Abstract
Without evidence for an organic framework, biological and biochemical processes observed during amelogenesis provided limited information on how extracellular matrix proteins control the development of the complex fibrous architecture of human enamel. Over a decade ago, amelogenin nanoribbons were first observed from recombinant proteins during in vitro mineralization experiments in our laboratory. In enamel from mice lacking the enzyme kallikrein 4 (KLK4), we later uncovered ribbon-like protein structures that matched the morphology, width, and thickness of the nanoribbons assembled by recombinant proteins. Interestingly, similar structures had already been described since the 1960s, when enamel sections from various mammals were demineralized and stained for transmission electron microscopy analysis. However, at that time, researchers were not aware of the ability of amelogenin to form nanoribbons and instead associated the filamentous nanostructures with possible imprints of mineral ribbons in the gel-like matrix of developing enamel. Further evidence for the significance of amelogenin nanoribbons for enamel development was stipulated when recent mineralization experiments succeeded in templating and orienting the growth of apatite ribbons along the protein nanoribbon framework. This article provides a brief historical review of the discovery of amelogenin nanoribbons in our laboratory in the context of reports by others on similar structures in the developing enamel matrix.
Keywords: enamel, biomineralization, protein nanoribbons, apatite, developing enamel matrix, extracellular matrix
Emergence of Recombinant Amelogenin Nanoribbons
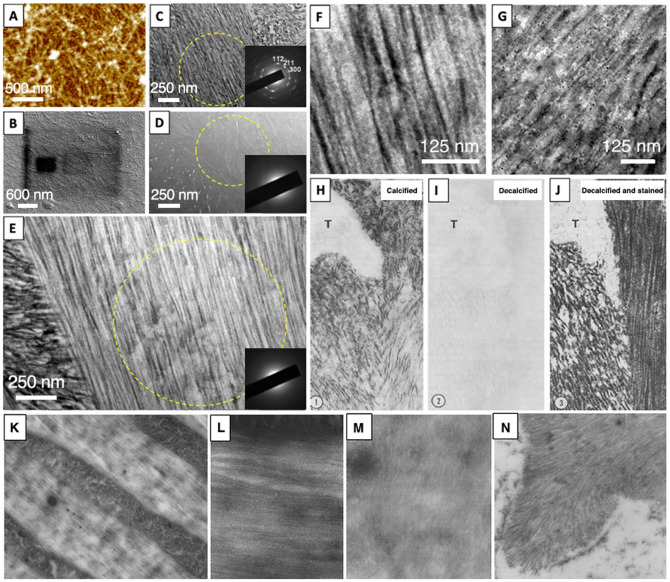
With most of the organic content being enzymatically degraded and inaccessible in mature enamel, expressing and studying recombinant amelogenin proteins in vitro became a feasible alternative approach for investigating amelogenin assembly structures and their role in biomineralization (Hu et al. 2005). In 2008, when performing constant composition experiments to induce mineral formation in the presence of recombinant full-length human amelogenin (rH174), we observed uniquely different precipitates in a few extracts of the solution when analyzed by atomic force microscopy on glass surfaces (Fig. A; Uskokovic et al. 2008). While we initially assumed that these structures were calcium phosphate minerals, it became clear that they were organic and were derived from amelogenin, as they dissolved in bleach and burned off by the electron beam in the scanning electron microscopy (Fig. B). These protein ribbons, however, only occasionally developed during the titration experiments with little reliability, and it took another 3 y of experimenting with oil emulsion systems, calcium phosphate concentrations, and pH optimization to develop protocols that ultimately allowed us to reproducibly form amelogenin nanoribbons in aqueous solutions (He et al. 2011; Martinez-Avila et al. 2012). Structure prediction revealed the presence of ß-sheets in amelogenin, and an N-terminal domain was identified as a critical driver for ribbon-like assembly and the development of an amyloid-like structure with cross-ß orientation (Sanii et al. 2014; Carneiro et al. 2016).
Figure.
(A) Atomic force microscopy image of nanoribbon structures from recombinant full-length amelogenin (rH174) observed for the first time in constant composition mineralization experiments in 2008. (B) Scanning electron microscopy image of rH174 nanoribbons obtained in 2008 shows nanoribbons on glass surface with damage from electron beam of areas imaged at higher magnification, indicating that the nanoribbons are organic and not mineral. (C) Transmission electron microscopy (TEM) image of secretory-stage enamel illustrates the presence of apatite mineral ribbons. As compared with mature enamel, secretory-stage apatite ribbons are thinner in width and lower in mineralization degrees. Inserted selected area electron diffraction pattern obtained from area within yellow circle: major reflections are indicated, including (112)–, (211)–, and (300)– plane, which show that the a-axis is oriented perpendicular to the mineral ribbon long axis and that the mineral phase is highly oriented. (D) TEM image of demineralized unstained secretory-stage enamel section. Only a vague pattern is visible, and detailed structures are undetectable. Insert: selected area electron diffraction data show that crystalline phases have been removed by demineralization. (E) TEM image of demineralized stained secretory-stage enamel section reveals a vast protein nanoribbon network with the signature enamel pattern. (F) Higher-magnification TEM image of 1 region in panel E, with filamentous nature of enamel protein matrix. (G) Immunostaining of demineralized wild type secretory-stage enamel section stained positive with amelogenin antibody. Section was incubated with primary amelogenin antibody from rabbit and secondary antibody from goat with 5-nm gold labeling in sequence and negatively stained. Other enamel proteins may be present in this section. (H–J) Series of TEM images of calcified, decalcified, and decalcified and stained young enamel tissue sections at area near Tomes’s process (labeled T; adapted from Bai and Warshawsky 1985; original magnification, 74,750×) with a very similar appearance to the sequence of images from this study (E–G). (K) Filamentous nature of enamel matrix was revealed in TEM (original magnification, 22,400×) after demineralization and without the use of tissue stains. (L) Ribbon-like structures were more pronounced after staining (adapted from Travis and Glimcher 1964; original magnification, 128,000×). Earliest known evidence for filamentous supramolecular structure in the developing enamel matrix was provided in 1962 by TEM analysis of developing human teeth: (M) demineralization of enamel tissue section by EDTA without staining (original magnification, 56,000×) and (N) human developing enamel matrix demineralized by EDTA and subsequent staining of section with uranyl acetate (from Ronnholm 1962; original magnification, 73,000×).
Uncovering Amelogenin Nanoribbons in Secretory-Stage Enamel
Numerous x-ray diffraction studies showed the presence of a cross-ß motif in the developing enamel matrix (DEM), which matched exactly the diffraction pattern obtained on nanoribbons from recombinant amelogenin and amelogenin peptides (Pautard 1961; Carneiro et al. 2016; Engelberth et al. 2018). In addition, staining of mouse Klk4−/− enamel or secretory-stage wild type enamel was positive for amyloid-like structures (Carneiro et al. 2016; Huang et al. 2021). Hence, we expected that the recombinant protein ribbons developed in vitro would also exist at some point in vivo during enamel development. But it was only recently that a study in our laboratory uncovered that supramolecular structure of the DEM (Bai et al. 2020). Using transmission electron microscopy (TEM) analysis of a Klk4−/− mouse revealed the characteristic architecture of mineral ribbons in murine enamel. Following a strict fixation protocol, a mild acid was applied to dissolve the apatite ribbons. Subsequent negative staining revealed the protein nanoribbons.
In this discovery article, we report that the same kind of nanoribbons exist in the DEM from wild type mice. In a standard TEM analysis, murine incisors were ultramicrotomed following the central line of the second molar to locate the secretory stage of enamel. TEM images of the mineralized tissue reveal the characteristic ribbon-like apatite crystals organized into the unique enamel texture (Fig. C) as previously reported in numerous studies. Selected area electron diffraction suggests that these apatite crystals are highly oriented (Fig. C insert). The translucency and width of the secretory-stage mineral appear lighter and thinner than the mature enamel apatite crystals. The mineral ribbons appear to cover about 80% of the TEM image but make up just one-third of the DEM by weight and <20% by volume.
On the contrary, matrix proteins should represent at least 40% by volume, which is not apparent from analysis of the mineralized enamel. Demineralization of the sections resulted in a largely featureless TEM image of the DEM (Fig. D). However, subsequent negative staining produced a TEM image (Fig. E) that was nearly identical to the features observed in the mineralized tissue sections. The protein ribbon dimensions of the demineralized enamel (Fig. F) matched well with the dimensions of the recombinant amelogenin nanoribbons developed in vitro (Fig. A, B), indicating a strong association between the two. Immunostaining (Fig. G) confirmed the nature of the nanoribbons as amelogenin protein: given that amelogenin represents >90% of all enamel proteins (Gibson 2011), no other matrix component is present at sufficient quantity in the DEM to resemble these expansive structures (Fig. E). These data are consistent with our previous observation of protein nanoribbons in Klk4−/− mouse incisors, further consolidating that amelogenin nanoribbons are indeed the structural templates in the DEM (Bai et al. 2020).
Crystal Ghosts Are Amelogenin Nanoribbons
While confirming the presence of nanoribbons in the DEM from wild type enamel without genetic modifications was exciting and novel to our group, we were surprised to learn that others had discovered similar structures already in the 1980s (Nanci et al. 1983; Bai and Warshawsky 1985; Hayashi et al. 1986) with some earlier reports reaching as far back as the 1960s (Ronnholm 1962; Travis and Glimcher 1964; Frank 1979). Following almost identical experimental protocols, these studies applied uranyl acetate, lead citrate, phosphotungstate, or other stains to demineralized secretory-stage enamel and revealed the presence of ribbon-like structures (Fig. H–J) matching our recent experiments and observations in an equivalent sequence (Fig. C–E). Some authors named these structures “crystal ghosts,” a term referred by Bonucci (1967) when structures reappeared after demineralization and staining of extracellular vesicles in cartilage, which resembled the dissolved mineral phase. According to the enamel literature, these structures revealed by staining were believed to be imprints of the enamel crystals into a proteinaceous gel that surrounded each mineral ribbon. A common hypothesis at that time claimed that enamel mineral develops in a gel-like protein substance that inhibits lateral growth of apatite mineral and allows growth along only the c-axis of the crystals (Nanci et al. 1983; Bai and Warshawsky 1985; Hayashi et al. 1986). According to this hypothesis, enamel proteins would accumulate at the enamel crystal surface and thus lead to an increased density of matrix along the crystal ribbons. After demineralization, staining would then reveal zones of higher protein density and therefore ribbon-like structures matching the dissolved crystals would make a ghost-like reappearance. However, Bonucci (2014) revised this initial assertion: “Electron microscope studies clearly exclude the possibility that these particles might be crystalline structures, as often believed, by showing that they are, instead, organic-inorganic hybrids, each comprising a filamentous organic component (the crystal ghost) made up of acidic proteins.”
It is nonetheless challenging to imagine how a protein gel without any structural resemblance to the filamentous mineral would be able to direct the growth direction so precisely as observed in developing prismatic enamel, with its uniform and highly organized mineral ribbons of defined width and thickness. In the 1970s, a number of investigators promoted the hypothesis that enamel crystallites and their growth determine the structure of the organic matrix in enamel (Fearnhead 1979; Nylen 1979). In conjunction with the observation that enamel crystals mostly grow perpendicular from the dentin surface, the argument was made that enamel crystals develop in an oriented fashion by nucleation on apatite mineral in dentin. That idea, however, seemed quite counterintuitive to some in the enamel research community, as laid out in a discussion transcript of the Enamel 3 meeting in 1978 (see appendix to Nylen 1979).
On the contrary, early TEM studies on enamel structure in the 1960s stimulated the idea that the matrix forms a proteinaceous template to mineralization through supramolecular assembly (Ronnholm 1962; Travis and Glimcher 1964; Frank 1979). Evidence for such structures was supported by fine, elongated structures often faintly visible after careful examination of demineralized developing enamel by TEM, without the application of heavy metal stains (Fig. K, M) and more pronounced after staining (Fig. L, N). Most studies identified these structures as filaments or ribbons, but some indications suggesting a tubular nature have been reported (Warshawsky 1989). According to our literature search, Ronnholm (1962) was first to demonstrate the filamentous matrix in unstained and stained TEM sections of developing murine enamel (Fig. N). That study additionally appears to be first in proposing that the filamentous nature of the organic matrix could template crystal growth of apatite ribbons forming along both sides of the protein filaments. A hypothesis was more clearly expressed by R.M. Frank (1979): “The great similarity of configuration, orientation and distribution with the growing ribbon-like apatite crystals suggested that these organic structures could be responsible for early oriented growth of the inorganic crystalline phase.”
The controversy between these models was not settled, mostly due to the ambiguity of the actual physiologic supramolecular structure of the main matrix component of the DEM—for example, the amelogenin protein and its enzymatic cleavage products. In the 1990s, novel in vitro amelogenin structures were proposed largely according to the morphology that the recombinant or extracted and purified amelogenin proteins adopted in various solutions. The nanosphere model—in which the amelogenin forms spheres of about 20 nm in diameter and acts as a spacer between existing enamel crystals—became the dominant paradigm and drew focus away from a template-based model with protein filaments (Fincham et al. 1994; Fincham et al. 1999). As the nanosphere model did not address the process of protein-guided mineral nucleation, a reformed model in 2011 hypothesized that oligomeric structures of the full-length amelogenin protein assemble into dodecahedral spheres, and it proposed that apatite mineral would develop from prenucleation clusters between 2 half spheres (Fang et al. 2011). Mineral ribbons extend in length as oligomeric spheres from full-length amelogenin line up and thus facilitate acicular growth of apatite mineral.
In the same year (He et al. 2011), our group published the first evidence for ribbon-like assemblies of recombinant amelogenin. Early on in our studies of amelogenin nanoribbons, we noticed the similarity of the organic structures to the enamel ribbons and proposed a protein template model in which growth of mineral ribbons is guided by amelogenin nanoribbons. Only recently did we realize that similar protein structures were described a half century earlier (Ronnholm 1962), later termed “crystal ghosts” (Bai and Warshawsky 1985), and similar models had already been proposed by others (Travis and Glimcher 1964; Frank 1979). Our recent model, in contrast to others, was able to elucidate phenotypical effects in the enamel of a series of existing mouse models, allowing us to advance the descriptive nature of earlier models into a mechanistic one (Bai et al. 2020; Habelitz and Bai 2021).
There Is More to Uncover
While the discovery of amelogenin nanoribbons and their ability to guide the formation of enamel mineral ribbons upon regulation by matrix metalloprotease 20 is a step toward a better understanding of how enamel’s dynamic matrix controls the synthesis of such an intricate architecture with nanometer precision of mineral growth (Habelitz and Bai 2021), new questions arise. Amorphous calcium phosphate (ACP) plays a distinct role as a transient precursor to bioapatite and has been detected in many biomineral systems from various phyla of the animal kingdom (Colfen 2008). This includes enamel mineralization, which starts as ACP, develops close to the secretion site of the Tomes’s process, and exists for about 8 to 12 µm at the mineralization front before transitioning into crystalline apatite ribbons in murine enamel (Shin et al. 2020). It is apparent that interaction of the organic framework with a charged, often phosphorylated protein that carries calcium and phosphate ions is critical for the formation of an amorphous mineral. The polymer-induced liquid precursor system (PILP method) with polyaspartate was applied to achieve templated growth of ACP on collagen fibrils and amelogenin nanoribbons (Gower 2008). None of the nonamelogenin proteins are as highly phosphorylated as phosphoproteins in bone and dentin. Nonetheless, enamelin is composed of domains rich in acidic residues, including aspartic acid, and the hydrophilic C-terminus of amelogenin, a cleavage product of the full-length protein, is a heavily charged molecule. Both are suitable candidates to interact with calcium and phosphate ions and to induce mineral formation on amelogenin nanoribbons through mechanisms similar but not identical to the PILP process that induces intrafibrillar mineralization of collagen.
In this regard, we like to emphasize that PILP mineralization is achieved through the formation of nanodroplets (15 to 20 nm) from polymer/protein aggregates with mineral ions (Olszta et al. 2007). Such nanodroplets would appear in conjunction with the amelogenin and mineral nanoribbons and may be related to the spherical structure and globular structure frequently described between mineral ribbons in developing enamel (Fincham et al. 1995). While the constituents, which fill the spaces between the protein and mineral ribbons in the DEM, must primarily be C- and N-terminal amelogenin cleavage products as well as nonamelogenin proteins immersed in enamel fluid, their structural configuration has not been defined. The matrix constituents between enamel ribbons will act as a spacer and inhibit lateral growth of apatite ribbons as previously hypothesized in the amelogenin nanosphere model (Fincham et al. 1999), but that space is not comprised by full-length amelogenin protein. In the nanoribbon model, the full-length amelogenin protein is critical to laying down a filamentous structure that templates ACP formation activated by matrix metalloprotease 20 hydrolysis. Subsequently, amelogenin cleavage products and nonamelogenin proteins aggregate around the mineral ribbons and inhibit the lateral growth of the apatite crystals, most likely by strong attachment of the hydrophilic C-terminus of amelogenin as described by others (Tarasevich et al. 2009).
In regard to future studies, the 3-dimensional molecular structure of the amelogenin nanoribbon itself has yet to be determined. Efforts are currently underway to resolve a near-atomic model of the amelogenin nanoribbons via cryoelectron microscopy with synchrotron radiation and promise to elucidate a series of critical information for our understanding of amelogenin-guided mineralization, including orientation of the hydrophilic C-terminus, enzymatic cleavage sites, and protein-mineral binding domains.
Other anticipated outcomes from the scientific exploration of the nanoribbon model system are improved ability to determine interaction of amelogenin domains with other protein components of the DEM. We expect interactions of amelogenin with enamelin and ameloblastin to differ when exposed to nanospherical or nanoribbon structures, as these superstructures have very different surfaces. In addition we expect to gain new insights of attachment mechanism between ameloblast membrane and the DEM. The protein matrix framework model has great potential to elucidate how the Tomes’s process engages with the DEM and is able to construct the remarkable hierarchical nano- and microstructure of prismatic enamel (Habelitz 2015). In this regard, too, a high-resolution structural model of amelogenin nanoribbons will be instrumental in answering many of these questions and is poised to reveal further details of the molecular mechanisms of enamel formation and its pathologies.
Author Contributions
Y. Bai, J. Bonde, K.M.M. Carneiro, Y. Zhang, W. Li, S. Habelitz, contributed to conception, design, data acquisition, analysis, or interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grant R01-DE025709 from the National Institute of Dental and Craniofacial Research, National Institutes of Health.
References
- Bai P, Warshawsky H. 1985. Morphological studies on the distribution of enamel matrix proteins using routine electron microscopy and freeze-fracture replicas in the rat incisor. Anat Rec. 212(1):1–16. [DOI] [PubMed] [Google Scholar]
- Bai Y, Yu Z, Ackerman L, Zhang Y, Bonde J, Li W, Cheng Y, Habelitz S. 2020. Protein nanoribbons template enamel mineralization. Proc Natl Acad Sci U S A. 117(32):19201–19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonucci E. 1967. Fine structure of early cartilage calcification. J Ultrastruct Res. 20(1):33–50. [DOI] [PubMed] [Google Scholar]
- Bonucci E. 2014. Understanding nanocalcification: a role suggested for crystal ghosts. Mar Drugs. 12(7):4231–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro KM, Zhai H, Zhu L, Horst JA, Sitlin M, Nguyen M, Wagner M, Simpliciano C, Milder M, Chen CL, et al. 2016. Amyloid-like ribbons of amelogenins in enamel mineralization. Sci Rep. 6:23105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colfen H. 2008. Single crystals with complex form via amorphous precursors. Angew Chem Int Ed Engl. 47(13):2351–2353. [DOI] [PubMed] [Google Scholar]
- Engelberth SA, Bacino MS, Sandhu S, Li W, Bonde J, Habelitz S. 2018. Progression of self-assembly of amelogenin protein supramolecular structures in simulated enamel fluid. Biomacromolecules. 19(10):3917–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang PA, Conway JF, Margolis HC, Simmer JP, Beniash E. 2011. Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc Natl Acad Sci U S A. 108(34):14097–14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead RW. 1979. Matrix-mineral relationships in enamel tissues. J Dent Res. 58(Spec Issue B):909–921. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P, Jr, Slavkin HC. 1995. Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J Struct Biol. 115(1):50–59. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP. 1999. The structural biology of the developing dental enamel matrix. J Struct Biol. 126(3):270–299. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. 1994. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol. 112(2):103–109. [DOI] [PubMed] [Google Scholar]
- Frank RM. 1979. Tooth enamel: current state of the art. J Dent Res. 58(Spec Issue B):684–694. [DOI] [PubMed] [Google Scholar]
- Gibson CW. 2011. The amelogenin proteins and enamel development in humans and mice. J Oral Biosci. 53(3):248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower LB. 2008. Biomimetic model systems for investigating the amorphous precursor pathway and its role in biomineralization. Chem Rev. 108(11):4551–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelitz S. 2015. Materials engineering by ameloblasts. J Dent Res. 94(6):759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelitz S, Bai Y. 2021. Mechanisms of enamel mineralization guided by amelogenin nanoribbons. J Dent Res. 100(13):1434–1443. doi: 10.1177/00220345211012925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Bianco P, Shimokawa H, Termine JD, Bonucci E. 1986. Organic-inorganic relationships, and immunohistochemical localization of amelogenins and enamelins in developing enamel. Basic Appl Histochem. 30(3):291–299. [PubMed] [Google Scholar]
- He X, Wu S, Martinez-Avila O, Cheng Y, Habelitz S. 2011. Self-aligning amelogenin nanoribbons in oil-water system. J Struct Biol. 174(1):203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, Yamakoshi Y, Yamakoshi F, Krebsbach PH, Simmer JP. 2005. Proteomics and genetics of dental enamel. Cells Tissues Organs. 181(3–4):219–231. [DOI] [PubMed] [Google Scholar]
- Huang Y, Bai Y, Chang C, Bacino M, Cheng IC, Li L, Habelitz S, Li W, Zhang Y. 2021. A N-terminus domain determines amelogenin’s stability to guide the development of mouse enamel matrix. J Bone Miner Res [epub ahead of print 6 May 2021] in press. doi: 10.1002/jbmr.4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Avila O, Wu S, Kim SJ, Cheng Y, Khan F, Samudrala R, Sali A, Horst JA, Habelitz S. 2012. Self-assembly of filamentous amelogenin requires calcium and phosphate: from dimers via nanoribbons to fibrils. Biomacromolecules. 13(11):3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanci A, Bai P, Warshawsky H. 1983. The effect of osmium postfixation and uranyl and lead staining on the ultrastructure of young enamel in the rat incisor. Anat Rec. 207(1):1–16. [DOI] [PubMed] [Google Scholar]
- Nylen MU. 1979. Matrix-mineral relationships—a morphologist’s viewpoint.J Dent Res. 58(Spec Issue B):922–929. [DOI] [PubMed] [Google Scholar]
- Olszta MJ, Cheng XG, Jee SS, Kumar R, Kim YY, Kaufman MJ, Douglas EP, Gower LB. 2007. Bone structure and formation: a new perspective. Mater Sci Eng R Rep. 58(3–5):77–116. [Google Scholar]
- Pautard FG. 1961. An x-ray diffraction pattern from human enamel matrix. Arch Oral Biol. 3:217–220. [DOI] [PubMed] [Google Scholar]
- Ronnholm E. 1962. III: the structure of the organic stroma of human enamel during amelogenesis. J Ultrastruct Res. 6:368–389. [DOI] [PubMed] [Google Scholar]
- Sanii B, Martinez-Avila O, Simpliciano C, Zuckermann RN, Habelitz S. 2014. Matching 4.7-Å XRD spacing in amelogenin nanoribbons and enamel matrix. J Dent Res. 93(9):918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin NY, Yamazaki H, Beniash E, Yang X, Margolis SS, Pugach MK, Simmer JP, Margolis HC. 2020. Amelogenin phosphorylation regulates tooth enamel formation by stabilizing a transient amorphous mineral precursor.J Biol Chem. 295(7):1943–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasevich BJ, Lea S, Bernt W, Engelhard MH, Shaw WJ. 2009. Changes in the quaternary structure of amelogenin when adsorbed onto surfaces. Biopolymers. 91(2):103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis DF, Glimcher MJ. 1964. The structure and organization of, and the relationship between the organic matrix and the inorganic crystals of embryonic bovine enamel. J Cell Biol. 23(3):447–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskokovic V, Kim MK, Li W, Habelitz S. 2008. Enzymatic processing of amelogenin during continuous crystallization of apatite. J Mater Res. 23(12):3184–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshawsky H. 1989. Organization of crystals in enamel. Anat Rec. 224(2): 242–262. [DOI] [PubMed] [Google Scholar]