Summary
Background
Osteoarthritis (OA) is the most common joint disease and is the leading cause of chronic disability among older people. Chondrocyte death and extracellular matrix (ECM) degradation was involved in OA pathogenesis. Ferroptosis was an iron-dependent cell death associated with peroxidation of lipids. Here, we proved that ferroptosis exists in OA and identified glutathione peroxidase 4 (GPX4) as an important regulator of OA.
Methods
Ferroptosis-related alterations were analyzed in human OA and undamaged cartilage. Expression of GPX4 was examined in 55 paired human OA samples. Ferrostatin-1 (Fer-1) and Deferoxamine (DFO) were used to treat OA, in vitro and in vivo. Alterations of GPX4-mediated signaling pathway were identified by RNA-seq analysis. AAV-Gpx4-shRNA were used to downregulate GPX4 expression in vivo.
Findings
Transcriptomic, biochemical, and microscopical analyses indicated that ferroptosis was closely associated with OA. Expression of GPX4 in the OA cartilage from 55 OA patients were significantly lower than undamaged cartilage. Fer-1 and DFO could protect OA in a necroptosis-independent manner, suggesting that ferroptosis exists in OA prog. Importantly, GPX4 downregulation could increase the sensitivity of chondrocytes to oxidative stress and aggravate ECM degradation through the MAPK/NFκB pathway. Furthermore, downregulation of GPX4 expression by AAV-Gpx4 shRNA aggravated OA in vivo.
Interpretation
Ferroptosis contributes to OA pathogenesis and GPX4 was the intersection of two mechanisms in regulating OA progression: ferroptosis and ECM degradation.
Keywords: Osteoarthritis, Ferroptosis, Chondrocyte, GPX4, Extracellular matrix
Research in context.
Evidence before this study
Ferroptosis is a recently discovered type of cell death initiated by lipid peroxidation and has been implicated in a number of diseases. However, there is still no evidence of ferroptosis existing in OA pathogenesis. The physiological and pathological roles of GPX4 (a pivotal regulator of ferroptosis) in cartilage have not yet been addressed.
Added value of this study
Our studies proved that ferroptosis was involved in human OA pathogenesis and contributed to OA progression by the evidence that (i) Ferroptosis-related alterations occurred in OA cartilage; (ii) Both a lipophilic antioxidant (Fer-1) and an iron chelator (DFO) could rescue OA progression; and (iii) Other cell death types were excluded. Moreover, GPX4 downregulation could not only increased the sensitivity of chondrocytes to oxidative stress, but also aggravated extracellular matrix (ECM) degradation through MAPK/NF-κB pathway.
Implications of all the available evidence
This is the first report to provide evidence that ferroptosis exists in human OA. It is pivotal to know whether one type of cell death occurs in the process of the disease, which can improve our understanding of OA pathogenic mechanisms. Remarkably, our results indicated dual function of GPX4 in OA: (i) regulating ferroptosis or oxidative stress; (ii) regulating ECM degradation. These data suggest that ferroptosis and GPX4 may stand for potential therapeutic regimens of OA.
Alt-text: Unlabelled box
Introduction
Osteoarthritis (OA) is the most common joint disease worldwide, and is the leading cause of chronic pain and disability among elderly people.1,2 OA is primarily characterized by joint tissue degradation caused by an imbalance between anabolic and catabolic processes of cartilage and other joint tissues,3,4 associated with a variety of pathological changes, including subchondral bone sclerosis, synovitis, and inflammation of joint capsule and tendon.5,6 OA progression is closely related to degradation of cartilage extracellular matrix (ECM) by catabolic matrix-degrading enzymes.7,8 Articular cartilage is a conjunctive tissue composed of only one cell type, chondrocytes, which are responsible for ECM production.9,10 The survival of chondrocytes is crucial for the maintenance of articular cartilage integrity and homeostasis.
A number of studies reported the presence of empty lacunae in cartilage with OA progression,11 suggesting that chondrocyte death is involved in OA pathogenesis.10,12,13 Various forms of chondrocyte death have been identified in the cartilage tissue from patients with OA and OA animal models, including apoptosis,14,15 necroptosis13,16 and autophagy.17 Although inhibiting apoptosis, necrosis, and autophagy can improve outcomes in animals subjected to experimental OA, there have been no successful clinical trials using any single cell-death inhibitor. These findings suggest that multiple forms of cell death may occur in OA and contribute collectively to cartilage deterioration. We hypothesized that additional forms of cell death, that mediate cartilage homeostasis in OA, likely remain to be discovered.
Recently, ferroptosis, an iron-dependent form of non-apoptotic cell death, has been recognized.18 Different from other forms of regulated cell death, such as necrosis, apoptosis or autophagy, ferroptosis has unique morphological, biochemical, genetic and immunological characteristics.18 Ferroptosis is caused by a redox imbalance between the production of oxidants and antioxidants, which is driven by the abnormal expression and activity of multiple redox-active enzymes that produce free radicals and lipid oxidation products.19 It is characterized by increased levels of lipid hydroperoxides and iron overload, leading to caspase- and necrosome-independent cell death.20 Previous studies have shown that ferroptosis is inhibited by lipid peroxidation inhibitor ferrostatin-1 (Fer-1) and iron chelator deferoxamine (DFO),19,21 but not all the cell death inhibited by Fer-1 or DFO was ferroptosis.19 In the process of ferroptosis, glutathione peroxidase 4 (GPX4) utilizes reduced glutathione (GSH) to convert phospholipid hydroperoxides to lipid alcohols and inhibits ferroptosis.22,23 Inactivation of GPX4 through depletion of GSH with erastin, or with the direct GPX4 inhibitor RSL3, would ultimately results in overwhelming lipid peroxidation that causes cell death.24 Up to now, ferroptosis is proved to be correlated with neurodegenerative disorders (such as Alzheimer's disease,25 Parkinson's diseases26), hemorrhagic stroke,27 and acute kidney injury (AKI).20 Recent studies reported that systemic iron overload and elevated intracellular iron uptake could induce and exacerbate OA in animal models,28, 29, 30 and Fer-1 could inhibit OA progression in a mouse model.31 However, these studies have not directly elucidated the changes related to ferroptosis in the natural course of human OA pathogenesis.
In the present studies, we investigated whether ferroptosis occurs in OA natural progression through specific detection of damaged cartilage from human OA patients, compared with relatively undamaged cartilage from the same patient. We also determined how GPX4 influenced OA pathogenesis and progression in vivo, using intra-articular injection of adeno-associated virus (AAV) carrying GPX4-specific short hairpin RNA (shRNA). In order to further confirm whether ferroptosis was involved in OA development, TBHP was used to induce chondrocyte oxidative damage to simulate OA in vitro and anterior cruciate ligament transection (ACLT)-induced OA mouse model in vivo. We demonstrated that ferroptosis inhibitors inhibited OA progression through different pathways from necroptosis in vivo and in vitro. Our studies confirmed that ferroptosis was involved in OA progression and we suggest that ferroptosis blockade may serve as an alternative therapeutic strategy for OA treatment.
Materials and methods
Ethics approval and consent to participate
This study was approved by the Ethics committee of Shanghai Sixth People's hospital and all aspects of the study comply with the criteria established by the Declaration of Helsinki. Human cartilage samples were obtained from individuals undergoing total knee arthroplasty, and were conducted in accordance with the informed consent of the patients and approval of ethics committee of Shanghai Sixth People's hospital (Approval Number: 2019-KY-007(K)). All animal experiments were approved by the Animal Care and Use Committee of Shanghai Sixth People's Hospital (DWSY2020-0203).
Patients and specimens
Articular samples were collected from 55 patients with knee OA who underwent knee arthroplasty surgery. Specimens that included all cartilage layers and subchondral bone were separately harvested, and divided into damaged area (designed as OA) and corresponding undamaged area (designed as Undamaged). All human studies were approved by the ethics committee of Shanghai Sixth People's hospital (Approval Number: 2019-KY-007(K)), and full written consents were obtained before the operative procedure. All the experiments described were carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). Clinical information was collected from patient records. The clinical and demographic characteristics of the study population were shown in Supplementary Table 1.
Experimental post-traumatic OA in mice
Ten-week-old male wildtype C57BL/6 mice underwent ACLT surgery of the right knee to induce mechanical instability and create an experimental OA model. Briefly, after anesthesia with intraperitoneally injection of 0.5% pentobarbital, the anterior cruciate ligament of the right knee was transected. A sham operation was performed without ligament transection. We randomly divided the mice into five groups: sham-operated treated with physiological saline (Sham), ACLT-operated treated with physiological saline (Vehicle), ACLT-operated treated with Fer-1(0.1mg/kg) (MedChemExpress), ACLT-operated treated with DFO (0.7mg/kg) (MedChemExpress), and ACLT-operated treated with Nec-1(0.3mg/kg) (Beyotime), respectively. Mice were given an intra-articular injection of Fer-1, DFO and Nec-1 or vehicle using 33-gauge needles (Hamilton Company) and 10 µl CASTIGHT syringes (Hamilton Company). The injection was repeated twice a week for 8 consecutive weeks. AAV-NC or AAV-shGpx4 mice also underwent ACLT surgery in ten-week-old. All mice were sacrificed at 8 weeks after the initial surgery. The right knee was harvested for further evaluation.
Ethical approval was received from the Animal Care and Use Committee of Shanghai Sixth People's Hospital (DWSY2020-0203). All animal experiments complied with the ARRIVE guidelines.
Isolation, culture of chondrocytes, and ATDC5 cells
Primary mouse chondrocytes (MCC) were isolated from cartilage fragments, which were dissected from femoral heads, femoral condyles and tibial plateau of C57BL/6 mice. Briefly, articular cartilage was firstly cut into small pieces and digested with 0.25% trypsin at 37°C for 30 min. After being washed 3 times by PBS, the pieces were fully digested using 0.25% collagenase II at 37°C for 8 h. Afterwards, the cell suspension was filtered using 70μm cell strainer and centrifuged (1000 rpm) for 5 min to collect primary chondrocytes. The cells were finally cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin cocktail. Chondrocytes at passage 3 were used in our study. ATDC5 cells were cultured in DMEM/F12 (Gibco) containing 5% FBS. ATDC5 cell line has been validated by STR profiling and recent mycoplasma testing has been performed. All cells were maintained in a humidified incubator containing 5% CO2 at 37°C.
GPX4 knockout chondrocytes and ATDC5 cells
To knock down specific target genes, 1 × 106 cells were transduced with the LV3 lentivirus carrying shRNA constructs (multiplicity of infection [MOI] of 100) overnight at 37°C with 5 μg/ml polybrene (GenePharma). Lentivirus packaging was provided by GenePharma Inc (Shanghai, China).
Cell viability assays and inhibitor studies
The cell counting kit‐8 (CCK‐8, Dojindo, Kumamoto, Japan) assay was used to measure the cell viability. Cells were seeded onto 96-well plates (1,0000 cells per well) and cultured with IL-1b and t-butyl hydroperoxide (TBHP) to simulate cell death in OA. Ferrostatin-1(Fer-1), Deferoxamine (DFO), Necrostatin-1 (Nec-1) and Z-VAD-FMK (Beyotime) were used to rescue cell death. Then, 90 µL of DMEM and 10 µL of CCK‐8 were mixed and added to each well after 24h. The absorbance of the wells at a wavelength of 450 nm was measured on a microplate reader (Mode 680, Bio‐Rad, Hercules, USA) after a 2 h incubation at 37°C.
Adeno-associated virus (AAV) and intra-articular administration
GPX4 short hairpin RNA (shRNA) fragment was cloned into adeno-associated virus (AAV) vector GV478 (U6-MCS-CAG-EGFP) (Shanghai Genechem Co., Ltd) to construct AAV-shGpx4. AAV-293 cells were co-transfected with recombinant AAV-shGpx4, pAAV-RC and pHelper for AAV packaging. AAV were collected from the AAV-293 cell supernatant, and then condensed purified.
Eight-week-old C57BL/6 mice were anesthetized with pentobarbital, and the skin above the articular joint was shaved. Mice were injected intraarticularly with 10μl of either 1.0 × 1010 v.g AAV-NC or AAV-shGpx4 using 33-gauge needles (Hamilton Company) and 10 µl CASTIGHT syringes (Hamilton Company).
Micro-CT analysis
Briefly, skin and muscles were removed, and the knee joints were obtained. The joint specimens were then fixed in 4% paraformaldehyde (PFA) and were stored in 70% ethanol. All samples were scanned at 9 µm resolution by a micro‐CT scanner (SkyScan 1176, Kontich, Belgium). Further analysis was performed with CTAn (Bruker MicroCT, Kontich, Belgium) and DataViewer (Bruker MicroCT, Kontich, Belgium). Three‐dimensional images were visualized using CTVox software (Bruker MicroCT, Kontich, Belgium). The transverse, coronal and sagittal images of the knee joints were used for analyses. The region of interest covering the surface area of tibia was collected.
Histology and immunohistochemical assay
Tissues were fixed in 4% buffered paraformaldehyde and subsequently decalcified with buffered EDTA (20% EDTA, pH 7.4). The tissues were embedded in paraffin, sectioned and stained with hematoxylin–eosin (HE) and safranin O/fast green. The cellularity and morphology of cartilage and subchondral bone were examined by another group of experienced histology researchers in a blinded manner using a microscope. The severity of cartilage degeneration of OA model was evaluated by OARSI scoring system.
For immunohistochemistry (IHC) staining, sections were heated at 95°C for 15 min, and then treated with 3% H2O2, 0.5% Triton X-100. Nonspecific binding sites was blocked by 10% bovine serum albumin for 1 h at room temperature. The sections were then incubated with the primary antibody (GPX4, 1:100, Abcam Cat# ab125066, RRID:AB_10973901) overnight at 4°C. Finally, the sections were incubated with biotinylated secondary antibody, counterstained with hematoxylin and visualized by DAB solution for IHC. The widely accepted German semi-quantitative scoring system were used to assess staining level.32,33 The final immunoreactive score was determined by multiplying the intensity and the extent scores, yielding a range from 0 to 12.
Iron ion detection in cartilage
Human OA cartilage Iron ion detection were performed using Iron Assay Kit (Dojindo, Kumamoto, Japan) according to the manufacturer's instructions. Briefly, 100mg fresh human OA cartilage tissue was placed in a sterile 2ml tube containing 1 ml assay buffer, and were then grinded using homogenizer on the ice at 4°C. Then the homogenate was centrifuged at 16,000 g for 10 min, and the supernatant was collected. Assay buffer was added in supernatant to test Fe2+ level, or reducer solution was added to test total Iron level. The absorbance of the wells at a wavelength of 593 nm was measured on a microplate reader after a 1 h incubation at 37°C. Then the Fe2+, Fe3+ and total Iron levels of OA cartilage and corresponding non-lesion cartilage were calculated.
Cellular and tissular glutathione peroxidase activity assay
The glutathione peroxidase (Gpx) activity was measured using a glutathione peroxidase assay kit (Beyotime, Jiangsu, China). Cells were seeded in a 6-well dish, treated with medium containing TBHP (30μM), TBHP+Fer-1 (1nM), TBHP/DFO (10nM), TBHP/Nec-1 (10nM) for 4 h at 37°C. For tissue, fresh human OA cartilage sample was placed in a sterile 2ml tube containing 1ml Assay buffer, and then grinded using a homogenizer on the ice at 4°C. The lysates were collected and measured according to the manufacturer's instructions. The absorbance at 340 nm was recorded every 5 min in a SpectraMax microplate reader with the temperature maintained at 25 °C, and then calculated according to the guideline.
Lipid peroxidation assessed by Liperfluo staining
For Liperfluo staining (Dojindo, Kumamoto, Japan), chondrocytes were seeded in a 48-well plate. Cells were treated as described above for 4 h, and then stained with Liperfluo (1μM) for 30 min at 37°C. After being washed by HBSS, the cells were observed immediately with a fluorescence microscope (Leica DMI8, Weztlar, German). The fluorescence intensity in FITC channel was monitored.
Determination of intracellular Fe2+ levels
Chondrocytes were seeded in a 48-well plate. As described above, cells were treated for 4 h, and then were washed 3 times in HBSS and stimulated for 20 min in HBSS at 37°C, 5% CO2. After that, cells were stained in 1μM FerroOrange (Dojindo, Kumamoto, Japan) in HBSS for exactly 30 min at 37°C, % CO2 and imaged immediately with a fluorescence microscope (Leica DMI8, Weztlar, German). Images were obtained with the Cy3 filter (ex 514nm, em 525–596).
Transmission electron microscopy of cartilage
Fresh human knee OA cartilage or mouse knee cartilage were collected and cut into 1mm3 pieces on ice immediately after the specimen was obtained. Then the cartilage tissue was fixed in 2.5% glutaraldehyde solution, post-fixed in 1% aqueous osmium tetraoxide, and dehydrated in gradual ethanol (30–100%) and acetone. The samples were prepared by gradient infiltration of anhydrous acetone and epoxy resin overnight, embedded in resin, and polymerized at 60 °C for 48 h. The embedded samples were cut into thick sections by ultra-microtome (Leica EM UC7). Ultrathin sections were collected onto copper grids, stained with uranium acetate and lead citrate, and examined by 120keV transmission electron microscopy (Talos L120C G2).
Measurement of GSH and GSSG
Chondrocytes were seeded in a 6-well plate. Cells were treated as described above for 4 h. For tissue sample, 500µl protein removal reagent was added to 30-50mg cartilage tissue. Subsequently, the tissue was homogenized with a homogenizer in ice bath, placed at 4 °C for 10 min, and centrifuged at 10,000g 4 °C for 10 min. The supernatant was collected, and the total GSH and GSSG levels were determined by GSH and GSSG Assay Kit (Beyotime, Jiangsu, China) following the manufacturer’ instruction.
Intracellular ROS detection
Intracellular ROS content was measured by the flow cytometry utilizing DCFH-DA. Chondrocytes were seeded in a 6-well plate. In the day of the experiment, cells were stained with 10 μM DCFH-DA at 37 °C for 30 min, then were treated as described above for 4 h. Cells were harvested and analyzed by the flow cytometry (Ex/Em = 488 nm/525 nm).
Malondialdehyde (MDA) assay
MDA content was measured by Lipid Peroxidation MDA Assay Kit (Beyotime, Jiangsu, China). Chondrocytes were seeded in a 6-well plate. Cells were treated as described above for 4 h. Protein lysates were harvested using Cell Lysis Buffer supplemented with Protease Inhibitor. 200μl MDA detection working buffer was added into 100μl samples or standard solution, which was followed by heat in 100 °C hot iron block for 15 min and water bath cooling to room temperature Then the tubes were centrifuged at 1000 x g at room temperature for 10 min and measured the absorbance of A532. The protein concentrations were quantified using a BCA Protein Assay Kit (Thermo, Waltham, USA)to normalize the MDA content.
Mitochondrial membrane potential detection
Mitochondrial membrane potential (ΔΨm) was measured by fluorescence microscopy of cells using JC-1 Mitochondrial Membrane Potential Assay Kit (FuShen, Shanghai, China). Briefly, chondrocytes were seeded in a 6-well plate, cultured overnight, and treated as described above for 4 h. The cells were then incubated with 1 × JC-1 staining solution for 20 min in the dark at 37 °C and analyzed with a fluorescent microscope (Leica DMI8, Weztlar, German).
Detection of synovial fluid
Synovial fluid samples were obtained from OA patients and centrifuged at 3000 g for 10 min within 4 h. The supernatant was stored at -80°C before the final analysis. The levels of 8-hydroxy-2 deoxyguanosine (8-OHdG), an important and reliable marker of oxidative stress, were measured using an ELISA kit (FuShen, Shanghai, China) and conducted according to the manufacturer's guidelines.
The iron levels were measured using Iron Assay Kit (Dojindo, Kumamoto, Japan) according to the manufacturer's instructions.
Quantitative reverse-transcription PCR (qRT-PCR)
Total RNA from ATDC5 cells and MCCs were extracted using EZ-press RNA Purification Kit (EZBioscience) and quantified by the NanoDrop 2000 spectrophotometer (Thermo, Waltham, USA). Complementary cDNA was synthesized through reverse transcription with the aid of a Reverse Transcription Master Mix (EZBioscience). The qPCR assay was performed using buffers from Vazyme on QuantStudio 7 (Thermo). Expression levels were normalized to β‐actin. The primers are listed in Supplementary table 2.
Western blotting
Cells were lysed with Cell Lysis Reagent (Sigma), separated by 12% SDS polyacrylamide gel electrophoresis (PAGE), and transferred to polyvinylidene fluoride (PVDF) membrane. PVDF membranes were incubated with primary antibodies overnight at 4°C, incubated with secondary antibodies at room temperature for 1 h, and visualized using the BIO-RAD ChemiDoc XRS + system. Proteins were analyzed with antibodies recognizing GPX4 (1:1000, Abcam Cat# ab125066, RRID:AB_10973901), MMP3 (1:1000, Santa Cruz Cat# sc-21732, RRID:AB_627958), MMP13 (1:1000, Santa Cruz Cat# sc-515284), ACAN (1:1000, Santa Cruz Cat# sc-33695, RRID:AB_626650), ACSL4 (1:1000, Santa Cruz Cat# sc-365230, RRID:AB_10843105), COL2A1 (1:1000, Abcam Cat# ab188570), Adamts5(1:1000, Affinity Cat# DF13268, RRID:AB_2846287), SLC3A2 (1:1000, Affinity Cat# DF7468, RRID:AB_2839405), SLC7A11(1:1000, Affinity Cat# DF12509, RRID:AB_2845314), FAK (1:1000, Cell Signaling Technology Cat# 3285, RRID:AB_2269034), MAPK 42/44 (1:1000, Cell Signaling Technology Cat# 4695, RRID:AB_390779), Phospho-MAPK 42/44 (1:1000, Cell Signaling Technology Cat# 4370, RRID:AB_2315112), NFκB (1:1000, Cell Signaling Technology Cat# 6956, RRID:AB_10828935), AKT (1:1000, Cell Signaling Technology Cat# 4685, RRID:AB_2225340), Phospho-AKT (1:1000, Cell Signaling Technology Cat# 4060, RRID:AB_2315049), PI3K (1:1000, Cell Signaling Technology Cat# 4249, RRID:AB_2165248), p38 MAPK (1:1000, Cell Signaling Technology Cat# 8690, RRID:AB_10999090), Phospho-p38 MAPK (1:1000, Cell Signaling Technology Cat# 4511, RRID:AB_2139682) and β-ACTIN (Servicebio Cat# GB11001, RRID:AB_2801259) (Supplementary Table 5). All the antibody used were commercial antibodies.
RNA extraction, library construction, and sequencing
Total RNA was extracted using the EZ-press RNA Purification Kit according to the manufacturer's protocol. RNA purity and quantification were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with RNA
Integrity Number (RIN) ≥ 7 were subjected to the subsequent analysis. Then the libraries were constructed using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. These libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten) and 125bp/150bp paired-end reads were generated. The transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China). Raw data (raw reads) were processed using Trimmomatic.34 The reads containing ploy-N and the low-quality reads were removed to obtain the clean reads. Then the clean reads were mapped to reference genome using hisat2.35 FPKM36 of each gene was calculated using Cufflinks,37 and the read counts of each gene were obtained by HTSeqcount.38 Expression analysis was performed using the DESeq R package. Hierarchical cluster analysis was performed to demonstrate the expression pattern of genes in different groups. GO enrichment and KEGG39 pathway enrichment analysis were performed respectively using R based on the hypergeometric distribution.
Gene set enrichment analysis (GSEA)
GSEA was performed using GSEA software (ver. 4.1.0; Broad Institute, MIT). Genes were ranked according to their expression; Gene set were searched from website (www.gsea-msigdb.org).
Statistical analysis
All data were expressed as mean± S.D. Statistical analyses were completed with Prism GraphPad. Unpaired Student's t test (for two groups), one-way or two-way ANOVA (for multiple groups) were used followed by the Tukey-Kramer test. Data based on ordinal grading systems were analyzed using non-parametric Mann-Whitney U tests. P<0.05 was considered statistically significant.
Role of funding source
The funders had no role in study design, data collection, interpretation and analysis, decision to publish or preparation of the manuscript.
Results
Ferroptosis occurred in osteoarthritic cartilage
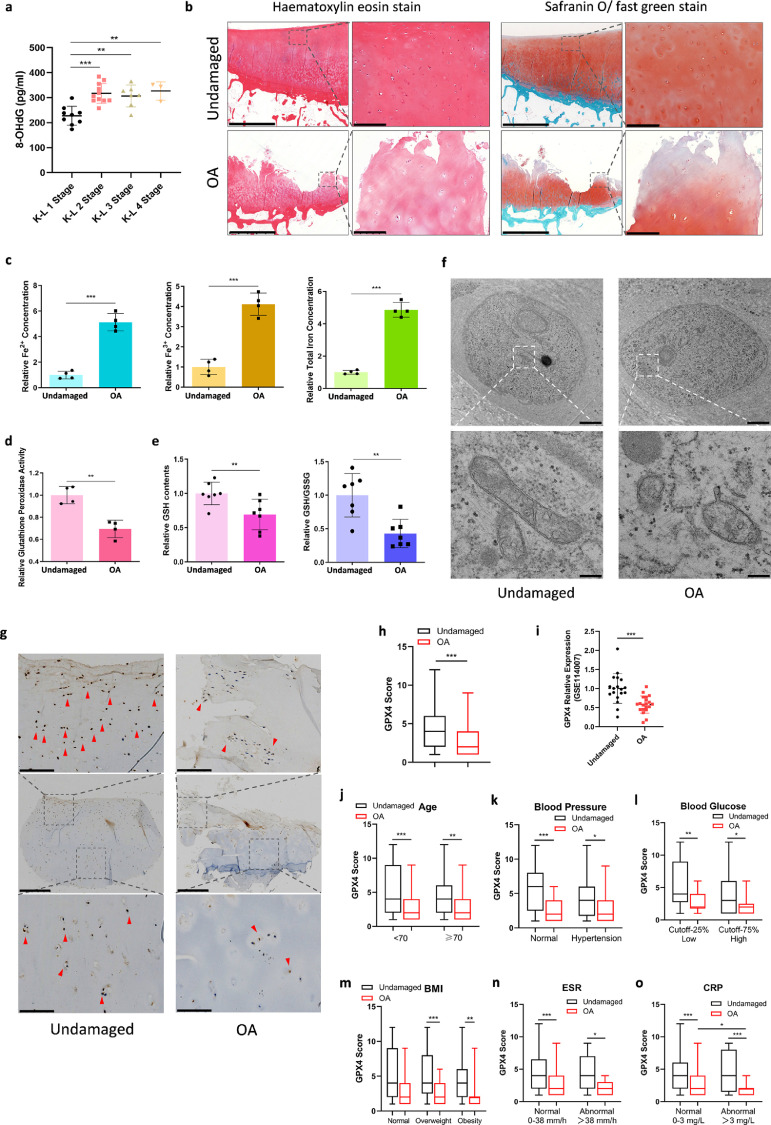
Genome-wide RNA-Seq data (GEO, https://www.ncbi.nlm.nih.gov/geo/, GEO accession number: GSE114007) were used to analyze the expression of ferroptosis-related genes (Supplementary Figure 1a, b, Supplementary Table 3 and 4) and the results showed that ferroptosis suppressor genes were downregulated in osteoarthritic cartilage. To explore whether ferroptosis was involved in the OA progression, we first detected 8-OHdG level in synovial fluid from OA patients to evaluate the oxidation level. The levels of 8-OHdG in synovial fluid from OA patients categorized as K-L (Kellgren-Lawrecne) Stage 2-4 were significantly higher than that in patients at Stage 1 (Figure 1a). Iron level in the synovial fluid was also assayed, and the data was similar to the results of 8-OHdG, showing that iron level in synovial fluid increased with the progression of OA (Supplementary Figure 1c). Then we detected ferroptosis-related alterations in OA cartilage and non-lesion counterparts. Hematoxylin–eosin (HE) and safranin O/fast green staining were performed in the cartilage specimens to assess the degradation level (Figure 1b). Fe2+, Fe3+ and total iron concentrations were all significantly higher in osteoarthritic cartilage than those in the undamaged counterparts (Figure 1c), indicating that there was iron accumulation in the cartilage during OA progression. Due to the fact that ferroptosis is dramatically modulated by pharmacological perturbation of lipid repair systems involving glutathione and GPX4,18 glutathione peroxidase activity was also detected in the cartilage specimens. Glutathione peroxidase could eliminate peroxides and plays a key role in the anti-peroxidant defense system, and GPX4 is one of the major functional enzymes. The results showed that glutathione peroxidase activity was decreased in OA cartilage (Figure 1d). Meanwhile, as the reducing substrate of GPX4 activity, the level of GSH was also examined (Figure 1e). The data revealed that there was GSH depletion in osteoarthritic cartilage, and GSH/GSSG level was also decreased in these samples. As previously reported, ferroptosis induced by erastin could lead to glutathione depletion and inactivation of the glutathione peroxidase.21,22 The variation of glutathione peroxidase activity and GSH in OA cartilage was consistent with that in ferroptosis. Moreover, as the characteristic change in ferroptosis, morphological changes of mitochondria were also observed by transmission electron microscopy (TEM). Ultrastructural analysis demonstrated that mitochondrial ridge was reduced or disappeared and rupture of outer mitochondrial membrane (OMM) occurred in chondrocytes embedded in OA cartilage (Figure 1f).
Figure 1.
Ferroptosis related to OA pathogenesis and GPX4 expression is downregulated in OA cartilage. (a) 8-OHdG level in synovial fluid from OA patients categorized as K-L stage 1–4. There were 9 samples in K-L 1 stage, 11 in K-L 2 stage, 7 in K-L 3 stage, 3 in K-L 4 stage. (b) Representative histopathological hematoxylin–eosin and safranin O/fast green staining of undamaged and OA cartilage tissues. Scale bar, left, 2.5 mm; right, 200 μm. (c) Fe2+, Fe3+ and total Iron level in OA cartilage and corresponding non-lesion cartilage (n = 4). (d) Glutathione peroxidase activity level were detected in OA and undamaged cartilage tissues (n = 4). (e) GSH contents and ratio of GSH/GSSG were measured in OA and undamaged cartilage tissues (n = 7). (f) Transmission electron micrographs showing outer mitochondrial membrane rupture in OA cartilage tissues. Scale bar, up, 2 μm; down, 200 nm. (g) Immunohistochemistry assay with anti-GPX4 in undamaged and OA cartilage tissues. Scale bar, middle, 400μm; up & down, 100μm. (h) The GPX4 scores in OA (n = 55) and corresponding undamaged (n = 55) cartilage tissues based on an immunohistochemistry assay were compared with the Mann–Whitney U-test. (i) Genome-wide RNA-Seq data of GPX4 expression level of OA cartilage and undamaged cartilage. Date was originated from GSE114007. (j) The GPX4 scores in OA patients: age<70 years old (n = 30) and ≥70 years old (n = 25). (k) The GPX4 scores in OA patients with (n = 29) or without hypertension (n = 26). (l) The GPX4 scores in OA patients: blood glucose 25% low group (n = 14) and 25% high group (n = 14). (m) The GPX4 scores in OA patients: BMI normal group (18<BMI<24, n = 7), overweight group (24≤BMI<32, n = 25) and obesity group (BMI≥32, n = 23). (n) The GPX4 scores in OA patients: normal ESR (Erythrocyte Sedimentation Rate) (n = 46) and high ESR (n = 9). (o) The GPX4 scores in OA patients with normal level of CRP (C-reactive protein) (n = 38) and high level of CRP (n = 17). All GPX4 scores were based on immunohistochemistry assay. Data are expressed as mean ± SD, * P< 0.05; ** P< 0.01; *** P< 0.001. Unpaired student's t-test were used for comparison between two groups in (a), (i). Paired non-parametric Mann-Whitney U tests were used in (c)–(e), (h), (j)–(o).
Decreased GPX4 levels in the cartilage of OA patients
To further explore the alterations of ferroptosis-related genes, we examined the expression levels of GPX4, a key regulator of ferroptosis, in 55 pairs of OA cartilage tissues and corresponding non-lesion samples. The expression of GPX4 protein was evaluated by immunohistochemical staining (Figure 1g). The results showed that GPX4 expression was significantly lower in OA cartilage than that in non-lesion tissues (Figure 1h). We also categorized patients into different groups according to age, hypertension, blood glucose, BMI, ESR and CRP level (Figure 1j–o). The results indicated that OA cartilage from patients with high CRP level exhibited significantly lower GPX4 level. In addition, patients with hypertension or high level of blood glucose had lower GPX4 level, in spite of no significant difference. Meanwhile, we analyzed RNA sequencing (RNA-seq) data of the transcriptome of OA cartilage and undamaged cartilage (GEO accession number: GSE114007) and revealed that mRNA level of GPX4 was lower in OA cartilage than that in undamaged cartilage (Figure 1i). Moreover, expression of system Xc− molecules, SLC3A2 and SLC7A11, were also analyzed in GSE114007, and we found that the expression level of SLC3A2 was significantly lower in OA cartilage (Supplementary Figure 1d). All these results suggested that ferroptosis is present during OA progression.
Fer-1 and DFO rescued TBHP-induced cell death in a necroptosis-independent manner
Due to the complexity of ferroptosis and lack of standard criteria and distinct biomarkers, we could not conclude that ferroptosis contributed to OA pathogenesis, despite our findings from OA patients which were consistent with ferroptosis. However, there are still some basic criteria to justify the involvement of ferroptosis except for the biological indicators as aforementioned: (i) ferroptosis should be suppressed by both an iron chelator (e.g., DFO) and a lipophilic antioxidant (e.g., ferrostatin-1, liproxstatin-1). (ii) Inhibitors of apoptosis or necroptosis should be examined to rule out these mechanisms.18 Therefore, the following experiments were undertaken to further examine the possibility of ferroptosis involvement in OA pathogenesis.
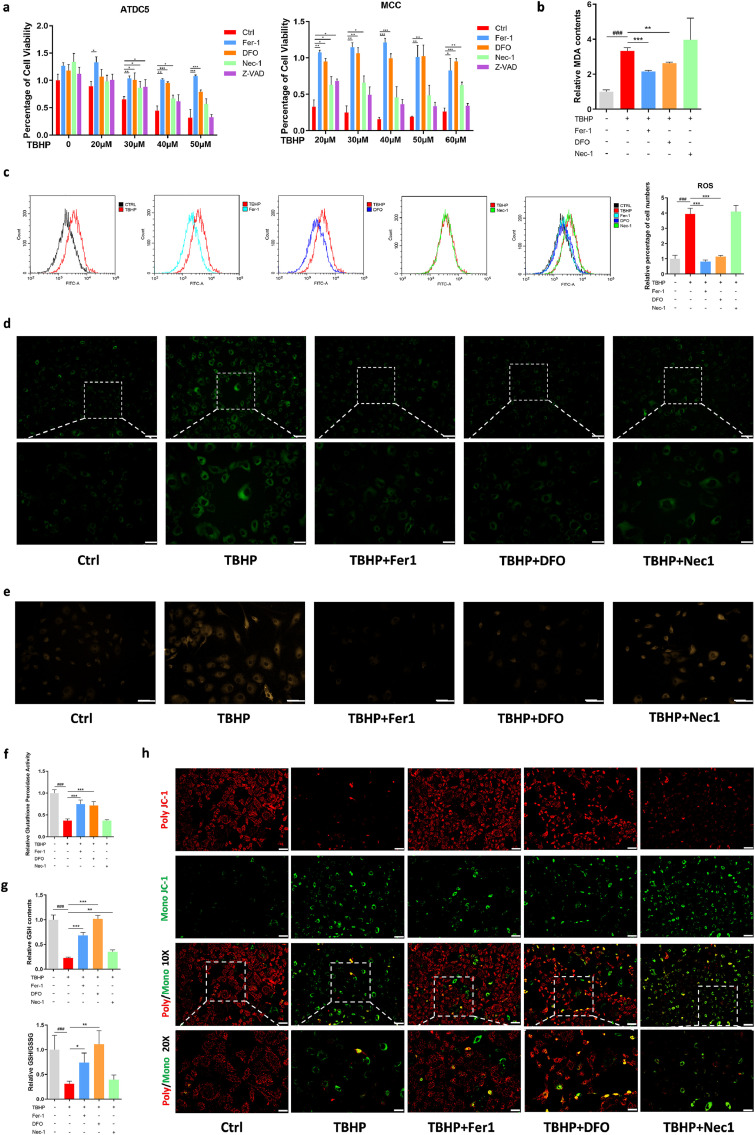
Researchers usually use IL-1β to induce inflammatory response and use TBHP to induce oxidative stress.40 Therefore, we examined effects of IL-1β and TBHP on inducing cell death (Supplementary Figure 2a, c). The results showed that IL-1β could not induce cell death effectively, while TBHP could obviously reduce cell viability. Because ferroptosis itself is related with oxidative stress, we induced the specific OA cell model by using TBHP, and treated cells with pan-caspase inhibitor z-VAD-FMK (zVAD), necroptosis inhibitor necrostatin-1 (Nec1), Fer1, and DFO, respectively. Traditionally, cell death induced by H2O2 was considered as necroptosis. We found that Fer-1 and DFO have a better protective effect than Nec-1, especially under high concentration of TBHP (Figure 2a). However, zVAD could protect cell death induced by low concentrations of TBHP, but had no obvious effects for high concentration of TBHP-induced cell death (Figure 2a). Thus, we ruled out the involvement of caspases in cell death induced by TBHP. This indicated that there may be more than one cell death modality triggered by TBHP, and ferroptosis and necroptosis might play a role in this process.
Figure 2.
Fer-1 and DFO protect TBHP induced MCC cell death in a necroptosis-independent manner. (a) Cell viability of ATDC5 cells and MCC treated with different concentration of TBHP and rescued via Fer-1, DFO, Nec-1 and Z-VAD for 24 h measure by CCK8. (b) MDA contents of MCC treated with TBHP and rescued via Fer-1, DFO, Nec-1 for 4 h. (c) Flow cytometry analyzed ROS level of MCC treated with TBHP and rescued via Fer-1, DFO, Nec-1, and relative ROS level performance. (d) Lipid ROS level was analyzed using Liperfluo fluorescence probe of MCC treated with TBHP and rescued via Fer-1, DFO, Nec-1. Scale bar, up, 100 μm; down, 50 μm. (e) Intracellular Fe2+ level was detected using FerroOrange Assay of MCC treated with TBHP and rescued via Fer-1, DFO, Nec-1. Scale bar, 75 μm. (f) Glutathione peroxidase activity level of MCC treated with TBHP and rescued via Fer-1, DFO, Nec-1. (g) GSH contents and ratio of GSH/GSSG of MCC treated with TBHP and rescued via Fer-1, DFO, Nec-1. (h) Mitochondrial membrane potential (ΔΨm) measured by fluorescence microscopy of MCC treated with TBHP and rescued via Fer-1, DFO, Nec-1 using JC-1 Mitochondrial Membrane Potential Assay, Scale bar, up, 100 μm; down, 50 μm. Data are expressed as mean ± SD, * P< 0.05; ** P< 0.01; *** P< 0.001. Student's t-test and one-way ANOVA were used for comparison between two groups and multiple groups, respectively.
Next, we addressed what was the difference between cells rescued by ferroptosis inhibitor and Nec-1. Flow cytometry was utilized to detect intracellular ROS contents. The results revealed that Fer-1 and DFO rescued the TBHP-induced increase of intracellular ROS levels, while Nec-1 had no effects on inhibiting ROS elevation (Figure 2c). Measurement of malondialdehyde (MDA) contents showed similar results: increased intracellular MDA levels could be restrained by Fer-1 and DFO, but not by Nec-1 (Figure 2b).
We then assessed changes in ferroptosis-related indicators when TBHP-induced cell death was rescued. Lipid peroxidation was detected by LiperFluo, which is a useful fluorescent probe in ferroptosis studies. The results showed that TBHP-triggered accumulation of lipid peroxidation was restrained by Fer-1 and DFO; in contrast, Nec-1 had no significant effect (Figure 2d). As ferroptotic cell death entails cellular Fe2+ accumulation, we also analyzed Fe2+ levels in this study. The results were similar to that detected by LiperFluo (Figure 2e). Glutathione peroxidase activity, GSH content and GSH/GSSG was obviously decreased when treated with TBHP, indicating that TBHP broke down the kinetic balance of GPX4/GSH and triggered ferroptosis (Figure 2f, g). In the rescue experiments, Fer-1 and DFO reversed the decline of glutathione peroxidase activity, GSH contents and GSH/GSSG, while Nec-1 had no significant effect. Because of the unique morphological and functional changes of mitochondria during ferroptosis, we also detected the cellular mitochondrial membrane potential (MMP). When cells were treated with TBHP, a large amount of poly JC-1 was dissociated into mono JC-1, indicating the decrease or loss of MMP. In Fer-1 and DFO group, red fluorescence (poly JC-1) was increased and green fluorescence (mono JC-1) was decreased (Figure 2h).
Furthermore, mRNA levels of anabolic and catabolic markers involved in chondrocyte metabolism were assessed. The protective effects of Fer-1, DFO and Nec-1 were analyzed by the reversal of TBHP-induced decrease of mRNA levels of anabolic genes (Supplementary Figure 3a, b), along with decrease of expression levels of catabolic (Supplementary Figure 3e–h) and inflammatory genes (Supplementary Figure 3c, d), compared with those in the TBHP-treated group. Moreover, the trends of variation of ferroptosis markers after TBHP treatment were consistent with those in classic ferroptosis (Supplementary Figure 3i–l). Fer-1 and DFO reversed the decrease of mRNA levels of GPX4 and Fth1 induced by TBHP, and decreased ACSL4 and PTGS2, while Nec-1 showed no effect on ferroptosis markers.
These data suggested that ferroptosis inhibitor Fer-1 and DFO could rescue TBHP-induced cell death through inhibiting ferroptosis, including reducing lipid peroxidation, decreasing iron accumulation and maintaining GPX4/GSH function, which was completely different from the function of Nec-1.
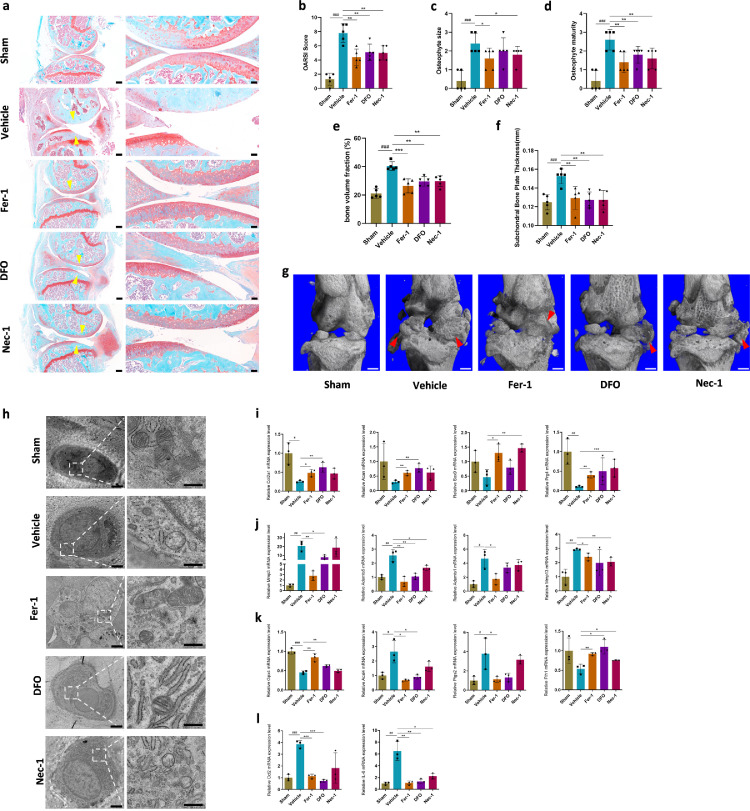
Both Fer-1 and DFO ameliorated OA progression
We performed an in vivo study to evaluate the effect of Fer-1 and DFO on OA development, and Nec-1 was also included as a control and performed ACLT surgery to induce OA. We found that mice with intra-articular injection of Fer-1 and DFO exhibited less severe OA phenotype to the extent similar to that of mice administered with Nec-1. This was demonstrated by analyzing histological changes (Figure 3a) and Osteoarthritis Research Society International (OARSI) scores (Figure 3b), osteophyte size (Figure 3c) and osteophyte maturity (Figure 3d). Micro-computerized tomography (μCT) analysis showed that significant osteophyte formation was observed 8 weeks after ACLT surgery (Figure 3g). Treatment with either Fer-1 or DFO significantly restricted osteophyte formation (Figure 3g). We also analyzed bone volume fraction of subchondral trabecular bone (STB) and thickness of subchondral bone plate (SBP). Results illustrated that both bone volume fraction of STB and SBP thickness was significantly increased after surgery (Figure 3e, f), and both ferroptosis inhibitors and necroptosis inhibitor significantly ameliorated ACLT-induced increases in bone volume fraction and SBP thickening (Figure 3 e, f).
Figure 3.
Fer-1 and DFO attenuated osteoarthritis (OA) development through inhibiting chondrocytes ferroptosis. (a) Representative safranin O-fast green images of osteoarthritic knee joints, that were collected 8 weeks after ACLT surgery. Yellow arrowheads indicate articular cartilage degradation. n = 5; Scale bar, left, 200 μm; right, 50 μm. (b) The severity of OA-like phenotype was analyzed using the Osteoarthritis Research Society International (OARSI) score system. n = 5. (c) and (d) Osteophytes were semi-quantified by evaluating the osteophyte formation score consisting of two domains, size (c) and maturity (d). n = 5. (e) Quantitative micro-CT analysis of tibial subchondral trabecular bone with bone volume fraction. n = 5. (f) Quantitative micro-CT analysis of tibial subchondral bone plate thickness. n = 5. (g) Three-dimensional models of mice knee joints. Red arrow shows osteophyte formation. Scale bar, 500μm. (h) Mitochondrial morphology of mice cartilage was observed using transmission electron microscopy. Scale bar, left, 2 μm; right, 500 nm. (i-l) Quantification of mRNA levels for cartilage anabolism related genes (Col2a1, Acan, Sox9, Prg4), catabolism related genes (Mmp3, Mmp13, Adamts1, Adamts5), ferroptosis related genes (Gpx4, Acsl4, Ptgs2, Fth1) and inflammatory related genes (Ccl2, IL-6) in articular cartilage obtained from sham, vehicle, and Fer-1, DFO, Nec-1 treated mice, respectively. Data are expressed as mean ± SD, * P< 0.05; ** P< 0.01; *** P< 0.001. Student's t-test and one-way ANOVA were used for comparison between two groups and multiple groups, respectively.
We next detected expression levels of OA-related biomarkers and ferroptosis core regulators from mice cartilage. The results showed that the expression levels of MMP3, MMP13, Adamts1, and Adamts5 were significantly upregulated in the cartilage from ACLT-induced OA mouse model (Figure 3j). The addition of Fer-1, DFO and Nec-1 could significantly suppress the increase of Adamts5 expression. Moreover, Fer-1 also effectively inhibited the expression of MMP3, MMP13, and Adamts1. Meanwhile, DFO inhibited MMP3 expression and Nec-1 inhibited Adamts1 expression. These findings suggested that ferroptosis is a potent pathway promoting catabolic responses. In addition to catabolic genes, Fer-1 also significantly reversed the downregulation of Col2a1, Aggrecan, Sox9 and Prg4 in OA cartilage (Figure 3i). DFO and Nec-1 exhibited similar effect in reversing the downregulated expression of anabolic genes. However, although Sox9 and Prg4 expressions in DFO-treated group had the reserved trend, there was no significant difference due to the large deviation, as well as Col2a1 and Acan expressions in Nec-1-treated group. Inflammatory biomarker Ccl2 and IL-6 were also suppressed by all these three inhibitors (Figure 3l). Ferroptosis core regulators, GPX4 and Fth1 was downregulated in the mice administered with vehicle, while Acsl4 and Ptgs2 were upregulated (Figure 3k). Fer-1 and DFO could reverse the expression of ferroptosis regulatory genes, while Nec-1 had no effect.
Fer-1 and DFO protected the rupture of outer mitochondrial membrane (OMM) of chondrocytes in the ACLT-induced OA mouse model
TEM was performed to observe ultrastructural alterations of the chondrocytes in the ACLT-induced OA mouse model. Ultrastructural analysis revealed that the chondrocytes in the OA model exhibited similar phenotype to the counterpart from human OA specimens, including condensed mitochondrial membrane densities, reduction or vanishing of mitochondria crista, as well as rupture of outer mitochondrial membrane (OMM) with normal nucleus (Figure 3h). Administration of Fer-1 and DFO could obviously prevent ultrastructural and morphological alterations of the chondrocytes in the OA model, while Nec-1 administration failed to protect these changes (Figure 3h). These in vivo results suggested that Fer-1 and DFO could play protective roles in OA progression by inhibiting ferroptosis, which was different from the effects of necroptosis inhibitor Nec-1. All the results above provided valid evidence that ferroptosis occurred in OA.
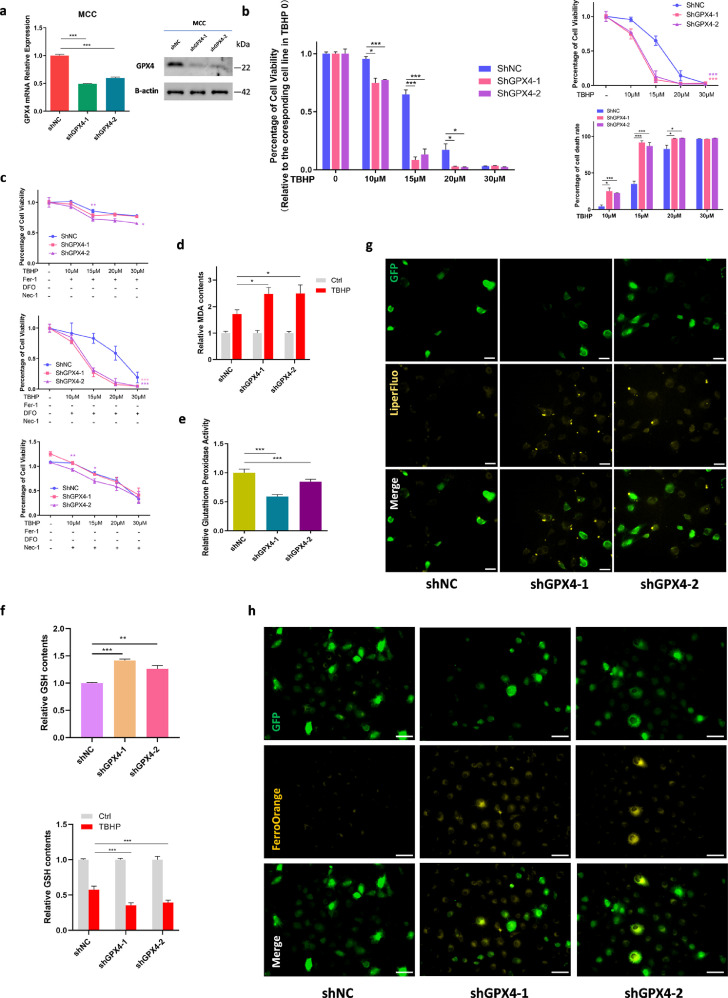
GPX4 knockdown increased the sensitivity of chondrocytes to oxidative stress
As the key regulator of ferroptosis, GPX4 is a selenoprotein, and selenium is crucial to the growth of cartilage.41 Therefore, we intended to explore the effect of GPX4 in chondrocyte. Given that deficiency of GPX4 would directly cause cell death, we established a stable GPX4-knockdown ATDC5 cells and primary mouse chondrocytes (MCCs) which simulate GPX4 expression level in OA cartilage (Figure 4a, Supplementary Figure 5a–c). We found that GPX4-knockdown significantly increased cell death rate when treated with the same concentration of TBHP (Figure 4b, Supplementary Figure 5d). Meanwhile, the effects of Fer-1 and DFO were impaired significantly (Figure 4c). Likewise, GPX4 knockdown increased the sensitivity of cells to TBHP, producing more MDA (Figure 4d). However, GPX4 knockdown did not affect the cellular MDA levels under normal circumstances (Supplementary Figure 5e). We also detected the glutathione peroxidase activity. Theoretically, glutathione peroxidase activity was related to the content of GPX4. Not surprisingly, after knockdown of GPX4, glutathione peroxidase activity decreased (Figure 4e). Nevertheless, the decrease rate of glutathione peroxidase activity exhibited no significant difference after TBHP treatment (Supplementary Figure 5f). It would be possible that the reduction of glutathione peroxidase activity is not related to the basic levels of GPX4, but more related to the treatment conditions. Interestingly, the levels of GPX4 substrate GSH was elevated in the GPX4-knockdown cells (Figure 4f). This might be attributed to the compensatory increase of GSH in order to balance the decrease of GPX4. Cellular GSH concentration is maintained by a complex homeostatic mechanism where the steady-state concentration is under dynamic control of specific enzymatic reactions.42 After treatment with TBHP, there was more reduction of GSH content in GPX4-knockdown cells (Figure 4f, Supplementary Figure 5g). This phenomenon might indicate that when TBHP breaks the balance of GPX4/GSH, GPX4/GSH is in a state of decompensation and GSH could be consumed dramatically. Liperfluo detection showed that GPX4 deficiency would lead to a slight increase of lipid peroxide levels (Figure 4g). Intracellular Fe2+ levels were enhanced obviously (Figure 4h). These results indicated that the decrease of GPX4 expression could reduce the tolerance of cells to external oxidative stimulation, and a slight decrease of GPX4 in chondrocytes in OA circumstances might render cells more sensitive to oxidative stress. However, beside of the anticipation in ferroptosis, the potential role of GPX4 in OA cartilage is still unknown.
Figure 4.
Gpx4 downregulation makes chondrocytes more sensitive to oxidative stress. (a) The efficacy of Gpx4 specific shRNA were measured by qRT-PCR and western blotting in MCC. (b) Cell viability and death rate of MCC-shNC and shGpx4 treated with different concentration TBHP for 24 h measure by CCK8. Data of TBHP 10μM, 15μM, 20μM, 30μM were relative to its corresponding cell line data in TBHP 0. (c) Cell viability of MCC-shNC and shGpx4 treated with different concentration TBHP and rescued via Fer-1, DFO and Nec-1, repectively. (d) MDA contents of MCC-shNC and shGpx4 and treated with TBHP 30μM for 4 h. Data of TBHP were relative to its corresponding cell line data of Ctrl. (e) Glutathione peroxidase activity level of MCC-shNC and shGpx4. (f) GSH contents of MCC-shNC and shGpx4 (left) and treated with TBHP 30μM for 4 h (right). Data of TBHP were relative to its corresponding cell line data of Ctrl. (g) Lipid ROS level was analyzed using Liperfluo fluorescence probe of MCC-shNC and shGpx4. Scale bar, 50 μm. GFP indicate lentivirus affected. (h) Intracellular Fe2+ level was detected using FerroOrange Assay of MCC-shNC and shGpx4. GFP indicate lentivirus affected. Scale bar, 75 μm. Data are expressed as mean ± SD, * P< 0.05; ** P< 0.01; *** P< 0.001. Student's t-test and one-way ANOVA were used for comparison between two groups and multiple groups, respectively.
GPX4 downregulation promoted extracellular matrix degradation through MAPK/NFκB pathway
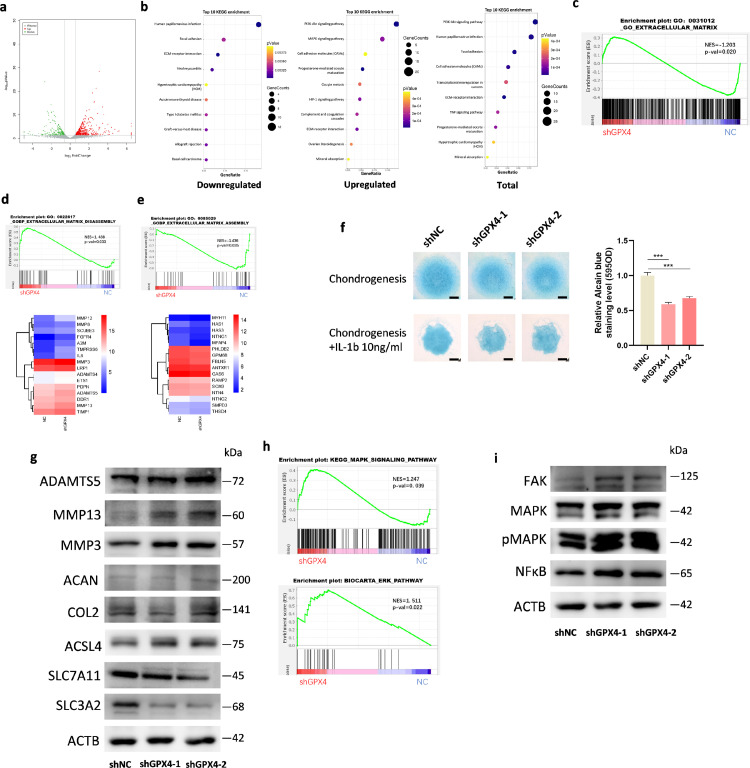
To further analyze the effect of GPX4 on gene expression, we conducted RNA-sequencing in mouse chondrocytes with or without GPX4 knockdown. GPX4 knockdown resulted in the upregulation of 426 genes and downregulation of 199 genes (Figure 5a). Gene ontology (GO) (Supplementary Figure 6a, b) analysis showed that GPX4 knockdown significantly affected extracellular matrix, extracellular region and extracellular space. In addition, KEGG pathway analysis showed inhibition of focal adhesion and ECM (extracellular matrix)-receptor interaction, and promotion of PI3K-Akt pathway and MAPK pathway in the GPX4-knockdown chondrocytes (Figure 5b). As the extracellular matrix category included both downregulated and upregulated genes, we performed GSEA analysis which categorized genes into those that promote and inhibit the degradation of ECM. The results showed that genes in extracellular matrix disassembly category (GO:0022617) were upregulated (Figure 5c, d) and genes in extracellular matrix assembly category (GO:0085029) were downregulated (Figure 5e). Taken together, these data revealed that ferroptosis pivotal regulator GPX4 plays an important role in articular cartilage homeostasis.
Figure 5.
GPX4 regulate ECM disassembly genes through MAPK/NFκB pathway. (a) A volcano plot illustrating differentially regulated gene expression from RNA-seq analysis between NC and shGpx4 MCCs. Genes upregulated and downregulated are shown in red and green, respectively. (b) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of downregulated, upregulated, and total genes in shGpx4 transcriptome. (Top 10) (c) GSEA plots evaluating the changes of extracellular matrix related genes. (d) GSEA plots evaluating the changes of extracellular matrix disassembly related genes and heatmap of representative genes. (e) GSEA plots evaluating the changes of extracellular matrix assembly related genes and heatmap of representative genes. (f) Alcian blue staining of ATDC5-shNC and shGpx4 cells after incubation in chondrogenesis media with or without IL-1β (10ng/ml). Left, representative image of staining at day 8 is shown. Scale bar, 0.25 mm. Right, quantification of staining by colorimetry. (g) Protein expression of Adamts5, MMP3, MMP13, COL2A1, ACAN and ACSL4,SLC7A11, SLC3A2 in chondrocytes infected with shNC or shGpx4 analyzed by Western blot. (h) GSEA plots evaluating the changes of MAPK pathway (up) and Erk pathway (down). (i) Protein expression of FAK, MAPK, pMAPK, and NFκB analyzed by Western blot. Data are expressed as mean ± SD, * P< 0.05; ** P< 0.01; *** P< 0.001. Student's t-test and one-way ANOVA were used for comparison between two groups and multiple groups, respectively.
To further determine the effects of GPX4 on extracellular matrix degradation, micromass culture of ATDC5 cells were induced to undergo chondrogenesis. Alcian blue staining displayed decreased proteoglycans in GPX4 knockdown cells compared to those in controls (Figure 5f). We next investigated whether GPX4 knockdown affects the expression of OA-related proteins. Expression of MMP3 and MMP13 was significantly upregulated, while there was no significant change of COL2A1 and ACAN in GPX4-knockdown MCCs (Figure 5g, Supplementary Figure 7a). Moreover, ferroptosis positive regulator ACSL4 was upregulated, and SLC3A2 was downregulated (Figure 5g, Supplementary Figure 7a). Based on KEGG analysis, focal adhesion, PI3K-AKT and MAPK pathways were significantly affected by GPX4. GSEA analysis also indicated similar gene expression pattern (Figure 5h, Supplementary Figure 7c, d). Immunoblot results showed that MAPK, pMAPK and NFκB were upregulated (Figure 5i, Supplementary Figure 7b), while AKT, pAKT and PI3K were not significantly altered in GPX4-knockdown MCCs (Supplementary Figure 7f, g). Expression of p38 MAPK and p-p38 MAPK was also not increased in GPX4-knockdown MCCs (Supplementary Figure 7 e–g), which is consistent with the GSEA analysis. These results indicated that cartilage catabolism-related proteins, such as MMP3 and MMP13, were activated through the MAPK/NFκB pathway to promote ECM degradation after GPX4 downregulation.
GPX4 modulated OA pathogenesis
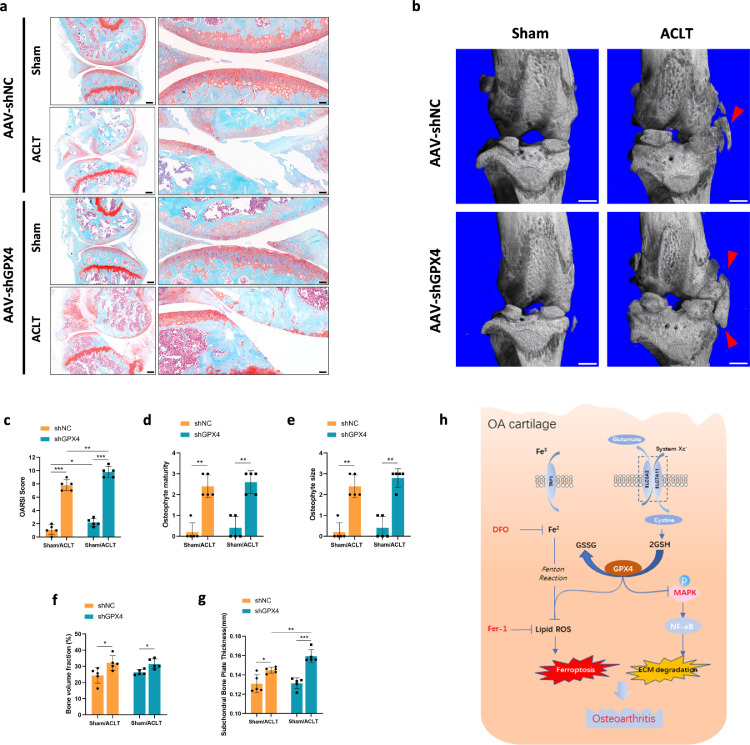
To clarify the contribution of GPX4 to OA disease onset or progression, 8-week-old male C57BL/6J mice underwent intra-articular injection of AAV expressing GPX4 specific shRNA. The efficiency of AAV expressing GPX4-specific shRNA in the joints was verified by immunofluorescence, animal image and qPCR (Supplementary Figure 8a–c). Two weeks after injection, we performed ACLT surgery in the knee joint in 10-week-old mice. Histological analysis including articular cartilage degradation, SBP deterioration and osteophyte formation were carried out to assess the severity of joint damage 10 weeks after ACLT surgery (Figure 6a). In Sham groups, mice injected with GPX4-specific shRNA showed slight cartilage degradation (Figure 6a, c). After ACLT surgery, cartilage degradation was obvious, evidenced by markedly higher OARSI scores (Figure 6c). Mice injected with GPX4-specific shRNA showed more severe cartilage degradation after ACLT surgery, compared to those injected with control shRNA. μCT also showed that osteophyte formation was significantly increased in mice undergoing ACLT surgery, and this effect was further enhanced by GPX4 knockdown (Figure 6b). SBP thickness was significantly increased after surgery (Figure 6g), and knockdown of GPX4 accelerated the increase of SBP thickness significantly. Similar results were observed for the osteophyte size and maturity, and bone volume fraction of subchondral trabecular bone (STB) (Figure 6d–f). Thus, GPX4 downregulation could increase ECM degradation and accelerate OA development.
Figure 6.
GPX4 downregulation accelerated OA progression. (a) Representative safranin O-fast green images of mice knee joints. One week before surgery, C57/BL6J mice (8 weeks old) were injected intra-articularly with AAV carrying GPX4-specific shRNA and analyzed 8 weeks after surgery. Scale bar, left, 500 μm; right, 50 μm. (b) Three-dimensional models of mice knee joints. Red arrow shows osteophyte formation. Scale bar, 500μm. (c) The severity of OA-like phenotype was analyzed using the Osteoarthritis Research Society International (OARSI) score system. n = 5. (d) and (e) Osteophytes were semi-quantified by evaluating the osteophyte formation score consisting of two domains, size (D) and maturity (E). n = 5. (f) Quantitative micro-CT analysis of tibial subchondral trabecular bone with bone volume fraction. n = 5. (g) Quantitative micro-CT analysis of tibial subchondral bone plate thickness. n = 5. (h) In OA cartilage, function of system Xc− was inhibited, result in content of GSH and GPX4 expression decreased. Downregulation of GPX4 not only increased the sensitivity of chondrocytes to oxidative stress, but also aggravated extracellular matrix (ECM) degradation potentially through MAPK/NFκB pathway. Data are expressed as mean ± SD, * P< 0.05; ** P< 0.01; *** P< 0.001. Student's t-test and one-way ANOVA were used for comparison between two groups and multiple groups, respectively.
Discussion
OA is the most common form of arthritis and the leading cause of chronic disability among older people.43 Targeting chondrocyte death in OA has been regarded as an important therapeutic approach. Cell death can be classified to apoptosis and the non-apoptotic processes, including necroptosis, pyroptosis, and ferroptosis.44,45 Recent reports suggested a possible link between ferroptosis and OA, based on the observations that OA progression is inhibited by Fer-1 and Iron overload induced chondrocyte ferroptosis in vivo.31 However, Fer-1 is essentially anti-lipid-oxidants and may inhibit other ROS-dependent forms of cell death; and the related molecular mechanisms in human OA are still unknown. In the current study, we systematically studied the role of ferroptosis, a recently discovered iron-dependent form of cell death, in human OA, in TBHP-induced OA-associated cell model and in ACLT-induced OA mouse model. Because the mechanisms of OA are complicated and multiple factors are involved in this process, including inflammation, extracellular matrix degradation, chondrocyte death and oxidative stress. All the existing OA cell models and animal models could not fully recapitulate OA process. Thus, it is essential to investigate OA in natural course of human patients in order to clarify whether ferroptosis is essentially involved in OA. We detected Fe2+ accumulation, decrease of GSH contents, glutathione peroxidase activity, and mitochondrial morphological alteration in damaged cartilage. We also analyzed the GEO database (GSE114007) and found that the most important antioxidant enzyme and a key regulator of ferroptosis, GPX4, and component of system Xc−, SLC3A2 were significantly downregulated in human OA cartilage. We also verified this observation in the damaged cartilage and compared with undamaged cartilage from the same patients by IHC. Combined with GEO database and IHC results, we showed that GPX4 expression was decreased by about 50% in OA cartilage compared with undamaged cartilage. Combined with the above results, we could conclude that ferroptosis was involved in OA progression.
Tang et al. suggested that researcher may not be able to conclude that excessive cell death by ferroptosis is involved in pathogenic processes just based on the preventive effects of ferrostatin-1 or liproxstatin-1 on tissue damage, because these compounds are essentially antioxidants and may suppress other ROS-dependent forms of cell death.19 Stockwell et al. also suggested that ferroptosis should be suppressed by both an iron chelator (e.g., DFO or CPX) and a lipophilic antioxidant (e.g., ferrostatin-1, liproxstatin-1) to evaluate the role of ferroptosis.18 Moreover, the verification of ferroptosis should generally rule out apoptosis and necroptosis.18 However, in the process of a specific disease, different types of cell death are usually involved at the same time. Hence, in vitro and in vivo experiments were designed to verify that Fer-1, DFO and Nec-1 to rescue OA process through different pathways. The results showed that non-necroptosis cell death type existed in OA development, thus ruling out the necroptosis mechanism. Collectively, ferroptosis was experimentally and logically proved to be involved in the OA pathogenic process in this study.
At present, there are still many unsolved problems in the field of ferroptosis research. First, there is still no golden standard for the detection of ferroptosis, especially in the disease. Therefore, we can only detect the indicators related to ferroptosis, as much as possible, and exclude other types of cell death to verify the existence of ferroptosis in the OA process. Second, the interaction and conversion between ferroptosis and other types of cell death is still a mystery. Different types of cell death usually share similar initial signals and molecular regulators. GPX4 is also involved in apoptosis15 and necroptosis16 in various injuries. The inhibition of ferroptosis may cause cells to engage in different lethal subroutines (instead of avoiding cell death as such).19 This may be the reason that there are no successful clinical trials using any single cell-death inhibitors to treat OA. It also comes to the question if we extend the experimental duration in vivo could still inhibit the OA progression when Fer-1, DFO or Nec-1 is used for OA treatment?
As the key regulator of ferroptosis, the expression of GPX4 is closely related with selenium. Pharmacological selenium augments GPX4 expression at the transcriptional level.46 Meanwhile, selenium deficiency impaired the growth of bone and cartilage tissue.41 However, the relation between GPX4 and OA has never been reported. According to the previous studies, GPX4 knockout could directly induce ferroptosis.20,22 However, in the OA process, the reduction of GPX4 expression may not induce ferroptosis directly. Thus, we evaluated GPX4 expression level in chondrocytes of OA cartilage. The results showed that about 50% of GPX4 knockdown did not directly induce cell death, but could increase the sensibility to oxidative stress. Meanwhile, its substrate GSH was increased, which was opposite to the results from OA cartilage (Figure 1d). Ferroptosis-inducing agents (FINs) have been classified into class I FINs that involve cellular GSH depletion, and class II FINs which trigger ferroptosis through inhibition of GPX4.20,22,47 These evidences suggested that ferroptosis in OA is similar to the ferroptosis induced by class I FINs, which involves GSH depletion through inhibiting system Xc−. In the meantime, expression of SLC3A2, the component of system Xc−, was also found to be downregulated in OA cartilage from GEO database, providing additional evidence to support our conclusion. Importantly, we discovered a previously unappreciated role of GPX4 in cartilage metabolism, in addition to the anticipation in ferroptosis. In the process of OA, knockdown of GPX4 could directly trigger ECM degradation through MAPK/NFκB signaling pathway. Thus, GPX4 was the intersection of two mechanisms in accelerating OA: ferroptosis and ECM degradation (Figure 6h). Further mechanistic studies are required to validate therapeutic potential of ferroptosis inhibitors and GPX4 inducers in OA in the future.
In summary, we provide comprehensive evidence showing that ferroptosis was involved in OA pathogenesis and development. Remarkably, our results indicated dual function of GPX4 in OA, (i) regulating ferroptosis or oxidative stress; (ii) regulating ECM degradation through MAPK/NFκB signaling pathway. With the development of detection techniques and methods, more patterns of cell death may be found in the future. It is pivotal to know whether one type of cell death occurs in the process of the disease. The current research laid a solid foundation for the future study of ferroptosis in OA and is meaningful for OA treatment strategy.
Data sharing statement
All reagents used in this work are available upon request and a brief statement describing the purpose for their use. RNA-Seq raw data reported in this paper have been deposited in the Gene Expression Omnibus database under accession number GSE190184.
Contributors
YM designed and conducted the in vitro and in vivo experiments, analyzed the data and wrote the manuscript. YC conducted some of the in vitro and in vivo experiments, MicroCT analysis and IHC analysis. KL and FX obtained and analyzed the data from the human OA patients. BZ and JY conducted in vivo experiments. JG and MZ conceived the study. CZ and GL designed and conducted the research, wrote the manuscript and directed the project. YM and GL have read and verified the underlying data. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by the Projects of International Cooperation and Exchanges of National Natural Science Funding of China (Grant no. 81820108020), the National Key Research and Development Project of China (Grant no. 2018YFC1106300) and the Basic Science Program of Shanghai Jiao Tong University Affiliated Sixth People's Hospital (Grant no. ynms202102).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103847.
Contributor Information
Changqing Zhang, Email: zhangcq@sjtu.edu.cn.
Guangyi Li, Email: guangyi.li@shsmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Glyn-Jones S., Palmer A.J., Agricola R., et al. Osteoarthritis. Lancet. 2015;386(9991):376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 2.Hunter D.J., Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 3.Kim J.H., Jeon J., Shin M., et al. Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell. 2014;156(4):730–743. doi: 10.1016/j.cell.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Chen D., Shen J., Zhao W., et al. Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 2017;5:16044. doi: 10.1038/boneres.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little C.B., Hunter D.J. Post-traumatic osteoarthritis: from mouse models to clinical trials. Nat Rev Rheumatol. 2013;9(8):485–497. doi: 10.1038/nrrheum.2013.72. [DOI] [PubMed] [Google Scholar]
- 6.Loeser R.F., Goldring S.R., Scanzello C.R., Goldring M.B. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheumatol. 2012;64(6):1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Husa M., Liu-Bryan R., Terkeltaub R. Shifting HIFs in osteoarthritis. Nat Med. 2010;16(6):641–644. doi: 10.1038/nm0610-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin A.C., Seeto B.L., Bartoszko J.M., et al. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med. 2009;15(12):1421–1425. doi: 10.1038/nm.2055. [DOI] [PubMed] [Google Scholar]
- 9.Carballo C.B., Nakagawa Y., Sekiya I., Rodeo SA. Basic science of articular cartilage. Clin Sports Med. 2017;36(3):413–425. doi: 10.1016/j.csm.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Charlier E., Relic B., Deroyer C., et al. Insights on molecular mechanisms of chondrocytes death in osteoarthritis. Int J Mol Sci. 2016;17(12) doi: 10.3390/ijms17122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu J.H., Shin Y., Huh Y.H., Yang S., Chun C.H., Chun J.S. Hypoxia-inducible factor-2alpha regulates Fas-mediated chondrocyte apoptosis during osteoarthritic cartilage destruction. Cell Death Differ. 2012;19(3):440–450. doi: 10.1038/cdd.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehana E.E., Khafaga A.F., El-Blehi SS. The role of matrix metalloproteinases in osteoarthritis pathogenesis: an updated review. Life Sci. 2019;234 doi: 10.1016/j.lfs.2019.116786. [DOI] [PubMed] [Google Scholar]
- 13.Jeon J., Noh H.J., Lee H., et al. TRIM24-RIP3 axis perturbation accelerates osteoarthritis pathogenesis. Ann Rheum Dis. 2020;79(12):1635–1643. doi: 10.1136/annrheumdis-2020-217904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park D.R., Kim J., Kim G.M., et al. Osteoclast-associated receptor blockade prevents articular cartilage destruction via chondrocyte apoptosis regulation. Nat Commun. 2020;11(1):4343. doi: 10.1038/s41467-020-18208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ran Q., Liang H., Gu M., et al. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J Biol Chem. 2004;279(53):55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 16.Canli O., Alankus Y.B., Grootjans S., et al. Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood. 2016;127(1):139–148. doi: 10.1182/blood-2015-06-654194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H., Wen Y., Zhang M., et al. MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint. Autophagy. 2020;16(2):271–288. doi: 10.1080/15548627.2019.1606647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockwell B.R., Friedmann Angeli J.P., Bayir H., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang D., Chen X., Kang R., Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31(2):107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedmann Angeli J.P., Schneider M., Proneth B., et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon S.J., Lemberg K.M., Lamprecht M.R., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W.S., SriRamaratnam R., Welsch M.E., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1-2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H., Zandkarimi F., Zhang Y., et al. Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat Cell Biol. 2020;22(2):225–234. doi: 10.1038/s41556-020-0461-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y., Hou W., Song X., et al. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derry P.J., Hegde M.L., Jackson G.R., et al. Revisiting the intersection of amyloid, pathologically modified tau and iron in Alzheimer's disease from a ferroptosis perspective. Prog Neurobiol. 2020;184 doi: 10.1016/j.pneurobio.2019.101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahoney-Sanchez L., Bouchaoui H., Ayton S., Devos D., Duce J.A., Devedjian JC. Ferroptosis and its potential role in the physiopathology of Parkinson's Disease. Prog Neurobiol. 2021;196 doi: 10.1016/j.pneurobio.2020.101890. [DOI] [PubMed] [Google Scholar]
- 27.Tuo Q.Z., Lei P., Jackman K.A., et al. Tau-mediated iron export prevents ferroptotic damage after ischemic stroke. Mol Psychiatry. 2017;22(11):1520–1530. doi: 10.1038/mp.2017.171. [DOI] [PubMed] [Google Scholar]
- 28.Burton L.H., Radakovich L.B., Marolf A.J., Santangelo KS. Systemic iron overload exacerbates osteoarthritis in the strain 13 guinea pig. Osteoarthr Cartil. 2020;28(9):1265–1275. doi: 10.1016/j.joca.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simao M., Gavaia P.J., Camacho A., et al. Intracellular iron uptake is favored in Hfe-KO mouse primary chondrocytes mimicking an osteoarthritis-related phenotype. Biofactors. 2019;45(4):583–597. doi: 10.1002/biof.1520. [DOI] [PubMed] [Google Scholar]
- 30.Jing X., Lin J., Du T., et al. Iron overload is associated with accelerated progression of osteoarthritis: the role of DMT1 mediated iron homeostasis. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.594509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao X., Sun K., Yu S., et al. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J Orthop Translat. 2021;27:33–43. doi: 10.1016/j.jot.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remmele W., Schicketanz K.H. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Computer-assisted image analysis (QIC score) vs. subjective grading (IRS) Pathol Res Pract. 1993;189(8):862–866. doi: 10.1016/S0344-0338(11)81095-2. [DOI] [PubMed] [Google Scholar]
- 33.Ji Q., Xu X., Kang L., et al. Hematopoietic PBX-interacting protein mediates cartilage degeneration during the pathogenesis of osteoarthritis. Nat Commun. 2019;10(1):313. doi: 10.1038/s41467-018-08277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C., Williams B.A., Pertea G., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S., Pyl P.T., Huber W. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Araki M., Goto S., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun K., Luo J., Jing X., et al. Astaxanthin protects against osteoarthritis via Nrf2: a guardian of cartilage homeostasis. Aging (Albany NY) 2019;11(22):10513–10531. doi: 10.18632/aging.102474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren F.L., Guo X., Zhang R.J., et al. Effects of selenium and iodine deficiency on bone, cartilage growth plate and chondrocyte differentiation in two generations of rats. Osteoarthr Cartil. 2007;15(10):1171–1177. doi: 10.1016/j.joca.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Ursini F., Maiorino M. Lipid peroxidation and ferroptosis: The role of GSH and GPx4. Free Radic Biol Med. 2020;152:175–185. doi: 10.1016/j.freeradbiomed.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Zhang B., Liu W.X., et al. Metformin limits osteoarthritis development and progression through activation of AMPK signalling. Ann Rheum Dis. 2020;79(5):635–645. doi: 10.1136/annrheumdis-2019-216713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galluzzi L., Vitale I., Aaronson S.A., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanden Berghe T., Linkermann A., Jouan-Lanhouet S., Walczak H., Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15(2):135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 46.Alim I., Caulfield J.T., Chen Y., et al. Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell. 2019;177(5):1262–1279. doi: 10.1016/j.cell.2019.03.032. e25. [DOI] [PubMed] [Google Scholar]
- 47.Dixon S.J., Stockwell B.R. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10(1):9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.