among Spike-specific MBCs, and (N) Spike-specific plasmablasts in the peripheral blood of vaccinated HCWs with breakthrough infections (n = 33). Samples were analyzed after the first positive NPS at the following time points: 0–3 days, 4–6 days, 7–10 days,11–14 days, and > 15days after the first positive NPS. Empty circles represent HCWs for which only one sample was collected at a known time point after the first positive NPS (n = 12). Antibody titer is shown on a log10 scale. Neutralizing antibodies are expressed as the reciprocal of the highest serum dilution inhibiting at least 90% of virus-induced cytopathic effect (MNA90) and values ≥10 were considered positive. Dashed line indicates the cut-off of the test (MNA90 < 10). Medians are indicated, and statistical significance was determined using unpaired Mann-Whitney t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We followed 21 HCWs for about 20 days after the first positive swab, whereas for 12 HCWs only one sample was obtained at a known time point after the first positive NPS.
In the serum, anti-Trimeric Spike and anti-RBD antibodies were already detectable 0–3 days from the first positive NPS because of the previous vaccination, immediately increased 4–6 days after, and reached the maximal level measurable by the standard assays performed in our Institute at 7 to 10 days after the first NPS (Figures 3G, 3H, S4A, and S4B). In contrast, anti-nucleoprotein (N) antibodies, which were absent at 0–3 days, became detectable in 4 cases 7–10 days after the first NPS, significantly increased at 11–14 days following the positive NPS, and were present in all individuals but one 15 days after the first positive NPS; this reflects the kinetics of the immune response to a novel antigen, which typically requires 2–3 weeks for the generation of specific antibodies (Figures 3I and S4C).
The rapid production of specific antibodies may be explained by the function of the already established population of MBCs that differentiate into antibody-producing cells once that virus has invaded the oropharynx.
In the peripheral blood, the frequency of Spike-specific MBCs started to increase at 4–6 days after the first positive NPS and became maximal 11–14 days after the first positive NPS (Figure 3L). Among MBCs, the RBD-specific fraction was always represented at a constant frequency of roughly 20%–25% (Figures 3M and S2 for the identification of the cell populations). Circulating Spike-specific antibody producing cells (plasmablasts) also increased with similar kinetics (Figures 3N and S2 for the identification of plasmablasts).
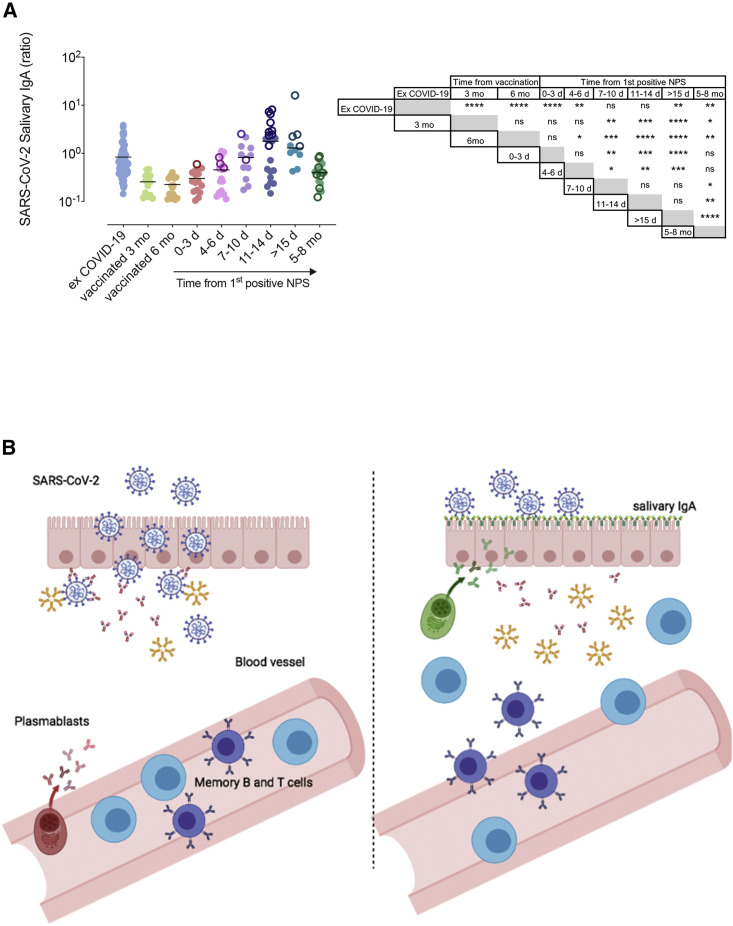
Salivary IgA is considered a reliable proxy of mucosal immunity (Brandtzaeg, 2013), and Spike-specific IgA has been detected in patients with COVID-19 (Sterlin et al., 2021) in response to the infection of the nasopharyngeal epithelium. Spike-specific salivary IgA was very low in the majority of vaccinated HCWs. In contrast, IgA against the Spike protein was still present in the saliva of individuals who had COVID-19 months before our study (Figure 4 A). Most importantly, salivary IgA was measurable in vaccinated HCWs who had a positive NPS after vaccination. Salivary IgA had already started to increase at 4–6 days and increased even more 7–10 days after the first positive NPS. Salivary IgA was augmented further at the following time points (Figure 4A). The rapid kinetics suggest that salivary IgA may be produced by MBCs migrated at the site of viral invasion. In the same HCWs with breakthrough infections, 5 to 8 months after the positive NPS, salivary IgA declined but remained significantly higher than in fully vaccinated HCWs who never experienced SARS-CoV-2 infection (Figure 4A).
Figure 4.
HCWs salivary IgA levels
(A) Detection of salivary IgA levels in 81 HCWs who previously had COVID-19, 34 vaccinated HCWs, and 33 HCWs with breakthrough infections measured at different time points after the first positive NPS (0–3 days, 4–6 days, 7–10 days, 11–14 days, >15 days, and 5–8 months). Empty circles refer to HCWs for which only one sample was collected at a known time point after the first positive NPS (n = 19). Statistical significances are reported in the table besides the graph. Medians are indicated, and statistical significance was determined using unpaired Mann-Whitney t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. (B) Graphical model of the possible mechanism by which SARS-CoV-2 infection turns vaccine-induced systemic immunity into local immunity. In vaccinated individuals, specific antibodies reach mucosal sites by transudation (on the left). After infection, memory T and B cells reach the site of viral entry. MBCs differentiate into plasma cells secreting large amounts of specific IgA (on the right).
Regarding the duration of SARS-CoV-2 PCR positivity in NPS, the median time from the first positive NPS to the first negative test was 28.3 ± 15 days in unvaccinated HCWs and 20 ± 9 days in the vaccinated group (p < 0.001). This finding confirms that vaccination-induced immune memory contributes to the control of viral infection in the nasopharynx, even now when the highly infectious and fast-replicating Delta variant has replaced the original Wuhan strain.
Discussion
The reason why vaccinated individuals may have breakthrough infections has been attributed to progressive decay of vaccine-induced immunity and, more specifically, to the reduction of specific antibodies in the serum (Bergwerk et al., 2021; Goldberg et al., 2021). In large population studies, the individual response cannot be studied in detail.
We studied two groups of HCWs, comparable for age, health and type of work, all without predisposing conditions associated with an increased risk of a positive SARS-CoV-2 test.
We asked the question of whether HCWs with breakthrough infections had a lower level of B cell immunity than their co-workers who received the same vaccine at the same time but never had a positive NPS.
Here, we show that in vaccinated HCWs, serum antibodies significantly declined between the peak level, recorded 1 week after the second dose, and 3 months later. The reduction of serum antibody slowly continued between 6 (Pape et al., 2021) and 9 months after vaccination (Figures 2A, 2B, and S1). The dramatic decrease of serum antibodies 3 months after vaccination is a physiological event occurring after every immunization and in each individual and is due to the programmed death of short-lived plasmablasts (Auner et al., 2010; Smith et al., 1996; Scott et al., 2021). Stable antibody levels are maintained by long-lived plasma cells homed to the bone marrow (Moser et al., 2006; Slifka et al., 1998; Brynjolfsson et al., 2018). We still do not know whether anti-SARS-CoV-2 antibody levels will remain unchanged or continue to decline in the next months.
In contrast to the reduction of serum antibodies, MBCs specific for the viral Spike protein significantly increased at all time points. Nine months after vaccination, the frequency of Spike-specific MBCs was 10-fold greater than immediately after the second dose (Figure 2C). Through the recirculation in the persistent germinal centers demonstrated in vaccinated individuals (Lederer et al., 2021; Turner et al., 2021b), MBCs may continuously be re-stimulated, thus explaining their increased numbers and progressively improved antigen affinity (Piano Mortari et al., 2021; Grimsholm et al., 2020; Wang et al., 2021a; Gaebler et al., 2021).
Two questions remain: 1) are serum antibodies and MBCs significantly reduced in HCWs with breakthrough infection compared to vaccinated HCWs who were never infected? And 2), are specific MBCs able to exert their function and be a rapid source of protective antibodies in the case of breakthrough infection?
In order to answer these questions, we compared the concentration of anti-RBD, anti-Trimeric Spike-specific and neutralizing antibodies of fully vaccinated HCWs infected 3 and 6 months after vaccination to the corresponding values of non-infected HCWs (Figures 3A–3C). We show, as already reported by others (Rovida et al., 2021), that at the time of the first positive NPS, vaccine-induced immunity had not waned in HCWs experiencing breakthrough infection because specific and neutralizing antibodies and MBCs were not reduced in comparison to the control HCWs. In addition, the rapid increase of Trimeric-Spike IgG and anti-RBD serum antibodies and the equally fast expansion of specific MBCs and plasmablasts strongly supports the hypothesis that vaccine-induced memory (Goel et al., 2021a), triggered by the virus, responds with the effect of limiting the infection and preventing symptoms (in 25/33; 75.7%) and severe disease (in all; 100%) and shortening the time of viral clearance. The role of established memory is also confirmed by the different kinetics of the response to the viral N protein that, for subjects never infected before, is a novel antigen. Anti-N antibodies become detectable after 2–3 weeks, as is expected by the germinal center response to a novel antigen (Gatto and Brink, 2010).
The increase of anti-Spike IgA in the saliva may have two different explanations. It may be the product of de novo generated mucosal response to the virus or represent the function of vaccine-induced memory B cells migrated to the site of viral invasion. The second hypothesis is supported by the rapid kinetics of IgA production in comparison to serum anti-N, against which no specific MBCs exist in vaccinated individuals before the infection.
In conclusion, the rapid response to the presence of the virus in the oropharynx demonstrates that, although serum antibodies decline, immunity induced by the vaccine does not wane.
Individuals convalescent from COVID-19 are rarely re-infected by SARS-CoV-2 (Hall et al., 2021; Omata et al., 2021; Turner et al., 2021a), whereas vaccinated individuals may have a positive NPS (Bergwerk et al., 2021), suggesting that viral infection and vaccination may generate different types of protective immunity.
The nasopharyngeal mucosa is the site of entry of SARS-CoV-2, and mucosal immunity has been demonstrated during the illness and in convalescence. Parenterally administered vaccines are not expected to generate mucosal immunity (Allie et al., 2019), and serum antibodies reach mucosal sites in small amounts by transudation (Sterlin et al., 2021). The reduced concentration of specific antibodies in the serum, and consequently in the tissues, may explain the observation that vaccine efficacy in prevention of infection is lower in individuals vaccinated at the beginning of the immunization campaign in comparison to the efficacy demonstrated in the recently vaccinated population (Rossman et al., 2021). Systemic protection induced by intramuscular vaccination can, however, be transformed into local protection. To do so, MBCs should be attracted to the site of infection and instructed to become resident MBCs (Allie et al., 2019). We suggest that breakthrough infections in vaccinated individuals may result from lack of local protection. Immune memory established by vaccination, however, prevents severe disease by the combined action of memory T and B cells that migrate to the infection site, thus turning systemic immunity into local protection (Figure 4B).
A third vaccine dose is necessary for all those individuals with primary and secondary immunodeficiency, immune-suppressive treatments, chemotherapy, bone marrow transplantation, or advanced age; these individuals may be unable to generate protective levels of antibodies and sufficient numbers of memory B and T cells (Salinas et al., 2021; Fernandez Salinas et al., 2021). The strength of our paper is the demonstration that B cell immunity does not wane and reacts as expected in vaccinated subjects independently from the time of immunization and the level of serum-specific antibodies at the time of infection. Our data also suggest that the ineffective mucosal protection represents the weak side of parenterally administered vaccines, including those containing mRNA. A corollary of our findings is that the measurement of serum antibodies is an insufficient indicator of established and functional immune memory to viral diseases.
In individuals with an intact immune system, the third vaccine dose will increase the level of Spike-specific antibodies and probably MBCs, similar to what is observed after breakthrough infections. The reduction of breakthrough infections demonstrated by the successful third-dose campaign in Israel (Bar-On et al., 2021; Barda et al., 2021) may be explained by the improvement of mucosal protection through the high concentration of serum antibodies induced by the booster dose and reaching the oropharynx by transudation. Serum antibodies will predictably decline a few months after the third dose, as happens in every type of vaccination and booster (Scott et al., 2021). We hope that the third dose will increase and stabilize the immunological memory, including MBCs, long-lived plasma cells, and of course memory T cells. Mucosal vaccines are in development (Mudgal et al., 2020; Lavelle and Ward, 2021; Lycke, 2012) and may be able to complete our protection with neutralizing secretory IgA posed on epithelial cells to stop viral invasion (Lavelle and Ward, 2021).
Our study has some limitations. First, we were unable to demonstrate that salivary IgA is produced by the same MBCs that migrate from the blood to the infection site. Comparison of immunoglobulin VH sequences in blood and salivary gland is necessary to demonstrate the clonal relationship. The difficulty is due to the facts that viable MBCs are not detectable in the saliva and that salivary gland biopsies cannot be performed in asymptomatic HCWs with a positive NPS. Further studies are necessary to demonstrate whether mucosal IgA specific for SARS-CoV-2 becomes a permanent protection and prevents re-infection. Additionally, we focused our analysis on a cohort of healthy HCWs, and our findings might not apply to the general population with a different exposure to the virus and various health conditions and age. Finally, the number of breakthrough infections reported in our sample was limited. Further studies will be crucial to better describe the dynamics of B cell immunity in time after vaccination for SARS-CoV-2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD19 BV786 (Clone SJ25C1) | Beckton Dickinson | Cat#563325; RRID:AB_2744314 |
| CD24 BV711 (Clone ML5) | Beckton Dickinson | Cat#56340; RRID: AB_2631261 |
| CD27 BV510 (Clone M-T271) | Beckton Dickinson | Cat#740167; RRID: AB_2739920 |
| CD38 BV421 (Clone HIT2) | Beckton Dickinson | Cat#562444; RRID: AB_11151894 |
| IgG BV650 (Clone G18-145) | Beckton Dickinson | Cat#740596; RRID: AB_2740297 |
| IgM APC (Polyclonal) | Jackson ImmunoResearch | Cat#709-136-073; RRID: AB_2340524 |
| Streptavidin PE | Beckton Dickinson | Cat#554061; RRID: AB_10053328 |
| Streptavidin BUV395 | Beckton Dickinson | Cat#564176; RRID AB_2869553 |
| Streptavidin FITC | Beckton Dickinson | Cat#554060; RRID: AB_10053373 |
| Streptavidin PE-Cy7 | Beckton Dickinson | Cat#557598; RRID: AB_10049577 |
| Biological samples | ||
| Human peripheral blood | This study, Bambino Gesù Children Hospital, Rome, Italy | N/A |
| Human saliva | This study, Bambino Gesù Children Hospital, Rome, Italy | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Biotinylated Recombinant SARS-CoV-2 Spike His-tag | R&D Systems | Cat#BT10549-050 |
| Biotinylated Receptor Binding Domain | This study, Takis, Rome, Italy | N/A |
| Critical commercial assays | ||
| Elecsys® Anti-SARS-CoV-2 | Roche Diagnostics | Cat#09 203 079 190 |
| Elecsys® Anti-SARS-CoV-2 S | Roche Diagnostics | Cat#09 289 275 190 |
| LIAISON® SARS-CoV-2 TrimericS IgG assay | Diasorin | Cat#311510D |
| ELISA Anti-SARS-CoV-2 IgA | Euroimmun | Cat#EI 2606-9601 A |
| SARS-CoV-2 isolate SARS-CoV2/Human/ITA/PAVIA10734/2020, clade G, D614G (S) | INMI | Cat#008V-04005 |
| EZ-LinkTM Sulfo-NHS-LC-Biotin reaction kit | ThermoScientific | Cat#21335 |
| QIAamp Viral RNA Mini Kit | QIAGEN | Cat# 52904 |
| Software and algorithms | ||
| FlowJo 10 | FLOWJO, LLC | https://www.flowjo.com |
| Prism 8 | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| RStudio | R Foundation for statistical computing | https://www.rstudio.com/ |
| Other | ||
| PANGOLIN v3.1.15 | Github | https://github.com/hCoV-2019/pangolin |
| Simple linear regression model code | This study | https://data.mendeley.com/datasets/3yxkps6msr/1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rita Carsetti (rita.carsetti@opbg.net).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Ethical approval
Ethics Committee of Bambino Gesù Children Hospital, Rome, Italy, approved the study (CE_291220). The study was performed in accordance with the Good Clinical Practice guidelines, the International Conference on Harmonization guidelines, and the most recent version of the Declaration of Helsinki.
Study design and population
We performed a monocentric observational study including both HCWs who received two doses of the BNT162b2 mRNA vaccine 21 days apart and developed SARS-CoV-2 breakthrough infections over 9 months of follow up, and fully vaccinated HCWs who never had a positive NPS (control group). All HCWs were fully vaccinated and recruited at the Bambino Gesù Children Hospital in Rome, Italy. Samples were collected from December 2020 to September 2021.
The population included in the study is described in Figure 1. The 34 HCWs included in the control group were randomly recruited among the 3511 HCWs of the Bambino Gesù Children Hospital (Rome, Italy), who received two doses of the BNT162b2 mRNA vaccine 21 days apart (Flowchart in Figure 1). For immunological studies, blood samples were collected at different time points: before vaccination (T0), 7 days after the second dose (1w) and, later, at 3, 6 and 9 months after vaccination. Before BNT162b2 mRNA administration, all HCWs had a negative SARS-CoV-2 status by molecular (Allplex2019-ncov, Seegene) and antibody assays (Elecsys Anti-N, Roche).
Among the 3511 HCWs who received two doses of the BNT162b2 mRNA vaccine, 38 (1.1%) had breakthrough infections 3-6 months after the second vaccine dose. We analyzed blood samples of 21 of them at different time points after the first positive NPS (0-3 days; 4-6 days; 7-10 days; 11-14 days; > 15 days). For 12 HCWs, we had a single sample at a known time point after the first positive NPS; the remaining 5 HCWs with breakthrough infections did not consent to participate to the study. None of the HCWs had predisposing conditions associated with an increased risk of a positive SARS-CoV-2 test. HCWs with breakthrough infections were compared to fully vaccinated HCWs matched for sex, age and interval from vaccination.
Saliva samples were obtained from 81 HCWs who had a SARS-CoV-2 positive NPS before vaccination, 34 vaccinated HCWs with a negative NPS at 3 and 6 months after vaccination and 33 vaccinated HCWs who had breakthrough infections. Of 21 HCWs with breakthrough infections, we collected saliva samples at different time points and from 12 HCWs only one sample was collected at a known time point after the first positive NPS.
In order to further evaluate the protective effects of vaccination, we compared the duration of SARS-CoV-2 PCR positivity of 299 unvaccinated HCWs who were infected from March 2020 to March 2021 to that of the 38 HCWs who had breakthrough infections.
Method details
RT-PCR assay
RNA detection and quantification of SARS-CoV-2 was carried out from NPS by using a fully automated magnetic bead platform Maelstrom 9600 (TANBead –Taiwan), followed by a multiplex real-time reverse transcription polymerase chain reaction (RT-PCR) PCR assay (Allplex SARS-CoV-2 Assay – Seegene South Korea) able to detect 4 target genes of SARS-CoV-2 (RdRP, S and N genes specific for SARS-CoV-2, and E gene for all of Sarbecovirus including SARS-CoV-2), in a single tube. Positive samples were run in a second Real Time PCR (COVID-19 Variant Catcher-Clonit Milano, Italy) for fast identification of well-known S gene mutations HV 69-70del, E484K and N501Y used for discrimination of SARS-CoV-2 Wuhan strain from SARS-CoV-2 variant strains B.1.1.7 (alpha variant), B.1.351 (beta variant) and P.1 (gamma variant).
Variant determination
SARS-CoV-2 sequences were available in 27 subjects in whom the amount of viral RNA was sufficient for whole genome sequencing. Total RNAs were extracted from NPS by using QIAamp Viral RNA Mini Kit (QIAGEN) and sequenced by using a multiplex approach (CleanPlex SARS-CoV-2 Research and Surveillance Panel, Paragon Genomics), and Illumina MiSeq platform (Illumina). Consensus sequence was obtained by in-house pipeline and lineages were assigned using PANGOLIN v3.1.15 (https://github.com/hCoV-2019/pangolin).
Quantitative determination of anti-N, anti-S, Trimeric Spike and RBD antibodies
Serum samples were tested by two different methods and analytical platforms. Qualitative detection of antibodies (IgA, IgM, IgG) direct against the nucleocapsid (N) protein and semiquantitative detection of total antibodies directed against the RBD of the virus Spike (S) protein of SARS-CoV-2 were tested by an electro-chemiluminescence sandwich immunoassay (ECLIA), using Elecsys-anti SARS-CoV-2 and Elecsys-anti SARS-CoV-2 S (Roche Diagnostics) test on a Cobas e801 analyzer following the manufacturer’s instructions. For anti-N antibodies, samples with a Cut off Index (COI; signal sample/cutoff) < 1.0 were considered as negative, those with a COI > 1.0 were considered as reactive (positive) and COI dynamic changes during infection were measured(Ko et al., 2020; Omata et al., 2021). Detection and quantification of anti-RBD antibodies were automatically calculated for each sample in U/mL, equivalent to the Binding Arbitrary Unit (BAU)/mL of the first WHO International Standard for anti-SARS-CoV-2 immunoglobulins. The quantitative determination of anti-Trimeric Spike protein specific-IgG antibodies to SARS-CoV-2 was run on LiaisonXL platform by a new generation of chemiluminescence immunoassay (CLIA) TrimericS IgG assay (DiaSorin).
Detection of neutralizing antibodies
Neutralizing antibodies were assessed by micro-neutralization assay (MNA) using live SARS-CoV-2 isolate. Briefly, seven two-fold serial dilutions (starting dilution 1:10) of heat-inactivated serum samples (56°C for 30 min) were titrated in duplicate, mixed with 100 TCID50 virus (SARS-CoV-2/Human/ITA/PAVIA10734/2020, clade G, D614G (S), Ref-SKU: 008V-04005, from EVAg portal) and incubated at 37°C for 30 min. Subsequently, virus-serum mixtures were added to Vero E6 cell monolayers in 96-well microplates and incubated at 37 °C, 5% CO2. After 48 h, microplates were examined by light microscope for the presence of virus-induced cytopathic effect (CPE). Neutralization titers were expressed as the reciprocal of the highest serum dilution inhibiting at least 90% (MNA90) of CPE. When 90% inhibition was not observed at the first dilution tested (1:10), the sample was considered not able to neutralize (neutralization titer < 1:10). Neutralization titer lower than 1:10 have been considered negative.
Detection of antigen-specific B cells
In order to detect SARS-CoV-2 specific B cells, biotinylated protein antigens were individually multimerized with fluorescently labeled streptavidin at 4°C for 1 h, as previously described(Piano Mortari et al., 2021). Briefly, recombinant biotinylated SARS-CoV-2 Spike (S1+S2; aa16-1211) was purchased from R&D systems (BT10549). RBD was generated in-house and biotinylation was performed using EZ-LinkTM Sulfo-NHS-LC-Biotin reaction kit (ThermoScientific) following the manufacturer’s standard protocol and dialyzed overnight against PBS. Separate aliquots of recombinant biotinylated Spike were mixed with streptavidin BUV395 or streptavidin PE (BD Bioscience) at 25:1 ratio and 20:1 ratio respectively. Streptavidin PE-Cy7 (BD Bioscience) was used as a decoy probe to gate out SARS-CoV-2 antigen non-specific streptavidin-binding B cells. The antigen probes individually prepared as above were then mixed in Brilliant Buffer (BD Bioscience). 4 × 106 previously frozen PBMC samples were prepared and stained with the antigen probe cocktail containing 100 ng of Spike per probe (total 200 ng), 27.5 ng of RBD and 2 ng of streptavidin PE-Cy7 at 4°C for 30 min to ensure maximal staining quality before surface staining with antibodies was performed in Brilliant Buffer at 4°C for 30 min.
B cell subsets were identified based on the expression of CD19, CD27, CD24 and CD38 markers by flow-cytometry. MBCs were defined as CD19+CD24+CD27+CD38- and plasmablasts were identified as CD19+CD27++CD24-CD38++. Stained PBMC samples were acquired on FACs LSRFortessa (BD Bioscience). At least 2 × 106 cells were acquired and analyzed using Flow-Jo10.7.1 (BD Bioscience). Phenotype analysis of antigen-specific B cells was only performed in subjects with at least 10 cells detected in the respective antigen-specific gate. The frequency of antigen-specific MBCs was expressed as a percentage of MBCs (CD19+CD24+CD27+CD38–, Pe-Cy7-, lymphocytes). Blank was determined in unexposed donors, before vaccination. LOB (limit of blank) was set as the mean of the blank + 1.645 X SD. LOD (limit of detection) as the mean of the blank + 3 × SD or the LOB + 1.645 × SD. LOS (limit of sensitivity) was set as the median + 2 × SD of the results in unexposed donors, before vaccination. The intra-subject biological coefficient of variation (CV) and the inter-subject biological coefficient of variation values were calculated on 10 replicates each. Percent of CV for intra-assay was 3.9% and for inter-assay was 4.9%.
ELISA for specific IgA detection in saliva samples
Saliva samples were collected aspirating with a sterile syringe, after removing the needle, from the oral cavity of all enrolled HCWs. Of note, saliva was collected 2-3 h after the last meal and/or drinking. All saliva samples were stored at −20°C until the time of testing. A semiquantitative determination of human IgA antibodies against the SARS-CoV-2 was performed on saliva samples, adapting the Anti-SARS-CoV-2 Spike ELISA (EUROIMMUN), according to the manufacturer’s instructions. Values were then normalized for comparison with a calibrator. Results were evaluated by calculating the ratio between the extinction of samples and the extinction of the calibrator. Results are reported as the ratio between OD samples and OD calibrator.
Quantification and statistical analysis
Demographics were summarized with descriptive statistics (median and IQR for continuous values). All statistical details can be found in figures and figure legends. Immunological, and clinical variables were compared between the different study times. A univariate analysis assessed the impact of variables of interest. Values were compared by the non-parametric Kruskal-Wallis test and, when appropriate, the Wilcoxon matched pair signed-rank test or the two-tailed Mann–Whitney U-test were used. Differences were deemed significant when p < 0.05. Statistical analyses were performed with GraphPad Prism 8.0 (GraphPad Software). The relationship between variables was studied using a simple linear regression model in the software R. https://www.R-project.org/
Acknowledgments
We thank all the HCWs of the Bambino Gesù Children’s Hospital who enthusiastically participated to the study. This work was funded by Italian Ministry of Health grant RF2013-02358960, Italian Ministry of Health grant COVID-2020-12371817, and grant “5 per mille, 2021” to Dr. Carsetti.
Author contributions
Drs. Terreri and Piano Mortari contributed equally to this work and are considered co-first authors. Drs. Zaffina and Carsetti contributed equally to this work and are considered co-last authors. Conceptualization, data analysis, validation: S.T., E.P.M., R.C. Investigation, methodology: S.T., E.P.M., M.R.V., C. Russo, C. Alteri, C. Albano, F.C., G.G., C. Agrati, G.L., L. Coltella, L. Colagrossi, G.D., M.S., R.B., V.C., A.S., N.M., R.S., G.R., E.P., A.M., A.V. Data curation: C. Albano, G.D. Statistical analysis: M.C.D.A., C. Rizzo. Visualization: M.R., F.L., C.F.P., S.Z., R.C. Writing – original draft: S.T., E.P.M., R.C. Writing – review & editing: S.T., E.P.M., C. Alteri, M.R., F.L., C.F.P., S.Z., R.C. Supervision, funding acquisition and project administration: C.F.P., S.Z., R.C.
Declaration of interests
All authors declare no competing interests.
Published: January 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.01.003.
Supplemental information
Data and code availability
All data has been included in main figures or supplementary information. All data reported in this paper will be shared by the lead contact upon reasonable request.
Consensus sequence are available at Github. The R code used to study the simple linear regression is deposited and available at Mendeley Data: https://data.mendeley.com/datasets/3yxkps6msr/1
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request
References
- Allie S.R., Bradley J.E., Mudunuru U., Schultz M.D., Graf B.A., Lund F.E., Randall T.D. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 2019;20:97–108. doi: 10.1038/s41590-018-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auner H.W., Beham-Schmid C., Dillon N., Sabbattini P. The life span of short-lived plasma cells is partly determined by a block on activation of apoptotic caspases acting in combination with endoplasmic reticulum stress. Blood. 2010;116:3445–3455. doi: 10.1182/blood-2009-10-250423. [DOI] [PubMed] [Google Scholar]
- Bar-On Y.M., Goldberg Y., Mandel M., Bodenheimer O., Freedman L., Kalkstein N., Mizrahi B., Alroy-Preis S., Ash N., Milo R., Huppert A. Protection of BNT162b2 Vaccine Booster against Covid-19 in Israel. N. Engl. J. Med. 2021;385:1393–1400. doi: 10.1056/NEJMoa2114255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barda N., Dagan N., Cohen C., Hernán M.A., Lipsitch M., Kohane I.S., Reis B.Y., Balicer R.D. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet. 2021;398:2093–2100. doi: 10.1016/S0140-6736(21)02249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandtzaeg P. Secretory immunity with special reference to the oral cavity. J. Oral Microbiol. 2013;5 doi: 10.3402/jom.v5i0.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynjolfsson S.F., Persson Berg L., Olsen Ekerhult T., Rimkute I., Wick M.-J., Mårtensson I.-L., Grimsholm O. Long-Lived Plasma Cells in Mice and Men. Front. Immunol. 2018;9:2673. doi: 10.3389/fimmu.2018.02673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsetti R., Zaffina S., Piano Mortari E., Terreri S., Corrente F., Capponi C., Palomba P., Mirabella M., Cascioli S., Palange P., et al. Different Innate and Adaptive Immune Responses to SARS-CoV-2 Infection of Asymptomatic, Mild, and Severe Cases. Front. Immunol. 2020;11:610300. doi: 10.3389/fimmu.2020.610300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Salinas A., Piano Mortari E., Terreri S., Milito C., Zaffina S., Perno C.F., Locatelli F., Quinti I., Carsetti R. Impaired memory B-cell response to the Pfizer-BioNTech COVID-19 vaccine in patients with common variable immunodeficiency. J Allergy Clin Immunol. 2021;149:76–77. doi: 10.1016/j.jaci.2021.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto D., Brink R. The germinal center reaction. J. Allergy Clin. Immunol. 2010;126:898–907, quiz 908–909. doi: 10.1016/j.jaci.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. UPenn COVID Processing Unit‡ mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Y., Mandel M., Bar-On Y.M., Bodenheimer O., Freedman L., Haas E.J., Milo R., Alroy-Preis S., Ash N., Huppert A. Waning Immunity after the BNT162b2 Vaccine in Israel. N. Engl. J. Med. 2021;385:e85. doi: 10.1056/NEJMoa2114228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsholm O., Piano Mortari E., Davydov A.N., Shugay M., Obraztsova A.S., Bocci C., Marasco E., Marcellini V., Aranburu A., Farroni C., et al. The Interplay between CD27dull and CD27bright B Cells Ensures the Flexibility, Stability, and Resilience of Human B Cell Memory. Cell Rep. 2020;30:2963–2977.e6. doi: 10.1016/j.celrep.2020.02.022. [DOI] [PubMed] [Google Scholar]
- Hall V.J., Foulkes S., Charlett A., Atti A., Monk E.J.M., Simmons R., Wellington E., Cole M.J., Saei A., Oguti B., et al. SIREN Study Group SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN) Lancet. 2021;397:1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Joo E.J., Park S.J., Baek J.Y., Kim W.D., Jee J., Kim C.J., Jeong C., Kim Y.J., Shon H.J., et al. Neutralizing antibody production in asymptomatic and mild COVID-19 patients, in comparison with pneumonic COVID-19 patients. J. Clin. Med. 2020;9:1–13. doi: 10.3390/jcm9072268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle E.C., Ward R.W. Mucosal vaccines - fortifying the frontiers. Nat. Rev. Immunol. 2021 doi: 10.1038/s41577-021-00583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Parvathaneni K., Painter M.M., Bettini E., Agarwal D., Lundgreen K.A., Weirick M., Goel R.R., Xu X., Drapeau E.M., et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. medRxiv. 2021 doi: 10.1101/2021.09.16.21263686. 2021.09.16.21263686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning Immune Humoral Response to BNT162b2 Covid-19 Vaccine over 6 Months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycke N. Recent progress in mucosal vaccine development: potential and limitations. Nat. Rev. Immunol. 2012;12:592–605. doi: 10.1038/nri3251. [DOI] [PubMed] [Google Scholar]
- Monto A.S. The Future of SARS-CoV-2 Vaccination - Lessons from Influenza. N. Engl. J. Med. 2021;385:1825–1827. doi: 10.1056/NEJMp2113403. [DOI] [PubMed] [Google Scholar]
- Moser K., Tokoyoda K., Radbruch A., MacLennan I., Manz R.A. Stromal niches, plasma cell differentiation and survival. Curr. Opin. Immunol. 2006;18:265–270. doi: 10.1016/j.coi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Mudgal R., Nehul S., Tomar S. Prospects for mucosal vaccine: shutting the door on SARS-CoV-2. Hum. Vaccin. Immunother. 2020;16:2921–2931. doi: 10.1080/21645515.2020.1805992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata M., Hirotsu Y., Sugiura H., Maejima M., Nagakubo Y., Amemiya K., Hayakawa M., Tsutsui T., Kakizaki Y., Mochizuki H., Miyashita Y. The dynamic change of antibody index against Covid-19 is a powerful diagnostic tool for the early phase of the infection and salvage PCR assay errors. J. Microbiol. Immunol. Infect. 2021;54:830–838. doi: 10.1016/j.jmii.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm A.E., Henry C. Remembrance of Things Past: Long-Term B Cell Memory After Infection and Vaccination. Front. Immunol. 2019;10:1787. doi: 10.3389/fimmu.2019.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape K.A., Dileepan T., Kabage A.J., Kozysa D., Batres R., Evert C., Matson M., Lopez S., Krueger P.D., Graiziger C., et al. High-affinity memory B cells induced by SARS-CoV-2 infection produce more plasmablasts and atypical memory B cells than those primed by mRNA vaccines. Cell Rep. 2021;37:109823. doi: 10.1016/j.celrep.2021.109823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano Mortari E., Russo C., Vinci M.R., Terreri S., Fernandez Salinas A., Piccioni L., Alteri C., Colagrossi L., Coltella L., Ranno S., et al. Highly Specific Memory B Cells Generation after the 2nd Dose of BNT162b2 Vaccine Compensate for the Decline of Serum Antibodies and Absence of Mucosal IgA. Cells. 2021;10:2541. doi: 10.3390/cells10102541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard A.J., Bijker E.M. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman H., Shilo S., Meir T., Gorfine M., Shalit U., Segal E. COVID-19 dynamics after a national immunization program in Israel. Nat. Med. 2021;27:1055–1061. doi: 10.1038/s41591-021-01337-2. [DOI] [PubMed] [Google Scholar]
- Rovida F., Cassaniti I., Paolucci S., Percivalle E., Sarasini A., Piralla A., Giardina F., Sammartino J.C., Ferrari A., Bergami F., et al. SARS-CoV-2 vaccine breakthrough infections with the alpha variant are asymptomatic or mildly symptomatic among health care workers. Nat. Commun. 2021;12:6032. doi: 10.1038/s41467-021-26154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas A.F., Mortari E.P., Terreri S., Quintarelli C., Pulvirenti F., Di Cecca S., Guercio M., Milito C., Bonanni L., Auria S., et al. SARS-CoV-2 Vaccine Induced Atypical Immune Responses in Antibody Defects: Everybody Does their Best. J. Clin. Immunol. 2021;41:1709–1722. doi: 10.1007/s10875-021-01133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J., Richterman A., Cevik M. Covid-19 vaccination: evidence of waning immunity is overstated. BMJ. 2021;374:n2320. doi: 10.1136/bmj.n2320. [DOI] [PubMed] [Google Scholar]
- Slifka M.K., Antia R., Whitmire J.K., Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8:363–372. doi: 10.1016/S1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- Smith K.G.C., Hewitson T.D., Nossal G.J., Tarlinton D.M. The phenotype and fate of the antibody-forming cells of the splenic foci. Eur. J. Immunol. 1996;26:444–448. doi: 10.1002/eji.1830260226. [DOI] [PubMed] [Google Scholar]
- Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:1–11. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., Kim W., Kalaidina E., Goss C.W., Rauseo A.M., Schmitz A.J., Hansen L., Haile A., Klebert M.K., Pusic I., et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature. 2021;595:421–425. doi: 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.-H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data has been included in main figures or supplementary information. All data reported in this paper will be shared by the lead contact upon reasonable request.
Consensus sequence are available at Github. The R code used to study the simple linear regression is deposited and available at Mendeley Data: https://data.mendeley.com/datasets/3yxkps6msr/1
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request