Abstract
The type 2 bradykinin receptor (B2R) is a G protein-coupled receptor (GPCR) in the cardiovascular system, and the dysfunction of B2R leads to inflammation, hereditary angioedema, and pain. Bradykinin and kallidin are both endogenous peptide agonists of B2R, acting as vasodilators to protect the cardiovascular system. Here we determine two cryo-electron microscopy (cryo-EM) structures of human B2R-Gq in complex with bradykinin and kallidin at 3.0 Å and 2.9 Å resolution, respectively. The ligand-binding pocket accommodates S-shaped peptides, with aspartic acids and glutamates as an anion trap. The phenylalanines at the tail of the peptides induce significant conformational changes in the toggle switch W2836.48, the conserved PIF, DRY, and NPxxY motifs, for the B2R activation. This further induces the extensive interactions of the intracellular loops ICL2/3 and helix 8 with Gq proteins. Our structures elucidate the molecular mechanisms for the ligand binding, receptor activation, and Gq proteins coupling of B2R.
Subject terms: Receptor pharmacology, Cryoelectron microscopy, G protein-coupled receptors
Type 2 bradykinin receptor (B2R) is essential in vasodilation and cardioprotection. Here the authors present two cryo-EM structures of human B2R-Gq in complex with bradykinin and kallidin to elucidate the mechanisms for ligand binding, receptor activation, and Gq proteins coupling.
Introduction
The kallikrein-kinin system (KKS) consists of kininogens, kallikrein enzymes, kinins, type 1 and type 2 bradykinin receptors (B1R and B2R), which plays important roles in vasodilation, inflammation, vascular permeability, cardioprotection, coagulation, and pain1–4. Kinins are short-lived peptides of various lengths and they are produced by enzymatic hydrolysis of precursor kininogens5. One derivative from the high molecular weight kininogen is called bradykinin (RPPGFSPFR), and another derivative from the low molecular weight kininogen is called kallidin (KRPPGFSPFR)6–8. Both kinins with only one N-terminal lysine difference are essential bioactive factors that regulate multiple physiological and pathological progress including blood circulation, smooth muscle contraction, and inflammatory responses. Increasing levels of kinins or dysfunction of their receptors lead to hereditary angioedema, an autosomal dominant disorder that is characterized by swelling of mucosal, submucosal tissue or subcutaneous9,10. The coronavirus disease (COVID-19) patients got pulmonary edema in the early stage mainly due to the increasing levels of B1R and B2R activation as well as their agonists stimulation11–14. And the B2R selective antagonist icatibant (D-Arg-Arg-Pro-Hyp-Gly-Thi-Ser-D-Tic-Oic-Arg) was suggested to relieve the symptoms15.
B1R and B2R belong to the γ-branch of class A G protein-coupled receptors (GPCRs) sharing the sequence identity of 32%. B2R is constitutively expressed in human tissues8. While, B1R is expressed robustly in the conditions of inflammation and oxidative stress16. Both B1R and B2R signal through Gq proteins pathway which promotes the phospholipase C activation and calcium mobilization17,18. Both the peptide ligands bradykinin and kallidin are endogenous full agonists of the human B2R19. The binding affinity of bradykinin is much higher for B2R than B1R. While, the endogenous B1R peptide agonist desArg10-kallidin (KRPPGFSPF) lacking the C-terminal arginine shows 100,000-fold higher affinity for B1R than B2R20,21.
Extensive efforts on developing new antagonists with higher potency and selectivity were hampered by the lacking of three-dimensional structures of B2R, and the molecular mechanisms for the ligand binding, receptor activation, and Gq proteins coupling were still elusive.
Here we determine two cryo-electron microscopy (cryo-EM) structures of B2R-Gq complexes in the presence of bradykinin and kallidin, respectively. Our structures reveal the critical interactions in the ligand-binding pocket and on the B2R-Gq interface, which shed light on a better understanding of the ligand selectivity and G proteins selectivity.
Results
Cryo-EM structure determination of B2R-Gq complexes
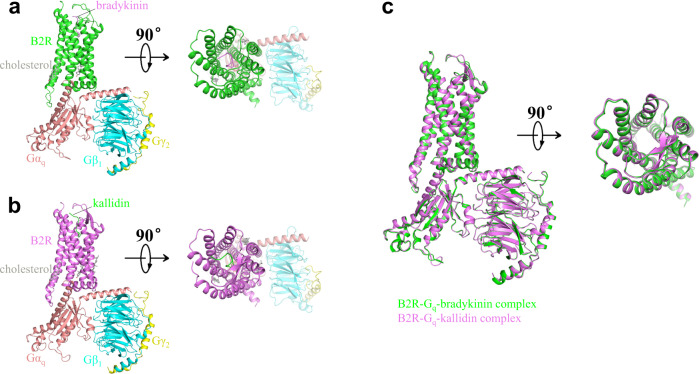
To obtain the active structures of B2R in complex with Gq heterotrimer, two B2R full agonists bradykinin and kallidin were used for the complex formation. The human B2R protein was engineered by replacing the N-terminal 28 amino acids with a maltose-binding protein (MBP) without any mutations (Supplementary Fig. 1). The engineered B2R used for the structure determination showed identical downstream calcium signaling to the wild-type (WT) B2R, indicating the N-terminal truncation and MBP fusion did not alter the B2R function (Supplementary Fig. 2a, Supplementary Table 1). The human Gαq protein was modified with two dominant-negative mutations R183Q and Q209L to stabilize the complexes22,23. The ability of the double-mutated Gq to activate the downstream calcium signaling was significantly decreased compared to the WT-Gq, since the R183Q mutation around the nucleotide-binding site decreased the affinity of GDP by disrupting the hydrogen bonds between R183 and GDP molecule23 (Supplementary Fig. 2a, Supplementary Table 1). The human Gβ1 and Gγ2 proteins were both wild types. GPCR-G proteins complexes were usually determined using antibodies or NanoBit tethering technology to stabilize the complexes24–26. However, our B2R-Gq complexes were found to be very stable during the purification and cryo-EM procedures without these stabilizing agents, and thus our B2R-Gq structures might represent more natural conformations. The B2R-Gq proteins were co-expressed in the insect cells and solubilized in lauryl maltose neopentyl glycol (LMNG) with agonists. During the cryo-EM data processing, 2D classification revealed averages with clear α-helices for the complex and 3D classification identified the desired particle partition showing well-defined features for the complex (Supplementary Figs. 3 and 4). The two structures of B2R-Gq in complex with bradykinin and kallidin were determined at the resolutions of 3.0 and 2.9 angstrom, respectively (Fig. 1; Supplementary Table 2). The overall structure of bradykinin-bound B2R was almost identical to that of kallidin-bound B2R, with Cα root mean square deviation (R.M.S.D.) values of 0.240 Å for the whole complex, 0.259 Å for B2R, 0.211 Å for Gαq, 0.214 Å for Gβ1, and 0.351 Å for Gγ2 (Fig. 1). We mainly focused on the B2R-Gq-kallidin complex for the following discussion unless otherwise indicated. From the cryo-EM density maps, the residues of transmembrane helices (TMs) of B2R could be clearly identified (Supplementary Fig. 5). However, the N-terminus of B2R (Leu29 to Lys46) and the α-helical domain (AHD) of Gq showed weak densities due to flexibility in these regions like many other GPCR-G protein complex structures. The densities of the orthosteric ligand-binding pocket and B2R-Gq interface were unambiguous, enabling near-atomic modeling building for the ligand binding and G protein coupling (Supplementary Fig. 5).
Fig. 1. Overall architectures of B2R-Gq complexes.
a, b Models for the B2R-Gq in complex with bradykinin (a, B2R, green; bradykinin, violet) and kallidin (b, B2R, violet; kallidin, green). c Structural comparison of bradykinin-bound (green) and kallidin-bound (violet) B2R.
Overall architectures of B2R-Gq complexes
B2R shares the sequence identity of 28% and 27% to the angiotensin receptors AT1R and AT2R, respectively, which are also GPCRs regulating the cardiovascular system and binding the endogenous peptide agonist angiotensin II (DRVYIHPF). Superposition of the Gq-bound active B2R structure with the inactive AT1R structure27 revealed a 9.0 Å outward movement of the transmembrane helix 6 (TM6) when measuring the Cα atoms of the residues at 6.31 (Ballesteros-Weinstein numbering)28, as shown in Supplementary Fig. 6b. Besides, at the cytoplasmic side, there was a 2.4 Å outward shift of TM1 and a 2.7 Å inward shift of TM7. Compared to the active AT1R structure29, a 6.3 Å inward movement of TM5 in B2R induced substantial compaction of the G protein binding pocket for signal transduction (Supplementary Fig. 6a). However, the transmembrane helices of both antagonist-bound30 and agonist-bound31 AT2R showed active conformations, similar to the active B2R structure (Supplementary Fig. 6d). The helices 8 of the active AT1R29 and other GPCR-G protein complexes, such as β2 adrenergic receptor (β2AR)-Gs32, M1 muscarinic acetylcholine receptor (M1R)-G1133, and histamine H1 receptor (H1R)-Gq24 lay proximately to the membrane, while the helix 8 of B2R adopted a ~25° downward rotation that was stabilized by the electrostatic interactions between R3388.49 of B2R and E355G.H5.22 of α5-helix of Gq (Supplementary Figs. 6a and 7). On the contrary, the helix 8 of antagonist-bound AT2R interacted with the intracellular tips of TM3, TM5, and TM6, functioning as a gatekeeper blocking the binding of G proteins (Supplementary Fig. 6d). At the extracellular side of B2R, except for the conserved disulfide bonds between C1303.25 and C211ECL2 bridging TM3 and extracellular loop 2 (ECL2), an additional disulfide bond linking the N-terminus and TM7 were observed between C47N-term and C3047.25. Similar disulfide bonds were also found in AT1R29 and AT2R31, while in other GPCRs they might bridge the residues between ECL1 and ECL234 or within ECL232 or ECL324,35 (Supplementary Fig. 8a). ECL2 of B2R formed a β-sheet which was a common feature in many peptide GPCRs such as AT1R, AT2R, and neurotensin receptor 1 (NTSR1)29,30,36. Interestingly, ECL2 of B2R on top of the ligand-binding pocket was stabilized by the hydrogen bonds with the residues D122ECL1, W123ECL1, and E1273.22 (Supplementary Fig. 8b). In the non-peptide GPCRs, ECL2 could form a short α-helix32 or interact with ECL135. As shown in Supplementary Fig. 9, three structured steroids (cholesterols) were found in the intracellular clefts between TM2-TM4, TM3-TM4, and TM6-TM7, with extensive hydrophobic interactions, which might help maintain the functional conformations of B2R and regulate the movements of transmembrane helices.
Molecular determinants of B2R-ligand binding
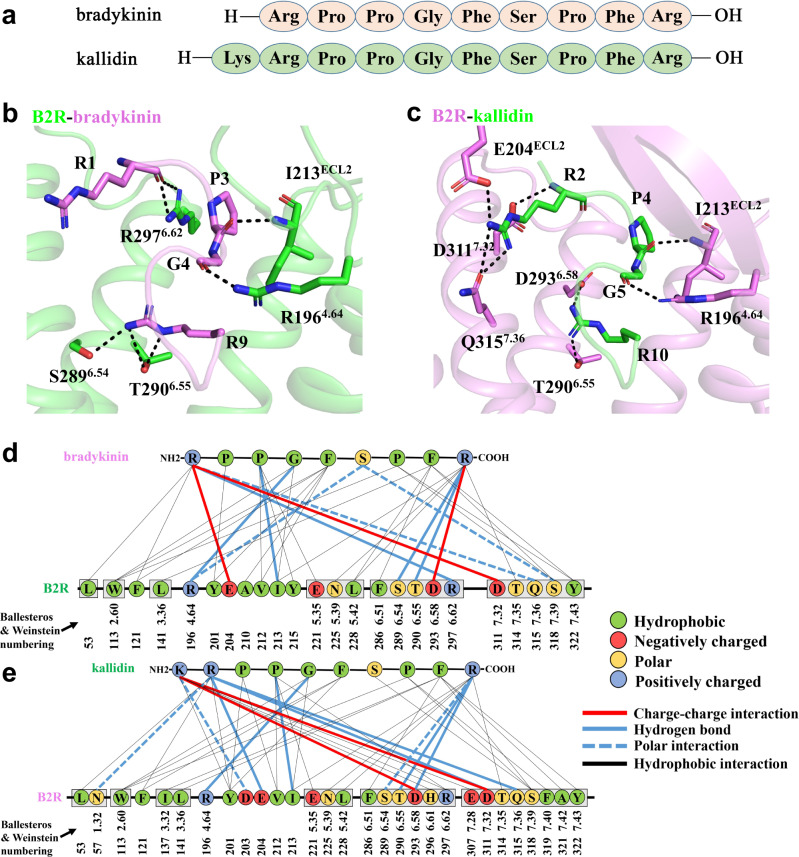
In our determined B2R-Gq structures, both ligands bradykinin and kallidin adopted S-shaped conformations with their C-termini inserting deeply into the orthosteric binding pocket and their N-termini extending to the extracellular side (Fig. 2). Extensive hydrogen bonds, polar, and hydrophobic interactions were found for the ligands binding to B2R (Fig. 2). The bradykinin and kallidin were both positively charged at the N and C-termini, where the arginines or lysine formed polar interactions with glutamates or aspartic acids of B2R. In the kallidin-bound structure, R2 and R10 of kallidin (referred to R2K and R10K, respectively) contributed most hydrogen bonds and polar interactions with B2R including N571.32, E204ECL2, D3117.32, and Q3157.36 for R2K and S2896.54, T2906.55, and D2936.58 for R10K. Pairs of P4K-I213ECL2 and G5K-R1964.64 were suggested to function as anchor points to avoid the conformational perturbation of kallidin and enhance its binding affinity. This was supported by our calcium mobilization assays that R1964.64A mutation completely abolished the kallidin-induced B2R activation to the downstream calcium signaling, and I213ECL2A mutation significantly decreased the kallidin-induced B2R activation with the EC50 value of 150-fold higher than the WT-B2R (Supplementary Fig. 2c, Supplementary Table 3). Bradykinin and kallidin shared similar interactions with B2R, except for the R1 of bradykinin (referred to R1B), which was at the equivalent position as R2K, formed the hydrogen bond with R2976.62 rather than Q3157.36, as well as the additional polar interactions and ionic interactions formed by K1K with D203ECL2, D2936.58, and D3117.32.
Fig. 2. Molecular determinants of B2R-ligand binding.
a Sequences of bradykinin and kallidin. b, c B2R-ligand interactions in the orthosteric ligand-binding pockets of the active B2R-Gq complexes with bradykinin (b, shown as violet sticks) and kallidin (c, shown as green sticks). Hydrogen bonds between B2R and agonists were labelled as dotted lines, and all residues participating in the interactions were shown as sticks. d, e Diagrams of the contacts between B2R and bradykinin (d) or kallidin (e).
Intriguingly, the charge distribution of the ligand-binding pocket determined the sigmoidal binding poses of the two peptides. Negatively charged residues such as glutamates and aspartic acids (D203ECL2, E204ECL2, E2215.35, D2936.58, E3077.28, and D3117.32) located at the entrance, while the bottom of the pocket was found to be mostly hydrophobic (Supplementary Fig. 10). Consequently, the positively charged R10K/R9B bent upwards to form electrostatic interactions with D2936.58, which was further stabilized by the interactions between P4K/P3B or G5K/G4B and B2R. Moreover, two intramolecular hydrogen bonds G4B-R9B/G5K-R10K and S6B-R9B/S7K-R10K maintained the S-shaped conformations of the peptides (Supplementary Fig. 11c). Surprisingly, the side chains of D203ECL2, E204ECL2, E2215.35, D2936.58, E3077.28, and D3117.32 covered the ligand-binding pocket by forming an anion trap preventing the ligand dissociation by the electrostatic interactions. Hydrophobic interactions and van der Waals forces also played important roles for the ligand binding to B2R, P3K/P2B with Y201ECL2, and P4K/P3B with V212ECL2/E2215.35, as well as extensive hydrophobic contacts among F6K/F5B with the residues from TM2, ECL1, and ECL2, and F9K/F8B with the residues from TM3, TM6, and TM7 (Fig. 2d, e, Supplementary Fig. 2b).
A recent study also reported the bradykinin-bound B2R structure, as well as the desArg10-kallidin-bound B1R structure, using the NanoBit strategy37. The two B2R structures exhibited identical conformations with R.M.S.D. values of 0.595 Å (for Cα atoms in receptors). Comparing the ligand-binding pockets of our determined bradykinin-bound B2R structure with the desArg10-kallidin-bound B1R structure revealed the molecular basis for kinin selectivity on B1R and B2R37. DesArg10-kallidin was a bradykinin derivative lacking the C-terminal arginine but with the N-terminal lysine. It was obvious that K1183.33 and R2025.38 of B1R could form electrostatic interactions with F9 of desArg10-kallidin (referred to F9DK), while the cognate residues in B2R were S1383.33 and T2245.38, which were unable to provide polar interactions to stabilize F8B (Supplementary Fig. 12b). Moreover, R9B could not fit into the narrow pocket consisting of R2025.38, Y2666.51, and E2736.58 of B1R, and there would be a severe steric hindrance between R9B and R2025.38 of B1R. On the contrary, the side chains of their cognate residues in B2R, T2245.38, F2866.51, and D2936.58, were small enough to provide space for R9B (Supplementary Fig. 12c). These differences provided the structural basis for the preference of bradykinin to B2R and desArg10-kallidin to B1R.
The structures of kallidin-bound B2R, AngII-bound AT1R, and AngII-bound AT2R were quite similar with R.M.S.D. values of 1.26 Å for B2R/AT1R and 1.07 Å for B2R/AT2R (for all Cα atoms in receptors)38–40. All the peptide agonists in these structures adopted their C-termini settling at the bottom of pockets and N-termini extending to the extracellular side. The AngII in AT1R exhibited anti-parallel β strands with the N-terminus and ECL2, compared to the unstructured N-termini of AngII in AT2R and kallidin in B2R (Supplementary Fig. 13b). Although these peptides showed diverse conformations at the N-termini, backbones of them converged in the middle and at the C-termini. The crooked backbone at F9K resulted in upward positioning of R10K, while it was absent in AngII. Intramolecular hydrogen bonds among G5K, S7K, and R10K in kallidin, as well as Y4 and F8 in AngII, stabilized the C-terminal shapes of peptides (Supplementary Figs. 11c and 13c). The pocket entrances of B2R, AT1R, and AT2R were similar, and the negatively charged D6.58 and D7.32 at the entrances were highly conserved, which interacted with the positively charged arginines of kallidin and AngII (Supplementary Fig. 11b and 13d). In the middle of kallidin, hydrophobic residues P4K and G5K formed hydrogen bonds with I213ECL2 and R1964.64. Similar interactions were also found in AngII-bound AT1R (Supplementary Fig. 11d). F9K in B2R and F8 in AT1R and AT2R were settled at the bottom of pockets forming extensive van der Waals forces to further stabilize the binding of peptides. Similar peptide agonist-binding patterns in the orthosteric pockets of B2R, AT1R, and AT2R indicated that they might share common activation mechanisms to propagate the extracellular signals to the intracellular G protein coupling.
Molecular mechanisms of B2R-Gq coupling
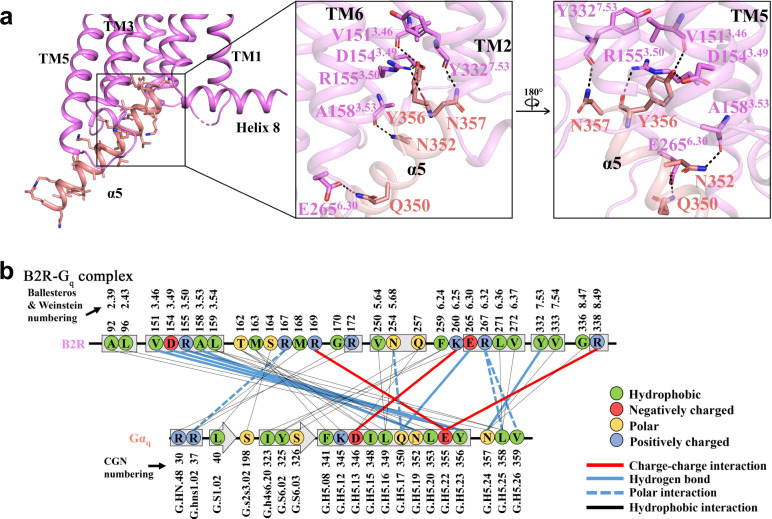
Extensive interactions from TMs2-3, TMs5-7, ICLs2-3, and helix 8 of B2R, with the α5-helix, αN-helix, and αN-β1 loop of Gq on the B2R-Gq interface were found to stabilize the complex (Fig. 3). Y356G.H5.23 (CGN numbering41) at the tail of α5-helix formed multiple hydrogen bonds with V1513.46, and D1543.49-R1553.50 of the conserved DRY motif. The α5-helix of Gq also formed hydrogen bonds with A1583.53, E2656.30, and Y3327.53 of the conserved NPxxY motif. Several polar interactions were found between R167ICL2-R37G.hns1.02, N2545.68-Q350G.H5.17, and R2676.32-L358G.H5.25/V359G.H5.26. The helix 8 of B2R was also involved in the Gq coupling by the interactions between R3388.49-E355G.H5.22 and G3368.47-N357G.H5.24 (Fig. 3b).
Fig. 3. Molecular mechanisms of B2R-Gq coupling.
a B2R-Gq interactions at the intracellular cleft of 7TMs and α5-helix of Gq. B2R was colored in violet and Gq was colored in salmon. Residues with hydrogen bonds were shown as sticks and dotted lines. b Diagram of the B2R-Gq contacts.
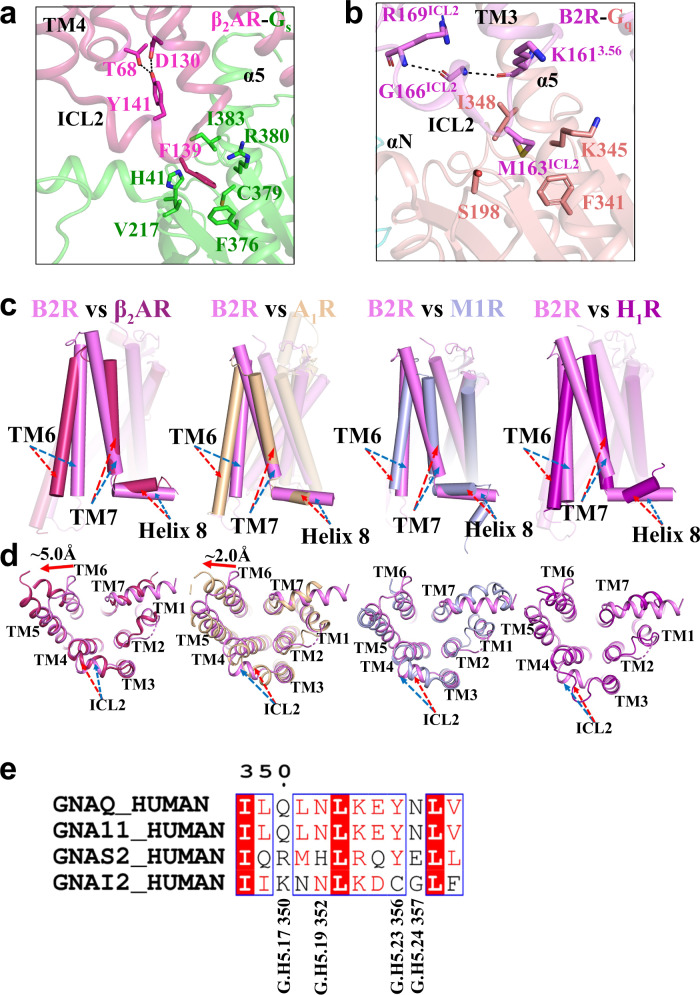
The ICL2 of B2R adopted a short α-helix like that in the β2AR-Gs structure. Y141ICL2 of β2AR formed hydrogen bonds with T682.39 and D1303.49 to stabilize the α-helical conformation of ICL2 (Fig. 4a)32. G166ICL2 of B2R formed two hydrogen bonds with R169ICL2 and K1613.56, while M163ICL2 interacted with F341G.H5.08, K345G.H5.12, and I348G.H5.15 in the α5-helix and S198G.s2s3.02 in the β2-β3 loop of Gq (Fig. 4b). In the β2AR-Gs structure, the intracellular tip of TM6 moved 14 Å outwards while only 9 Å in the B2R-Gq structures (Fig. 4c, d). It was suggested that the residues tyrosine and leucine at the C terminus of Gs required more space to accommodate the bulky side chains, compared to the smaller side chain of valine at the C terminus of Gq. Additionally, a ~15° rotation of α5-helix towards TM1 was found in the B2R-Gq structure, compared to the β2AR-Gs structure (Supplementary Fig. 7).
Fig. 4. Comparison of various GPCR-G protein complexes.
a In the β2AR-Gs complex, T68 and D130 formed hydrogen bonds with Y141 of ICL2 to stabilize the short α-helix. F139 of β2AR inserted into a hydrophobic pocket on the Gs. b In the B2R-Gq complex, G166 formed hydrogen bonds with K161 and R169, and the α-helical conformation of ICL2 positioned M163 into a hydrophobic pocket on the surface of Gq. The pocket was consisted of residues from α5-helix (F341, K345, I348) and β2-β3 loop (S198). c, d Lateral (c) and intracellular (d) views of the B2R-Gq (violet) superposed with the β2AR-Gs (warm-pink, PDB ID: 3SN6), A1R-Gi (wheat, PDB ID: 6D9H), M1R-G11 (light-blue, PDB ID: 6OIJ), and H1R-Gq (purple, PDB ID: 7DFL). e Sequence alignment of human Gq, Gs2, Gi2, and G11 proteins.
Diverse interactions for Gq/11-coupling were observed by comparing the B2R-Gq structures with the M1R-G1133 and H1R-Gq structures24. All α5-helices in these structures inserted into the intracellular cavities and tilted toward TM1 and TM7 (Supplementary Fig. 7). However, some additional contacts at the Gq and helix 8 interface were found in the B2R-Gq structures (Fig. 3b). The QG.H5.17 and NG.H5.24 were highly conserved in the Gq/11 family proteins and critical for the polar interactions with B2R, which might determine the Gq coupling of B2R. However, it was suggested that B2R could also couple to Gi42–44, in which the common NG.H5.19 in the Gi2/11/q might form hydrogen bonds with B2R (Fig. 4e).
Molecular insights of B2R activation
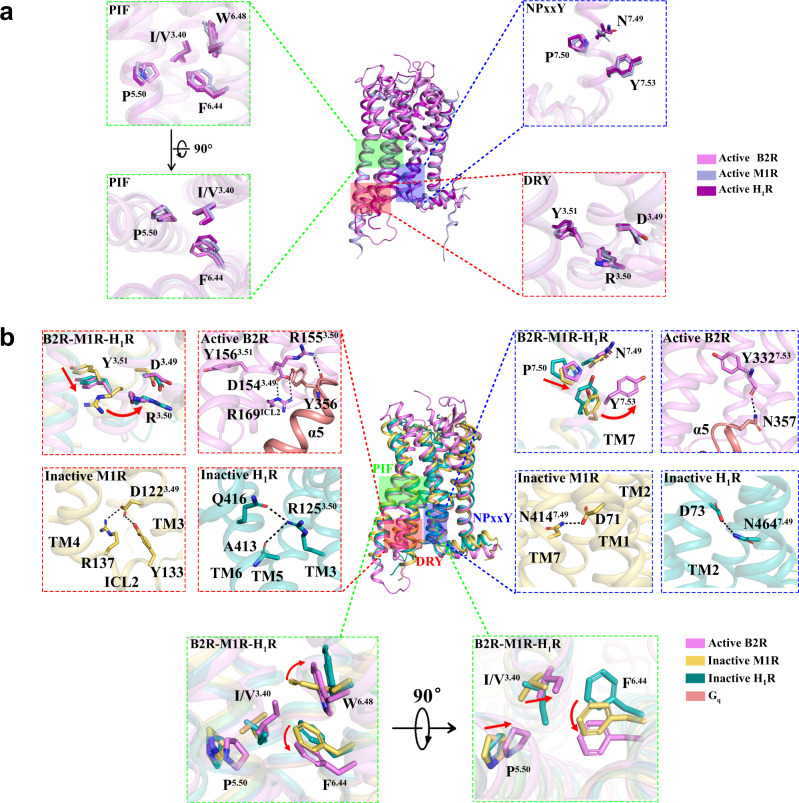
Superposition of our determined active B2R structures with the doxepin-bound inactive H1R45, histamine-bound active H1R24, tiotropium-bound inactive M1R46, and iperoxo-bound active M1R33 structures provided the molecular insights into the activation mechanisms of B2R. In the inactive H1R and M1R structures, R3.50 of the conserved DRY motif formed hydrogen bonds with the residues in TM6, and locked the receptor in an inactive state (Fig. 5b). Additionally, N7.49 of the conserved NPxxY motif formed hydrogen bonds with aspartic acids in TM2. While in the active structures, the DRY, NPxxY, and PIF motifs exhibited similar conformations, indicating the conserved activation mechanism across the class A GPCRs (Fig. 5a).
Fig. 5. Molecular insights of B2R activation.
a Structural comparison of active B2R (violet) with active M1R (light-blue, PDB ID: 6OIJ) and active H1R (purple, PDB ID: 7DFL). DRY motif, NPxxY motif, PIF motif, and toggle switch were highlighted. b Structural comparison of active B2R with inactive M1R (yellow-orange, PDB ID: 5CXV), and inactive H1R (teal, PDB ID: 3RZE). DRY motif, NPxxY motif, PIF motif, and toggle switch were highlighted with the interactions labelled by dotted lines.
The B2R antagonist icatibant showed a different chemical structure from bradykinin and kallidin in the P4K/P3B, F6K/F5B, P8K/P7B, and F9K/F8B. Especially, F9K/F8B in bradykinin and kallidin induced the side chain displacement and rotation of the conserved toggle switch W2836.48 (Supplementary Fig. 6c) thus influencing the P5.50I3.40F6.44 motif, which was found in many GPCRs as an allosteric bridge to coordinate the communications between the ligand-binding pockets and G protein-coupling interfaces47. It was suggested that the translocation of F6.44 in the PIF motif induced the intracellular side of TM6 extending outward from the TMs core. Upon TM6 displacement, hydrogen bonds within DRY-TM6/ICL2 and NPxxY-TM2 were destructed to allow these two motifs interacting with G proteins. In our determined B2R structure, the hydrogen bonds of D1543.49-Y356G.H5.23 and R1553.50-Y356G.H5.23 were found between DRY motif and α5-helix. Besides, the additional hydrogen bonds were observed between D1543.49 and R169ICL2 to stabilize the helical structure of ICL2, as well as between NPxxY motif and α5-helix (Y3327.53-N357G.H5.24) (Fig. 5b). Therefore, rearrangements of these important microswitches, as well as displacements of intracellular helices and loops, might facilitate B2R activation and Gq coupling. The C-terminal F8 of AngII was also important for AT1R and AT2R activation since it triggered a series of conformational changes38,39. The side chain of F8 in AngII formed a hydrogen bond with K5.42 of AT1R and AT2R, while the side chain of F9K in kallidin orientated to the opposite direction with no hydrogen bond formed (Supplementary Fig. 13e). The bulky side chain of F8 in AngII pushed W2536.48 and Y2927.43 of AT1R downward to avoid steric clashes, and consequently promoted conformational changes at the toggle switch and PIF motif, which further induced TM6 outward displacement. Upon activation, the internal lock between N1113.35 and N2957.46 that stabilized the inactive AT1R was disrupted29,39,40. AT2R shared a similar activation process except for the absence of an internal lock between N1273.35 and S3117.46 of AT2R29,38. Therefore, the C-terminal phenylalanines of the peptides bradykinin and AngII triggered the B2R and AT1R/AT2R activation with conserved mechanisms.
Discussion
Our determined B2R-Gq complex structures provided deep insights into the molecular mechanisms of the ligand binding, receptor activation, and G protein coupling of the human bradykinin receptor. The peptide agonists bradykinin and kallidin adopted the S-shaped binding poses. The N/C-termini of the peptides fell into the anion trap located at the entrance of the orthosteric ligand-binding pocket and were stabilized by the electrostatic interactions. To accommodate the hydrophobic residues in the middle of peptides, B2R formed hydrogen bonds with proline and glycine of the ligands. These two residues could act as the anchor points to immobilize the backbones of agonists. Divergences of the peptide-receptor interactions between B2R and B1R mainly focused on the C-termini of peptides. In B1R37, the carboxyl group of F9DK was negatively charged and fell into a cationic pocket formed by K3.33 and R5.38. However, F8B was electroneutral and only hydrophobic interactions could be found (Supplementary Fig. 12b). Additionally, the pocket consisting of R5.38, Y6.51, and E6.58 in B1R was infeasible to accommodate R9B, because of the steric hindrance with R5.38 (Supplementary Fig. 12c).
Gq formed substantial interactions with ICL2, ICL3, and helix 8 of B2R. ICL3 was usually truncated to increase the thermostability of GPCR-G protein complex for structure determination48, which made it difficult to investigate the roles of ICL3 in the G protein coupling. In our structures, ICL3 was found to grasp the α5-helix through polar interaction of N2545.68-Q350G.H5.17 and electrostatic force of K2606.25-D346G.H5.13 as well as hydrophobic interactions (Fig. 3b). Helix 8-Gq interactions were unusual in the previously determined GPCR-Gq/11 structures24,49, which might further induce inward movement of TM7 at the cytoplasmic side during activation.
Our structures of B2R-Gq complexes also indicated the activation mechanisms of B2R. Upon bradykinin or kallidin binding to the bulky orthosteric binding pocket, F8B/F9K interacted with the conserved toggle switch W2836.48, and then induced outward movement of TM6 at the cytoplasmic side and F2796.44 in the PIF motif. The rearrangement of PIF motif destabilized the inactive state of the DRY and NPxxY motifs, opened the intracellular cleft for the α5-helix insertion of Gq. The active B2R-Gq complex structures we determined in this study are thus expected to pave the way to the structure-based drug design of B2R ligands for the treatment of cardiovascular diseases and COVID-19.
Methods
Constructs
To increase the yields and stability of the complex, the first 28 residues of human B2R were replaced by a maltose-binding protein (MBP) and the gene was cloned into pFastbac1 that contained a haemagglutinin (HA) tag, a FLAG tag (DYKDDDDK), a 10xHis tag, and a TEV protease cleavage site before the N-terminus of MBP. A DNGαq was generated by introducing the mutations R183Q and Q209L, to decrease the binding affinity of GDP to Gq, while the human Gβ1 and Gγ2 proteins were both wild types22,23. There was a 10xHis tag at the N-terminus of each subunit.
Expression and purification of B2R-Gq complexes
The human B2R, DNGαq, Gβ1 and Gγ2 were co-expressed in sf9 insect cells. Cell cultures were grown to a density of 2 million cells per ml in ESF921 serum-free media (Expression Systems). Then the cells were infected by adding the baculoviruses of B2R, DNGαq-Ric8A and Gβ1γ2 at the ratio of 1:1:1. After 48 h incubation at 27 °C, shaking at 125 rpm, cells were harvested by centrifugation and stored at −80 °C until use.
Cell pellets infected with B2R and Gq heterotrimer were thawed at room temperature and suspended by dounce homogenization in 20 mM HEPES pH 7.5, 50 mM NaCl, 2 mM MgCl2, 500 μM AEBSF, 1 μM E-64, 1 μM Leupeptin, 150 nM Aprotinin. The complex was formed on membranes in the presence of 10 μM bradykinin (Sangon Biotech) or 5 μM kallidin (Alomone), and was treated with apyrase (100 mU/ml, NEB) and β-mercaptoethanol (β-ME, 2 mM), followed by 3 h incubation at room temperature. Membranes were collected by centrifugation at 30,000 × g for 30 min. The washed membranes were resuspended and solubilized in 60 mM HEPES pH 7.5, 100 mM NaCl, 0.5% lauryl maltose neopentyl glycol (LMNG, Anatrace), 0.05% cholesteryl hemisuccinate Tris salt (CHS, Anatrace), 10 mM imidazole, 2 mM MgCl2, 50 mU/ml apyrase, 1 mM β-ME supplemented with 5 μM bradykinin or 2.5 μM kallidin. The membranes were incubated at 4 °C for 4 h. Insoluble material was removed by centrifugation at 58,000 × g for 1 h and the supernatant was incubated with pre-equilibrated Talon IMAC resin overnight. Then the resin was packed into a gravity column (Bio-Rad) and washed with 15 column volumes of 50 mM HEPES pH 7.5, 100 mM NaCl, 0.05% LMNG, 0.005% CHS, 20 mM imidazole, 2 mM MgCl2, 5 μM for bradykinin or 2.5 μM for kallidin, and 15 column volumes of 20 mM HEPES pH 7.5, 100 mM NaCl, 0.002% LMNG, 0.0002% CHS, 40 mM imidazole, 2 mM MgCl2, 5 μM for bradykinin or 2.5 μM for kallidin. The protein was eluted in 5 column volumes of 20 mM HEPES pH 7.5, 100 mM NaCl, 0.002% LMNG, 0.0002% CHS, 300 mM imidazole, 2 mM MgCl2, 5 μM for bradykinin or 2.5 μM for kallidin. The eluted material was incubated with 100 mU/ml apyrase and 2 mM β-ME for 2 h at 4 °C followed by incubation with pre-equilibrated amylose resin (NEB) for 4 h at 4 °C. The resin was loaded onto a gravity column and washed with 10 column volumes of 20 mM HEPES pH 7.5, 100 mM NaCl, 0.002% LMNG, 0.0002% CHS, 2 mM MgCl2, 5 μM bradykinin or kallidin. The protein was eluted in 6 column volumes of 20 mM HEPES pH 7.5, 100 mM NaCl, 0.002% LMNG, 0.0002% CHS, 2 mM MgCl2, 5 μM bradykinin or kallidin, and 10 mM maltose. Eluted protein was concentrated using a Vivaspin Turbo Ultrafiltration Units (MWCO 50 kDa) and subjected to size-exclusion chromatography on a Superdex 200 Increase 10/300 column (GE Healthcare) pre-equilibrated with 20 mM HEPES pH 7.5, 100 mM NaCl, 0.002% LMNG, 0.0002% CHS, 2 mM MgCl2, 0.5 μM bradykinin or kallidin. Peak fractions containing B2R-Gq complex were pooled and concentrated to ~4 mg/ml for electron microscopy studies. The final yields of the purified complexes were ~0.15 mg/L insect cell culture.
Cryo-EM grid preparation and data collection
For cryo-EM grids preparation, 3 μL B2R-Gq complex solution was applied to glow-discharged holey carbon EM grids (C-flat 300 Cu mesh R1.2/1.3) using Vitrobot (FEI Vitrobot Mark IV). The grid was plunge-freezing into liquid ethane and stored in liquid nitrogen for further data collection.
Cryo-EM data collection was performed on a Titan Krios (ThermoFisher) electron microscope operated at 300 kV equipped with a K2 Summit direct electron detector (Gatan) at a magnification of 29,000× in the Center of Cryo-Electron Microscopy, Zhejiang University (Hangzhou, China), corresponding to a nominal pixel size of 1.014 Å. Movies were recorded using SerialEM software in counting mode at a dose rate of 8.0 e/Å2/s with a defocus range of −0.5 to −2.0 μm. The total exposure time was 8 s and a total of 40 frames per micrograph. A total of 4093 and 3425 movies were collected for B2R-Gq-bradykinin and B2R-Gq-kallidin, respectively.
Cryo-EM data processing and structure determination
The flow charts of data processing were presented in Supplementary Figs. 3 and 4. For three batches of movie stacks, global motion correction was performed using the MotionCorr2 program50, and contrast transfer function (CTF) parameters were estimated using CTFFIND451. The remaining image processing steps were carried out using RELION 3.052.
For the B2R-Gq-bradykinin complex, 2,630,394 particles were picked from 4,093 movies using the Laplacian-of-Gaussian-based auto-picking method and binned three times before 2D classification. The iterative 2D classification caused 1,169,656 well-qualified particles, which were then selected for further 3D classification. Using TT-OAD2–GLP-1R–Gs complex (EMDB-20179)53 as a reference, the 3D classification resulted in 1,056,238 well-defined particles, which were then re-extracted using the original pixel size of 1.014 Å and used for 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 2.93 Å at a Fourier shell correlation of 0.143.
For the B2R-Gq-kallidin complex, the Laplacian-of-Gaussian-based auto-picking produced 2,042,659 particles, which were binned 3 times and subjected to integrative 2D classifications. Particles from qualified 2D averages were then selected for further 3D analysis using the map of B2R-Gq-bradykinin complex as the reference, resulting in a well-defined subset with 1,388,486 particles. The particles were re-extracted using the original pixel size of 1.014 Å and subjected to 3D refinement, particle polishing and CTF refinement. The final refinement yielded a map with a resolution of 2.76 Å at FSC = 0.143.
Cryo-EM structure models were built using the cryo-EM structure of M1R-G11 (PDB ID: 6OIJ)33 as the initial model. The model was docked into the EM density maps using Chimera and manually rebuilt using COOT54. Realspace refinement was performed using Phenix55. The final refinement statistics were validated using Phenix and shown in Supplementary Table 2. All the figures were prepared using PyMol56 and UCSF Chimera57.
Calcium mobilization assays
CHO cells transfected with wild-type or mutated B2R and wild-type or mutated Gq were seeded into 96-well black plates at a density of 30,000 cells per well and incubated for 24 h. Then cells were loaded with reagents from Calcium-5 Assay Kit (Molecular Devices) for 45 min at 37 °C in 5% CO2 according to the manufacturer’s protocol. Cells were treated with varying concentrations of kallidin and detected with Flexstation 3 Multi-Mode Microplate Reader (Molecular Devices) with excitation at 485 nm and emission at 525 nm. Data were analyzed by GraphPad Prism 5 and presented as Mean ± S.E.M. from three independent experiments.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
H.Z. is supported by National Key R&D Program of China (2018YFA0508100), National Natural Science Foundation of China (81722044, 91753115, 21778049, 81861148018), and National Science and Technology Major Project of China (2018ZX09711002). We thank S.C. and X.Z. in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University for their technical assistance on Cryo-EM. We thank W.L. and M.L. in the Shanghai Yuyao Biotech Ltd for the assistance on the calcium mobilization assays.
Source data
Author contributions
J.S. and D.Z. designed the constructs. J.S. and Y.F. expressed and purified the complexes. D.Z. prepared the cryogenic samples, collected, and processed the cryo-EM data. A.C. collected the cryo-EM data. X.Y. performed the calcium mobilization assays. H.Z. conceived and supervised the project, and determined the structures. J.S. and H.Z. wrote the manuscript with input from all other authors.
Peer review
Peer review information
Nature Communications thanks So Iwata and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes: EMD-31480 (B2R-Gq-bradykinin) and EMD-31481 (B2R-Gq-kallidin). The atomic coordinates have been deposited in the Protein Data Bank under accession codes: 7F6H (B2R-Gq-bradykinin) and 7F6I (B2R-Gq-kallidin). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-28399-1.
References
- 1.Chatzikyriakidou A, Voulgari PV, Drosos AA. An extensive study of the functional polymorphisms of kinin-kallikrein system in rheumatoid arthritis susceptibility. Arch. Rheumatol. 2018;33:33–38. doi: 10.5606/ArchRheumatol.2018.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dendorfer A, Wolfrum S, Dominiak P. Pharmacology and cardiovascular implications of the kinin-kallikrein system. Jpn. J. Pharmacol. 1999;79:403–426. doi: 10.1254/jjp.79.403. [DOI] [PubMed] [Google Scholar]
- 3.Kashuba E, Bailey J, Allsup D, Cawkwell L. The kinin-kallikrein system: physiological roles, pathophysiology and its relationship to cancer biomarkers. Biomarkers. 2013;18:279–296. doi: 10.3109/1354750X.2013.787544. [DOI] [PubMed] [Google Scholar]
- 4.Webb JG. The kallikrein/kinin system in ocular function. J. Ocul. Pharmacol. Ther. 2011;27:539–543. doi: 10.1089/jop.2011.0187. [DOI] [PubMed] [Google Scholar]
- 5.Marceau F, Regoli D. Bradykinin receptor ligands: therapeutic perspectives. Nat. Rev. Drug Discov. 2004;3:845–852. doi: 10.1038/nrd1522. [DOI] [PubMed] [Google Scholar]
- 6.Lau J, Rousseau J, Kwon D, Benard F, Lin KS. A systematic review of molecular imaging agents targeting bradykinin B1 and B2 receptors. Pharmaceuticals. 2020;13:199. doi: 10.3390/ph13080199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahdi F, et al. Mapping the interaction between high molecular mass kininogen and the urokinase plasminogen activator receptor. J. Biol. Chem. 2004;279:16621–16628. doi: 10.1074/jbc.M313850200. [DOI] [PubMed] [Google Scholar]
- 8.da Costa PLN, Sirois P, Tannock IF, Chammas R. The role of kinin receptors in cancer and therapeutic opportunities. Cancer Lett. 2014;345:27–38. doi: 10.1016/j.canlet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Aldayini NA, et al. Hereditary angioedema in pediatric age: an overview. Arch. Pharm. Pract. 2020;11:130–133. [Google Scholar]
- 10.Lumry WR, Settipane RA. Hereditary angioedema: epidemiology and burden of disease. Allergy Asthma Proc. 2020;41:S8–S13. doi: 10.2500/aap.2020.41.200050. [DOI] [PubMed] [Google Scholar]
- 11.Garvin MR, et al. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;9:e59177. doi: 10.7554/eLife.59177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rameshrad M, Ghafoori M, Mohammadpour AH, Nayeri MJD, Hosseinzadeh H. A comprehensive review on drug repositioning against coronavirus disease 2019 (COVID19) Naunyn Schmiedebergs Arch. Pharmacol. 2020;393:1137–1152. doi: 10.1007/s00210-020-01901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Razeghian-Jahromi I, et al. Angiotensin-converting enzyme 2: a double-edged sword in COVID-19 patients with an increased risk of heart failure. Heart Fail Rev. 2021;26:371–380. doi: 10.1007/s10741-020-10016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roche JA, Roche R. A hypothesized role for dysregulated bradykinin signaling in COVID-19 respiratory complications. FASEB J. 2020;34:7265–7269. doi: 10.1096/fj.202000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cicardi M, et al. Icatibant, a new bradykinin-receptor antagonist, in hereditary angioedema. N. Engl. J. Med. 2010;363:532–541. doi: 10.1056/NEJMoa0906393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parekh RU, Robidoux J, Sriramula S. Kinin B1 receptor blockade prevents angiotensin II-induced neuroinflammation and oxidative stress in primary hypothalamic neurons. Cell Mol. Neurobiol. 2020;40:845–857. doi: 10.1007/s10571-019-00778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman JA, Willits AB, Bellemer A. G alpha q and Phospholipase C beta signaling regulate nociceptor sensitivity in Drosophila melanogaster larvae. PeerJ. 2018;6:e5632. doi: 10.7717/peerj.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, et al. Bradykinin-mediated Ca2+ signalling regulates cell growth and mobility in human cardiac c-Kit(+) progenitor cells. J. Cell Mol. Med. 2018;22:4688–4699. doi: 10.1111/jcmm.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess J, et al. Differential pharmacology of cloned human and mouse B2 bradykinin receptors. Mol. Pharmacol. 1994;45:1–8. [PubMed] [Google Scholar]
- 20.Austin CE, et al. Stable expression of the human kinin B1 receptor in Chinese hamster ovary cells. J. Biol. Chem. 1997;272:11420–11425. doi: 10.1074/jbc.272.17.11420. [DOI] [PubMed] [Google Scholar]
- 21.Leeb-Lundberg LMF, Marceau F, Müller-Esterl W, Pettibone DJ, Zuraw BL. International Union of Pharmacology. XLV. Classification of the kinin receptor family: from molecular mechanisms to pathophysiological consequences. Pharmacol. Rev. 2005;57:27–77. doi: 10.1124/pr.57.1.2. [DOI] [PubMed] [Google Scholar]
- 22.Fan G, Ballou LM, Lin RZ. Phospholipase C-independent activation of glycogen synthase kinase-3β and C-terminal Src kinase by Gαq. J. Biol. Chem. 2003;278:52432–52436. doi: 10.1074/jbc.M310982200. [DOI] [PubMed] [Google Scholar]
- 23.Martins L, et al. Computational analysis for GNAQ mutations: new insights on the molecular etiology of Sturge-Weber syndrome. J. Mol. Graph. Model. 2017;76:429–440. doi: 10.1016/j.jmgm.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Xia R, et al. Cryo-EM structure of the human histamine H1 receptor/Gq complex. Nat. Commun. 2021;12:2086. doi: 10.1038/s41467-021-22427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang Y, et al. Structure of formylpeptide receptor 2-Gi complex reveals insights into ligand recognition and signaling. Nat. Commun. 2020;11:885. doi: 10.1038/s41467-020-14728-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto HH, et al. Cryo-EM structure of the human MT1–Gi signaling complex. Nat. Struct. Mol. Biol. 2021;28:694–701. doi: 10.1038/s41594-021-00634-1. [DOI] [PubMed] [Google Scholar]
- 27.Zhang HT, et al. Structural basis for ligand recognition and functional selectivity at angiotensin receptor. J. Biol. Chem. 2015;290:29127–29139. doi: 10.1074/jbc.M115.689000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 29.Wingler LM, McMahon C, Staus DP, Lefkowitz RJ, Kruse AC. Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell. 2019;176:479–490. doi: 10.1016/j.cell.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang HT, et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017;544:327–332. doi: 10.1038/nature22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asada H, et al. Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog. Nat. Struct. Mol. Biol. 2018;25:570–576. doi: 10.1038/s41594-018-0079-8. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda S, Qu Q, Robertson MJ, Skiniotis G, Kobilka BK. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science. 2019;364:552–557. doi: 10.1126/science.aaw5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bous J, et al. Cryo-electron microscopy structure of the antidiuretic hormone arginine-vasopressin V2 receptor signaling complex. Sci. Adv. 2021;7:eabg5628. doi: 10.1126/sciadv.abg5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Draper-Joyce CJ, et al. Structure of the adenosine-bound human adenosine A1 receptor–Gi complex. Nature. 2018;558:559–563. doi: 10.1038/s41586-018-0236-6. [DOI] [PubMed] [Google Scholar]
- 36.White JF, et al. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin YL, et al. Molecular basis for kinin selectivity and activation of the human bradykinin receptors. Nat. Struct. Mol. Biol. 2021;28:755–761. doi: 10.1038/s41594-021-00645-y. [DOI] [PubMed] [Google Scholar]
- 38.Asada H, et al. The crystal structure of angiotensin II type 2 receptor with endogenous peptide hormone. Structure. 2020;28:418–425. doi: 10.1016/j.str.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Wingler LM, et al. Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR. Science. 2020;367:888–892. doi: 10.1126/science.aay9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, et al. Structural insights into ligand recognition and activation of angiotensin receptors. Trends Pharmacol. Sci. 2021;42:577–587. doi: 10.1016/j.tips.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flock T, et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blaukat A, Barac A, Cross MJ, Offermanns S, Dikic I. G protein-coupled receptor-mediated mitogen-activated protein kinase activation through cooperation of G alpha(q), and G alpha(i) signals. Mol. Cell Biol. 2000;20:6837–6848. doi: 10.1128/mcb.20.18.6837-6848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.deWeerd WFC, LeebLundberg LMF. Bradykinin sequesters B2 bradykinin receptors and the receptor-coupled G alpha subunits G alpha(q) and G alpha(i) in caveolae in DDT1 MF-2 smooth muscle cells. J. Biol. Chem. 1997;272:17858–17866. doi: 10.1074/jbc.272.28.17858. [DOI] [PubMed] [Google Scholar]
- 44.Liao JK, Homcy CJ. The G-proteins of the G-alpha(I) and G-alpha(Q) family coupled the bradykinin receptor to the release of endothelium-derived relaxing factor. J. Clin. Invest. 1993;92:2168–2172. doi: 10.1172/JCI116818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimamura T, et al. Structure of the human histamine H-1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thal DM, et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan D, et al. Activation of the alpha2B adrenoceptor by the sedative sympatholytic dexmedetomidine. Nat. Chem. Biol. 2020;16:507–512. doi: 10.1038/s41589-020-0492-2. [DOI] [PubMed] [Google Scholar]
- 48.Kato HE, et al. Conformational transitions of a neurotensin receptor 1-Gi1 complex. Nature. 2019;572:80–85. doi: 10.1038/s41586-019-1337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim K, et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell. 2020;182:1574–1588. doi: 10.1016/j.cell.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao P, et al. Activation of the GLP-1 receptor by a non-peptidic agonist. Nature. 2020;577:432–436. doi: 10.1038/s41586-019-1902-z. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janson G, Zhang CX, Prado MG, Paiardini A. PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics. 2017;33:444–446. doi: 10.1093/bioinformatics/btw638. [DOI] [PubMed] [Google Scholar]
- 57.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes: EMD-31480 (B2R-Gq-bradykinin) and EMD-31481 (B2R-Gq-kallidin). The atomic coordinates have been deposited in the Protein Data Bank under accession codes: 7F6H (B2R-Gq-bradykinin) and 7F6I (B2R-Gq-kallidin). Source data are provided with this paper.