Abstract
Background SARS-CoV-2 claimed 5,209,104 lives, infected 260,997,910 individuals, globally. Infection is caused due to exposure or susceptibility; deaths occur due to age,comorbidity,higher-viral-load, immuno-suppression, inflammation, and multi-organ failure. Theaflavin-gallate, the major black tea component, showed previous evidence to inhibit HIV-1. Purpose As theaflavin-gallate prevents experimental rat-lung injury, the study of inhibitory effects of theaflavin-gallate was done, on SARS-CoV-2proteins and various host proteins related to some adverse effects in COVID-19 patients. Study Design Currently, some prospective phytochemical, black-tea (Camellia-sinensis) extract (BTE) was initially tested in vivo in strong oxidant-mutagen arsenic-induced model rat lung injury similar to that of COVID-19 manifestations like severe inflammation, oxidative stress, lung tissue degenerations, and apoptotic death. In silico, extensive bioinformatics and molecular docking experiments were performed on all catechin or theaflavin derivatives of C. sinensis, and finally theaflavin-3′-O-gallate (TFMG) were screened for blocking or inactivation of several proteins of SARS CoV-2 and host adversely-acting proteins or factors. Methods In vivo experiments in DNA stability (ladder, comet assay), free radicals attack (malondialdehyde; MDA, superoxide dismutase SOD, catalase gel-zymogram assay), extra cellular matrix damage (matrix metalloprotease; MMP2and9 zymogram assay) and inflammation (TNF-α, ELISA). In silico experiments- molecular docking by AutoDock-Patchdock analysis, Surface Topology Calculation by CASTp, Grid-value calculation, and Ramachandran Plot study. Results The BTE showed strong protection of lung DNA and cell-matrix by decreasing MMPs, TNF-α, and free radicals, the same factors affecting COVID-19 patients. In silico data suggest that TFMG significantly blocked the entry, exit, and amino acids at catalytic active-site of more than thirty proteins including viral (nsp1,nsp2,Mainpro,∼-9.0 kcal/mol) and host inflammatory, oxidants, apoptotic, vaso-destabilizer molecules (FAS, CD40R, BCL2, TLR2, ∼ -10 and ACE1or2 ∼ -9.0 and AT1or2∼ -7.5 kcal/mol and more). When the binding energy of TFMG ranged from -7 to -11.7 kcal/mol(average -9.0) the same for hydroxy‑Chloroquine ranged (-2.5 to -7 average -4.5) and dexamethasone (-3.0 to -6.0, average -4.0 kcal/mol). Conclusions TFMG has some novel physicochemical or structural properties like (ACE values of binding to all tested proteins, -300 to -625), (for TFMG H-bond acceptor or donor: 15/10, for TFDG 20/13). Their topological-polar-surface-area (264Ų and 351Ų) and travel depth/height; 17.0/9.6 Å and 15.4/11.3 Å, respectively) were more potent than other compounds. Conclusively, the efficacy of TFMG may be further verified.
Keywords: Global pandemic and SARS CoV-2, Inflammation and vaso-destabilizing proteins, Theaflavin mono gallate, Active site docking, Therapeutic intervention
Graphical abstract
Abbreviations
- ACE
Angiotensin-converting enzyme
- ACE value
Atomic Contact Energy value
- ALP
Alkaline phosphatase
- BTE
Black-tea extract
- CASTp
Computed-Atlas-of-Surface-Topography
- COVID-19
Coronavirus disease 2019
- CQ
Chloroquine
- CS
Camellia-sinensis
- DEX
Dexamethasone
- EGCG
Epigallocatechin-gallate
- ELISA
Enzyme-linked immunosorbent assay
- HCQ
Hydroxy-Chloroquine
- HCV
Hepatitis-C-virus
- HSV
Herpes-simplex-virus
- MDA
Malondialdehyde
- MMP
Matrix metalloprotease
- REM
Remdesivir
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SGOT
Serum glutamic oxaloacetic transaminase
- SGPT
Serum glutamic pyruvic transaminase
- SOD
Superoxide dismutase
- TBA
Thiobarbituric acid
- TFDG
Theaflavin-3,3′-digallate
- TFMG
Theaflavin-3′-O-gallate
- TNF-α
Tumor necrosis factor-α
- WHO
World Health Organization
Introduction
The global pandemic by SARS CoV-2 virus claimed and infected millions of lives. This virus was extremely spreadable, highly proliferating and generated its most stable form than its earlier variant SARS CoV (van Doremalen et al., 2020). High rate of mutability and fidelity to capture human lung ACE2 make this virus tremendously powerful. Screenings of new drugs and repurposing old drugs are the current strategies. Chloroquine (CQ), hydroxy-Chloroquine (HCQ), remdesivir (REM), and the recently proposed dexamethasone (DEX) were being used with inconclusive results (Molina et al., 2020; WHO, 2021).
The epidemiological data suggest that (1) high rate of viral reproducibility (2) immuno-suppression (3) inflammation and (4) impairment in the vaso-vascular state were the major causes of pathogenicity (Ackermann et al., 2020). Presently, we screened a number of drugs and we foundtheaflavin-3′-O-gallate (TFMG) as the most potent active-sites binder of a total 31-proteins related to above mentioned factors like host immune- and vasoregulatory proteins, inflammatory markers and some viral proteins for host lung cell interaction, viral entry, replication and immune-invasion proteins. Antiviral effects of TFMG and epigallocatechin-gallate (EGCG) have been previously shown (Mhatre et al., 2021). Here, TFMG showed consistently higher binding and molecular-docking affinity than other drugs i.e. EGCG, HCQ, DEX, theaflavin-di-gallate (TFDG), hesperidin, quercetin and kaempferol. TFMG can block the COVID-19 RNA polymerase active site (Banerjee et al., 2021). Theaflavin (and its gallate-form) was the major component of black tea (0.27 to 1.48 mg/g of tea). A total 78% of the tea produced in the world (in India, USA etc.) is consumed as black-tea. Theaflavin, TFMG and TFDG were produced as catechin oxidation products (Takemoto and Takemoto, 2018).
Polyphenol can bind to diverse protein structures. They can inhibit the SARS-CoV-2 M protease (Bhardwaj et al., 2021). The binding of Theaflavin to Bovine serum albumin (BSA), a model-protein has been extensively studied (Lei et al., 2017).TFMG decreased inflammatory-cytokine IL-6 during viral-infection (Zu et al., 2012) and inhibited influenza-A virus and other subtype H1N1.TFDG completely inactivated herpes-simplex-virus (HSV) and HSV-1 viral particles in Vero and A549 cells (Mhatre et al., 2020). Theaflavin also inhibited the entry of hepatitis-C-virus (HCV) to the target-cells (Mhatre et al., 2020) and it also neutralized bovine Rota virus and bovine-coronavirus infections. Tea-polyphenols especially with galloyl-moiety significantly prevented zika-virus NS3-NS2B and HIV-1 replications (Mhatre et al., 2020). Resveratrol, genistein and curcumin have also demonstrated to inhibit viral-infections (Liu et al., 2007).Another recent finding suggests that theaflavin gallate can block the central-channel of SARS CoV-2 spike-protein which hampers spike-ACE2 binding (Mhatre et al., 2021). Recent evidence suggests some drugs like lopinavir, amodiaquine, and TFDG were capable to block protease-Mpro of SARS-CoV-2 (pdb id: 6LU7) (Ghosh et al., 2020). Not only pathogen, induced tissue injury, a large number of toxic substances and environmental pollutants like arsenic can cause lungs and other organ damages by similar mechanisms like oxidative stress, inflammation and apoptosis (Maiti et al., 2017).Arsenic induced similar mode of tissue damages and its protection by tea extract has been demonstrated earlier (Acharyya et al., 2014; Acharyya et al., 2015). All these previous reports suggest the multi-potential protective effects of tea and its constituents against several types of tissue damages.
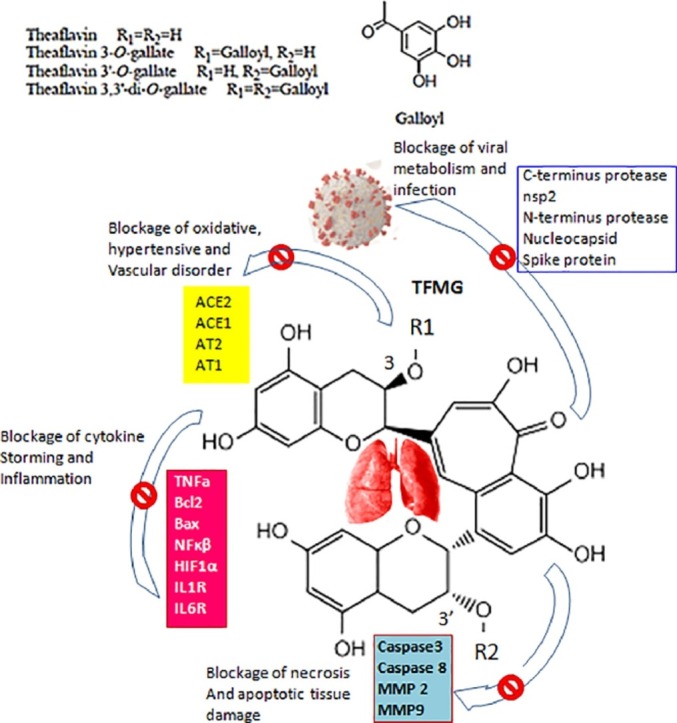
Based on this background, we hypothesized that blocking viral-proteins and host hostile-proteins can efficiently manage COVID-19. After extensive searching, we found from molecular-docking study that TFMG, due to its unique physico-chemical properties, could block several proteins of viral and host sources. In brief, from the rat model experimental data we have shown that stress-induced lung damages occur by inflammations, oxidative stress and immuno-modulations. We have further demonstrated that black tea which is enriched with TFMG is highly protective against these damaging steps.
Materials and methods
Protein structure retrieval
A total of 31-proteins were finally selected for reporting on 6 COVID-19 functional-proteins like; C-terminus protease (Zhang Lab ID: QHD43415_5), nsp2 (Zhang Lab ID: QHD43415_2), N-terminus protease (Zhang Lab ID: QHD43415_3), nucleocapsid (Zhang Lab ID: QHD43423), COVID-19 main protease (PDB ID: 6LU7), nCov2 Spike (PDB ID: 6svb)and 25 human-proteins i.e. Apoptotic regulators: Bcl2 (4ieh), Bax (4s0o), NFκβ (1nfk), monomeric TLR2 (2Z80), dimeric TLR2 (2Z80), Monomeric TLR4 (3fxi), MultimericTLR4 (3fxi), Caspase3 (1 gfw), Caspase 8 (1f9e), MMP 2 (3ayu) and MMP9 (2ow0), FAS_D (3tje),proinflammatory regulators: CD40 (6pe9), CD40_R (1aly), IL1R (1itb), IL6R (1p9m), IFNϒ (1fg9), TNFα (1tnf), immuno-inflammation: TNFR1 (1ncf), TNFR2 (3alq),vaso-regulator proteins: ACE1 (6en5), ACE2 (4aph), AT1 (6os1), AT2 (5xjm), Stress regulator: HIF1α (4ajy)etc.(Table 1 ), and retrieved from RCSB PDB (https://www.rcsb.org/) and Zhang Lab (https://zhanglab.ccmb.med.umich.edu/COVID-19/). Their 3D structures were collected (.pdb-format) fromX-ray-crystallography, NMR or Electron-Microscopy data (Maiti et al., 2017: Acharyya et al., 2014).
Table 1.
Patchdock analysis data on TFMG docking on 31 proteins are presented.
| Name of the molecules | PDB/Uniprot ID | Best PatchDock Atomic Contact Energy Value (ACE) | Surface area |
|---|---|---|---|
| Viral proteins | |||
| C-terminus protease | QHD43415_5 | −531.35, −458.28, −436.10 | 733.30, 708.90, 800.00 |
| nsp2 | QHD43415_2 | −498.50, −487.90, −477.94 | 878.20, 758.80, 807.30 |
| N-terminus protease | QHD43415_3 | −383.73, −378.93, −376.85 | 744.30, 713.90, 770.70 |
| nucleocapsid | QHD43423 | −556.35, −273.85, −266.78 | 868.10, 706.00, 624.50 |
| COVID-19 main protease | 6LU7 | −383.73, −378.93, −376.85 | 744.30, 713.90, 770.70 |
| nCov2 Spike | 6svb | −434.42, −397.12, −392.53 | 906.20, 969.10, 911.40 |
| Human proteins | |||
| FAS_D | 3tje | −467.22, −400.62, −400.04 | 878.40, 797.50, 773.60 |
| CD40 | 6pe9 | −506.45, −475.73, −473.95 | 856.80, 933.20, 869.10 |
| CD40_R | 1aly | −353.37, −282.93, −244.56 | 696.80, 479.90, 588.90 |
| IL1R | 1itb | −333.00, −329.21, −303.53 | 761.80, 663.20, 763.20 |
| IL6R | 1p9m | −413.76, −355.92, −342.27 | 777.50, 672.30, 698.70 |
| TNFR1 | 1ncf | −412.25, −351.30, −324.34 | 660.80, 673.50, 646.60 |
| TNFR2 | 3alq | −199.59, −190.95, −153.60 | 732.60, 666.30, 668.00 |
| IFNϒ | 1fg9 | −467.46, −430.45, −389.77 | 830.50, 861.00, 701.90 |
| TNFa | 1tnf | −456.27, −323.00, −317.63 | 729.20, 810.70, 650.80 |
| Bcl2 | 4ieh | −309.63, −265.62, −258.02 | 712.50, 784.60, 629.50 |
| Bax | 4s0o | −474.43, −377.74, −346.51 | 773.40, 686.40, 814.80 |
| NFκβ | 1nfk | −397.99, −373.50, −312.99 | 780.50, 782.40, 753.30 |
| HIF1α | 4ajy | −446.61, −363.67, −342.04 | 609.80, 700.80, 530.80 |
| monomeric TLR2 | 2Z80 | −455.90, −321.99, −320.23 | 686.00, 525.40, 676.40 |
| dimeric TLR2 | 2Z80 | −155.51, −93.60, −91.630 | 936.60, 891.60, 794.00 |
| Monomeric TLR4 | 3fxi | −203.49, −137.48, −130.44 | 776.10, 690.50, 794.20 |
| MultimericTLR4 | 3fxi | −489.31, −456.83, −389.56 | 808.50, 740.20, 724.10 |
| Caspase3 | 1 gfw | −373.91, −356.51, −338.93 | 681.30, 780.90, 592.70 |
| Caspase 8 | 1f9e | −299.09, −239.61, −239.35 | 720.90, 738.00, 804.70 |
| MMP 2 | 3ayu | −616.95, −593.93, −583.02 | 734.70, 780.50, 736.00 |
| MMP9 | 2ow0 | −625.14, −614.45, −591.09 | 880.30, 948.20, 890.20 |
| ACE2 | 4aph | −319.12, −248.17, −229.89 | 911.90, 785.50, 891.90 |
| ACE1 | 6en5 | −351.19, −269.59, −226.69 | 717.00, 721.70, 724.10 |
| AT2 | 5xjm | −360.15, −343.08, −334.87 | 819.20, 700.90, 683.20 |
| AT1 | 6os1 | −378.52, −285.57, −282.28 | 741.40, 736.90, 853.90 |
Ligand structure retrieval
The 3D-structures (PubChem as .SDF-file) of ligand-molecules were by AutoCAD Map-3D, Autodesk-GIS and MapGuide. We have selected TFMG or Theaflavin-5-oxo-8-[(2R,3R)−3,5,7-trihydroxy-3,4-dihydro-2H-chromen-2-yl]benzo[7]annulen-1-yl]−3,4-dihydro-2H-chromen-3-yl] for our study (Mhatre et al., 2021). The canonical SMILES of the molecule was
C1C(C(OC2=CC(=CC(=C21)O)O)C3=CC4=C(C(=C(C = C4C5C(CC6=C(C = C(C = C6O5)O)O)OC(=O)C7=CC(=C(C(=C7)O)O)O)O)O)C(=O)C(=C3)O)O(MW 716.6 g/mol; Molecular-formula C36H28O16). TFMG is a group of catechins having two enantiomers (+ &-) and plays a significant role as a metabolite.
Ligand and protein structures preparation
The PyMol software was used as molecular-visualization system. The structures were saved as a .pdb format. Ligand molecules were retrieved in .sdf format (Table 1).
Surface-topology calculation of proteins
Solvent accessibility factor defines two properties of protein; pocket, where water enters and cavity, where does not. The CASTp: Computed-Atlas-of-Surface-Topography of Protein (http://sts.bioe.uic.edu/castp/index.html?j_5e8c7bec25090) was used to define pocket and cavity.
Grid-value calculation and optimization
During the grid calculation system of Autogrid, the energy of each and every atom of the ligand was being calculated [Table S3]. Then the receptor protein molecule was placed at the center of a three-dimensional grid box having adjustable x, y and z axis. During molecular docking the free movement of receptor and ligand within the grid box generates different energy parameters.
Molecular-docking
The molecular-docking of TFMG with selected proteins were individually performed using Patchdock Beta 1.3 Version (https://bioinfo3d.cs.tau.ac.il/PatchDock/) and Autodock version 1.5.6. Sep_17_14 software (Trott and Olson, 2010). It generates Atomic-Contact-Energy (ACE-value) and corresponding area for each probable position of ligand. Among different parameters we have discussed the binding-energy, inhibitory-constant (Ki) and ligand-efficiency-value in TFMG inhibition.
Preparation of tea extract and arsenic solution
The raw matured leaves (without any preservatives, pesticides or contaminants) of Camellia sinensis were collected locally and dried/dehydrated in an incubator for several days at 37 °C. Those were grinded in moisture free condition to make dust. Extraction was done by taking the freshly prepared Camellia sinensis leaf dust of 1 g% in distilled water. To achieve the optimum extraction of the bioactive phytochemicals, antioxidants and flavonoids, catechin and theaflavin products higher temperature (100 °C) should be avoided. Longer duration of extractions (∼24 h) may also yield fewer amounts of bioactive compounds (Banerjee and Chatterjee, 2015). In the current study, 4–5 h of extraction process was continued at 70 °C. When the color of the aqueous extract became dark brown, it was collected for the experiment and stored after lyophilization. This condition was termed as optimum and it was reported earlier (Acharyya et al., 2014). The total antioxidant capacity of the extract solution was found to be the maximum in the present set up of the experiment. Sodium-meta-arsenite (NaAsO2) from Sigma-Aldrich (St Louis, MO, USA), Chemicals was dissolved in distilled water to prepare 0.6 ppm final concentration. We used this dose because it was not lethal for the animal but generated tissue toxicity after a moderate time of its exposure (Acharyya et al., 2014).
Treatment and group distribution of rat model
For this experiment, male albino rats weighing 150 g ± 10 were acclimatized for 7 days at 12 hour light-dark cycle, 25 ± 2⁰C temperature and humidity 50–70٪. Standards pellet diet (Hindustan Lever, Mumbai, India) was given to them. Rats were purchased from rodent Government accredited firm house (CPCSEA-Committee for the Purpose of Control and Supervision of Experiments on Animals: Reg. no 1A2A/PO/BT/S/15/CPCSEA. <http://cpcsea.nic.in/Auth/index.aspx>) organization under the Dept of Animal Husbandry and Dairy, Ministry of Agriculture and Farmer's Welfare, Govt. of India. Animal experiments were performed in the OIST Animal Resource Facilities. Institutional ethical concerns Oriental Institute of Science and Technology) Review Board; OIST-IRB, reference no. oist/EC/sAM/20_2) were maintained throughout the investigation. Rats were randomly selected into 3 different groups having four in each. Group distribution for drug application was as follows, Control or group I; fed only drinking water, Arsenic or group-II; fed0.5 ml of arsenic solution (0.6 ppm), Arsenic+ black tea extract (BTE) or group-III; 0.5 ml arsenic (0.6 ppm) + 0.5 ml lyophilized tea extract (Acharyya et al., 2014, 2015). In a different set of experiment, one group of animals (n = 4) was fed only drinking water (Control group) and other group (n = 4) was fed with 0.5 ml lyophilized tea extract (1 g%). All treatment was continued for 21 days
Evaluation of toxicity
On the day 22, animals were exposed to halothane and after some time, blood was collected using disposable syringe (21-gage needle) and the serum was separated. By sacrificing the rats, experimental lung tissues were collected and homogenized in cold PBS with protease inhibitors cocktail and their cytosols were preserved at −40⁰Cfor testing within 3 to 4 days. Fresh tissues were used for DNA fragmentation and comet assay.
Serum TNF-α assay
Serum TNF- level was measured and quantified by ELISA using a suitable antibody (BioLegend) following the supplier's and published protocol (Maiti et al., 2017).
Malondialdehyde (MDA) assay
Tissue homogenates were treated with the same volume of 5% TCA, centrifuged at 9000 x g. After discarding pellet, supernatant was treated with 3% TBA (thiobarbituric acid) solution, incubated at 80⁰C in water bath for 30 min and cooled at RT for 20 min. The color intensity was measured in spectrophotometer and MDA concentration was calculated in the different groups utilizing the molar extinction coefficient of MDA (1.56 × 105 cm2/mmol) (Acharyya et al., 2014).
Gel zymogram assay of catalase and SOD activities
Catalase activity assay was done in gel-zymogram by using 8% native gel. The substrate 0.003% H2O2 (30% solution) was added in this enzymatic reaction. Then the gel was stained by using 2% ferric chloride and 2% potassium ferrocyanide. A transparent band showing enzyme activity was developed in a dark green-blue background (Maiti et al., 2014).Superoxide dismutase (SOD) enzyme was assessed in 12.5%a non-denaturing PAGE. The gel was left in NBT (nitro-bluetetrazolium) solution in shaking condition. After incubation the gel was soaked in a solution containing potassium phosphate, Tetramethylethylenediamine (TEMED) and riboflavin for approximately 15 min. The gel was illuminated in UV source. The transparent SOD activity bands were visible after some time (Nazmeen et al., 2020).
Gel zymogram assay of matrix metalloproteinase (MMP2 and MMP9)
Samples were run in SDS-polyacrylamide gel (8%) electrophoresis. During gel preparation gelatin solution (1%) was added. Gel was run at 110 V (Bio-Rad apparatus, USA). After removing the gel from the gel apparatus, it was incubated in zymographic development buffer (50 mM Tris pH-7.4, 1 mM CaCl2, 0.02% NaN3, sterile diH2O) at 37⁰C for 42 h. The gel was stained by Coommassie Brilliant Blue R-250 solution at room temperature on a rotary shaker. Staining was followed by the distaining solution (ethanol, acetic acid, dH2O) until the clear MMP activity bands were observed (Zhao et al., 2019).
Comet assay and DNA stability
The alkaline Comet assay was conducted according to the method of Singh and colleagues’ with some modifications (Maiti et al., 2017; Singh et al., 1988). Low melting-point agarose (0.6%) in PBS at 37 °C was added to a cell suspension (105 cells) and dropped onto a microscope slide pre-coated with 1% agarose, covered by a coverslip. After The ice-cold lysis buffer (2.5 mM NaCl, 85 mM EDTA, 10 mM Trizma base, 1% Triton X-100, 10% DMSO and 1% sodium lauryl sarcosinate, adjusted to pH 10) was added (1 hr at 4 °C) on the after removal of the coverslip. Slides were washed three times in PBS, 50 ml of buffer (control) or T4 endo V (epicentre) (4 U/slide) in buffer was transferred to the slides. Coverslips were placed on and the slides were incubated at 37 °C for 45 min. After careful washing, slides were placed in a submarine gel electrophoresis chamber (Bio-Rad) filled with alkaline electrophoresis buffer (0.3 M NaOH and 1 mM EDTA) for 25 min. After incubation, electrophoresis was performed for 30 min at 25 V and the current was adjusted to 300 mA by raising the buffer level. Slides were neutralized with PBS, stained with 10 mg/ml ethidium bromide for 5 min and then washed. Slides were read using a fluorescence microscope (Nikon, Eclipse LV100 POL), with the VisComet (ImpulsBild analyze) software. A total of 100 comets per slide were read for each experiment. Lung tissue was treated with buffer (50 mM Tris pH 8.0, 20 mM EDTA, 10 mM NaCl, 1% SDS) and centrifuged at 12,000 × g for 30 min. The supernatant was extracted with a 1:1 mixture of phenol: chloroform and then precipitated with2 equivalents of cold-ethanol and 1/10 equivalents of sodium-acetate. After spinning the precipitate was re- suspended in TE buffer and 5 µl of loading-buffer. The 0.8% agarose gel was run. The band densities were estimated by a gel documentation system. The DNA ladder (EZ Load 500 bp MW Ruler #1,708,354, Life Sciences, Bio-Rad) was run to estimate the DNA damage pattern.
Results
Bioinformatics data analysis
Tables and figures are provided in this manuscript as integrated and multi-paneled as representative image but the detail images, but the details Figures and Tables are provided in the Supplementary Tables (Tables S1 to S7) and Figures (Figs. S1 to S24).
Binding of TFMG to viral proteins
COVID-19 main protease
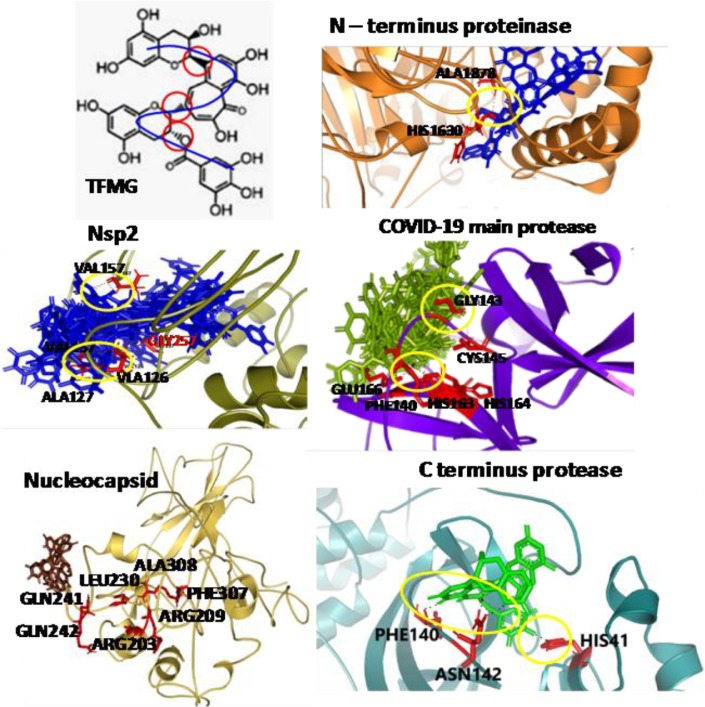
An inhibitor α-ketoamide was found as active site blocker of this enzyme. It was interacted with the amino acids like GLU166/PHE140/GLY143/CYS145/HIS164. In our study TFMG also interacted with GLU166/PHE140/HIS163/HIS164/CYS145/GLY143. According to Patchdock result, TFMG formed covalent bonds with ASN142/THR25 and THE24 of C-terminus protease at the active site (Fig. S2).So, all the common amino acids were blocked with high affinity. At those positions the Ki values of two molecules of TFMG were 8.11 µM and 12.58 µM. The ligand efficiency values were −0.13 for each (Tables 1, S1, Figs. 1 and S1).
Fig. 1.
AutoDock and/or Patchdock binding images of TFMG with viral proteins are presented. Yellow circle designate H- bonding. Amino acids demarcated in the active site locations and those participate with TFMG binding. Refer to supplementary-file.
C-terminus protease
According to ACE value of −531.35, TFMG showed highest affinity to C-terminus protease. Among the 20 different positions of TFMG, only one has the ACE value of −46.74. All others were significantly higher. TFMG showed the cascade like interaction with C-terminus protease. And in the active site it interacts with the amino acids THR24/THR25/ASN142 (Tables 1, S1, Figs. 1 and S2). Very low Ki values i.e. 1.29 µM and 1.35 µM for these binding sites strongly suggests higher affinity of TFMG which blocked the pockets of C-terminus possibly influencing this enzymatic function.
N-terminus protease
COVID-19 N-terminus protease cleaves the N-terminus of replicase polyprotein. It also works in de-ubiquitination from different cellular molecules, and positively associated with the viral replication with the help of Nsp4 (https://zhanglab.ccmb.med.umich.edu/COVID-19/). During molecular docking with TFMG it was found that an entry pocket of COVID-19 N-terminus protease was blocked. The highest ACE value of TFMG- N-terminus protease was −380.07 where it blocked the pocket by forming H-bond with HIS1630. (Tables 1, S1, Figs. 1 and S3). In Fig. S4, TFMG interacted with COVID-19 Nucleocapsid. It blocked the amino acids VAL157, GLY257, THR1590, and ALA127, VAL126 on one side and PHE156, ASN130, SER128, PRO129 on another side. This substrate entry pocket blocked by TFMG can inhibit viral Nsp2 function.
TFMG binding to vaso-regulator proteins
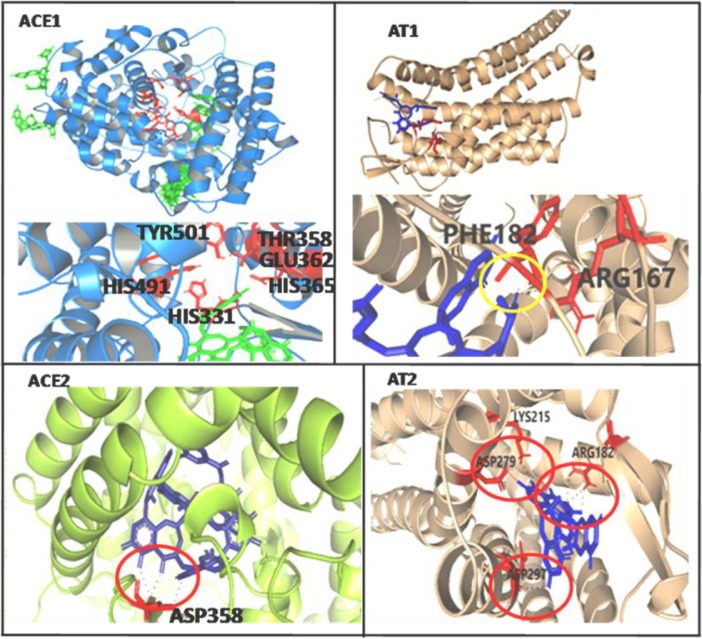
ACE2 and ACE1
TFMG showed higher affinity to Angiotensin Converting Enzyme 2 (ACE2) rather than ACE1. According to AutoDock result,−8.77/−8.33/−8.31/−7.41/−7.4 Kcal/mol binding energy values were found in TFMG-ACE2 interaction at the active site. Here the lowest Ki value was 3.67 µM for binding energy value of −7.41 Kcal/mol. On the other hand, only one position was observed at the active site of ACE1 with binding energy value of −7.54 Kcal/mol and Ki value of 2.99 µM (Table 1, Table S1, Figs. 2 and S6).ACE value −319.12 was the highest value for TFMG-ACE2 interaction but the frequency was higher than ACE1. On the other hand −351.19 was the highest value of TFMG-ACE1 interaction but the frequency of getting higher negative value was low in comparison to ACE2.
Fig. 2.
AutoDock and/or Patchdock binding images of TFMG with host vasoactive proteins are presented. Yellow/red circle designate H- bonding. Amino acids demarcated in the active site locations and those participate with TFMG binding. Refer to supplementary file.
AT1 andAT2
For both the proteins AT1/AT2, TFMG interacted with the angiotensin-binding-site or active-sites. The highest ACE value of −378.52 was found for TFMG-AT1 interaction. And, −360.15 values were observed for TFMG-AT2 interaction. The affinity was near about same for AT1 & AT2. TFMG-AT1 showed higher interaction affinity with another location just opposite of the active site pocket (Tables 1, S1, Figs. 2 and S7, S8).At active site of AT1, H-bond was formed with PHE182 and ARG167. Moreover, several bonding was observed at AT2 active site with the amino acids LYS215, ASP279, ARG182 and ASP297.
TFMG binding with proinflammatory regulators
TNFα
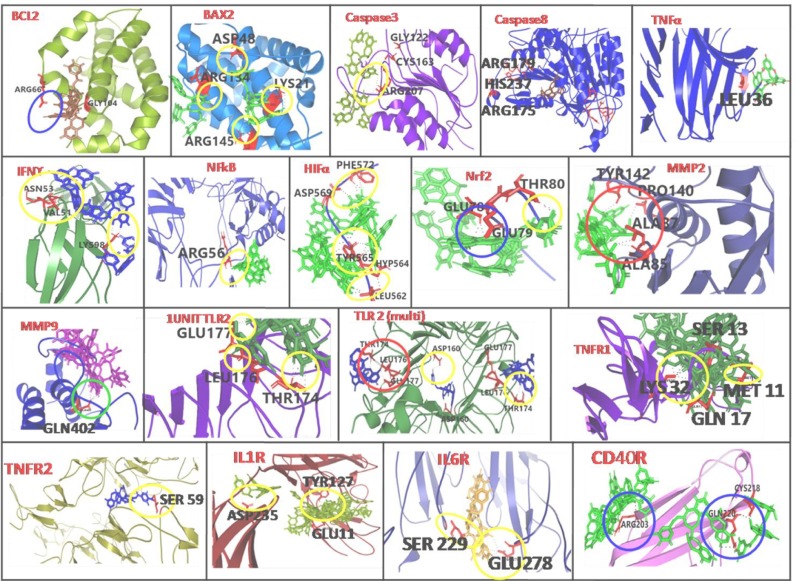
The TNFα has three subunits (Nazmeen et al., 2020). Among them different polypeptide ranges (I: 32–34, II: 84–91, III: 117–119 and IV: 143–148) were reported to maintain the biological activities as confirmed through the mutagenesis study. In our study TFMG was found to block PRO100, SER99 and GLU116 from one β-barrel and ARG103, CYS101, PRO100, LYS98 GLU116 from another β-barrel. ACE value at this location was −323.00 (Tables 1, S2, Figs. 3 and S9). In AutoDock study it showed highest binding energy value of −9.55, −8.13 Kcal/mol with Ki value of 99.59 µM and 1.1 µM, respectively. Among others LEU36 was mutation also showed functional inactivation (Ostade et al., 1991).Which was also blocked by TFMG molecule in our study (Table 1, Table 2 , Fig. 3 and Fig. S9).
Fig. 3.
AutoDock and/or Patchdock binding images of TFMG with host immuno- suppressive and inflammatory stress proteins are presented. Yellow/red/blue circle designate H- bonding. Amino acids demarcated in the active site locations and those participate with TFMG binding. Refer to supplementary file.
Table 2.
Different molecular parameter analysis of used ligand molecules to predict their druggability.
| Name | Pubchem ID | Molecular Weight (g/mol) | H-bond | Rotatable Bond count | Topological Polar Surface Area (Ų) | Depth (Å) | Height (Å) | |
|---|---|---|---|---|---|---|---|---|
| Acceptor | Donor | |||||||
| Theaflavin-3′-O-gallate | 71,307,578 | 704.6 | 15 | 10 | 5 | 264 | 17.0 | 9.6 |
| Theaflavin 3,3′-digallate | 135,403,795 | 868.7 | 20 | 13 | 8 | 351 | 15.4 | 11.3 |
| (-)- Gallocatechin gallate | 199,472 | 458.4 | 11 | 8 | 4 | 197 | 14.5 | 8.3 |
| (-)-Epigallocatechin gallate | 65,064 | 458.4 | 11 | 8 | 4 | 197 | 11.5 | 8.3 |
| Epigallocatechin | 72,277 | 306.27 | 7 | 6 | 1 | 131 | 11.5 | 1.4 |
| Epigallocatechin-(4beta->8)-epicatechin-3-O-gallate | 442,678 | 746.6 | 17 | 13 | 6 | 308 | 10.4 | 8.3 |
| Hydroxychloroquine | 3652 | 335.9 | 4 | 2 | 9 | 48.4 | 8.9 | 6.8 |
| Dexamethasone | 5743 | 392.5 | 6 | 3 | 2 | 94.8 | 12.1 | 4.1 |
IFNγ
IFNγ is a kind of homodimeric cytokines which exerts its different activities, i.e., development of innate and adaptive immunity against viral, some bacterial and protozoal infections, induces MHC class II complex and macrophages also (Zhao et al., 2019). At the binding location, TFMG showed higher affinity, with binding energy of −6.84, −6.48 and −6.36 Kcal/mol with the Ki value of 9.72, 17.84 µM and 21.77 µM. (Tables 1, S2, Fig. 3).Here Ki value decreases with the decrease of binding energy. And the higher affinity of TFMG to the base location indicated that it can significantly inhibit the IFN γ interaction with its receptor
IL1R and IL6R
Interleukin - 1(IL1) is the molecular mediator of inflammatory diseases and it mediates its function through the interaction with IL-1α, IL-1β and IL-1R.TheIL-1β interacts with IL-1R at the location in loop 1 and 2 including the specific amino acids. According to AutoDock results, that loops region was also blocked by TFMG (Tables 1, S2, Figs. 3 and S10). The affinity of TFMG towards the active site was high according to ACE value of −333.00 where the amino acid occupied were GLU11, LYS12, ILE13, TYR127 and PRO26
IL-6 interaction was activated when one unit of gp130 is attached with the one unit of IL-6R.The pocket in between the two units (gp130 and IL-6R) is the IL-6 binding site. A number of H-bond formed with GLU278, MET250, VAL230, LYS281 and ASN224. The ACE value was −184.01. But the highest ACE value of −413.76 and −355.92 positions were found at the extra arm on gp130. This can also block the heterodimeric unit formation. At the active site location, the TFMG binding energy was −5.52 and −5.24 Kcal/mol (Tables 1, S2, Figs. 3 and S11).
CD40 and CD40R
CD40 is expressed in macrophages, dendritic cells and B cells. CD40 has a direct involvment in T-helper cell-stimulation, resulting in proinflamatory cytokine production. Over activation of CD40 inducecscytokine level which augment the inflamatory-responses. According to ACE value (−353.37) TFMG showed moderate affinity to CD40R (Tables 1, S2, Figs. 3, S19). Active site interaction was not found for this interaction. But TFMG was found to block a particular location at the β-barral structure of CD40R. TFMG formed rigid bond (ACE value: −159.35) with CYS218, ALA130 and GLN246. Hydrogen bonds were formed with VAL247 and CYS178.
TFMG binding to stress regulated HIF1α
TFMG was found to interact with amino acids PHE572, ASP569, TYR565, HYP564 and LEU562. At this location affinity of TFMG was high enough, showing the binding energy value ranged from −6.47 to −5.11 Kcal/mol. Among 10 postures 6 were found at that location (Tables1, S2, and Fig. 3). The lowest Ki value was 18.16 µM. The PatchDock analysis showed that ACE value also high at this location i.e. −446.61. Most of the values were lower than −200 indicating higher affinity of TFMG at active site forming H-bonds.
TFMG binding to apoptotic regulators
Bcl2
According to Touré et al. (2013) saccharin analogue 6 was a potent Bcl-2 inhibitor which forms electrostatic interaction with TYR67 and also blocked ARG66 residue in Bcl-2. In our study TFMG was also found to interact with ARG66 and have blocked the channel similar to the inhibitor. At this site a strong binding energy of TFMG (−7.23, −9.92, −7.02 and −6.43 Kcal/mol) was observed with a lower Ki value of 5.04 µM. According to ACE value −309.63 in PatchDock analyses, indicated the interaction affinity of TFMG with Bcl-2. At that position TFMG formed rigid bond with both TYR67 and ASP70. Hydrogen bonds were also observed with GLT104 and ALA108 (Tables 1, S2, Figs. 3 and S12).
Bax
To inhibit apoptosis, BAX is inserted into the mitochondrial membrane. Among the 9 α-helix present in BAX molecule (Fig. 3). Where ASP48 was present in the center part of long loop region, LYS21 was in α1 and ARG145 was present in α6 (Tables 1, S2, Figs. 3 and S13).
MMP 2 and MMP9
Matrix-metalloproteinases (MMPs), zinc-dependent endopeptidase degrades extracellular proteins and acts in morphogenesis, angiogenesis, tissue-repair and tumor-invasion. It is known that active-site-inhibitor interacts with amino acids TYR112/TYR142/ILE141and others. In our study TFMG was found to interact with same cleft and with some common amino acids like TYR142/PRO140/ALA87/ALA85 (Fig. 3). Hydroxamate inhibitor interacts with ALA191/GLU402/LEU188/LEU187/GLY186/TYR423 and PRO421. In our study TFMG was also found to block the active-site, mainly PRO421/TYR423 (Tables 1, S2, Figs. 3 and S14). It showed active site binding energy value of −6.61 Kcal/mol and showing strong hydrogen bond energy of −6.857 kcal/mol and the Ki value was 14.29 µM. On the other hand affinity of TFMG in the form of Atomic Contact Energy (ACE) value was too much significant as it ranged from −625.14 to −258.52.
Caspase3 and caspase 8
Inhibition of caspase 3 and 8can block apoptosis (Brentnall et al., 2013) and can terminate osteoarthritis and other diseases. Report reveals inhibitor complex interact with the amino acids HIS121/CYS163/TYR204/TRP206 and PHE256. In our study TFMG was also found to interact with those exact same amino acids HIS121/CYS163/TYR204. It also formed rigid bonds with ASP253, THR255, PHE256, TYR204 and SER205. It also formed H-bonds with SER120, ARG64 and ARG207. That pocket was completely blocked by one molecule of TFMG. It showed the ACE value of −356.51 with surface area occupancy of 780.90. In this location binding energy of TFMG were −6.71 and −5.52 with Ki value of 12.1 and 89.21 µM, respectively. Caspase 8 inhibits the FAS-mediated pathway to control caspase 3 target. TFMG binding was observed at the nearby position of active site location containing amino acids Cys285/His237/Arg341 (Tables 1, S2, Figs. 3 and S15).
Monomeric and dimeric TLR2
TLR2 has a significant function in recognition of microbial lipoprotein in combination with TLR1 or TLR6 (Oliveira-Nascimento et al., 2012). TFMG interact best with the dimeric form of TLR2 with the highest binding energy of −9.77/−8.77/−7.17 Kcal/mol at the active site. Both in mono and dimeric TLR2, amino acids GLU177, LEU176 and THR174 were found to form H-bond with TFMG. In addition to that, ASP160 from both units (dimeric TLR2) were found H-bonded with TFMG (Tables 1, S2, and Fig. 3).
Homodimeric and heterodimeric TLR4
TLR4 is the major component of innate immune responses, which recognizes the lipopolysaccharide (LPS) at the surface of Gram negative bacteria] (Park et al., 2009). TFMG binds to the homodimer attachment site where it formed rigid bonds with MET557 and LYS660. Nevertheless, it formed H-bonds with THR558/SER534/GLU509/GLN510. And GLY480/GLN505/HIS456/ASN433/THR459 and ASN481 formed H-bonds with TFMG (Tables 1, S2, Figs. 3 and S16, S17).
NF-κβ
NF-κβ is a transcription factor with 10-base pair κβ site of immunoglobulin κ light chain. Structurally, NF-κβ is a homodimer and each tertiary unit has two domains (1 & 2). The junction of two domains in two tertiary units together forms the active site of DNA binding. The complete passage was blocked by TFMG during molecular docking analysis in our study. (Tables 1, S2, Figs. 3, S18). TFMG formed rigid bond with HIS64, ASN136 and LYS49. Hydrogen bonds were formed with other amino acids like LEU137 and GLY52. ARG56 was blocked by one TFMG molecule binding. The highest TFMG binding energy was −8.23 Kcal/mol. But at the active site location three binding energies were −6.76 and −6.29 with respective Ki values of 5.2, 11 and 24.43 µM.
Data from in vivo experiments
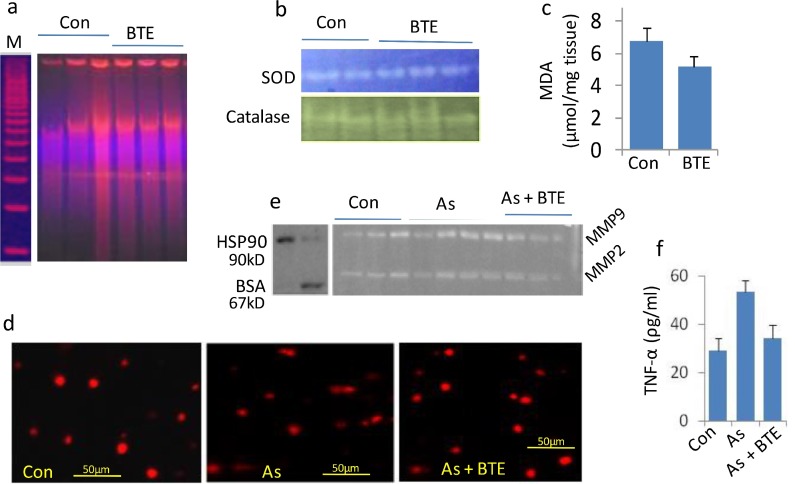
Present result suggests that BTE has multi-protective roles for different cellular components. And this protection was implicated from biochemical, cellular and molecular levels. It can protect DNA and cell matrix structures and their stability from the emerging damages raised by arsenic. Another major cell damaging (both apoptotic and necrotic) MMPs were inhibited by BTE. The inflammatory components like TNF-α and NFκβ introduce adverse cellular events. BTE significantly decreased TNF-α level in the serum sample of the arsenic intoxicated rats. It activates catalase and SOD possibly resulting in minimizing free-radical burden. And that effect is reflected in serum MDA level, DNA stability in BTE alone or BTE+As group. Low DNA ladder and less comet tails were observed in BTE treated groups. Other physiological effects were also verified by liver function testing (SGOT, SGPT, ALP etc.), kidney function testing (urea and creatinine) and found the present dose is very much safe (Maiti et al., 2017; Acharyya et al., 2014, 2015).
Discussion
Our in vivo data suggest the possible versatile interaction ability of the black tea extract (BTE) to different cellular components. That is reflected from its protective effects at cellular and molecular level. Catechin and theaflavin are the major flavonoid components in BTE. Theaflavin level is comparatively higher in black tea than the green tea. Strong anti-inflammatory and anti-degenerative efficacy of BTE was observed from its ability in decreasing TNF and MMPs levels (Fig. 4 ). Our bioinformatics and AutoDock modeling data suggest direct binding effects of TFMG with MMPs, TNF and TNFR1and2 correlating tissue protective effects. In our bioinformatics study we evaluated the docking and binding effects of all catechin and theaflavin derivatives and observed the best outcome from TFMG. The docking results were also better than that of EGCG which is a known antioxidant and anti-inflammatory agent. This versatile binding feature of TFMG has been critically analyzed from our present in silico data and unique physico-chemical properties of this flavonoid.
Fig. 4.
Protective effects of black tea extract (BTE) against arsenic induced rat lung injuries have been demonstrated. Black tea extract only (BTE) showed better Protective effects of DNA stability than that of control (a). BTE showed protection and/or strong restoration of SOD, catalase (b) and decreased free radical damage as MDA level (c). In arsenic induced model rat lung injury, it showed protection in DNA protection as lower number of comets (d). Matrix damaging MMP2and MMP9 was also decreased in BTE supplemented group than that of arsenic treated group (e). BTE showed strong anti-inflammatory effects by restoring the TNF-α compared to the arsenic treated group (f).
Compared with the known and currently used immune-supporting prophylactic-drugs like HCQ (Table S1), DEX (Table S5) or EGCG (Table S6), TFMG showed the best binding-energy and lower Ki-values. TFMG has also been extensively compared with few more drugs or pharmacologically active agents like catechin derivatives, EGCG, theaflavin-digallate, hesperidin, quercetin, kaempferol and observed that TFMG generated 3–4 fold better results (data not shown). The hydroxyl‑groups of the benzocycloheptene 1 ring were important for TF function (Ohba et al., 2017). The inhibition by TF of Akt and nuclear factor kappaB (NFκβ) with a lower COX-2 was explained by TF-ability to dock in NFκβ structure (Fig. 3). Eventually, NFκβ-mediated inflammation (enhanced in COVID-19 infection) was diminished. Beside the transcriptional control, regulations at cell-signaling level by Fas with Fas-ligand and TNF-α with TNFR1and2 interaction are also important. As a result, SARS-CoV-2 initiated severe inflammation and could directly infect T lymphocytes leading to lymphopenia (Li et al., 2020); this can be blocked by TFMG (Tables S1 and S3 and Figs. 3, S20). Lowering the serum TNF level was also observed in our experimental rat results.
The signaling of CD40-CD40L correlates with SARS-CoV-2 infected lung. The IL-1orIL-6 signaling pathways (their receptors were blocked by TFMG, Fig. 3, Table 1 and Fig. S1–S24) were also involved (Wu et al., 2020). Patients with chronic inflammatory-diseases have adverse impact from IL1or6 leading to intrinsic immune-dysfunction (Wu et al., 2020).TFMG blocking of NFκβ-TNFα-CD40 could prevent both canonical and non-canonical inflammatory responses. These types of signaling responses were associated with IL1 regulations (Wu et al., 2020). TNF-α receptors, TNFR1 and 2 were blocked by TFMG (Table 1, Figs. 3 and S21). During COVID-19 viral sepsis TNF (macrophages, DCs, endothelium, lymphocytes, myocytes), and its interaction to TNFR1and2 were significantly up-regulated (Mangalmurti and Hunter, 2020). TFMG binding to Bcl-2 and BAX and possible modulation of their functions may resist lung cellular-apoptosis and viral-propagation (Cheong et al., 2020).
One of the major targets of COVID-19 is the vasovascular system. The angiotensin is the key regulator of this system. Nevertheless, stress-induced factor HIF1α has a direct influence on angiotensin associated AT1 (angiotensin-II receptor) and IL1, IL6-mediated inflammatory-responses. Under physiological conditions, there is an equilibrium between ACE-1 (catalyzes angiotensin-I to angiotensin-II) and ACE-2 (catalyzes angiotensin II to angiotensin1–7) function. However, during the hypoxemia in COVID-19, the ACE-1 was up-regulated by the hypoxia-inducible factor-1 (HIF-1); meanwhile the expression of ACE-2 was markedly decreased (Afsar et al., 2020). It has been suggested that increased levels of ACE-2 were positively associated with COVID-19 infection (Arias-Reyes et al., 2020). Thus, both hypoxemia and related ACE-2 up-regulation worsen clinical outcomes in COVID-19. Our result suggests that TFMG blocks both ACE1 and ACE2 and down-regulates angiotensin function. Moreover, it blocks the HIF1α and AT1 and AT2 also. Report revealed that HIF-1αstabilizers may dampen both hypoxia and adverse outcomes in COVID-19 (Arias-Reyes et al., 2020). Indeed, in a recent study, COVID-19 infection was found less prevalent in high altitudes (+2500 m above sea-level) possibly due to physiological acclimatization to hypoxia and down-regulation of ACE-2. Other investigator also proposed that HIF-1α stabilizers may prevent the hypoxia and related adverse outcomes in COVID-19 (Arias-Reyes et al., 2020).
TLR members can up-regulate anti-viral and pro-inflammatory mediators through the activation of NFκβ resulting in a "cytokine storm" (Mangalmurti and Hunter, 2020).TFMG has been shown to block the active sites of TLR2 and TLR4 (Figs. 3, S16, S17 and S22). When SARS CoV-2 infects the upper and lower respiratory tract (acute respiratory syndrome) and bind with TLR, it causes the release of pro-IL-1β which was cleaved by a caspase enzyme. The Caspases are involved in the generation of Fas-dependant apoptotic stimuli. However, TFMG blocks caspase 3and8 proteins suggesting possible termination of TLR dependant IL-1β maturation. Modulator effects of tea flavonoids on TLR and caspase have been demonstrated earlier (Marinovic et al., 2015).
Increase in the level of circulating MMP-9 in COVID-19 patient results in respiratory failure (Ueland et al., 2020). The blocking and inactivation of matrix metalloproteinases (MMPs) by TFMG may terminate this cycle and the pivotal events of MMP in the degradation of tissue matrix. TFMG inhibitory efficacy of MMPs has been shown in our in vivo results and by other research group also (Sazuka et al., 1997). Our previous report on the similarities in the angiotensin binding proteins i.e. ACE, ACE2, AT1, AT2has explained this issues (Maiti et al., 2021). In this investigation TFMG has been shown to bind some common amino acids in the active sites of angiotensin regulating proteins (Fig. 2). As a result, angiotensin production was minimized, inactivation of ACE2 may be discouraging to SARS-CoV-2 and finally AT1 mediated IL1 activation was restricted in low angiotensin II condition. In addition to the antiviral therapy, potential strategies of ACE2 and AT1-R inhibition and novel anti-inflammatory potential which have been demonstrated here by TFMG, could be extremely beneficial (Liu et al., 2020).
Theaflavin's benzotropolone moiety was produced after the condensation between a catechol-type B-ring of EC and a pyrogallol-type B-ring of EGC. Beside B-rings, galloyl esters have significant reactivity also. This justifies better interaction of TFMG with proteins by forming hydrophobic and hydrogen-bonds (Lei et al., 2017). TFs contain a bisflavan substituted10, 20-dihydroxy-3, 4-benzptroplone moiety, resulting in better solubility in more non-polar solvent. So, lipid rich bi-layer of the membrane transportation was facilitated and bioavailability was increased. Though, both of EGCG and theaflavin interacts with BSA (standard protein for drug docking study) but EGCG distort a greater extent of BSA secondary structures (wavelength shift from 342 to 346 nm). However, theaflavin only caused a concentration-dependent quenching of the intrinsic fluorescence intensity of the protein without changing its structure. Flavonoids help in quenching the aromatic amino acid buried in that protein hydrophobic-pocket (Xu et al., 2015). TF exerts dose-duration dependent both static and dynamic quenching increasing its interaction as observed in the current study. Van-der-Waals and hydrophobic-forces favors theaflavins binding to proteins spontaneously, suggested based on positive ∆H and ∆S values. TFMG decreased α-helix and increased the β-sheet with more unfolding in protein secondary structure possibly for optimal binding adjustment (Xu et al., 2015). COVID-19 increases the levels of large number of proteins of both viral and host origin. Host proteins include vaso-active, tissue damaging, oxidative stress inducing and inflammatory proteins. More adverse proteomic environment (both qualitative and quantitative) dominates over sustainable and controlled proteomic environment. Our data suggest that very high levels of adversely over populated proteins may become good targets for TF binding.
Proteins have cavity (enclosed-space) and pocket (disclosed-space) and pockets are more interactive due to solvent-ligand accessibility. Generally, pocket shares important from pharmacological perspective. The binding-affinities were observed significant of TFMG to the selected-proteins. In this study we have used theaflavin 3′-O-gallate. From the stereo chemical point of view; it occupies four different plains with 5 rotatable bonds which are more favorable than in other gallate and catechin (Fig. S1–S24, Table S3). Topological-polar-surface-area, hydrogen-bonding capacity determines thedruggabilityof a ligand by its polar-pocket-accessibility in a protein (Sirk et al., 2011). In Table 4 TFMG showed most compatible to polar-surface-area (264 Ų) and travel-depth (17.2 Å) and height in a protein-pocket (Table S3& S4).TFMG occupied variable molecular-posture in same or different locations of proteins (Figs. S22 and S24). Free bond rotation within a conical-bond-vector in different plains develops a drastic stereochemical effect in the ligand helping it searching its proper-pocket (Bronowska et al., 2000). The more the rotatable bond the more will be the ligand-receptor association. In TFMG, different posture occupancies were observed for the same and different locations (Fig. S22, S23, S24, respectively).
Conclusions
In conclusion, TFMG showed to bind and block a large number of viral and host protein molecules which are associated with several adverse effects. Proteomics data from severely infected COVID-19patients suggests that these proteins (Table 1) expressed 5 to 10fold higher than the uninfected patients (due to higher rate of viral proliferation, inflammatory ‘cytokine storming’ and vaso-activation) (Bojkova et al., 2020). In an urgent quest for searching some suitable drugs, we found TFMG to be a good target for further analysis.
Data availability statement
All data are available if required. All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy
Authors' agreement
We the undersigned declare that this manuscript is original, has not been published before and is not currently being considered for publication elsewhere.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We understand that the Corresponding Author is the sole contact for the Editorial process. He is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs.
CRediT authorship contribution statement
Smarajit Maiti: Visualization, Investigation, Writing – original draft. Amrita Banerjee: Investigation, Writing – original draft. Mehak Kanwar: Investigation.
Declaration of Competing Interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Acknowledgement
Partially by West Bengal State DST (WB/2016/05)
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.phyplu.2022.100237.
Appendix. Supplementary materials
References
- Acharyya N., Chattopadhyay S., Maiti S. Chemoprevention against arsenic-induced mutagenic DNA breakage and apoptotic liver damage in rat via antioxidant and SOD1 upregulation by green tea (Camellia sinensis) which recovers broken DNA resulted from arsenic-H2O2 related in vitro oxidant stress. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2014;32:338–361. doi: 10.1080/10590501.2014.967061. [DOI] [PubMed] [Google Scholar]
- Acharyya N., Sajed, Ali S., Deb B., Chattopadhyay S., Maiti S. Green tea (Camellia sinensis) alleviates arsenic-induced damages to DNA and intestinal tissues in rat and in situ intestinal loop by reinforcing antioxidant system. Environ. Toxicol. 2015;30:1033–1044. doi: 10.1002/tox.21977. [DOI] [PubMed] [Google Scholar]
- Ackermann M., Verleden S.E., Kuehnel M., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afsar B., Kanbay M., Afsar R.E. Hypoxia inducible factor-1 protects against COVID-19: a hypothesis. Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.109857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Reyes C., Zubieta-DeUrioste N., Poma-Machicao L., Aliaga-Raduan F., Carvajal-Rodriguez F., Dutschmann M., Schneider-Gasser E.M., Zubieta-Calleja G., Soliz J. Does the pathogenesis of SARS-CoV-2 virus decrease at high-altitude? Respir. Physiol. Neurobiol. 2020;277 doi: 10.1016/j.resp.2020.103443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Chatterjee J. Efficient extraction strategies of tea (Camellia sinensis) biomolecules. J. Food Sci. Technol. 2015;52(6):3158–3168. doi: 10.1007/s13197-014-1487-3. JunEpub 2014 Aug 6. PMID: 26028699; PMCID: PMC4444893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banarjee A., Kanwar M., Maiti S. Theaflavin-3’-O-gallate a black-tea constituent blocked SARS CoV-2 RNA dependent RNA polymerase active-site with better docking result than remdesivir. Drug Res. 2021;71:462–472. doi: 10.1055/a-1467-5828. [DOI] [PubMed] [Google Scholar]
- Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentnall M., Rodriguez-Menocal L., De Guevara R.L., Cepero E., Boise L.H. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013;14:32. doi: 10.1186/1471-2121-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronowska A., Sylte I., Edvardsen Ø., Østensen R., Leś A., Chilmonczyk Z. Theoretical models of interactions between buspirone analogues and 5-HT1A and 5-HT2A serotonin receptor subtypes. Acta Pol. Pharm. 2000;57:40–45. [PubMed] [Google Scholar]
- Cheong D.H.J., Tan D.W.S., Wong F.W.S., Tran T. Anti-malarial drug, artemisinin and its derivatives for the treatment of respiratory diseases [published online ahead of print. Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020;22:1–13. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S., Xu D., Saeeduddin M., Riaz A., Zeng X. Characterization of molecular structures of theaflavins and the interactions with bovine serum albumin. J. Food Sci. Technol. 2017;54:3421–3432. doi: 10.1007/s13197-017-2791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu L., Zhang D., et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Wang T., Zhou Y., Zhao Y., Zhang Y., Li J. Potential role of ACE2 in coronavirus disease 2019 (COVID-19) prevention and management. J. Transl. Int. Med. 2020;8(1):9–19. doi: 10.2478/jtim-2020-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Lu H., Zhao Q., et al. Theaflavin derivatives in black tea and catechin derivatives in green tea inhibit HIV-1 entry by targeting gp41. Biochim. Biophys. Acta. 2007;1770:312. doi: 10.1016/j.bbagen.2005.02.012. [published correction appears in. [DOI] [PubMed] [Google Scholar]
- Maiti S., Acharyya N., Ghosh T.K., Ali S.S., Manna E., Nazmeen A., Sinha N.K. Green tea (Camellia sinensis) protects against arsenic neurotoxicity via antioxidative mechanism and activation of superoxide dismutase activity. Cent. Nerv. Syst. Agents Med. Chem. 2017;17:187–195. doi: 10.2174/1871524917666170201145102. [DOI] [PubMed] [Google Scholar]
- Maiti S., Banerjee A., Kanwar M. In siliconigellidine (N. sativa) bind to viral spike/active-sites of ACE1/2, AT1/2 to prevent COVID-19 induced vaso-tumult/vascular-damage/comorbidity. Vascul. Pharmacol. 2021;138 doi: 10.1016/j.vph.2021.106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020 doi: 10.1016/j.immuni.2020.06.017. [published online ahead of print. S1074-7613r 30272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinovic M.P., Morandi A.C., Otton R. Green tea catechins alone or in combination alter functional parameters of human neutrophils via suppressing the activation of TLR-4/NFκB p65 signal pathway. Toxicol. In Vitro. 2015;29:1766–1778. doi: 10.1016/j.tiv.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Mhatre S., Srivastava T., Naik S., Patravale V. Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: a review. Phytomedicine. 2020;17 doi: 10.1016/j.phymed.2020.153286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre S., Naik S., Patravale V. A molecular docking study of EGCG and theaflavin digallate with the druggable targets of SARS-CoV-2. Comp. Biol. Med. 2021;129 doi: 10.1016/j.compbiomed.2020.104137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J.M., Delaugerre C., Le Goff J., et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 2020;50:384. doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazmeen A., Chen G., Ghosh T.K., Maiti S. Breast cancer pathogenesis is linked to the intra-tumoral estrogen sulfotransferase (hSULT1E1) expressions regulated by cellular redox dependent Nrf-2/NFκβ interplay. Cancer Cell. Int. 2020;20:70. doi: 10.1186/s12935-020-1153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba M., Oka T., Ando T., et al. Antiviral effect of theaflavins against caliciviruses. J. Antibiot. 2017;70:443–447. doi: 10.1038/ja.2016.128. [DOI] [PubMed] [Google Scholar]
- Oliveira-Nascimento L., Massari P., Wetzler L.M. The role of TLR2 in infection and immunity. Front. Immunol. 2012;3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostade X.V., Tavernier J., Prange T. Localization of the active site of human tumour necrosis factor (hTNF) by mutational analysis. EMBO J. 1991;10:827–836. doi: 10.1002/j.1460-2075.1991.tb08015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.S., Song D.H., Kim H.M. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature. 2009;458:1191–1196. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Sazuka M., Imazawa H., Shoji Y., Mita T., Hara Y., Isemura M. Inhibition of collagenases from mouse lung carcinoma cells by green tea catechins and black tea theaflavins. Biosci. Biotechnol. Biochem. 1997;61:1504–1506. doi: 10.1271/bbb.61.1504. [DOI] [PubMed] [Google Scholar]
- Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation 598 of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Sirk T.W., Friedman M., Brown E.F. Molecular binding of black tea theaflavins to biological membranes: relationship to bioactivities. J. Agric. Food Chem. 2011;59:3780–3787. doi: 10.1021/jf2006547. [DOI] [PubMed] [Google Scholar]
- Takemoto M., Takemoto H. Synthesis of theaflavins and their functions. Molecules. 2018;23:918. doi: 10.3390/molecules23040918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueland T., Holter J.C., Holten A.R., Müller K.E., Lind A., Bekken G.K., Dudman S., Aukrust P., Dyrhol-Riise A.M., Heggelund L. Distinct and early increase in circulating MMP-9 in COVID-19 patients with respiratory failure. J. Infect. 2020;81:e41–e43. doi: 10.1016/j.jinf.2020.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doremalen N., Bushmaker T., Morris D.H., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.Y., Wang S.H., Wu C.H., Yen L.C., Lai H.F., Ho C.L., Chiu Y.L. In silico immune infiltration profiling combined with functional enrichment analysis reveals a potential role for naïve B cells as a trigger for severe immune responses in the lungs of COVID-19 patients. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0242900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Wang Q., Zhang W., Bing H., Li Z., Zeng X.X., Sun Y. Inhibitory activities of caffeoylquinic acid derivatives from Ilexkudingcha C. J. Tseng ona-glucosidase from Saccharomyces cerevisiae. J. Agric. Food Chem. 2015;63:3694–3703. doi: 10.1021/acs.jafc.5b00420. [DOI] [PubMed] [Google Scholar]
- Zhao L., Niu H., Liu Y., Wang L., Zhang N., Zhang G., Liu R., Han M. LOX inhibition downregulates MMP-2 and MMP-9 in gastric cancer tissues and cells. J. Cancer. 2019;10:6481–6490. doi: 10.7150/jca.33223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu M., Yang F., Zhou W., Liu A., Du G., Zheng L. In vitro anti-influenza virus and anti-inflammatory activities of theaflavin derivatives. Antiviral Res. 2012;94:217–224. doi: 10.1016/j.antiviral.2012.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available if required. All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy