Abstract
A total of 221 isolates of multidrug-resistant Salmonella enterica serovar Typhimurium in Japan were characterized in the present study. The results revealed that clonal serovar Typhimurium definitive phage type 104 strains prevailed and that these strains had drug resistance patterns, integron types, and pulsed-field gel electrophoresis patterns similar to those predominant among isolates in Western countries.
Infection with Salmonella enterica has been recognized as a major public health concern in developed countries. Recently, a multidrug-resistant Salmonella enterica serovar Typhimurium strain with definitive phage type 104 (DT104) has emerged and spread over Western countries (2, 6, 13, 15). The dominant resistance type of such strains is ACSSuT; that is, they are resistant to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline, and the resistance is encoded by chromosomally located genes containing class 1 integron structures (3).
Although we have reported on the isolation of multidrug-resistant serovar Typhimurium strains in Japan (8), in the study described here we further analyzed the strains by pulsed-field gel electrophoresis (PFGE) and PCR for detection of integrons as well as by bacteriophage typing and antimicrobial susceptibility testing to investigate the relationships of S. enterica serovar Typhimurium DT104 strains and other isolates.
A total of 221 isolates of S. enterica serovar Typhimurium resistant to more than two antimicrobial agents were used in the present study. One hundred forty-seven isolates were from human sources and were involved in 14 outbreaks and 133 sporadic cases of infection; they were isolated between 1980 and 2000 and had no epidemiological relationship to each other. Seventy-four isolates were from nonhuman sources including cattle, poultry, pigs, and the environment and were isolated between 1981 and 2000.
Antimicrobial susceptibility tests were done by the disk diffusion method on Mueller-Hinton II agar (Becton Dickinson Microbiology Systems, Cockeysville, Md.) by the standard methods outlined by the National Committee for Clinical Laboratory Standards (11). Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as quality control strains. Disks with the following antibiotics (abbreviations; diffusible amounts) were provided from a commercial source (Becton Dickinson Microbiology Systems): ampicillin (A; 10 μg), chloramphenicol (C; 30 μg), streptomycin (S; 10 μg), tetracycline (T; 30 μg), ciprofloxacin (Cip; 5 μg), kanamycin (K; 30 μg), cefotaxime (Ctx; 30 μg), sulfamethoxazole-trimethoprim (Sx; 23.75 and 1.25 μg, respectively), trimethoprim (Tm; 5 μg), gentamicin (G; 10 μg), nalidixic acid (N; 30 μg), and sulfisoxazole (Su; 250 μg). Resistance was determined according to reference zone diameter interpretive standards (11). The results are summarized in Table 1. The ACSSuT type of resistance, the major type in DT104 strains, was the most predominant (n = 93), followed by ACSSuTK (n = 26), ACSSuTN (n = 13), and ACSSuTKSxTmG (n = 10).
TABLE 1.
Resistance types and PTs of the S. enterica serovar Typhimurium isolates in the present study
| Drug resistance no. | Resistance type | No. of isolates of the following PT:
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 12 | 29 | 36 | 49 | 52A | 99 | 104 | 104B | 104C | 104L | 106 | 110B | 116 | 126 | 130 | 136 | 188 | 193 | 193A | 194 | 208 | U302 | U288 | RDNC | UT | Total | ||
| 3 | ACK | 1 | 1 | |||||||||||||||||||||||||
| ASK | 1 | 1 | ||||||||||||||||||||||||||
| CSSu | 1 | 1 | ||||||||||||||||||||||||||
| CTK | 2 | 2 | ||||||||||||||||||||||||||
| SuTK | 1 | 1 | ||||||||||||||||||||||||||
| SSuT | 1 | 1 | 1 | 3 | ||||||||||||||||||||||||
| TKN | 1 | 1 | ||||||||||||||||||||||||||
| 4 | ACSN | 1 | 1 | |||||||||||||||||||||||||
| ACST | 1a | 2 | 4 | 7 | ||||||||||||||||||||||||
| ACTK | 1 | 1 | 2 | 4 | ||||||||||||||||||||||||
| ASSuT | 1 | 2 | 1 | 4 | ||||||||||||||||||||||||
| ASTK | 1 | 1 | 2 | |||||||||||||||||||||||||
| CSTK | 1 | 1 | ||||||||||||||||||||||||||
| SSuTK | 2 | 1 | 3 | |||||||||||||||||||||||||
| 5 | ACSTK | 2 | 1 | 1 | 1 | 5 | ||||||||||||||||||||||
| ACSSuK | 1 | 1 | ||||||||||||||||||||||||||
| ACSSuT | 1 | 1 | 78a | 6a | 1a | 1a | 4a | 1 | 93 | |||||||||||||||||||
| ACSuTK | 1 | 1 | ||||||||||||||||||||||||||
| ASSuTK | 7 | 1 | 1 | 1 | 1 | 2 | 13 | |||||||||||||||||||||
| ASuTSxTm | 1 | 1 | ||||||||||||||||||||||||||
| CSuTKG | 1 | 1 | ||||||||||||||||||||||||||
| 6 | ACSSuTK | 2 | 2 | 2a | 1 | 4 | 1 | 5(2a) | 3 | 6 | 26 | |||||||||||||||||
| ACSSuTN | 7a | 2a | 1 | 1 | 1 | 1 | 13 | |||||||||||||||||||||
| CSSuKSxTm | 1 | 1 | ||||||||||||||||||||||||||
| 7 | ACSSuTKN | 2 | 3 | 5 | ||||||||||||||||||||||||
| ACSuTKGN | 2 | 2 | ||||||||||||||||||||||||||
| ACSSuTSxTm | 1 | 1a | 2 | |||||||||||||||||||||||||
| CSSuTKGN | 1 | 1 | ||||||||||||||||||||||||||
| 8 | ACSSuKSxTmG | 1 | 1 | |||||||||||||||||||||||||
| ACSSuTKTmN | 1 | 1 | ||||||||||||||||||||||||||
| ACSSuTKSxTm | 4 | 1 | 5 | |||||||||||||||||||||||||
| ACSSuTSxTmN | 1 | 1 | 2 | |||||||||||||||||||||||||
| 9 | ACSSuTKSxTmG | 7 | 1 | 2 | 10 | |||||||||||||||||||||||
| ACSSuTKSxTmN | 1 | 1 | ||||||||||||||||||||||||||
| ACSSuTSxTmGN | 1 | 1 | ||||||||||||||||||||||||||
| 10 | ACSSuTKSxTmGN | 1 | 1 | 1 | 3 | |||||||||||||||||||||||
| Total | 1 | 1 | 4 | 1 | 1 | 1 | 13 | 88 | 8 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 24 | 2 | 15 | 3 | 16 | 1 | 5 | 27 | 221 | |
Number of isolates positive for DT104-like integrons.
Bacteriophage typing was performed in accordance with the methods of the Public Health Laboratory Service (PHLS), London, United Kingdom (1). The typing phages and scheme were kindly provided by PHLS. The pattern of lysis produced by infection with typing phages was recorded and designated in accordance with the standard scheme. UT indicates untypeable, which means that a tested strain was not lysed by any of the typing phages. RDNC indicates that a tested strain reacted with some of the typing phages but did not conform to any of the schemes. The results are summarized in Table 1. Eighty-eight isolates were identified as DT104. DT104B, DT104C, and DT104L, which are DT104-related types, were also identified (n = 8, 2, and 1 isolates, respectively). Sixty-four of them were from human sources involved in five outbreaks and 19 sporadic cases of infection and were recovered between the years 1986 and 2000, and 35 were from nonhuman sources and were recovered between the years 1990 and 2000. Among the other isolates tested, DT193 (n = 24 isolates, 20 from human sources) was predominant, as was UT (n = 27 isolates, 14 from human sources). DT194 and phage type (PT) U302 were also identified (n = 15 and 16 isolates, respectively) from both human sources (n = 8 and 13 isolates, respectively) and nonhuman sources (n = 7 and 3 isolates, respectively).
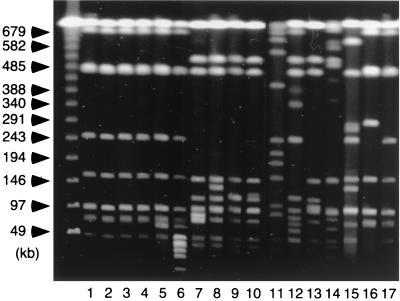
To obtain clues as to the genetic backgrounds of the isolates, PFGE and PCR for detection of class 1 integrons were applied. PFGE was carried out as described elsewhere (9). All the isolates were analyzed by using restriction endonuclease BlnI (Takara, Shiga, Japan). Electrophoresis was performed with a 1% agarose gel by using a CHEF-DR II apparatus (Bio-Rad Laboratories, Hercules, Calif.) in 0.5× TBE (Tris-borate-EDTA) buffer at 6 V/cm and 4°C. A linearly ramped switching time from 5 to 50 s was applied for 21 h. DT104-related isolates showed PFGE patterns similar to each other, even though they were from various sources in terms of years and locations of isolation. Sixty-four and 26 isolates showed patterns quite similar to those depicted in Fig. 1, lanes 17 and 16, respectively, of which 53 and 23, respectively, showed patterns indistinguishable from those in Fig. 1, lanes 17 and 16, respectively. U.S. strains, kindly provided by the Centers for Disease Control and Prevention (Atlanta, Ga.), also showed patterns indistinguishable from that in Fig. 1, lane 16 (data not shown). On the contrary, the isolates of other PTs showed multiple patterns different from those of the DT104-related isolates, which might reflect variations in their origins.
FIG. 1.
PFGE patterns of PT U302 and representative DT104 isolates from Japan. Total DNAs of PT U302 (lanes 1 to 15) and DT104 (lanes 16 and 17) isolates were digested with BlnI and separated in a 1% agarose gel. Lanes 1 to 6, isolates positive for DT104-like integrons with resistance type ACSSuT (lanes 1 to 4) or ACSSuTK (lanes 5 and 6); lanes 7 to 15, isolates negative for DT104-like integrons with resistance type ACSSuTK (lanes 7 to 9), ACSSuK (lane 10), ASSuTK (lane 11), ACSSuTSxTmN (lane 12), ACSSuTSxTmGN (lane 13), ASTK (lane 14), or SuTK (lane 15); lanes 16 and 17, DT104 isolates with the ACSSuT resistance type. The sizes of the molecular size markers, a bacteriophage λ ladder, are shown on the left.
For detection of class 1 integrons, primers whose sequences comprised the consensus sequences of class 1 integrons were used (primers intI-F [5′-GGCATCCAAGCAGCAAGC-3′] and intI-R [5′-AAGCAGACTTGACCTGAT-3′]) (4, 7). Amplification was carried out in 25-μl reaction mixture with a boiled bacterial suspension with 200 μM (each) deoxynucleotide triphosphate, the primer pairs at concentrations (each) of 0.4 μM, Taq buffer (Qiagen, Hilden, Germany), and 1 U of Taq DNA polymerase (Qiagen). The PCR was run at 93°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 35 cycles. The reaction mixture was separated on a 1% agarose gel in 1× TAE (Tris-acetate-EDTA) buffer. Isolates harboring the typical DT104-like integrons are expected to show bands with sizes of approximately 1.0 and 1.2 kb (3). All the DT104, DT104B, DT104C, and DT104L isolates were positive for DT104-like integrons (Table 1 and data not shown). Furthermore, 6 of 16 PT U302 isolates were positive, whereas 4 were from humans with sporadic infections and 2 were from nonhuman sources. They were considered DT104 related since they had PFGE patterns similar to those of DT104 strains, especially in bands more than 50 kb in size (Fig. 1, lanes 1 to 6) as well as resistance types, as described below, and since PT U302 strains were previously referred to as being DT104 related (3). In contrast, isolates of all of the other PTs were negative for DT104-like integrons. These results suggest that DT104-related isolates from various years and areas in Japan harbor the resistance locus typical for the DT104 strain, while the others did not.
The resistance types of 105 DT104-related isolates were characteristic; ACSSuT, the typical DT104 resistance type, was the most predominant (n = 90 isolates, including 4 PT U302 isolates; 60 were from human sources), followed by ACSSuTN (n = 9 isolates, isolated from 1997 to 1999; 7 were from human sources) and ACSSuTK (n = 4 isolates, isolated in 1995, 1996, and 2000, including 2 PT U302 isolates and 1 DT104 isolate from human sources). All of the isolates with the ACSSuTN resistance type and three of the isolates with the ACSSuTK resistance type had PFGE patterns quite similar to or indistinguishable from those shown in Fig. 1, lanes 16 and 17, despite differences in their resistance types (Fig. 1 and data not shown). We found that the resistance to these additional antibiotics, nalidixic acid or kanamycin, was associated with point mutations (S83F, n = 3; D87G, n = 5; and D87N, n = 1) in the gyrA gene, which encodes the A subunit of gyrase and which is responsible for quinolone resistance (5, 16), or plasmids (approximately 2 kb [n = 2] and 70 kb [n = 2]), respectively; such mutations might be acquired during spreading of the isolates (data not shown).
Among the other resistance types, types ACSSuTK (n = 22 isolates), ASSuTK (n = 13 isolates), and ACSSuTKSxTmG (n = 10 isolates) were frequently detected. Fifty-three of the 116 isolates not related to DT104 had resistance types that included ACSSuT. However, they were negative for the DT104-like integron (Table 1) and showed multiple PFGE patterns different from those of the DT104-related isolates (Fig. 1 and data not shown). Furthermore, a variety of plasmid profiles and other kinds of class 1 integrons could be detected in preliminary experiments, which might account for the resistance types of these isolates.
In Japan, nearly 200 isolates of S. enterica serovar Typhimurium are recovered each year, with these isolates accounting for approximately 3% of Salmonella isolates detected from human sources at local public health institutes. Although the number of cases of S. enterica serovar Typhimurium infection has decreased and is much less than the number of cases of S. enterica serovar Enteritidis infection, the latter of which accounts for 45 to 60% of Salmonella infections in Japan, serovar Typhimurium still ranks high as a cause of Salmonella infections (12). The number of the cases of infection caused by S. enterica serovar Typhimurium DT104 is low at present; however, the frequency of identification of DT104 seemed to increase gradually during the 1990s (10, 14). As the resistance types and PFGE patterns of the DT104-related isolates tested in the present study are quite similar to each other, clonal serovar Typhimurium DT104 strains appear to be spreading to Japan, acquiring resistance to additional antibiotics such as quinolones or kanamycin. On the other hand, a variety of multidrug-resistant strains of serovar Typhimurium other than DT104, which seem to have origins different from those of DT104 strains, have prevailed in Japan. It would be important to survey further the occurrence of resistance types, PTs, and genotypes of serovar Typhimurium strains in Japan as well as other countries in order to learn how they change.
Acknowledgments
We thank all the municipal and prefectural public health institutes for providing us with S. enterica serovar Typhimurium isolates. We thank the Public Health Laboratory Service for providing us with the typing phages. We thank N. Zaitsu, M. Miura, M. Ishikawa, and N. Sudo for assistance.
This work was supported by a grant from the Ministry of Health, Labor and Welfare of Japan.
REFERENCES
- 1.Anderson E S, Ward L R, de Saxe M J, de Sa J D. Bacteriophage typing designation of Salmonella typhimurium. J Hyg. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baggesen D L, Aarestrup F M. Characterisation of recently emerged multiple antibiotic-resistant Salmonella enterica serovar typhimurium DT104 and other multiresistant phage types from Danish pig herds. Vet Rec. 1998;143:95–97. doi: 10.1136/vr.143.4.95. [DOI] [PubMed] [Google Scholar]
- 3.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraud E, Brisabois A, Martel J L, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43:2131–2137. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glynn M K, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 7.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination crossover point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 8.Izumiya H, Tamura K, Terajima J, Watanabe H. Salmonella enterica serovar Typhimurium phage type DT104 and other multidrug resistant strains in Japan. Jpn J Infect Dis. 1999;52:133. [PubMed] [Google Scholar]
- 9.Izumiya H, Terajima J, Wada A, Inagaki Y, Itoh K, Tamura K, Watanabe H. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J Clin Microbiol. 1997;35:1675–1680. doi: 10.1128/jcm.35.7.1675-1680.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushita S, Konishi N, Arimatsu M, Kai A, Yamada S, Morozumi S, Izumiya H, Terajima J, Watanabe H. Drug-resistance and definitive type 104 of Salmonella serovar typhimurium isolated from sporadic cases in Tokyo, 1980–1998. Kansenshogaku Zasshi. 1999;73:1087–1094. doi: 10.11150/kansenshogakuzasshi1970.73.1087. . (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests; approved standard, 7th ed. NCCLS document M2–A7. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 12.National Institute of Infectious Diseases. Salmonellosis in Japan as of June 2000. Infect Agents Surveillance Rep. 2000;21:162. [Google Scholar]
- 13.Ng L K, Khakhria R, Woodward D, Johnson W. National laboratory surveillance of enteric pathogens. Can J Infect Dis. 1997;8:133–136. [Google Scholar]
- 14.Sameshima T, Akiba M, Izumiya H, Terajima J, Tamura K, Watanabe H, Nakazawa M. Salmonella typhimurium DT104 from livestock in Japan. Jpn J Infect Dis. 2000;53:15–16. [PubMed] [Google Scholar]
- 15.Threlfall E J, Frost J A, Ward L R, Rowe B. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet Rec. 1994;134:577. doi: 10.1136/vr.134.22.577. [DOI] [PubMed] [Google Scholar]
- 16.Walker R A, Saunders N, Lawson A J, Lindsay E A, Dassama M, Ward L R, Woodward M J, Davies R H, Liebana E, Threlfall E J. Use of a LightCycler gyrA mutation assay for rapid identification of mutations conferring decreased susceptibility to ciprofloxacin in multiresistant Salmonella enterica serotype Typhimurium DT104 isolates. J Clin Microbiol. 2001;39:1443–1448. doi: 10.1128/JCM.39.4.1443-1448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]