Abstract
To define relationships between Listeria monocytogenes genetic lineages, ribotypes, and serotypes, 235 L. monocytogenes isolates were characterized by serotyping and automated EcoRI ribotyping. Genetic lineage predicted the following serovar clusters: lineage I, comprising serotypes 1/2b, 3b, 3c, and 4b; lineage II, comprising serotypes 1/2a, 1/2c, and 3a; and lineage III, comprising serotypes 4a and 4c. Some EcoRI ribotypes contained multiple serotypes; a subset of these isolates was further differentiated with PvuII ribotyping. Of the 12 resultant EcoRI-PvuII combination types, only 4 contained multiple serotypes, demonstrating the potential of ribotyping for serotype prediction.
Listeria monocytogenes is a food-borne pathogen associated with severe invasive diseases in humans and animals that is estimated to cause approximately 2,500 cases of human illness and 500 deaths annually in the United States (11). Accurate, reliable, and standardized subtyping methods provide epidemiological markers, which are critical to disease outbreak investigations.
A variety of subtyping methods have been used to differentiate L. monocytogenes beyond the species level (19). Although serotyping is not very discriminatory, it is a universal technique sometimes used as a prerequisite for other subtyping methods (18). Serology divides L. monocytogenes into 13 serotypes on the basis of somatic and flagellar antigens; this technique relies on high-quality, specific sera prepared with standardized strains and is currently performed in only a small number of reference laboratories. The vast majority of human listeriosis cases are caused by three serotypes (1/2a, 1/2b, and 4b), rendering this method minimally useful in epidemiologic investigations (19). The goal in using molecular methods for subtyping of L. monocytogenes is to attain greater discrimination of strains than is achieved by serotyping (19).
Ribotyping, a sensitive and reproducible subtyping method, is based on rRNA gene restriction fragment polymorphisms. Previous work has shown that EcoRI ribotyping can differentiate L. monocytogenes from other Listeria spp., a large database of L. monocytogenes ribotype patterns already exists, and a fully automated ribotyping system is commercially available (3, 10, 12). As traditional phenotypic methods are now being used in conjunction with or have been replaced by molecular subtyping for L. monocytogenes surveillance, we must define the relationships between subtypes determined by different methods. Genotypic analyses have consistently grouped L. monocytogenes into two major lineages. Multilocus enzyme electrophoresis divides the species into two primary subgroups, division I (serotypes 1/2b, 4a, and 4b) and division II (serotypes 1/2a and 1/2c) (14). Pulsed-field gel electrophoresis also yields a binary division into group I (serotypes 1/2b, 3b, 4b, 4d, and 4e) and group II (serotypes 1/2a, 3a, 1/2c, and 3c) (2). Corresponding genetic subdivisions were also found with ribotyping; one group (RTα) contained serotypes 1/2a, 1/2c, and 3a, while a second group (RTβ) contained serotypes 1/2b, 3b, 4b, and 4ab (8). Ribotyping and virulence gene allelic analysis have been shown to subdivide L. monocytogenes into three lineages that may differ in pathogenic potential (10, 15, 21). Ribotyping is commonly used for subtyping of L. monocytogenes and has played an important role in outbreak detection (1, 3, 8). Hence, there is a need to define associations between L. monocytogenes serotypes and genetic lineages and ribotypes to further improve the diagnostic utility of this subtyping method. Understanding of correlations between ribotypes and genetic lineages with serotypes may also provide insight into the evolution of L. monocytogenes subtypes. We describe here the relationships among serotypes, ribotypes, and genetic lineages in a set of L. monocytogenes isolates predominantly from humans and animals.
Bacterial isolates.
A total of 235 L. monocytogenes isolates from humans (n = 161), animals (n = 72), and foods (n = 2) were selected from the Cornell University Listeria Collection for inclusion in this study. Some of the isolates had previously been characterized by EcoRI ribotyping (10). All of the isolates were stored in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) with 15% glycerol at −80°C.
Automated ribotyping.
Bacterial isolates were streaked onto brain heart infusion agar plates and incubated at 37°C for 24 h, after which the plates were submitted for automated ribotyping. Ribotyping was performed with the RiboPrinter Microbial Characterization System (Qualicon, Wilmington, Del.) as previously described (3, 9). All isolates were ribotyped by using EcoRI chromosomal digests, and a subset was ribotyped by using PvuII (n = 32). Isolates were assigned to genetic lineage I, II, or III based on EcoRI ribotypes as previously described (21).
Serotyping.
All isolates were serotyped in accordance with the scheme for routine serodiagnosis of L. monocytogenes (20). Serotypes were designated based on agglutination reactions with factor antisera. All serodiagnoses were performed blinded.
Correlations between serotypes, EcoRI ribotypes, and lineages.
We identified eight serotypes among the 235 isolates tested; the majority of isolates were characterized as serotype 1/2a (33%), 1/2b (17%), or 4b (40%). EcoRI ribotyping differentiated these isolates into 24 distinct ribotypes; 8 fell within lineage I, 10 were in lineage II, and 6 were in lineage III (Table 1). Consistent with previous studies, genetic lineages predicted serovar clusters. Lineage I contained serotypes 1/2b, 3b, 3c, and 4b; lineage II contained serotypes 1/2a, 1/2c, and 3a; and lineage III included serotypes 4a and 4c. There also is a specific correlation between single antigens and lineages. Lineages II and III contain flagellar antigens a and c, while lineage I predominantly contains antigen b.
TABLE 1.
Distribution of serotypes among L. monocytogenes EcoRI ribotypes
| Lineage | EcoRI ribotype | No. of isolates of serotype:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1/2a | 1/2b | 1/2c | 3a | 3b | 3c | 4a | 4b | 4c | ||
| I | DUP-1038 | 41 | ||||||||
| I | DUP-1042 | 26 | 1 | 1 | 19 | 1 | ||||
| I | DUP-1044 | 1 | 31 | |||||||
| I | DUP-1052 | 6 | 1 | 1 | ||||||
| I | DUP-1026 | 1 | ||||||||
| I | DUP-1024 | 5 | 2 | |||||||
| I | DUP-1043 | 1 | ||||||||
| I | DUP-1027 | 2 | ||||||||
| II | DUP-1062 | 8 | ||||||||
| II | DUP-1030 | 30 | 7 | 1 | ||||||
| II | DUP-1039 | 9 | 1 | 1 | ||||||
| II | DUP-1045 | 13 | ||||||||
| II | DUP-1053 | 4 | ||||||||
| II | DUP-1054 | 1 | ||||||||
| II | DUP-1056 | 3 | ||||||||
| II | DUP-1029 | 2 | ||||||||
| II | DUP-1035 | 1 | ||||||||
| II | DUP-1047 | 4 | ||||||||
| III | DUP-1061 | 1 | 1 | |||||||
| III | DUP-1059 | 4 | 1 | |||||||
| III | DUP-10146 | 1 | ||||||||
| III | DUP-10147 | 1 | ||||||||
| III | DUP-10145 | 1 | ||||||||
Two atypical lineage I isolates (one of ribotype DUP-1042 and one of ribotype DUP-1044) were of serotypes 4c and 1/2a, respectively. One atypical lineage III isolate was of serotype 1/2a. While it has previously been shown that serotyping may not always be highly reproducible (18), the occurrence of these atypical combinations of serotypes and molecular subtypes allows speculation that horizontal gene transfer may occur among L. monocytogenes or that point mutations could result in phenotypic shifts detectable by serodiagnosis.
Lineages I and II correspond to the primary divisions of L. monocytogenes uncovered by multilocus enzyme electrophoresis and pulsed-field gel electrophoresis, whereas lineage III represents a distinct taxonomic unit that has been proposed to represent at least one new subspecies (15, 21). Our results are consistent with previous studies in grouping different L. monocytogenes serotypes into lineages I and II and further confirm that these two lineages represent distinct subgroups. Serotype 4a is unique to lineage III, and serotype 4c occurred only once outside lineage III. Our findings corroborate the conclusions of others that lineage III strains represent a distinct subset of L. monocytogenes (13, 17). Furthermore, previous studies have shown that lineage III strains are isolated significantly less frequently from humans (0.8%) than from animals (10.5%), indicating that lineage III may rarely cause human disease (10, 21). Other studies have also indicated that only 0 to 2% of human cases are caused by lineage III (12, 21) or serotype 4a and 4c strains (7). Similar findings led to the definition of Listeria serotype 5 strains (which predominantly causes disease in animals) as a new species, L. ivanovii, in 1985 (16). Further analyses, including total genomic DNA-DNA homology studies, are required, however, to clarify the taxonomic status of lineage III.
Lineage I isolates characterized as EcoRI ribotypes DUP-1042, DUP-1052, DUP-1024, and DUP-1044 contained more than one serotype. Also, two lineage II ribotypes (DUP-1030 and DUP-1039) contained multiple serotypes (1/2a, 1/2c, and 3a). Two of the lineage III ribotypes (DUP-1061 and DUP-1059) accommodated multiple serotypes (4a and 4c). Overall, we found four distinct genetic groups (two lineage II ribotypes and two lineage III ribotypes), each of which contains both a and c flagellar antigen groups. This observation indicates the importance of considering the distribution of single antigens among genetic subtypes or lineages, in addition to serotypes as a whole. The fact that both a and c flagellar antigens are present within closely related genetic groups possibly indicates that a single genetic event or, less likely, horizontal gene transfer could lead to conversion from a to c or vice versa. Currently, we have little knowledge of the molecular basis for the serotypes of L. monocytogenes. Understanding of the genetic determinants of flagellar and somatic antigenic groups would allow further probing of this issue.
Correlations between serotypes and PvuII ribotypes.
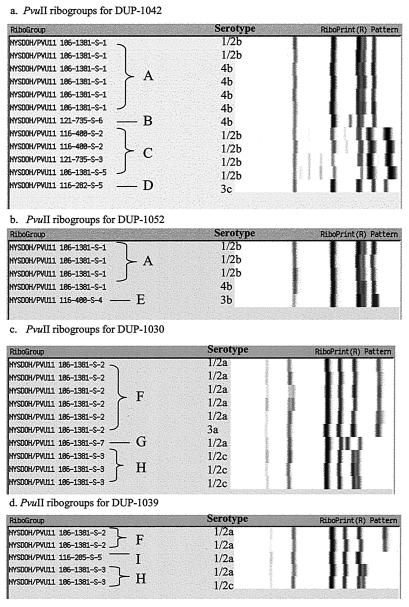
Eighteen EcoRI ribotypes appear to be predictive of a specific serotype, while four EcoRI ribotypes in lineage I and two ribotypes each in lineages II and III contain multiple serotypes. PvuII ribotyping has previously been shown to improve the discriminatory power of ribotype-based differentiation of L. monocytogenes strains over EcoRI ribotyping (6). Thus, a subset of isolates representative of EcoRI ribotypes that contained more than one serotype (32 isolates; DUP-1042, DUP-1052, DUP-1030, and DUP-1039) were further characterized by PvuII ribotyping. These isolates gave rise to nine PvuII patterns (A through I; Fig. 1), resulting in 12 combination types. The combination of EcoRI and PvuII ribotypes improves serotype prediction, as only 4 of the 12 combination types contained multiple serotypes (DUP-1042/A, DUP-1052/A, DUP-1030/F, and DUP-1039/H). Specifically, within lineage I, DUP-1042 separated into four PvuII ribogroups (A, B, C, and D) and DUP-1052 split into two groups (A and E; Fig. 1a and b). One PvuII ribogroup (A) was common to both DUP-1042 and DUP-1052. Isolates of serotypes 3b and 3c each had unique PvuII ribogroup patterns, whereas PvuII ribogroups generally did not differentiate serotypes 1/2b and 4b, with the exception of group B (serotype 4b) and group C (exclusively serotype 1/2b). Within lineage II, PvuII ribotyping separated DUP-1030 and DUP-1039 into three groups each (F, G, and H and F, I, and H, respectively), two of which (F and H) were common to both (Fig. 1c and d). PvuII ribotyping differentiated serotype 1/2a and 1/2c isolates, with the exception of one DUP-1039 serotype 1/2a isolate. Specifically, PvuII types G and I contained only serotype 1/2a isolates, PvuII type F contained five serotype 1/2a isolates and one 3a isolate, and PvuII type H contained four serotype 1/2c isolates and one 1/2a isolate. Our results provide further evidence that ribotyping with two enzymes allows finer discrimination of strains and improved prediction of L. monocytogenes serotypes. Nevertheless, even when two restriction enzymes are used, a limited number of ribotypes still contain multiple serotypes.
FIG. 1.
PvuII ribotype patterns found within EcoRI ribogroups, which contain multiple serotypes. (a) PvuII ribogroups for DUP-1042 isolates. (b) PvuII ribogroups for DUP-1052 isolates. (c) PvuII ribogroups for DUP-1030 isolates. (d) PvuII ribogroups for DUP-1039 isolates. The ribotype patterns shown represent normalized data, and gels were run from left to right. Ribotype patterns within each letter grouping (A to I) are considered the same.
Conclusion. Although both molecular subtyping methods and serotyping are valuable techniques for studying the epidemiology of bacterial pathogens, the connection between molecular subtypes and serotypes has yet to be defined for many organisms. Like listeriosis, outbreaks of cholera are caused primarily by specific serogroups of Vibrio cholerae and an analysis of 103 clinical V. cholerae strains yielded a low degree of correlation between ribotypes and serotypes (5). However, within group A Streptococcus isolates, serotypes correlate with ribotype patterns (with two restriction enzymes), although genetic heterogeneity has been demonstrated among certain Streptococcus pyogenes serotypes (4). Our results demonstrate the potential of ribotyping for the prediction of L. monocytogenes serotypes, although a small number of subtypes differentiated by EcoRI and PvuII ribotyping contain more than one serotype. A complete understanding of the genetic determinants of flagellar and somatic antigenic groups is required to elucidate the complex relationships between subtyping methods and to provide further insight into the evolution of the serotypes of this food-borne pathogen.
Acknowledgments
This paper is a result of research funded partially by the USDA National Research Initiative under award 99-35201-8074 and partially by National Oceanic and Atmospheric Administration award NA86RG0056 to the Research Foundation of the State University of New York for New York Sea Grant.
Ribotyping was performed by Qualicon, Inc., and at the Laboratory for Molecular Typing at Cornell University. We thank Mary Bodis for kind assistance with the RiboPrinter system.
REFERENCES
- 1.Anonymous. Multistate outbreak of listeriosis—United States, 1998. Morb Mortal Wkly Rep. 1998;47:1085–1086. [PubMed] [Google Scholar]
- 2.Brosch R, Chen J, Luchansky J B. Pulsed-field fingerprinting of listeriae: identification of genomic divisions of Listeria monocytogenes and their correlation with serovar. Appl Environ Microbiol. 1994;60:2584–2592. doi: 10.1128/aem.60.7.2584-2592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce J L, Hubner R J, Cole E M, McDowell C I, Webster J A. Sets of EcoRI fragments containing ribosomal RNA sequences are conserved among different strains of Listeria monocytogenes. Proc Natl Acad Sci USA. 1995;92:5229–5233. doi: 10.1073/pnas.92.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruneau S, DeMontclos H, Drouet E, Denoyel G A. rRNA gene restriction patterns of Streptococcus pyogenes: epidemiological applications and relation to serotypes. J Clin Microbiol. 1994;32:2953–2958. doi: 10.1128/jcm.32.12.2953-2958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalsgaard A, Forslund A, Mortensen H F, Shimada T. Ribotypes of clinical Vibrio cholerae non-O1 non-O139 strains in relation to O-serotypes. Epidemiol Infect. 1998;121:535–545. doi: 10.1017/s0950268898001654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gendel S M, Ulaszek J. Ribotype analysis of strain distribution in Listeria monocytogenes. J Food Prot. 2000;63:179–185. doi: 10.4315/0362-028x-63.2.179. [DOI] [PubMed] [Google Scholar]
- 7.Gilot P, Genicot A, Andre P. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J Clin Microbiol. 1996;34:1007–1010. doi: 10.1128/jcm.34.4.1007-1010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graves L M, Swaminathan B, Reeves M W, Hunter S B, Weaver R E, Plikaytis B D, Schuchat A. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J Clin Microbiol. 1994;32:2936–2943. doi: 10.1128/jcm.32.12.2936-2943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubner R J, Cole E M, Bruce J L, McDowell C I, Webster J A. Predicted types of Listeria monocytogenes created by the positions of EcoRI cleavage sites relative to rRNA sequences. Proc Natl Acad Sci USA. 1995;92:5234–5238. doi: 10.1073/pnas.92.11.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffers G T, Bruce J L, McDonough P L, Scarlett J, Boor K J, Wiedmann M. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology. 2001;147:1095–1104. doi: 10.1099/00221287-147-5-1095. [DOI] [PubMed] [Google Scholar]
- 11.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norton D M, Scarlett J M, Horton K, Sue D, Thimothe J, Boor K J, Wiedmann M. Characterization and pathogenic potential of Listeria monocytogenes isolates from the smoked fish industry. Appl Environ Microbiol. 2001;67:646–653. doi: 10.1128/AEM.67.2.646-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notermans S, Chakraborty T, Leimeister-Waechter M, Dufrenne J, Heuvelman K J, Maas H, Jansen W, Wernars K, Guinee P. Specific gene probe for detection of biotyped and serotyped Listeria strains. Appl Environ Microbiol. 1989;55:902–906. doi: 10.1128/aem.55.4.902-906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piffaretti J-C, Kressebuch H, Aeschenbacher M, Bille J, Bannerman E, Musser J M, Selander R K, Rocourt J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci USA. 1989;86:3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen O F, Skouboe P, Dons L, Rossen L, Olsen J E. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology. 1995;141:2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- 16.Rocourt J. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification. In: Ryser E T, Marth E H, editors. Listeria, listeriosis, and food safety. New York, N.Y: Marcel Dekker, Inc; 1999. pp. 1–20. [Google Scholar]
- 17.Rocourt J, Grimont F, Grimont P A D, Seeliger H P R. DNA relatedness among serovars of Listeria monocytogenes sensu lato. Curr Microbiol. 1982;7:383–388. [Google Scholar]
- 18.Schönberg A, Bannerman E, Courtieu A L, Kiss R, McLauchlin J, Shah S, Wilhelms D. Serotyping of 80 strains from the W. H. O. multicentre international typing study of Listeria monocytogenes. Int J Food Microbiol. 1996;32:279–287. doi: 10.1016/s0168-1605(96)01142-7. [DOI] [PubMed] [Google Scholar]
- 19.Schuchat A, Swaminathan B, Broome C. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeliger H P R, Höhne K. Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 1979;13:31–49. [Google Scholar]
- 21.Wiedmann M, Bruce J L, Keating C, Johnson A E, McDonough P L, Batt C A. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]