Abstract
Lung cancer is one of the most common malignancy worldwide and causes estimated 1.6 million deaths each year. Cancer immunosurveillance has been found to play an important role in lung cancer and may be related with its prognosis. KLRK1, encoding NKG2D, is a homodimeric lectin-like receptor. However, there has not been one research of KLRK1 as a biomarker in lung cancer. Data including patients` clinical characteristics and RNAseq information of KLRK1 from TCGA were downloaded. A total of 1019 patients with lung cancer were included in this study, among which 407 patients were female and 611 patients were male. Evaluations of mRNA expression, diagnostic value by ROC (receiver operating characteristic) curves and prognostic value by survival curve, Cox model and subgroup analysis were performed. The level of KLRK1 expression in lung adenocarcinoma cancer tissues and normal lung tissues was detected by qRT-PCR. The CCK-8 assay investigated the proliferation rate and the wound healing assay assessed the migratory ability in vitro. The expression of KLRK1 in tumor was lower than that in normal tissue. KLRK1 expression was associated with gender, histologic grade, stage, T classification and vital status. Patients with high KLRK1 expression presented an improved overall survival (P = 0.0036) and relapse free survival (P = 0.0031). KLRK1 was found to have significant prognostic value in lung adenocarcinoma (P = 0.015), stage I/II (P = 0.03), older patients (P = 0.0052), and male (P = 0.0047) by subgroup overall survival analysis, and in lung adenocarcinoma (P = 0.0094), stage I/II (P = 0.0076), older patients (P = 0.0072), and male (P = 0.0033) by subgroup relapse free survival analysis. Lung adenocarcinoma cancer patients with high KLRK1 expression presented an improved overall survival (P = 0.015) and relapse free survival (P = 0.0094). In vitro studies indicated that KLRK1 inhibited tumor cell proliferation and migration. KLRK1 was an independent prognostic factor and high KLRK1 expression indicated a better overall and relapse free survival. KLRK1 may be a prognostic biomarker for lung adenocarcinoma cancer.
Subject terms: Cancer, Lung cancer
Introduction
Lung cancer is one of the most common malignancy worldwide and causes estimated 1.6 million deaths each year1,2. Depending on stage and regional differences, the 5-year survival rate of lung cancer varied from 4 to 17%3. Lung cancer is classified into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). SCLC only accounts for 15% of lung cancer cases, while the rest approximately 85% are NSCLC, among which lung adenocarcinoma and lung squamous cell carcinoma are the most common histological types1,2.
Cancer immunosurveillance has been found to play an important role in lung cancer and may be related with its prognosis. Recently, the role of KLRK1 in cancer immunosurveillance and immune escape is widely studied4–6. KLRK1, encoding NKG2D, is a homodimeric lectin-like receptor7. As a cytotoxic and co-stimulatory molecule on NK cells and T cells, KLRK1 is unique because it does not have any inhibitory isoforms8,9. KLRK1 has two kinds of ligands, which belong to MIC and RAET1 gene family10. The ligands of KLRK1 are frequently expressed on primary tumor cells and KLRK1 was reported to play a role in the control of tumor and infection4,9. Some studies have demonstrated the antitumor function of KLRK1, while some researchers argue that KLRK1 contributes to tumor growth in a model of inflammation-driven liver cancer11,12. As Neal et al. reviewed about developing biomarker-specific end points in lung cancer clinical trials, the exploration of biomarkers for a better diagnosis and prognosis is of great importance13. However, there has not been one research of KLRK1 as a biomarker in lung cancer.
In this study, expression of KLRK1 in patients with lung cancer was first evaluated. First, the relationship of KLRK1 and clinical features was examined. Then, the receiver operating characteristic (ROC) curves was plotted for analyzing the diagnostic value of KLRK1. To study the prognostic value of KLRK1, the survival package in R was used and Cox model was established, followed by evaluation of subgroups including different genders, different ages, and different stages.
Methods
Data mining
To collect original data from patients suffering from lung cancer, data mining was carried out. Specifically, we download data including patients’ clinical characteristics and RNAseq information of KLRK1 from TCGA (The Cancer Genome Atlas) database by UCSC Xena. Given that all the data were open to public, no ethical approval was needed. A total of 1019 patients with lung cancer were included in this study, among which 407 patients were female and 611 patients were male.
The RNA expression of KLRK1 was shown in boxplots as center line represents median, top line and bottom line of the box represents upper and lower quartiles and vertical lines represents 95% confidence intervals (95% CI).
To study the diagnostic value of KLRK1 in patients with lung cancer, ROC (Receiver operating characteristic) curves was plotted by pROC package14. The calculated area under curves (AUC) indicated the diagnostic value. Furthermore, according to the identified threshold level of KLRK1, the patients were grouped into high expression group and low expression.
To study the prognostic value of KLRK1 in patients with lung cancer, the survival package in R was used and Cox model was established15. Evaluation of subgroups was performed as well.
Wound healing assay
Wound-healing assays were performed as previously described16. The migration of cells toward the wound was photographed under a Nikon fluorescence microscope.
Immunohistochemical staining in HPA database and qRT-PCR
The Human Protein Atlas (HPA, http://www.proteinatlas.org/) online database was explored to validate the KLRK1 protein expression in lung cancer by immunohistochemical (IHC) staining by CAB021896 antibody17. Ten pairs of lung adenocarcinoma cancer tissues and normal lung tissues were obtained from primary adenocarcinoma cancer patients at the Affiliated Hospital of Weifang Medical University. According to the manufacturer’s instructions, total RNA was extracted using TRIzol reagent, cDNA was synthesized and the qRT-PCR was performed and calculated by means of 2−ΔΔCt methods. The related primer of KLRK1 were displayed as following: F: 5ʹ-TGGATTCGTGGTCGGAGGTCTC-3ʹ, R: 5ʹ-GGACATCTTTGCTTTTGCCATCGTG-3ʹ.
Cell culture and cell transfection
A549 cell lines were purchased from American Tissue Culture Collection, and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37 °C with 5% CO2.
The KLRK1 sequence was amplified and inserted into pCMV vector (Beyotime). The transfection of KLRK1 over-expression or control plasmid was performed using Lipofectamine 3000 (Invitrogen).
Cell proliferation assay
The cells were treated with plasmids and cultured for 24 h. Then, 10 μL of CCK-8 reagent was added and cultured for 20 min. A microplate reader was used to measure the absorbance at 490 nm. The cell viability was calculated relative to the untreated control.
To evaluate the cell viability, co-staining of calcein AM and PI was performed. The cells were seeded and incubated for 16 h. After different treatments for 24 h, the cells were stained and observed using a Nikon fluorescence microscope.
Wound healing assay
Wound-healing assays were performed as previously described16. The migration of cells toward the wound was photographed under a Nikon fluorescence microscope.
Statistical analysis
R version 3.5.2 packages (https://www.R-project.org) was used for bioinformatics analysis18. Data were presented using the ggplot2 package in R19. The Wilcoxon rank-sum test was used for comparison between two groups, and the Kruskal–Wallis test was used for comparison among three or more groups. For assessment of associations between KLRK1 expression and clinical parameters, the chi-squared test was used, and corrected by Fisher’s exact test. Data from in vitro and in vivo experiments were analyzed by the Student’s t-test (unpaired, two-tailed). P < 0.05 was statistical significance.
Ethical approval
This study was approved by the ethics committee of the Affiliated Hospital of Weifang Medical University and conducted in strict accordance with the National Institutes of Health guidelines.
Results
Characteristics of patients with lung cancer
Clinical characteristics of the patients with lung cancer, including age, gender, histological type, stage, T classification, N classification, M classification, radiation therapy, residual tumor, vital status, sample type, KLRK1 expression, were shown in Table 1. The percentage of two histological types, lung adenocarcinoma (50.74%) and lung squamous cell carcinoma (49.26%), was close. About half of the patients were in stage I (51.13%). As for T, N, M classification, T2 (56.04%), N0 (63.98%), and M0 (74.39%) were highest among each classification. Most patients (99.80%) were primary tumor.
Table 1.
Clinical characteristics of the patients with lung cancer.
| Characteristics | Number (%) |
|---|---|
| Age | |
| < 55 | 109 (10.70) |
| ≥ 55 | 881 (86.46) |
| NA | 29 (2.84) |
| Gender | |
| Female | 407 (39.94) |
| Male | 611 (59.96) |
| NA | 1 (0.10) |
| Histological type | |
| Lung adenocarcinoma | 517 (50.74) |
| Lung squamous cell carcinoma | 502 (49.26) |
| Stage | |
| I | 521 (51.13) |
| II | 284 (27.87) |
| III | 168 (16.49) |
| IV | 33 (3.24) |
| NA | 13 (1.28) |
| T classification | |
| T1 | 284 (27.87) |
| T2 | 571 (56.04) |
| T3 | 118 (11.58) |
| T4 | 42 (4.12) |
| TX | 3 (0.29) |
| NA | 1 (0.10) |
| N classification | |
| N0 | 652 (63.98) |
| N1 | 227 (22.28) |
| N2 | 114 (11.19) |
| N3 | 7 (0.69) |
| NX | 17 (1.67) |
| NA | 2 (0.20) |
| M classification | |
| M0 | 758 (74.39) |
| M1 | 32 (3.14) |
| MX | 220 (21.59) |
| NA | 9 (0.88) |
| Radiation therapy | |
| NO | 782 (76.74) |
| YES | 110 (10.79) |
| NA | 127 (12.46) |
| Residual tumor | |
| R0 | 743 (72.91) |
| R1 | 25 (2.45) |
| R2 | 8 (0.79) |
| RX | 48 (4.71) |
| NA | 195 (19.14) |
| Vital status | |
| Deceased | 404 (39.65) |
| Living | 614 (60.26) |
| NA | 1 (0.10) |
| Sample type | |
| Primary tumor | 1017 (99.80) |
| Recurrent tumor | 2 (0.20) |
| KLRK1 expression | |
| High | 701 (68.79) |
| Low | 318 (31.21) |
NA not available, X represents uncertain.
Low KLRK1 expression in tumor
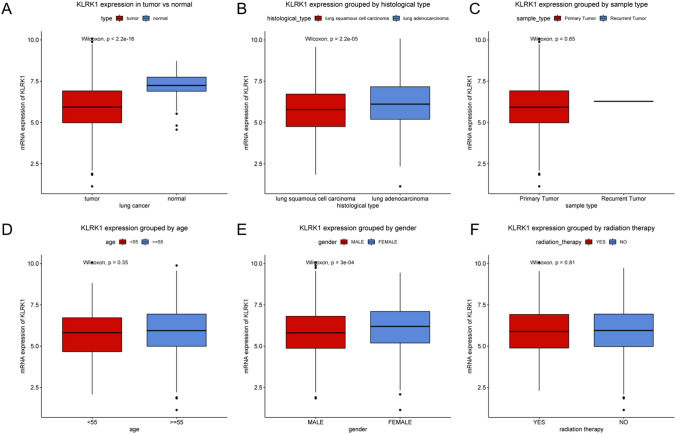
As shown in Fig. 1, the expression of KLRK1 in tumor was lower than that in normal tissue (P < 2.2e−16). The association of KLRK1 expression with clinical features, including histological type, sample type, age, gender, radiation therapy, was further evaluated. KLRK1 expression in lung squamous cell carcinoma was lower than that in lung adenocarcinoma (P = 2.2e−05). KLRK1 expression in male was lower than that in female (P = 3e−04). Other clinical features did not show statistical differences (P > 0.05).
Figure 1.
KLRK1 expression in lung cancer. Expression of KLRK1 grouped by (A) tumor vs. normal tissue, (B) histological type, (C) sample type, (D) age, (E) gender, and (F) radiation therapy.
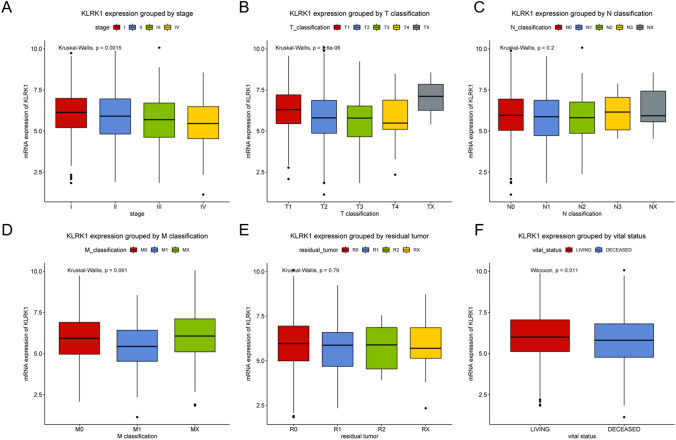
Association of KLRK1 expression with stage, T classification, N classification, M classification, residual tumor and vital status was evaluated (Fig. 2). The KLRK1 expression got decreased progressively with higher stages (P = 0.0015) and T classification (P = 2.6e−06). No significances were observed in N classification (P = 0.200), M classification (P = 0.091) and residual tumor (P = 0.790). The KLRK1 expression in living patients was a little higher than that in deceased patients (P = 0.011).
Figure 2.
KLRK1 expression in lung cancer. Expression of KLRK1 grouped by (A) stage, (B) T classification, (C) N classification, (D) M classification, (E) residual tumor and (F) vital status.
Diagnostic value of KLRK1 for lung cancer
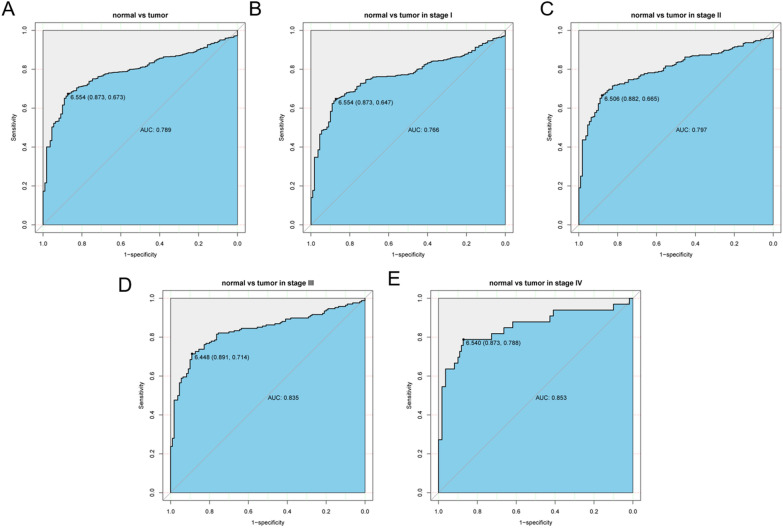
As shown in Fig. 2, the ROC analysis was first performed in all lung cancer patients, indicating a modest diagnostic value with AUC of 0.789. Moreover, different stages of lung cancer were analyzed. It was suggested that the diagnostic value was increasing with stage getting higher from stage I (AUC = 0.766) to stage II (AUC = 0.797) to stage III (AUC = 0.835) finally to stage IV (AUC = 0.853) (Fig. 3).
Figure 3.
ROC (Receiver operating characteristic) curves of KLRK1 expression in patients with lung cancer. (A) Normal vs. overall, (B) stage I, (C) stage II, (D) stage III and (E) stage IV tumor.
Correlation of KLRK1 expression with clinical features
According to the threshold value ascertained by ROC analysis, patients were divided to two subgroups, high (n = 701) and low (n = 318) KLRK1 expression group. The relationship between the clinical features and KLRK1 expression in patients with lung cancer was shown in Table 2. KLRK1 expression was associated with gender (P = 0.005), histologic grade (P = 0.001), stage (P = 0.001), T classification (P < 0.001) and vital status (P = 0.001). No correlation was found between KLRK1 expression and age (P = 0.431), N classification (P = 0.218), M classification (P = 0.383), radiation therapy (P = 0.399), residual tumor (P = 0.336), and sample type (P = 0.850).
Table 2.
Relationship between the clinical features and KLRK1 expression in patients with lung cancer.
| Clinical characteristics | Variable | Number of patients |
KLRK1 | χ2 | P value | |||
|---|---|---|---|---|---|---|---|---|
| High | % | Low | % | |||||
| Age | < 55 | 109 | 71 | (10.41) | 38 | (12.34) | 0.620 | 0.431 |
| ≥ 55 | 881 | 611 | (89.59) | 270 | (87.66) | |||
| Gender | Female | 407 | 301 | (42.94) | 106 | (33.44) | 7.819 | 0.005 |
| Male | 611 | 400 | (57.06) | 211 | (66.56) | |||
| Histological type | Lung adenocarcinoma | 517 | 381 | (54.35) | 136 | (42.77) | 11.285 | 0.001 |
| Lung squamous cell carcinoma | 502 | 320 | (45.65) | 182 | (57.23) | |||
| Stage | I | 521 | 386 | (55.86) | 135 | (42.86) | 15.336 | 0.001 |
| II | 284 | 181 | (26.19) | 103 | (32.70) | |||
| III | 168 | 102 | (14.76) | 66 | (20.95) | |||
| IV | 33 | 22 | (3.18) | 11 | (3.49) | |||
| T classification | T1 | 284 | 229 | (32.67) | 55 | (17.35) | 29.391 | < 0.001 |
| T2 | 571 | 373 | (53.21) | 198 | (62.46) | |||
| T3 | 118 | 69 | (9.84) | 49 | (15.46) | |||
| T4 | 42 | 27 | (3.85) | 15 | (4.73) | |||
| TX | 3 | 3 | (0.43) | 0 | (0) | |||
| N classification | N0 | 652 | 461 | (65.86) | 191 | (60.25) | 5.610 | 0.218 |
| N1 | 227 | 146 | (20.86) | 81 | (25.55) | |||
| N2 | 114 | 75 | (10.71) | 39 | (12.30) | |||
| N3 | 7 | 4 | (0.57) | 3 | (0.95) | |||
| NX | 17 | 14 | (2.00) | 3 | (0.95) | |||
| M classification | M0 | 758 | 516 | (74.03) | 242 | (77.32) | 1.904 | 0.383 |
| M1 | 32 | 21 | (3.01) | 11 | (3.51) | |||
| MX | 220 | 160 | (22.96) | 60 | (19.17) | |||
| Radiation therapy | No | 782 | 540 | (88.38) | 242 | (86.12) | 0.711 | 0.399 |
| Yes | 110 | 71 | (11.62) | 39 | (13.88) | |||
| Residual tumor | R0 | 743 | 513 | (90.80) | 230 | (88.8) | 3.305 | 0.336 |
| R1 | 25 | 14 | (2.48) | 11 | (4.25) | |||
| R2 | 8 | 4 | (0.71) | 4 | (1.54) | |||
| RX | 48 | 34 | (6.02) | 14 | (5.41) | |||
| Vital status | Deceased | 404 | 253 | (36.09) | 151 | (47.63) | 11.673 | 0.001 |
| Living | 614 | 448 | (63.91) | 166 | (52.37) | |||
| Sample type | Primary tumor | 1017 | 699 | (99.71) | 318 | (100) | 0.036 | 0.850 |
| Recurrent tumor | 2 | 2 | (0.29) | 0 | (0) | |||
X represents uncertain.
IHC and qRT-PCR result
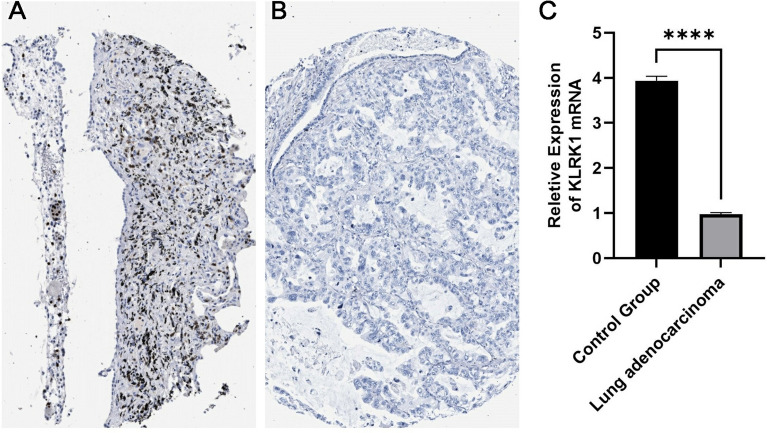
Comparing the IHC experimental pictures obtained in the HPA database, it can be seen that the expression of KLRK1 in lung adenocarcinoma cancer tissues was significantly lower than that in normal lung tissues. As shown in Fig. 4, we utilized qRT-PCR to validate the KLRK1 expression in lung adenocarcinoma cancer tissues and found the KLRK1 expression was down-regulated in the lung adenocarcinoma cancer (N = 10) compared with normal lung tissues (N = 10; P < 0.001).
Figure 4.
IHC of KLRK1 expression in normal lung tissue (A) and lung adenocarcinoma tissue (B), qRT-PCR of KLRK1 in two tissues (C).
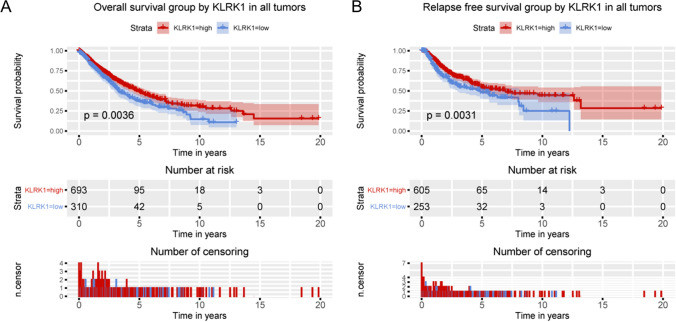
Overall survival and relapse free survival of KLRK1 for lung cancer
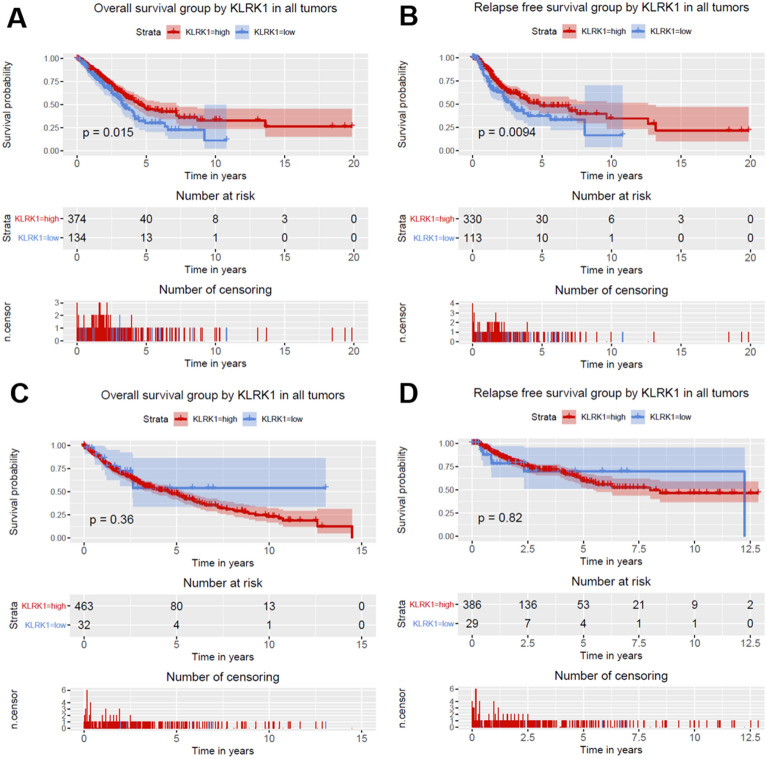
Given that KLRK1 was correlated with survival, the prognostic value of KLRK1 was further studied. As shown in Fig. 5, lung cancer patients with high KLRK1 expression presented an improved overall survival (P = 0.0036) and relapse free survival (P = 0.0031). As shown in Fig. 6, lung adenocarcinoma cancer patients with high KLRK1 expression presented an improved overall survival (P = 0.015) and relapse free survival (P = 0.0094). But no significances were found in lung squamous cell cancer patients with high KLRK1 expression.
Figure 5.
Relationship of KLRK1 expression with (A) overall and (B) relapse free survival in all patients with lung cancer.
Figure 6.
Relationship of KLRK1 expression with (A) overall and (B) relapse free survival in patients with lung adenocarcinoma cancer, and (C) overall and (D) relapse free survival in patients with lung squamous cell cancer.
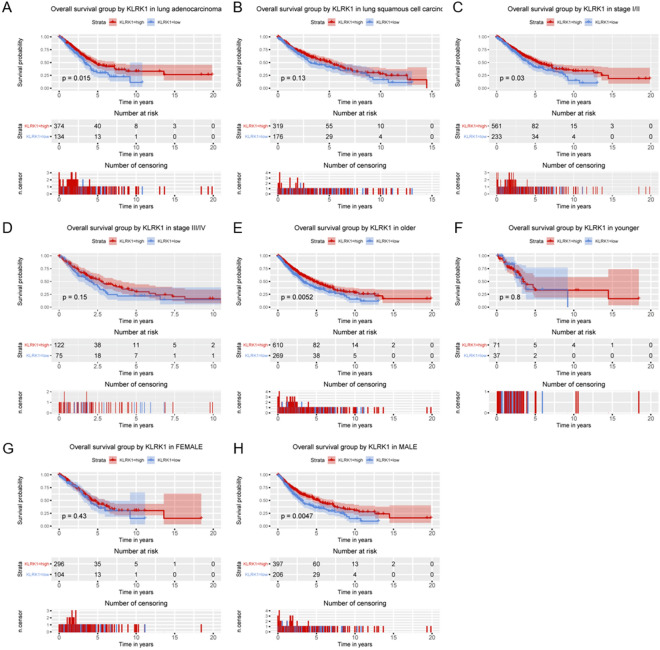
Subgroup analysis of KLRK1 for lung cancer’s overall survival
By overall survival analysis of subgroups (Fig. 7, Figs. S1, S2), KLRK1 was found to have significant prognostic value in lung adenocarcinoma (P = 0.015), stage I/II (P = 0.03), older patients (P = 0.0052), and male (P = 0.0047). For lung adenocarcinoma cancer, KLRK1 was found to have significant prognostic value in stage I/II (P = 0.049), older patients (P = 0.0064), and male (P = 0.013).
Figure 7.
Relationship of KLRK1 expression with overall survival in all patients with lung cancer in (A) lung adenocarcinoma, (B) lung squamous cell carcinoma, (C) stage I/II, (D) stage III/IV, (E) older patients, (F) younger patients, (G) female and (H) male.
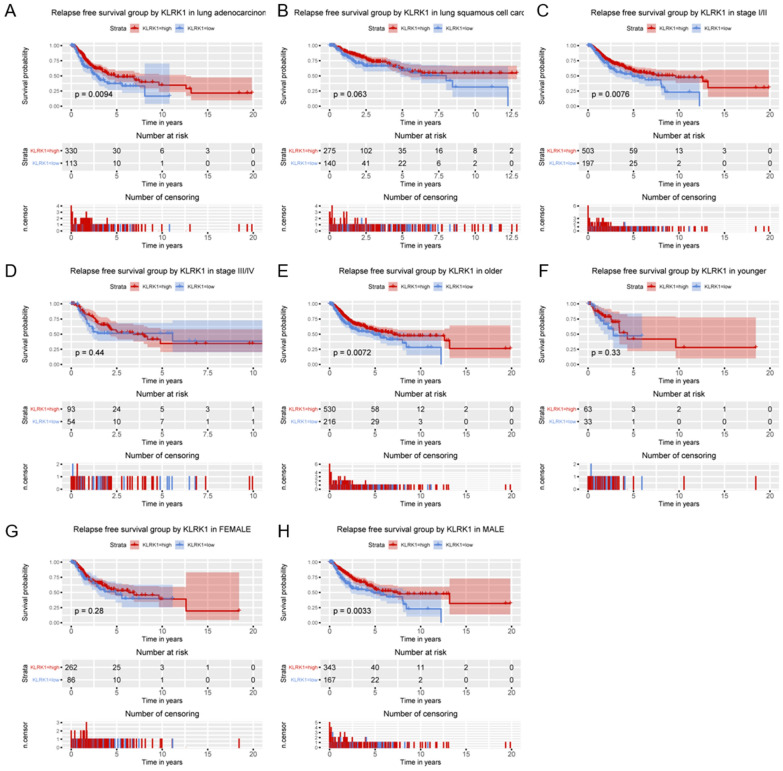
Subgroup analysis of KLRK1 for lung cancer’s relapse free survival
By relapse free survival analysis of subgroups (Fig. 8, Figs. S3, S4), KLRK1 was found to have significant prognostic value in lung adenocarcinoma (P = 0.0094), stage I/II (P = 0.0076), older patients (P = 0.0072), and male (P = 0.0033). For lung adenocarcinoma cancer, KLRK1 was found to have significant prognostic value in stage I/II (P = 0.0025), and older patients (P = 0.012).
Figure 8.
Relationship of KLRK1 expression with relapse free survival in all patients with lung cancer in (A) lung adenocarcinoma, (B) lung squamous cell carcinoma, (C) stage I/II, (D) stage III/IV, (E) older patients, (F) younger patients, (G) female and (H) male.
KLRK1 is an independent risk factor for lung cancer’s overall survival
The univariate Cox model of overall and relapse free survival in patients with lung cancer was established (Tables 3, 4, Table S1), and the multivariate analysis of overall and relapse free survival in patients with lung cancer was further performed (Tables 5, 6, Table S2).
Table 3.
Univariate analysis of overall and relapse free survival in patients with lung cancer.
| Overall survival | Relapse free survival | |||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | |
| Age | 1.03 | 0.74–1.43 | 0.867 | 0.95 | 0.63–1.43 | 0.819 |
| Gender | 1.14 | 0.93–1.40 | 0.201 | 0.94 | 0.73–1.20 | 0.613 |
| Histological type | 1.12 | 0.91–1.36 | 0.282 | 0.63 | 0.49–0.82 | < 0.001 |
| T classification | 1.42 | 1.25–1.61 | < 0.001 | 1.35 | 1.14–1.59 | < 0.001 |
| N classification | 1.39 | 1.22–1.57 | < 0.001 | 1.41 | 1.20–1.65 | < 0.001 |
| M classification | 1.16 | 0.91–1.49 | 0.220 | 0.83 | 0.62–1.11 | 0.210 |
| Radiation therapy | 1.00 | 0.79–1.27 | 0.983 | 1.76 | 1.32–2.35 | < 0.001 |
| Residual tumor | 1.17 | 1.03–1.33 | 0.016 | 1.29 | 1.11–1.49 | 0.001 |
| Stage | 1.48 | 1.33–1.64 | < 0.001 | 1.41 | 1.23–1.62 | < 0.001 |
| KLRK1 expression | 0.74 | 0.60–0.91 | 0.004 | 0.68 | 0.52–0.88 | 0.003 |
Table 4.
Univariate analysis of overall and relapse free survival in patients with lung adenocarcinoma cancer.
| Overall survival | Relapse free survival | |||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | |
| Age | 0.82 | 0.55–1.22 | 0.322 | – | – | – |
| Gender | 1.05 | 0.78–1.4 | 0.760 | – | – | – |
| T classification | 1.52 | 1.26–1.82 | < 0.001 | 1.16 | 0.96–1.4 | 0.132 |
| N classification | 1.67 | 1.41–1.97 | < 0.001 | 1.15 | 0.92–1.44 | 0.228 |
| M classification | 1.4 | 1.04–1.9 | 0.029 | 0.93 | 0.68–1.28 | 0.662 |
| Radiation therapy | 1.25 | 0.89–1.75 | 0.199 | – | – | – |
| Residual tumor | 1.2 | 1.01–1.42 | 0.037 | 1.14 | 0.96–1.35 | 0.150 |
| Stage | 1.68 | 1.47–1.93 | < 0.001 | 1.51 | 1.21–1.89 | < 0.001 |
| KLRK1 | 0.69 | 0.51–0.93 | 0.016 | 0.78 | 0.57–1.07 | 0.129 |
Table 5.
Multivariate analysis of overall survival and relapse free survival in patients with lung cancer.
| Overall survival | Relapse-free survival | |||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | |
| Histological type | – | – | – | 0.58 | 0.45–0.76 | < 0.001 |
| T classification | 1.18 | 1.02–1.36 | 0.025 | 1.22 | 1.01–1.47 | 0.040 |
| N classification | – | – | – | 1.11 | 0.89–1.4 | 0.343 |
| Radiation therapy | – | – | – | 1.42 | 1.06–1.91 | 0.018 |
| Residual tumor | 1.13 | 0.99–1.28 | 0.067 | 1.23 | 1.07–1.42 | 0.004 |
| Stage | 1.34 | 1.14–1.57 | < 0.001 | 1.16 | 0.93–1.43 | 0.183 |
| KLRK1 | 0.79 | 0.64–0.97 | 0.025 | 0.74 | 0.57–0.96 | 0.022 |
Table 6.
Multivariate analysis of overall survival and relapse free survival in patients with lung adenocarcinoma cancer.
| Overall survival | Relapse free survival | |||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | |
| T classification | 1.39 | 1.13–1.71 | 0.002 | 1.18 | 0.94–1.49 | 0.155 |
| N classification | 1.45 | 1.2–1.76 | < 0.001 | 1.12 | 0.85–1.48 | 0.410 |
| Radiation therapy | 1.97 | 1.37–2.82 | < 0.001 | 1.62 | 1.12–2.35 | 0.010 |
| Residual tumor | 1.20 | 1.01–1.43 | 0.035 | 1.16 | 0.98–1.38 | 0.088 |
| Stage | 1.44 | 1.23–1.7 | < 0.001 | 1.17 | 0.91–1.51 | 0.209 |
| KLRK1 | 0.64 | 0.45–0.9 | 0.010 | 0.74 | 0.52–1.06 | 0.099 |
As shown in Table 3, no obvious differences were observed in age (P = 0.867), gender (P = 0.201), histological type (P = 0.282), M classification (P = 0.220), and radiation therapy (P = 0.983). T classification (P < 0.001), N classification (P < 0.001), residual tumor (P = 0.016), stage (P < 0.001) and KLRK1 expression (P = 0.004) showed significant differences for lung cancer. As shown in Table 4, T classification (P < 0.001), N classification (P < 0.001), M classification (P = 0.029), residual tumor (P = 0.037), stage (P < 0.001) and KLRK1 expression (P = 0.016) showed significant differences for lung adenocarcinoma cancer. As shown in Table 5, T classification (HR 1.18; 95% CI 1.02–1.36; P = 0.025) and stage (HR 1.34; 95% CI 1.14–1.57; P < 0.001) were risk factors for lung cancer.
KLRK1 is an independent risk factor for lung cancer’s relapse free survival
As shown in Table 3, no obvious differences were observed in age (P = 0.819), gender (P = 0.613), and M classification (P = 0.210). Histological type (P < 0.001), T classification (P < 0.001), N classification (P < 0.001), radiation therapy (P < 0.001), residual tumor (0.001), stage (P < 0.001) and KLRK1 expression (P = 0.003) showed significant differences. As shown in Table 4, stage (P < 0.001) showed significant differences for lung adenocarcinoma cancer. As shown in Table 5, T classification (HR 1.22; 95% CI 1.01–1.47; P = 0.040), radiation therapy (HR 1.42; 95% CI 1.06–1.91; P = 0.018) and residual tumor (HR 1.23; 95% CI 1.07–1.42; P = 0.004) were risk factors for lung cancer.
KLRK1 inhibits lung cancer cell proliferation and migration in vitro
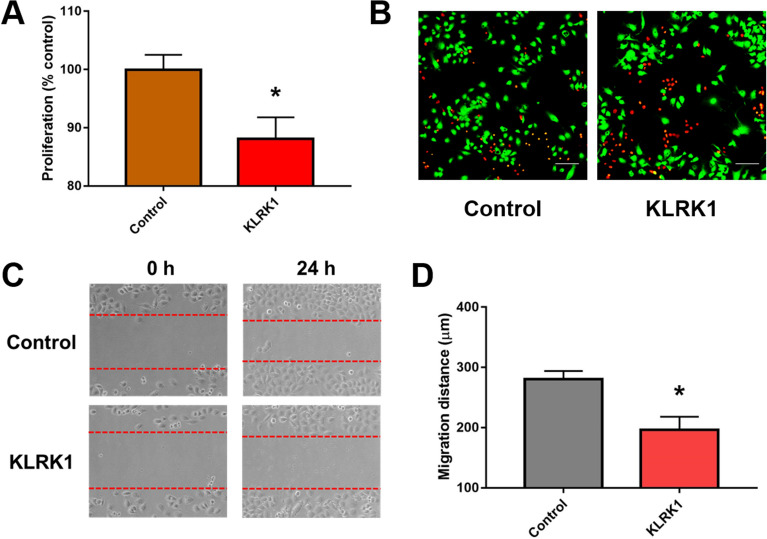
We further studied the effect of KLRK1 on cells by overexpressing KLRK1 in lung cancer A549 cell lines. The CCK-8 assay showed that KLRK1 upregulation significantly inhibited cell proliferation (P < 0.05; Fig. 9A). Compared with control group, KLRK1 increased the percentage of dead cells by live/dead staining (Fig. 9B). KLRK1 decreased the migration distance of cancer cell (P < 0.05; Fig. 9C,D).
Figure 9.
The effect of KLRK1 in lung cancer A549 cell lines. (A) The CCK-8 assay; (B) live/dead staining; (C) the migration and (D) migration distance.
Discussion
Lung cancer is one of the most common malignancy with poor prognosis worldwide. Many researches have working on mining novel prognostic biomarker in different types of cancer20–36. However, little is done in the field of lung cancer. In this bioinformatics study, KLRK1 was found to lower expressed in lung cancer compared with normal lung tissue. KLRK1 expression was associated with gender, histologic grade, stage, T classification and vital status. Moreover, KLRK1 presented a moderate diagnostic value for lung cancer.
Recently, research about biomarkers by data mining is popular20,21,23,24,27,30,36–40. Although some protein blood biomarkers have already been put into clinical practice, the ability of their diagnosis and prognosis is limited and he exploration of biomarkers for lung cancer is ongoing13. CEA (carcinoembryonic antigen) and CTC (Circulating tumor cells) are used in some lung cancers41,42. Also, there are studies reporting some novel biomarkers. Jiang et al. found thymidine kinase 1 combined with CEA, CYFRA21-1 and NSE improved its diagnostic value for lung cancer43. Yang et al. compared the different value of combination in CEA, CA125 (carbohydrate antigen 125), CY211 (cytokeratin 19 fragment), NSE (neuron-specific enolase), and SCC (squamous cell carcinoma antigen)44. Tang et al. identified HSP70 (circulating heat shock protein 70) as a novel biomarker for early diagnosis of lung cancer45. In this study, we first found KLRK1 as a prognostic biomarker for lung cancer.
Researches of biomarkers for lung cancer are not only limited in prognosis but also in diagnosis. Wu et al. found UCK2 (Uridine‐cytidine kinase 2) as a potential diagnostic and prognostic biomarker for lung cancer, and identified UCK2 highly expressed in stage IA lung cancer with AUC > 0.946. Jiang et al. reported a diagnostic value of circulating lncRNA XLOC_009167 with AUC of 0.739847. Similarly, we found the KLRK1 presented a promising diagnostic value in lung cancer, especially advanced stages. In comparison of lung cancer vs. tumor, the AUC could get into 0.789, indicating a relatively high diagnostic value of KLRK1 as a biomarker for lung cancer.
KLRK1 is an immunoreceptor binding to a variety of cell surface glycoproteins distantly related to MHC class I molecules48. A recent study conducted by Shi et al. reported the up-regulation of KLRK1-activating receptors that recognize lung cancer could facilitate the clearance of lung cancer49. Their results confirmed the role of KLRK1 in changing state of NK cells, leading to the control of lung cancer through immunosurveillance. Paczulla et al. reported the absence of KLRK1 ligand mediated the immune evasion of leukaemia stem cells30. Dong et al. found T cells expressing KLRK1 chimeric antigen receptors could efficiently eliminate glioblastoma and cancer stem cells34. Moreover, blockade drugs for EGFR and PD-1 can enhance the effective of NKG2D, encoded by KLRK1, lead to cancer cell recognition and killing by NK effector cells50,51. From our results, high KLRK1 expression is associated with a better overall and relapse free survival, which may attributes to the immunosurveillance of KLRK1 therefore suppressing the proliferation and metastasis of lung cancer4. Of note, our results showed the significances of KLRK1 in both overall survival and relapse free survival in lung cancer.
Our research first suggested the diagnostic and prognostic value of KLRK1 for lung cancer. However, the major limitation is that this study analyzed the data from a single database by data mining. Further verifications in different areas and populations are required. Besides, in vivo function experiments and exploration of its molecular mechanism would further illuminate the role of KLRK1 in lung cancer.
Conclusions
In conclusion, KLRK1 was lower expressed in lung cancer in comparison with normal lung tissue. KLRK1 expression was associated with gender, histologic grade, stage, T classification and vital status. KLRK1 had a diagnostic value for lung adenocarcinoma cancer. KLRK1 was an independent prognostic factor and high KLRK1 expression indicated a better overall and relapse free survival. KLRK1 may be a novel biomarker for lung adenocarcinoma cancer.
Supplementary Information
Author contributions
Y.Z. contributed to the study design and wrote the manuscript. Z.C. contributed to the data evaluation and revised the manuscript. A.J. provided experimental specimens and assisted in the experiment. G.G. supervised the study and extensively revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Aifang Jiang and Guanqi Gao.
Contributor Information
Aifang Jiang, Email: wfjaf@sina.com.
Guanqi Gao, Email: lygqgao@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-05997-z.
References
- 1.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 2.Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat. Rev. Dis. Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 3.Fred R, Hirsch GVS, Mulshine JL, Kwon R, Curran WJ, Wu Y-L, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.Ljunggren HG. Cancer immunosurveillance: NKG2D breaks cover. Immunity. 2008;28(4):492–494. doi: 10.1016/j.immuni.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Schmiedel D, Mandelboim O. NKG2D ligands-critical targets for cancer immune escape and therapy. Front. Immunol. 2018;9:2040. doi: 10.3389/fimmu.2018.02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding H, Yang X, Wei Y. Fusion proteins of NKG2D/NKG2DL in cancer immunotherapy. Int. J. Mol. Sci. 2018;19(1):177. doi: 10.3390/ijms19010177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanier LL. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008;9(5):495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antoun A, Vekaria D, Salama RA, et al. The genotype of RAET1L (ULBP6), a ligand for human NKG2D (KLRK1), markedly influences the clinical outcome of allogeneic stem cell transplantation. Br. J. Haematol. 2012;159(5):589–598. doi: 10.1111/bjh.12072. [DOI] [PubMed] [Google Scholar]
- 9.Jelencic V, Sestan M, Kavazovic I, et al. NK cell receptor NKG2D sets activation threshold for the NCR1 receptor early in NK cell development. Nat. Immunol. 2018;19(10):1083–1092. doi: 10.1038/s41590-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David Cosman MI, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. ULBPs, novel MHC class I–related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 11.Sheppard S, Ferry A, Guedes J, Guerra N. The paradoxical role of NKG2D in cancer immunity. Front. Immunol. 2018;9:1808. doi: 10.3389/fimmu.2018.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan S, Guo W, Xu Z, et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol. Cancer. 2019;18(1):29. doi: 10.1186/s12943-019-0956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neal JW, Gainor JF, Shaw AT. Developing biomarker-specific end points in lung cancer clinical trials. Nat. Rev. Clin. Oncol. 2015;12(3):135–146. doi: 10.1038/nrclinonc.2014.222. [DOI] [PubMed] [Google Scholar]
- 14.Robin X, Turck N, Hainard A, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Betancur M, Sadaoui H, Piffaretti C, Rey G. Survival analysis with multiple causes of death: Extending the competing risks model. Epidemiology. 2017;28(1):12–19. doi: 10.1097/EDE.0000000000000531. [DOI] [PubMed] [Google Scholar]
- 16.Yan F, Shen N, Pang J, Zhao N, Deng B, Li B. A regulatory circuit composed of DNA methyltransferases and receptor tyrosine kinases controls lung cancer cell aggressiveness. Oncogene. 2017;36(50):6919–6928. doi: 10.1038/onc.2017.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 18.Team RDCJC. R: A Language and Environment for Statistical Computing, Vol. 14, 12–21 (R Foundation for Statistical Computing, 2009).
- 19.Wickham H. Ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. 2011;174(1):245–246. [Google Scholar]
- 20.Cai H, Jiao Y, Li Y, Yang Z, He M, Liu Y. Low CYP24A1 mRNA expression and its role in prognosis of breast cancer. Sci. Rep. 2019;9(1):13714. doi: 10.1038/s41598-019-50214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui Y, Jiao Y, Wang K, He M, Yang Z. A new prognostic factor of breast cancer: High carboxyl ester lipase expression related to poor survival. Cancer Genet. 2019;239:54–61. doi: 10.1016/j.cancergen.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Hou L, Zhang X, Jiao Y, et al. ATP binding cassette subfamily B member 9 (ABCB9) is a prognostic indicator of overall survival in ovarian cancer. Medicine (Baltimore) 2019;98(19):e15698. doi: 10.1097/MD.0000000000015698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao Y, Fu Z, Li Y, Meng L, Liu Y. High EIF2B5 mRNA expression and its prognostic significance in liver cancer: A study based on the TCGA and GEO database. Cancer Manage. Res. 2018;10:6003–6014. doi: 10.2147/CMAR.S185459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao Y, Fu Z, Li Y, Zhang W, Liu Y. Aberrant FAM64A mRNA expression is an independent predictor of poor survival in pancreatic cancer. PLoS ONE. 2019;14(1):e0211291. doi: 10.1371/journal.pone.0211291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao Y, Li Y, Fu Z, et al. OGDHL expression as a prognostic biomarker for liver cancer patients. Dis. Mark. 2019;2019:9037131. doi: 10.1155/2019/9037131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiao Y, Li Y, Ji B, Cai H, Liu Y. Clinical value of lncRNA LUCAT1 expression in liver cancer and its potential pathways. J. Gastrointest. Liver Dis. 2019;28(4):439–447. doi: 10.15403/jgld-356. [DOI] [PubMed] [Google Scholar]
- 27.Jiao Y, Li Y, Jiang P, Han W, Liu Y. PGM5: A novel diagnostic and prognostic biomarker for liver cancer. PeerJ. 2019;7:e7070. doi: 10.7717/peerj.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao Y, Li Y, Liu S, Chen Q, Liu Y. ITGA3 serves as a diagnostic and prognostic biomarker for pancreatic cancer. OncoTargets Ther. 2019;12:4141–4152. doi: 10.2147/OTT.S201675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao Y, Li Y, Lu Z, Liu Y. High trophinin-associated protein expression is an independent predictor of poor survival in liver cancer. Dig. Dis. Sci. 2019;64(1):137–143. doi: 10.1007/s10620-018-5315-x. [DOI] [PubMed] [Google Scholar]
- 30.Paczulla AM, Rothfelder K, Raffel S, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572(7768):254–259. doi: 10.1038/s41586-019-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Jiao Y, Li Y, Liu Y. Expression of la ribonucleoprotein domain family member 4B (LARP4B) in liver cancer and their clinical and prognostic significance. Dis. Mark. 2019;2019:1569049. doi: 10.1155/2019/1569049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Jiao Y, Luo Z, Li Y, Liu Y. High peroxidasin-like expression is a potential and independent prognostic biomarker in breast cancer. Medicine (Baltimore) 2019;98(44):e17703. doi: 10.1097/MD.0000000000017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie Y, Jiao Y, Li Y, Li W. Investigation of the clinical significance and prognostic value of the lncRNA ACVR2B-As1 in liver cancer. Biomed. Res. Int. 2019;2019:13. doi: 10.1155/2019/4602371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang D, Sun B, Dai H, et al. T cells expressing NKG2D chimeric antigen receptors efficiently eliminate glioblastoma and cancer stem cells. J. Immunother. Cancer. 2019;7(1):171. doi: 10.1186/s40425-019-0642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Cui Y, He M, Jiao Y, Yang Z. Lipocalin-1 expression as a prognosticator marker of survival in breast cancer patients. Breast Care. 2019;15:272. doi: 10.1159/000503168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao YC, Jiao Y, Li YQ, Fu Z, Yang ZY, He M. Elevated high mobility group A2 expression in liver cancer predicts poor patient survival. Rev. Esp. Enferm. Dig. 2019 doi: 10.17235/reed.2019.6365/2019. [DOI] [PubMed] [Google Scholar]
- 37.Fu Z, Jiao Y, Li YQ, et al. PES1 in liver cancer: A prognostic biomarker with tumorigenic roles. Cancer Manage. Res. 2019;11:9641–9653. doi: 10.2147/CMAR.S226471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiao Y, Li Y, Fu Z, et al. OGDHL expression as a prognostic biomarker for liver cancer patients. J. Dis. Mark. 2019;2019:9. doi: 10.1155/2019/9037131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Jiao Y, Luo Z, Li Y, Liu Y. High peroxidasin-like expression is a potential and independent prognostic biomarker in breast cancer. Medicine. 2019;98(44):e17703. doi: 10.1097/MD.0000000000017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou LL, Jiao Y, Chen HM, et al. Differentially expressed long noncoding RNAs and regulatory mechanism of LINC02407 in human gastric adenocarcinoma. World J. Gastroenterol. 2019;25(39):5973–5990. doi: 10.3748/wjg.v25.i39.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011;29(12):1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 42.Grunnet M, Sorensen JB. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76(2):138–143. doi: 10.1016/j.lungcan.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Jiang ZF, Wang M, Xu JL. Thymidine kinase 1 combined with CEA, CYFRA21-1 and NSE improved its diagnostic value for lung cancer. Life Sci. 2018;194:1–6. doi: 10.1016/j.lfs.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 44.Yang Q, Zhang P, Wu R, Lu K, Zhou H. Identifying the best marker combination in CEA, CA125, CY211, NSE, and SCC for lung cancer screening by combining ROC curve and logistic regression analyses: Is it feasible? Dis. Mark. 2018;2018:2082840. doi: 10.1155/2018/2082840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang T, Yang C, Brown HE, Huang J. Circulating Heat shock protein 70 is a novel biomarker for early diagnosis of lung cancer. Dis. Mark. 2018;2018:6184162. doi: 10.1155/2018/6184162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y, Jamal M, Xie T, et al. Uridine-cytidine kinase 2 (UCK2): A potential diagnostic and prognostic biomarker for lung cancer. Cancer Sci. 2019;110(9):2734–2747. doi: 10.1111/cas.14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang N, Meng X, Mi H, et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin. Chim. Acta Int. J. Clin. Chem. 2018;486:26–33. doi: 10.1016/j.cca.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 48.Lazarova M, Steinle A. The NKG2D axis: An emerging target in cancer immunotherapy. Expert Opin. Therap. Targets. 2019;23(4):281–294. doi: 10.1080/14728222.2019.1580693. [DOI] [PubMed] [Google Scholar]
- 49.Lei Shi KL, Guo Y, Banerjee A, Wang Q, Lorenzc UM, Parlak M, Sullivane LC, Onyem OO, Arefanianf S, Stelowg EB, Brautigang DL, Bullockcg TNJ, Brownch MG, Krupnick AS. Modulation of NKG2D, NKp46, and Ly49C/I facilitates natural killer cell-mediated control of lung cancer. PNAS. 2018;115(46):11808–11813. doi: 10.1073/pnas.1804931115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tallerico R, Garofalo C, Carbone E. A new biological feature of natural killer cells: The recognition of solid tumor-derived cancer stem cells. Front. Immunol. 2016;7:179. doi: 10.3389/fimmu.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao L, Wang S, Kang G, et al. PD-1 blockade improves the anti-tumor potency of exhausted CD3 (+)CD56 (+) NKT-like cells in patients with primary hepatocellular carcinoma. Oncoimmunology. 2021;10(1):2002068. doi: 10.1080/2162402X.2021.2002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.