ABSTRACT
Rickettsia species (spp.) are strict obligate intracellular bacteria, some of which are pathogenic in their mammalian host, including humans. One critical feature of these stealthy group of pathogens is their ability to manipulate hostile cytosolic environments to their benefits. Although our understanding of Rickettsia cell biology and pathogenesis is evolving, the mechanisms by which pathogenic Rickettsia spp. evade host innate immune detection remain elusive. Here, we show that disease severity in wild-type (WT) C57BL/6J mice infected with Rickettsia typhi (the etiologic agent of murine typhus) and Rickettsia rickettsii (the etiologic agent of Rocky Mountain spotted fever), but not with the nonpathogenic species Rickettsia montanensis, correlated with levels of bacterial burden as detected in the spleens of mice, as well as the serum concentrations of proinflammatory cytokine interleukin-1α (IL-1α) and, to a lesser extent, IL-1β. Antibody-mediated neutralization of IL-1α confirmed a key role in controlling mortality rates and bacterial burdens of rickettsia-infected WT mice. As macrophages are a primary source of both IL-1α and IL-1β cytokines, we determined the mechanism of the antirickettsial activities using bone marrow-derived macrophages. We found that pathogenic R. typhi and R. rickettsii, but not nonpathogenic R. montanensis, eluded pro-IL-1α induction and benefited predominantly from the reduced IL-1α secretion, via a caspase-11–gasdermin D (Gsdmd)-dependent pathway, to facilitate intracytosolic replication. Adoptive transfer experiments identified that IL-1α secretion by macrophages was critical for controlling rickettsiosis in WT mice. In sum, we identified a previously unappreciated pathway by which pathogenic, unlike nonpathogenic, rickettsiae preferentially target the caspase-11–Gsdmd–IL-1α signaling axis in macrophages, thus supporting their replication within the host.
KEYWORDS: R. typhi, R. rickettsii Sheila Smith, R. montanensis, IL-1α, IL-1β, caspase-1, caspase-11, macrophages, inflammasomes
INTRODUCTION
Invasive cytosolic bacteria, including Listeria, Shigella, Burkholderia, Francisella, Orientia, and Rickettsia species, have developed strategies to induce their own uptake by phagocytosis and to circumvent host innate immune defenses for their intracellular survival (1, 2). Human infection with Rickettsia spp. occurs via infected hematophagous arthropods such as fleas, ticks, and human body lice (3), either through the bite or deposited as infected feces on skin and mucosal surfaces. Upon entry, Rickettsia spp. encounter tissue-resident immune cells, like macrophages (Mϕ). Activated Mϕ play a crucial role in either terminating an infection at an early stage, which commonly is the fate of nonpathogenic Rickettsia spp., or succumbing to bacterial replication and pathogen colonization as well as host dissemination to distant organs (3). After internalization into host cells, Rickettsia spp. escape from phagosomes and subvert host cytosolic defense mechanisms (i.e., autophagy and inflammasomes) to establish an intracytosolic replication niche. Recently, we reported that pathogenic Rickettsia spp. secret effectors to promote host colonization by modulating endoplasmic reticulum structures or by hijacking the autophagic defense pathway (4–9). Subversion of autolysosomal destruction to colonize the host cytosol exposes Rickettsia spp. to another cytosolic host sensor-regulated defense mechanism, the inflammasomes (1, 10). Inflammasomes are immune signaling complexes categorized into canonical (caspase-1 [Casp-1]) and noncanonical (murine Casp-11 or human Casp-4/5) inflammasomes. The inflammasome complex assembly involves the adaptor protein ASC and upstream sensors, including NLRP1, NLRP3, NLRC4, AIM2, and pyrin, which are primed by exogenous pathogen-associated molecular pattern molecules (PAMPs) and activated through endogenous damage-associated molecular pattern molecules (DAMPs). Initiation of the canonical inflammasome results in the activation of Casp-1. Active Casp-1 leads to the maturation of proinflammatory cytokines interleukin-1β (IL-1β) and IL-18 and the activation of gasdermin D (Gsdmd), the executor of pyroptosis (11, 12). Of note, recent findings have also suggested that active Casp-11 is capable of activating Gsdmd (11, 12). Although IL-1β is released by both canonical and noncanonical inflammasome pathways, IL-1α is preferentially released by the noncanonical inflammasome pathway (13–15). IL-1α is expressed by a wide range of hematopoietic and nonhematopoietic cell types, whereas IL-1β is primarily produced by myeloid cells (15). Importantly, although IL-1α and IL-1β signal through the same receptor, IL-1R, these two cytokines are not completely functionally redundant (15). Given the importance for both canonical and noncanonical inflammasome-mediated IL-1 signaling in limiting pathogen colonization, many bacteria have evolved strategies to block their activation (16–21). In fact, various pathogenic intracellular bacteria utilize their own effector repertoire to evade these pathways to successfully colonize and disseminate in their host cells (10, 22).
In the case of Rickettsia, our understanding of the role of inflammasomes in controlling host colonization is only now emerging (23–25). Specifically, how pathogenic Rickettsia spp. manipulate immune defenses to replicate within the host cytosol not only relies almost exclusively on data from tick-transmitted rickettsiae (e.g., members of the spotted fever group [SFG] or transition group [TRG]), but also shows that these pathogens likely employ species-specific strategies to evade host cytosolic defense mechanisms. For instance, Rickettsia australis, a pathogenic TRG member, benefited from ATG5-mediated autophagy induction and suppression of inflammasome-dependent IL-1β production to colonize both bone marrow-derived macrophages (BMDMs) (23) and mice (26). In contrast, Rickettsia parkeri, a mildly pathogenic member of SFG (see Fig. 1A below), utilizes its surface cell antigen Sca5 (OmpB) for protection against autophagic recognition and consequently benefits from inflammasome-mediated host cell death that antagonizes the action of type I interferon (IFN) in BMDMs and mice (24, 27). In contrast, our recent report on the flea-transmitted Rickettsia typhi (a pathogenic member of the typhus group [TG]), showed that R. typhi is ubiquitinated upon host entry and escapes autolysosomal fusion to establish an intracytosolic niche in nonphagocytic cells (9). These unexpected findings on how members of SFG, TRG, and TG Rickettsia differentially promote intracytosolic host survival prompted us to explore the underlying mechanism(s) by which pathogenic, but not nonpathogenic, Rickettsia spp. block immune defense responses to establish a replication niche in phagocytic cells, like Mϕ. Specifically, we sought to test the hypothesis that pathogenic, but not nonpathogenic, Rickettsia spp. reduce inflammasome-mediated IL-1 responses, thereby promoting their intracytosolic replication within host cells.
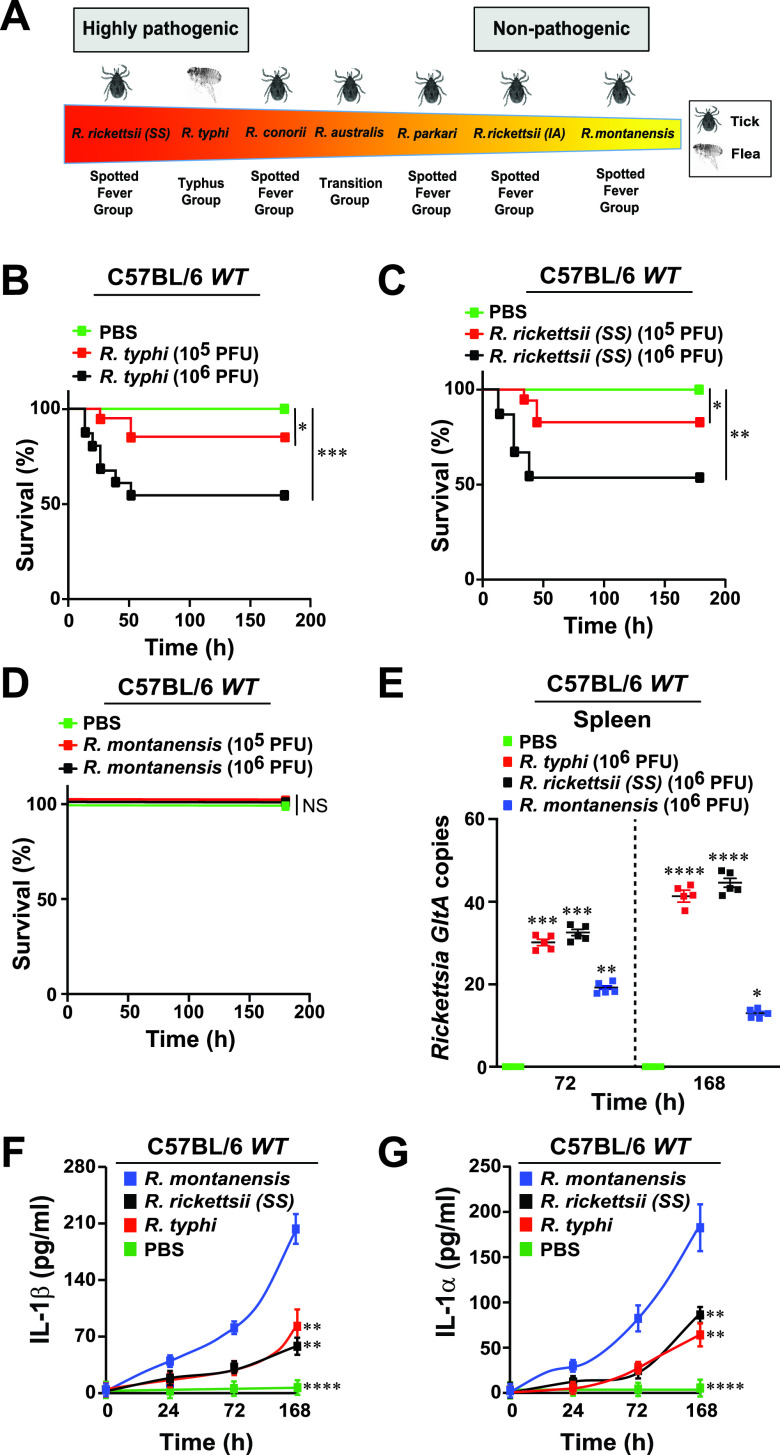
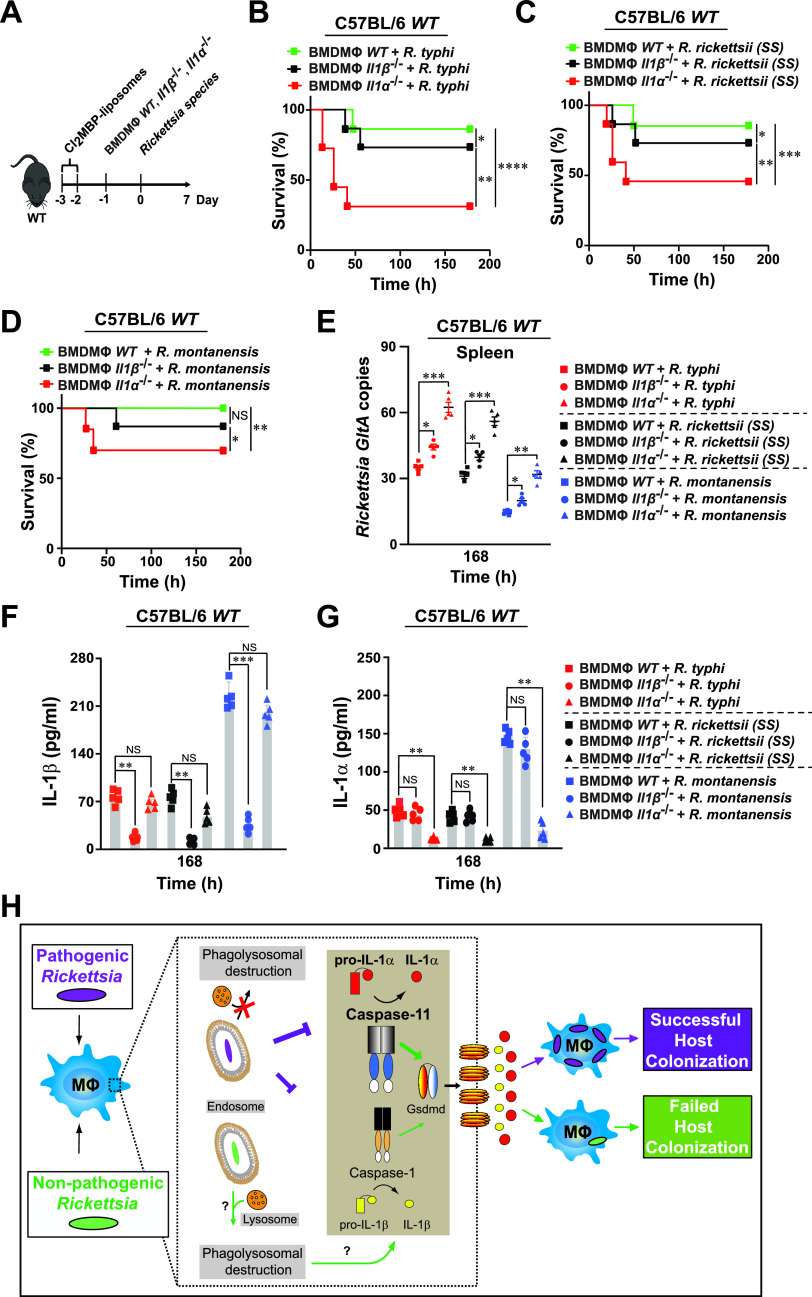
FIG 1.
In vivo models of rickettsiosis. (A) Rickettsia spp. and their level of pathogenicity to humans (3, 44). (B to D) Establishment of a model of rickettsiosis (LD25 and LD50) for R. typhi (B), R. rickettsii (C) or R. montanensis (D) in C57BL/6J WT mice. Animals were injected via tail vein (i.v.) with different doses (105 to 106 PFU) of R. typhi, R. rickettsii, R. montanensis, or PBS (n = 12 for each treatment). Survival was monitored for 7 days. (E) Bacterial burden was tested in spleens of R. typhi-, R. rickettsii-, R. montanensis-, or PBS-injected WT mice (n = 5 per each treatment) shown in panels B to D by GltA RT-qPCR at days 3 and 7 (n = 5 for each treatment) using the host housekeeping gene Gapdh for normalization. (F and G) Serum samples from Rickettsia-infected mice described in panels B to D were analyzed for IL-1β (F) and IL-1α (G) production using the Legendplex kits (BioLegend), followed by flow cytometry analysis. Error bars in panels E to G represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001.
RESULTS
In vivo models of rickettsiosis.
At present, more than 30 Rickettsia spp. have been described globally, but less than a dozen is known to cause disease in humans, with some being notoriously pathogenic and associated with high morbidity and mortality, while others exert limited or no pathogenicity (Fig. 1A). We sought to test our hypothesis that pathogenic, but not nonpathogenic, Rickettsia spp. evade immune responses in host defense cells, like Mϕ, to replicate and disseminate. We simultaneously evaluated the cytosolic host defense responses between pathogenic (R. rickettsii strain Sheila Smith and R. typhi Wilmington) and nonpathogenic (Rickettsia montanensis) strains in vivo. We first established a mouse model of mild (approximate 25% lethal dose [∼LD25]) or more severe (∼LD50) rickettsiosis in C57BL/6J wild-type (WT) mice. For both R. rickettsii and R. typhi, LD25 or LD50 were achieved with doses of 105 or 106 PFU, respectively (Fig. 1B and C); however, R. montanensis-infected mice showed no signs of lethality at either 105 or 106 PFU (Fig. 1D). Bacterial burdens in spleens from infected C57BL/6J WT mice (only the dose of 106 PFU is shown) confirmed successful infection with all three Rickettsia spp. at day 3 postinfection, while R. typhi- and R. rickettsii-infected WT mice displayed a significantly higher bacterial burden in the spleens compared to splenic tissues from R. montanensis-infected mice at day 7 (Fig. 1E). This correlated with the observed differences in the spleen sizes and weights of the infected animals (see Fig. S1 in the supplemental material). Given the earlier findings from other laboratories and ours (9, 23–28), we hypothesized that the observed dissimilarities in pathogenicity among Rickettsia spp. are likely linked to differences in host defense responses. Recent findings further suggest that the intracytosolic survival of different Rickettsia spp. is either supported or suppressed by immune defense responses (e.g., IFN-I, tumor necrosis factor alpha [TNF-α], or IL-1β) (23–28), thus leaving the precise mechanism to be determined. Therefore, we first sought to evaluate immune defense responses at the level of IL-1β and IL-1α cytokine secretion in the sera of R. typhi-, R. rickettsii-, and R. montanensis-infected animals. The increase in mortality and elevated bacterial burden correlated with reduced serum levels of both IL-1β and IL-1α cytokines (Fig. 1F and G), suggesting that reduced activation of both IL-1 signaling responses is a potential mechanism for lethality and survival of pathogenic Rickettsia spp.
Splenic data during infection of pathogenic and nonpathogenic Rickettsia species. (A) Representative images of spleens (at day 7) from C57BL/6J WT mice injected i.v. with R. typhi, R. rickettsii, R. montanensis, or PBS (dose of 106 PFU). (B) Spleen weight from injected animals was evaluated at day 7 day (n = 5). Error bars in panel B represent means ± SEM from five independent experiments. *, P ≤ 0.05; ***, P ≤ 0.005. Download FIG S1, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
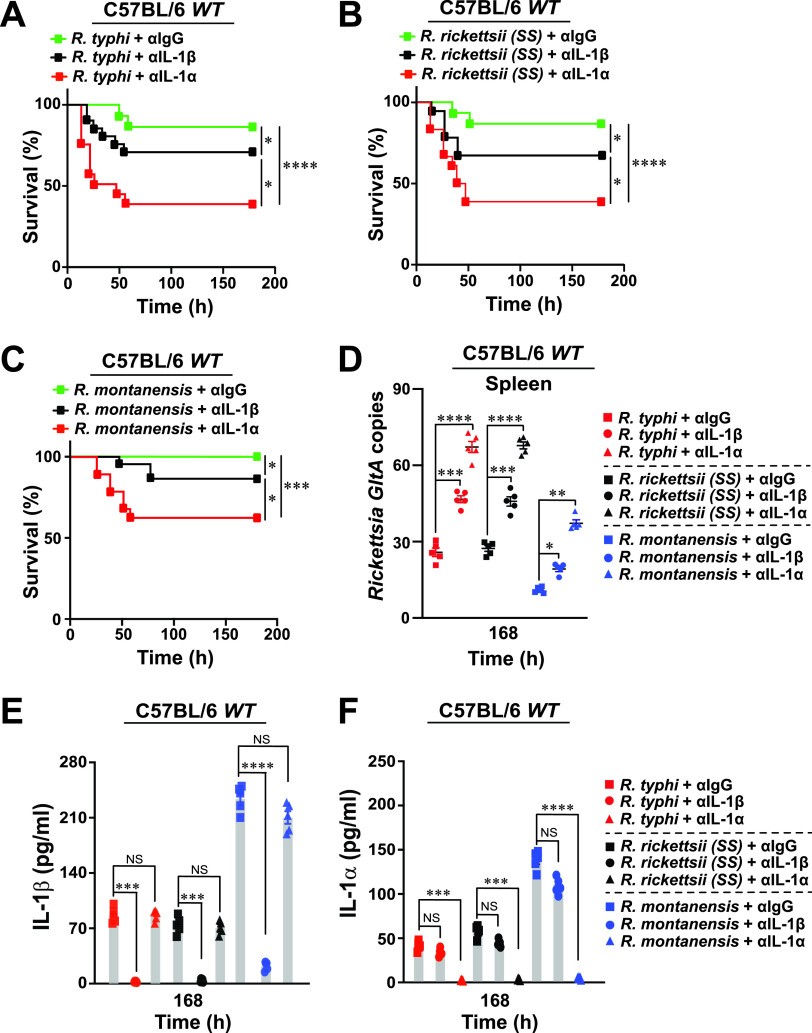
Antirickettsial activity of IL-1α is involved in restricting Rickettsia infection.
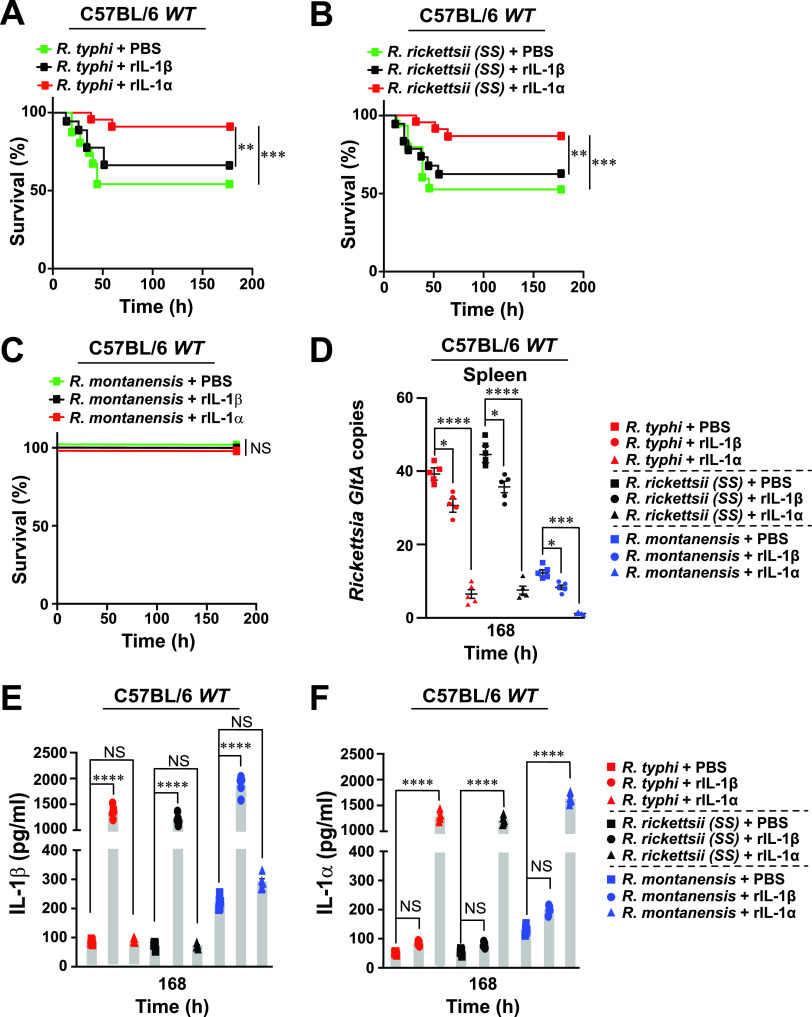
To characterize further the role of IL-1α and IL-1β cytokines in restricting nonpathogenic and pathogenic Rickettsia spp. in vivo, IL-1α or IL-1β function was neutralized via tail vein (intravenous [i.v.]) injection with anti-IL-1α, anti-IL-1β, or anti-IgG-isotype control antibodies (Abs) into C57BL/6J WT mice in our established model of mild rickettsiosis (LD25; ∼105 PFU) (Fig. 1B to D). Neutralization of IL-1α, and to a much lesser extent IL-1β, was associated with a significant increase in the mortality of R. typhi-, R. rickettsii-, and R. montanensis-infected mice (Fig. 2A to C) and resulted in the development of splenomegaly (see Fig. S2 in the supplemental material), which correlated with an increase in bacterial loads in the spleen (Fig. 2D). The efficiency of Ab-mediated blocking was confirmed by measuring the levels of IL-1β and IL-1α cytokine in the sera of the rickettsia-infected mice (Fig. 2E and F). Next, we sought to determine the effect of administering recombinant IL-1α (rIL-1α) or rIL-1β proteins on Rickettsia colonization in vivo. Accordingly, we administered (i.v.) endotoxin-free rIL-1α and rIL-1β proteins following infection with R. typhi, R. rickettsii, and R. montanensis. Pretreatment of mice with rIL-1α and, to a lesser extent, rIL-1β protected C57BL/6J WT mice from pathogenic Rickettsia-induced lethality (Fig. 3A to C), with both reduced splenomegaly (see Fig. S3 in the supplemental material) and decreased splenic bacterial burdens (Fig. 3D). Moreover, the observed phenotypes correlated with the measured IL-1α and IL-1β serum concentrations (Fig. 3E and F). Collectively, these findings suggest that IL-1α and, to a lesser extent, IL-1β are involved in restricting the replication and colonization of nonpathogenic and pathogenic Rickettsia spp. in C57BL/6J WT mice.
FIG 2.
Neutralization of IL-1α activity augments mortality of pathogenic and nonpathogenic Rickettsia-induced rickettsiosis. (A to C) C57BL/6J WT mice were injected via tail vein (i.v.) with 105 PFU of R. typhi (A), R. rickettsii (B), or R. montanensis (C) (A to C; n = 12 for each treatment), followed by a subsequent injection (i.v.) with anti-IL-1β, anti-IL-1α, or anti-IgG isotype control antibody (Ab) (250 μg Ab/mouse). Survival was monitored for 7 days. (D) Bacterial burden was tested in the spleens of the Ab-treated R. typhi-, R. rickettsii-, and R. montanensis-injected WT mice shown in panels A to C by GltA RT-qPCR at day 7 (n = 5 for each treatment) using the host housekeeping gene Gapdh for normalization. (E and F) Serum samples from mice described in panels A to C were analyzed for IL-1β (E) and IL-1α (F) production at day 7 (n = 5 for each treatment) using the Legendplex kits (BioLegend), followed by flow cytometry analysis. Error bars in panels D to F represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001.
FIG 3.
Administration of recombinant IL-1α rescues fatal Rickettsia-induced rickettsiosis. (A to C) C57BL/6J WT mice were injected i.v. with rIL-1β or rIL-1α protein (500 ng/mouse), followed by infection (24 h post-protein injection) with 106 PFU of R. typhi (A), R. rickettsii (B), R. montanensis (C), or PBS (A to C; n = 15 for each treatment). Survival was monitored for 7 days. (D) Bacterial burden was tested in the spleens of the protein-treated R. typhi-, R. rickettsii-, R. montanensis-, or PBS-injected WT mice shown in panels A to C by GltA RT-qPCR at day 7 (n = 5 for each treatment), using the housekeeping gene Gapdh for normalization. (E and F) Serum samples from the mice described in panels A to C were analyzed for IL-1β (E) and IL-1α (F) production at day 7 (n = 5 for each treatment) using the Legendplex kits (BioLegend), followed by flow cytometry analysis. Error bars in panels D to F represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001.
Splenic data after IL-1 signaling neutralization during infection of pathogenic and nonpathogenic Rickettsia species. (A) Representative images of spleens (day 7) from C57BL/6J WT mice injected i.v. with R. typhi, R. rickettsii, or R. montanensis (dose of 105 PFU), followed by administration of anti-IL-1β, anti-IL-1α, or IgG isotype control antibody (Ab) (250 μg Ab/mouse) (24 h post-Ab injection). (B) Spleen weight from injected animals was evaluated at day 7 (n = 5). Error bars in panel B represent means ± SEM from five independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. Download FIG S2, EPS file, 3.2 MB (3.2MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Splenic data after administration of recombinant IL-1α and IL-1β proteins during infection of pathogenic and nonpathogenic Rickettsia species. (A) Representative images of spleens (day 7) from C57BL/6J WT mice injected i.v. with rIL-1β or rIL-1α protein, followed by administration of R. typhi, R. rickettsii, R. montanensis, or PBS (dose of 106 PFU) at day 7. (B) Spleen weight from injected animals was evaluated at day 7 day (n = 5). Error bars in panel B represent means ± SEM from five independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. Download FIG S3, EPS file, 4.0 MB (4MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pathogenic, but not nonpathogenic, Rickettsia species block IL-1α secretion and avoid pro-IL-1α induction to establish a replication niche in macrophages.
As Mϕ are one of the cell types first encountered during infection by Rickettsia spp. and are considered to play a crucial role in either terminating the infection early at the skin site or allow initial pathogen colonization and subsequent dissemination within the infected host (3), we tested the hypothesis that pathogenic, but not nonpathogenic, rickettsiae suppress Mϕ immune responses. In this effort, we determine the importance of IL-1α and IL-1β in restricting the replication of Rickettsia spp. by infecting BMDMs derived from WT, Il-1β−/−, or Il-1α−/− mice with pathogenic R. typhi and R. rickettsii or nonpathogenic R. montanensis and assessed the cytokine levels of IL-1β and IL-1α in cultured supernatants as well as bacterial burdens. Infection of Il-1β−/− or Il-1α−/− BMDMs with Rickettsia spp. did not result in the secretion of either IL-1β or IL-1α, respectively, compared to infected WT BMDMs (Fig. 4A to F). Moreover, infections with R. montanensis resulted in overall higher IL-1 responses and lower bacterial burden compared to infections performed with R. typhi or R. rickettsii in WT, Il-1β−/−, or Il-1α−/− BMDMs (Fig. 4A to G). Of note, the bacterial burden of all three Rickettsia spp. in infected Il-1α−/− BMDMs was higher than the levels detected in Il-1β−/− or WT BMDMs (Fig. 4G), suggesting that IL-1α and, to significantly lesser extent, IL-1β play a role in restricting rickettsia survival. In agreement with our in vivo data, infection assays using BMDMs also displayed an overall higher bacterial burden of both pathogenic Rickettsia spp., compared to the nonpathogenic Rickettsia strain (Fig. 1E and Fig. 4G). We further tested the protein expression levels of pro-IL-1β and pro-IL-1α upon bacterial-infection of WT BMDMs and showed that pro-IL-1β levels were induced by all three Rickettsia spp. to similar levels (∼5-fold) compared to uninfected WT BMDMs (Fig. 4H). Intriguingly, only R. montanensis-infected WT BMDMs produced significantly higher levels of pro-IL-1α than R. typhi- or R. rickettsii-infected Mϕ (Fig. 4H).
FIG 4.
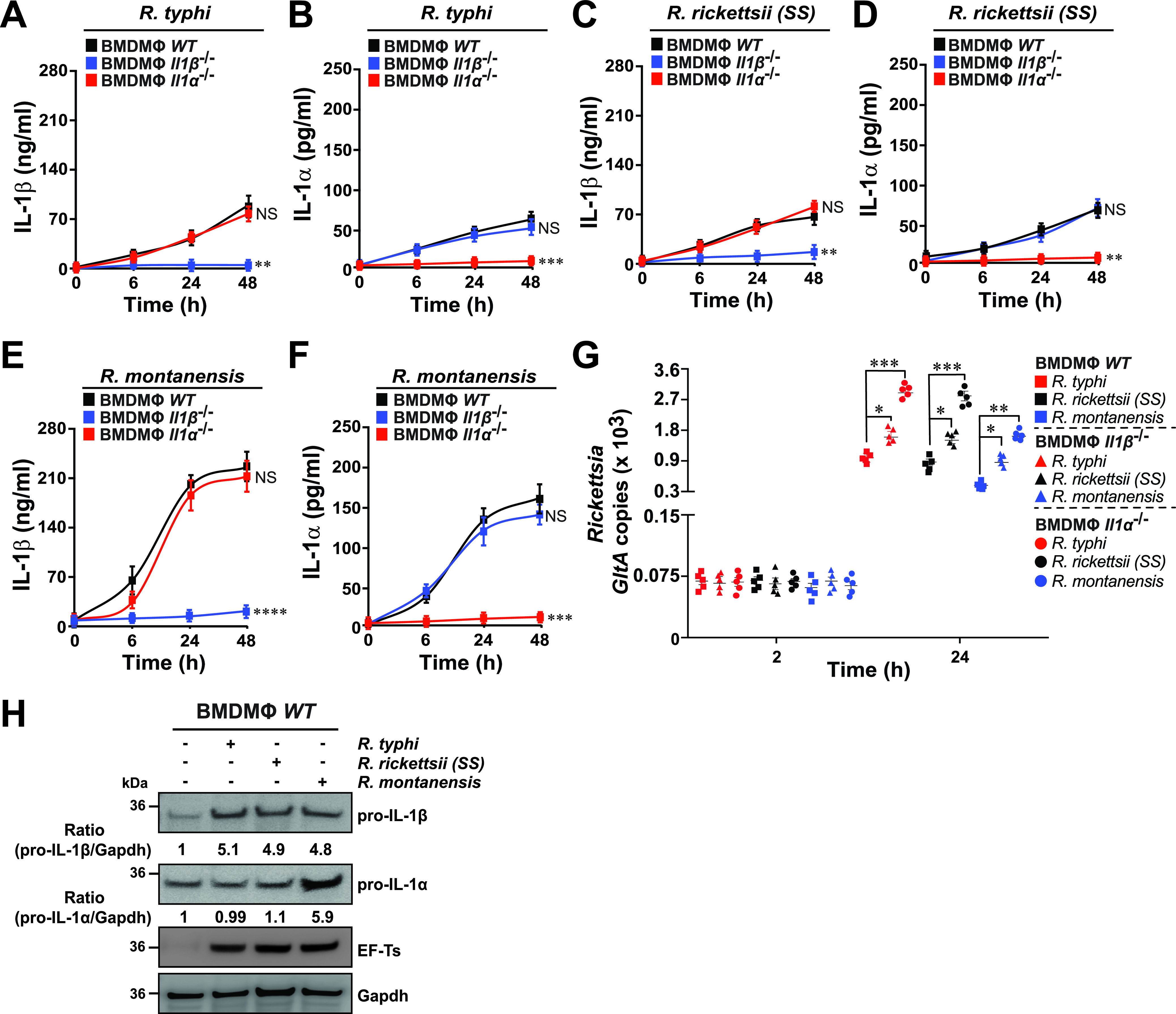
IL-1α, but not IL-1β, contributes to the survival of Rickettsia species in macrophages. (A to F) BMDMs from WT, Il-1β−/−, or Il-1α−/− mice were infected with R. typhi, R. rickettsii, or R. montanensis (MOI = 50) for 0, 6, 24, and 48 h. Culture supernatants were analyzed for production of IL-1β (A, C, and E) and IL-1α (B, D, and F) using Legendplex kits (BioLegend), followed by flow cytometry. (G) Bacterial burden in Rickettsia-infected BMDMs from WT, Il-1β−/−, or Il-1α−/− mice was evaluated at 2 and 24 h postinfection by GltA RT-qPCR. Expression of the housekeeping gene Gapdh was used for normalization. (H) BMDMs from WT mice were either left uninfected (−) or were infected with R. typhi, R. rickettsii, or R. montanensis (MOI = 50) for 24 h. Lysates were immunoblotted with anti-IL-1α, anti-IL-1β, anti-ET-Ts, and anti-Gapdh Abs. Densitometry was performed using Fiji software, and data representing the fold change ratios of pro-IL-1β/Gapdh or pro-IL-1α/Gapdh between uninfected and infected cells are shown. Immunoblot data are representative of three independent experiments. Error bars in panels A to G represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001.
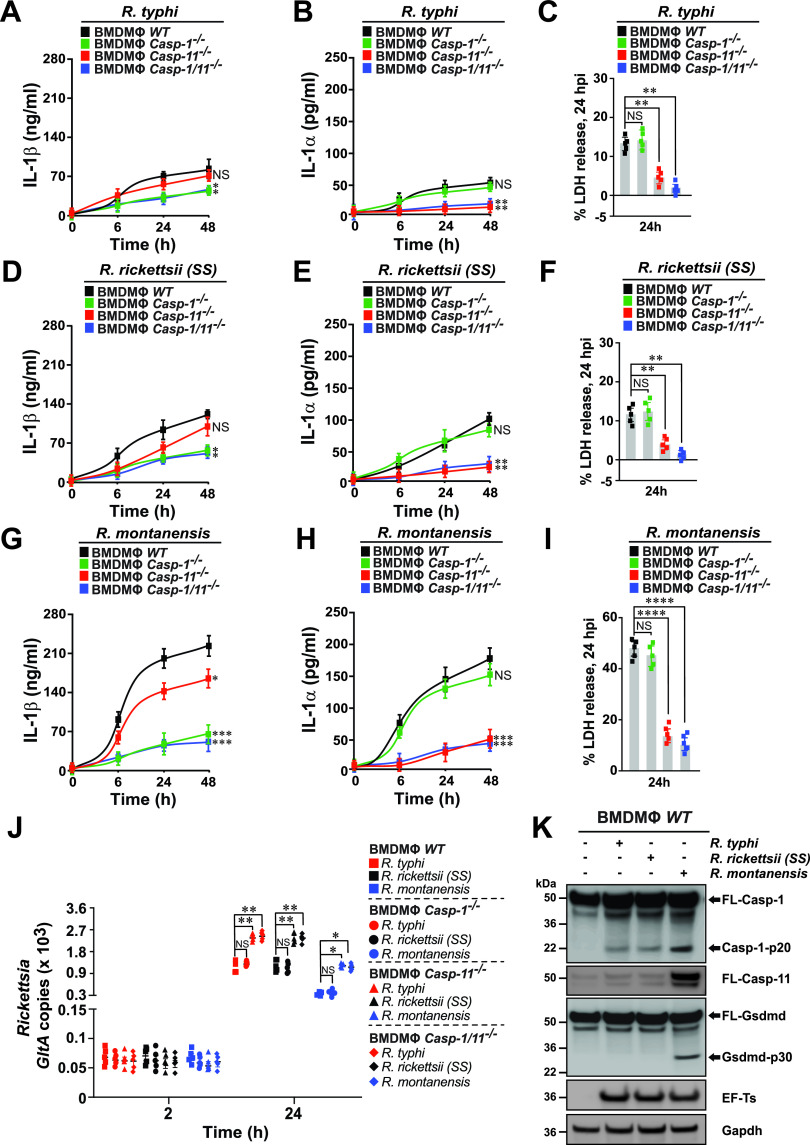
Intracytosolic replication of pathogenic Rickettsia species in macrophages depends on the inhibition of IL-1 cytokine secretion via a caspase-11–Gsdmd-dependent pathway.
As our findings suggest that pathogenic, but not nonpathogenic, Rickettsia spp. prevent the activation of signaling pathways required for IL-1α and IL-1β production and release, we explored the mechanism of IL-1 signaling in greater detail. As IL-1 signaling responses commonly involve the canonical and noncanonical inflammasome pathways, which in turn involves the proteolytic processing of both cytokines by activated caspase-1 (Casp-1 [canonical]) and Casp-11 [noncanonical]), respectively, we assessed their potential role in regulating replication of nonpathogenic versus pathogenic Rickettsia in BMDMs. In this effort, BMDMs derived from WT, Casp-1−/−, Casp-11−/−, and Casp-1/11−/− mice were infected with R. typhi, R. rickettsii, and R. montanensis, and the levels of IL-1β and IL-1α and cell death, as well as bacterial burdens, were evaluated over the course of infection. Our assays revealed that Casp-11 was involved in the secretion of cleaved IL-1α upon infection of BMDMs with nonpathogenic and pathogenic Rickettsia spp. (Fig. 5A, B, D, E, G, and H). In addition, Casp-11 deficiency resulted in a significant decrease in host cell death (Fig. 5C, F, and I). Casp-1 deficiency (Casp-1−/−) caused a significant decrease in IL-1β production (Fig. 5A, D, and G, green lines), while IL-1α secretion as well as the level of cell death remained unaffected during infection with all three Rickettsia spp. (Fig. 5B, C, E, F, H, and I, green lines and symbols). Analysis of bacterial burdens further revealed a prominent role for Casp-11, but not for Casp-1, in restricting the replication of both pathogenic and nonpathogenic Rickettsia spp. (Fig. 5J; WT [squares], Casp-1−/− [circles], Casp-11−/− [triangles], and Casp-1/11−/− [diamonds]). As our findings indicate that infections with R. typhi and R. rickettsii resulted in a significant reduction of IL-1α secretion, likely via a Casp-11-dependent mechanism, we assessed the expression and activation status of Casp-1 and Casp-11 via Western blot analyses. R. montanensis infection resulted in a robust activation of Casp-1, as indicative of the detection of the Casp-1–p20 fragment (Fig. 5K). In contrast, infection with R. typhi and R. rickettsii spp. resulted in a lower activation of Casp-1 (∼5-fold) (Fig. 5K). Intriguingly, only infection with R. montanensis resulted in a robust induction of Casp-11 (∼8-fold) compared to infection data using both pathogenic Rickettsia spp. (Fig. 5K). To test if IL-1 cytokine secretion is dependent on the bacterial load, we heat inactivated both pathogenic and nonpathogenic Rickettsia spp. and showed that IL-1β and IL-1α release was significantly impaired compared to that in infections using viable Rickettsia spp., a phenotype more strongly observed in infections using R. montanensis (see Fig. S4 in the supplemental material). As IL-1 cytokine secretion is dependent on Gsdmd, the pore-forming executor of pyroptosis (29–32), we assessed the proteolytic processing of Gsdmd and showed that only R. montanensis infection resulted in a robust cleavage of Gsdmd (∼8-fold), as indicative by the detection of the Gsdmd-p30 fragment (Fig. 5K). In support of our findings, we showed that infection of Gsdmd−/− BMDMs with nonpathogenic and pathogenic Rickettsia spp. released significantly lower levels of IL-1β and IL-1α than infection of WT BMDMs (see Fig. S5A and B in the supplemental material). Furthermore, analysis of bacterial burdens provided additional evidence that Gsdmd plays a role in restricting the replication of R. typhi, R. rickettsii, and R. montanensis (Fig. S5C). These findings suggest that pathogenic, unlike nonpathogenic, Rickettsia spp. suppress IL-1 cytokine secretion via a Casp-11–Gsdmd-dependent pathway to support an intracytosolic replication in Mϕ.
FIG 5.
Pathogenic, but not nonpathogenic, Rickettsia spp. limit caspase-1- and caspase-11-dependent IL-1 signaling to facilitate their intracellular replication in macrophages. (A to I) BMDMs from WT, Casp-1−/−, Casp-11−/−, or Casp-1/11−/− mice were infected with R. typhi (A to C), R. rickettsii (D to F), or R. montanensis (G to I) (MOI = 50) for 0, 6, 24, and 48 h. Culture supernatants were analyzed for production of IL-1β (A, D, and G) and IL-1α (B, E, and H) using Legendplex kits (BioLegend), followed by flow cytometry. BMDM cell death at 24 h postinfection was measured by lactate dehydrogenase (LDH) release assay (C, F, and I). (J) Bacterial burdens in infected BMDMs were evaluated 2 and 24 h postinfection by GltA RT-qPCR. Expression of the host housekeeping gene Gapdh was used for normalization. (K) Western analysis of Casp-1, Casp-11, and Gsdmd induction and processing at 24 h postinfection with R. typhi, R. rickettsii, or R. montanensis using anti-Casp-1, anti-Casp-11, and anti-Gsdmd Abs. Reblotting with Rickettsia-specific anti-EF-Ts and host-cell-specific anti-Gapdh Abs served as infection and equal loading controls, respectively. Densitometry was performed using Fiji software, and data representing the fold change ratios of Casp-1-p20/FL-Casp-1, Gsdmd-p30/FL-Gsdmd, or Casp-11/Gapdh between uninfected and infected cells are shown. Error bars in panels A to J represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001. Immunoblot data are representative of three independent experiments.
Heat inactivation of Rickettsia species resulted in a reduced IL-1α and IL-1β cytokine release by macrophages. (A and B) BMDMs from WT mice were infected with heat-inactivated or non-heat-treated R. typhi, R. rickettsii, or R. montanensis (MOI = 50) for 24 h. Culture supernatants were analyzed for production of IL-1β (A) and IL-1α (B) using Legendplex kits (BioLegend), followed by flow cytometry. (C) Bacterial burden in Rickettsia-infected BMDMs from WT mice was evaluated at 2 and 24 h postinfection by GltA RT-qPCR. Expression of the housekeeping gene Gapdh was used for normalization. Error bars in panels A to C represent means ± SEM from five independent experiments. NS, nonsignificant; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001. Download FIG S4, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gsdmd is involved in the release of IL-1α and IL-1β by macrophages upon Rickettsia infection. (A and B) BMDMs from WT and Gsdmd−/− mice were infected with R. typhi, R. rickettsii, or R. montanensis (MOI = 50) for 24 h. Culture supernatants were analyzed for production of IL-1β (A) and IL-1α (B) using Legendplex kits (BioLegend), followed by flow cytometry. (C) Bacterial burden in Rickettsia-infected BMDMs from WT and Gsdmd−/− mice was evaluated at 2 and 24 h postinfection by GltA RT-qPCR. Expression of the housekeeping gene Gapdh was used for normalization. Error bars in panels A to C represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. Download FIG S5, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Secretion of IL-1α by macrophages is crucial in restricting the replication of pathogenic and nonpathogenic Rickettsia species in vivo.
To examine whether secretion of IL-1 by Mϕ limits the replication of Rickettsia spp. in vivo, we first injected (i.v.) WT mice with phosphate-buffered saline (PBS)- or dichloromethylene biphosphate (Cl2MBP)-liposomes to deplete endogenous macrophages as described previously (33). Next, PBS- or Cl2MBP-liposome-treated WT mice were injected (i.v.) with BMDMs isolated from WT, Il-1β−/−, or Il-1α−/− mice prior to infection with R. typhi, R. rickettsii, or R. montanensis. Strikingly, adoptive transfer of Il-1α−/− BMDMs, but not Il-1β−/− or WT Mϕ, significantly increased the mortality of Cl2MBP- but not PBS-treated WT mice injected with all three Rickettsia spp., reaching levels similar to the survival percentages observed in IL-1α Ab neutralization studies (Fig. 2 and Fig. 6A to D; see Fig. S6 in the supplemental material). Moreover, transfer of Il-1α−/− BMDMs resulted in the development of splenomegaly (see Fig. S7 in the supplemental material) and an increase in bacterial burden in the spleens of Cl2MBP-treated WT mice (Fig. 6E), which correlated with a decrease in IL-1α serum concentrations without affecting IL-1β serum levels (Fig. 6F and G). In contrast, transfer of Il-1β−/− BMDMs had an overall lesser effect on the severity of rickettsiosis, as evidenced by a lower mortality rate, smaller spleen size, and lower bacterial burden in Cl2MBP-treated WT mice (Fig. 6A to E; see Fig. S7 in the supplemental material), which is in agreement with our IL-1β Ab neutralization data (Fig. 2). In addition, transfer of Il-1β−/− BMDMs resulted in a decrease in IL-1β serum concentrations without affecting IL-1α serum levels (Fig. 6F and G). Collectively, these data suggest that modulation of expression and secretion of IL-1α by macrophages is important to limit the replication of Rickettsia spp. in vivo.
FIG 6.
Macrophage-dependent secretion of IL-1α contributes more than IL-1β for controlling the survival and colonization of pathogenic and nonpathogenic Rickettsia species. (A to D) Dichloromethylene biphosphate (Cl2MBP)-treated C57BL/6J WT mice were injected (i.v.) with WT, Il-1β−/−, or Il-1α−/− BMDMs (5 × 106 cells/mouse), followed by infection (24 h post-Mϕ transfer) with R. typhi (B), R. rickettsii (C), or R. montanensis (D) (dose 105 PFU) (B to D; n = 12 for each treatment). Survival was monitored for 7 days. (E) Bacterial burden was tested in spleens from mice described in panels B to D by GltA RT-qPCR at day 7 (n = 5 for each treatment), using the housekeeping gene Gapdh for normalization. (F to G) Serum samples from mice described in panels B to D were analyzed for IL-1β (F) and IL-1α (G) production at day 7 (n = 5 for each treatment) using the Legendplex kits (BioLegend), followed by flow cytometry analysis. Error bars in panels E to G represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001. (H) Proposed working model on how pathogenic Rickettsia spp. suppress Casp-1- and Casp-11-dependent IL-1 signaling responses to establish a replication niche in vitro and in vivo. Of note, the majority of nonpathogenic Rickettsia spp. are likely destroyed by phagolysosomal fusion, while a subpopulation may escape lysosomal fusion, ultimately allowing for the induction of inflammasome-mediated IL-1 responses.
Survival data after adoptive transfer of WT, Il-1β−/−, or Il-1α−/− macrophages and infection of pathogenic and nonpathogenic Rickettsia species into PBS-liposome-treated C57BL/6J WT mice. (A to D) PBS-treated C57BL/6J WT mice were injected i.v. with WT, Il-1β−/−, or Il-1α−/− BMDMs (5 × 106 cells/mouse), followed by infection (24 h post-Mϕ transfer) with R. typhi (B), R. rickettsii (C), R. montanensis (D), or PBS (dose of 105 PFU) (B to D; n = 12 for each treatment). Survival was monitored for 7 days. Download FIG S6, EPS file, 1.6 MB (1.6MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Splenic data after adoptive transfer of WT, Il-1β−/−, or Il-1α−/− macrophages and infection of pathogenic and nonpathogenic Rickettsia species into dichloromethylene biphosphate (Cl2MBP)-treated C57BL/6J WT mice. (A) Representative images of spleens (day 7) from Cl2MBP-treated C57BL/6J WT mice injected i.v. with WT, Il-1β−/−, or Il-1α−/− BMDMs (5 × 106 cells/mouse), followed by administration (24 h post-Mϕ transfer) of R. typhi, R. rickettsii, R. montanensis, or PBS (dose of 105 PFU). (B) Spleen weight from injected animals was evaluated at day 7 (n = 5). Error bars in panels B represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01. Download FIG S7, EPS file, 3.8 MB (3.8MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Obligate intracellular bacterial pathogens, which successfully reside and replicate within the host cell, overcome responses of innate immune defense surveillance (e.g., inflammasomes and autophagy). However, in the case of strict obligate intracytosolic Rickettsia spp., the roles for both inflammasome and autophagy to restrict their replication in endothelial cells and immune cells, like Mϕ, is only now emerging, although without consistent mechanistical insights (23–28, 34). Given these knowledge gaps and our current lack of understanding on how TG Rickettsia spp. evade immune defense responses to facilitate host colonization, we established an animal model of rickettsiosis using C57BL/6J WT mice by comparing development of both mild (∼LD25) and more severe (∼LD50) disease for two pathogenic spp., R. rickettsii and R. typhi. In fact, data of disease severity correlated with the bacterial burdens detected in the mice spleens. We also observed that infections with pathogenic, but not nonpathogenic, Rickettsia spp. resulted in a reduced serum response of both proinflammatory cytokines, IL-1β and IL-1α, which is likely the result of pathogenic Rickettsia spp. to evade canonical and noncanonical inflammasome-dependent defense sensing, of which the former is in agreement with recent studies using R. australis (26). Thus, our data suggest that inhibition of both canonical and noncanonical inflammasome-dependent signaling contributes to the enhanced survival and colonization of pathogenic Rickettsia spp. in our in vivo experiments.
Our data further suggest that IL-1α and, to a significantly lesser extent, IL-1β play a role in limiting rickettsial infection in vivo. By testing the putative antirickettsial capabilities of these cytokines through employing Ab neutralization and recombinant protein assays, we showed that IL-1α and, to a much lesser extent, IL-1β was able to restrict the replication and colonization of both nonpathogenic and pathogenic Rickettsia spp. in vivo. Our current understanding by which Rickettsia spp. evade host-induced antibacterial activities, and in particular, how cytosolic rickettsiae overcome host immune surveillance in defense cells like Mϕ, is primarily based on reports that are not aligned with one another (23–25, 27). Thus, we sought to address the hypothesis that pathogenic R. typhi and R. rickettsii spp., but not the nonpathogenic species R. montanensis, evade innate immune defense responses in order to establish an intracytosolic replication niche in Mϕ. In agreement with our in vivo infection models, R. montanensis-infected WT BMDMs produced higher levels of IL-1α and IL-1β cytokines and displayed reduced bacterial loads during the course of infection, compared to R. typhi- or R. rickettsii-infected BMDMs. These data support the notion that nonpathogenic, but not pathogenic, Rickettsia spp. are more efficiently cleared by Mϕ, a mechanism that further supports the previously published findings with SFG Rickettsia using THP-1 cells, a human macrophage-like cell line (35). Collectively, our presented data strengthen our hypothesis that pathogenic, but not nonpathogenic, Rickettsia spp. suppress antirickettsial inflammasome-dependent IL-1 cytokine responses to establish an intracytosolic replication niche in Mϕ.
Given that IL-1 signaling is modulated through inflammasome-dependent Casp-1, Casp-11, and Gsdmd activation, we tested the role of both caspases as well as Gsdmd, and found that nonpathogenic, but not pathogenic, Rickettsia spp. ensured Casp-1 activation and Casp-11 induction, which ultimately resulted in the proteolytic processing of Gsdmd and release of IL-1α and IL-1β cytokines. Intriguingly, the lack of Casp-11 induction by pathogenic, but not nonpathogenic, Rickettsia spp. suggests that the membrane-bound lipopolysaccharide (LPS) of R. typhi and R. rickettsii spp. is likely less immunogenic than that of R. montanensis, which is further supported by our recent reports (36, 37). Our findings further suggest that pathogenic, compared to nonpathogenic, Rickettsia spp. benefit from evasion of the Casp-11–Gsdmd–IL-1α signaling axis to establish a replication niche in Mϕ, as evidenced by the increased replication in Mϕ from Casp-11−/−, Casp-1/11−/−, or Gsdmd−/− mice compared to WT and Casp-1−/− BMDMs. Finally, we sought to determine the role of IL-1 cytokine responses by Mϕ in restricting the replication of Rickettsia spp. and showed that transfer of Il-1α−/− BMDMs and, to much lesser extent, the administration of Il-1β−/− BMDMs exacerbated the disease progression in WT mice injected with either pathogenic or nonpathogenic Rickettsia spp. It is worth noting that the observed differences in IL-1α release could be partially attributed to alternative mechanisms, including the retainment of IL-1α in the cytosol or nucleus, the dependence of IL-1β, and/or the association with the decoy receptor of IL-1 (IL-1R2), and future experiments are under way to address these possibilities (15, 29, 33, 38–40). Also, IL-1α is produced by other immune cells, such as neutrophils (15). Although our study did not evaluate a potential contributing role of neutrophils, preceding findings suggest that neutrophils did not alter the course of rickettsiosis or contribute to the restriction of bacterial growth (28).
Importantly, preceding findings suggest that intracellular pathogens, like rickettsiae, not only encounter inflammasome-dependent defense mechanisms but also are confronted by another cytosolic defense pathway, autophagy (23, 25). Both responses not only are key to mount the appropriate host defense responses (16, 18), but also are functionally interconnected. In fact, recent reports indicated that autophagy acts on intracellular microbes upstream of the inflammasome and thereby functions as a negative regulator by degrading inflammasome components (10, 16, 18, 22). In the case of rickettsiae, however, the role of autophagy in regulating inflammasome responses to facilitate their host colonization remains inconclusive. For instance, R. australis, a pathogenic TRG member, benefited from ATG5-dependent autophagy induction and suppression of inflammasome-dependent IL-1β production to colonize Mϕ (23, 25). In contrast, R. parkeri, a mildly pathogenic member of SFG, demonstrated that its surface protein OmpB is critical for protecting against autophagic recognition, while evasion of autophagy was critical for invasion of BMDMs and WT mice by R. parkeri (24, 27). Intriguingly, our recent report on R. typhi showed that R. typhi is ubiquitinated upon host entry, induces autophagy, but escapes autophagolysosomal maturation for intracellular colonization in nonphagocytic cells (9). Given these reports by others and our recent findings, it is tempting to speculate that pathogenic, but not nonpathogenic, Rickettsia spp. induce autophagy to downregulate inflammasome-dependent IL-1β and IL-1α cytokine responses to establish an intracytosolic replication niche in vitro and in vivo, and our future research will address this possibility.
Overall, our findings present a previously unappreciated model of host invasion by which pathogenic, but not nonpathogenic, Rickettsia spp. avoid the activation of signaling pathways required for IL-1α production and release—likely via the suppression of the Casp-11–Gsdmd signaling pathway—to facilitate their intracytosolic replication in Mϕ and ultimately cause host colonization (Fig. 6H).
MATERIALS AND METHODS
Animals.
All experiments were conducted in fully AAALAC-accredited program using 8- to 10-week-old female C57BL/6J WT mice in a specific-pathogen-free environment according to the University of Maryland School of Medicine Institutional Animal Care and Use Committee (IACUC) in compliance with the National Institutes of Health guide (41).
Antibodies and reagents.
Anti-IL-1α (clone ALF-161), anti-IL-1β (clone B122), and an isotype control IgG (Armenian hamster IgG) antibody (Ab) were purchased from BioXCell. Anticaspase (anti-Casp-1) Ab was purchased from Adipogen, while anti-Casp-11 (clone EPR18628) and anti-Gsdmd (clone EPR19828) Abs were obtained from Abcam. Elongation factor Ts (EF-Ts) Ab was obtained from Primm Biotech as previously described (9), while the anti-Gapdh (FL-335) Ab was purchased from Santa Cruz Biotechnology. Halt protease and phosphatase inhibitor cocktail were obtained from Thermo Fisher Scientific. Endotoxin-free recombinant mouse IL-1α and IL-1β proteins were purchased from BioLegend.
Bacterial strains, cell culture, and infection.
Vero76 cells (an African green monkey kidney line; ATCC, RL-1587) were maintained in minimal Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37°C with 5% CO2. R. montanensis strain M5/6 and R. rickettsia strain Sheila Smith were obtained from Ted Hackstadt (Rocky Mountain Laboratories, NIH, MT, USA), and R. typhi strain Wilmington was obtained from the CDC. All Rickettsia strains were propagated in Vero76 cells grown in DMEM supplemented with 5% FBS at 34°C with 5% CO2. All Rickettsia cells were purified as previously described (7). For infection of BMDMs, purified Rickettsia spp. were used at a multiplicity of infection (MOI) of 50, to ensure the presence of enough bacteria at early stage of infection, for host response (5, 8, 9). For infection using heat-inactivated bacteria, purified Rickettsia spp. were heated at 90°C for 20 min (42). Rickettsiosis in mice was induced by tail vein injection (i.v.) of purified Rickettsia cells (105 to 106 PFU) resuspended in PBS. At days 1, 3, and 7 after administration, blood was collected, and serum cytokine levels were measured by flow cytometry. Splenic tissue specimens were collected at the indicated times and used for bacterial burden analysis by quantitative PCR (qPCR) as described below.
Differentiation of bone marrow-derived macrophages.
Bone marrow cells were isolated from femurs and tibias of WT, Il-1β−/−, Il-1α−/−, Gsdmd−/−, Casp-1−/−, Casp-11−/−, and Casp-1/11−/− mice. Femurs from Casp-1−/−, Casp-11−/−, and Casp-1/11−/− mice were kindly provided by Amal Amer (The Ohio State University, OH, USA), while bones from Il-1β−/− or Il-1α−/− mice were obtained from Thirumala-Devi Kanneganti (St. Jude Children’s Research Hospital, TN, USA). Femurs from Gsdmd−/− were kindly provided by Matthew Welch (University of California, Berkeley, CA, USA). Differentiation was induced by culturing bone marrow cells in RPMI 1640 medium supplemented with 10% FBS and 30% L929-conditioned medium (a source of macrophage colony-stimulating factor) with culture for 7 days as described previously (43).
Measurement of cytokines and chemokines.
IL-1 cytokine concentrations in the sera of mice or supernatants from cultured BMDMs were assessed using the Legendplex mouse inflammation kit (BioLegend) following the manufacturer’s instructions as described previously (43).
RNA isolation and quantitative real-time PCR.
BMDM samples were collected at 2, 6, 24, and 48 h postinfection, while spleens were collected at day 3 or 7 postinfection. RNA was extracted from 1 × 106 BMDMs or 100 μL of organ homogenate using the Quick-RNA miniprep kit (ZymoResearch). The iScript reverse transcription supermix kit (Bio-Rad; 1708841) was used to synthesize cDNAs from 200 ng of RNA according to the manufacturer's instructions. Quantitative real-time PCR (qRT-PCR) was performed using SYBR green (Thermo Fisher Scientific) and 2 μL cDNA, and for the rickettsial citrate synthase gene (GltA), 1 μM each oligonucleotides 5′-CATAATAGCCATAGGATGAG-3′ (forward [F]) and 5′-ATGATTTATGGGGAACTACC-3′ (reverse [R]) were used and the results analyzed as described previously (25). Oligonucleotides for Gapdh were obtained from Qiagen.
Extract preparation and Western blot analysis.
Rickettsia-infected BMDM cells were lysed for 2 h at 4°C in ice-cold lysis buffer (50 mM HEPES [pH 7.4], 137 mM NaCl, 10% glycerol, 1 mM EDTA, and 0.5% NP-40, supplemented with protease and phosphatase inhibitory cocktails) as described previously (43). Equal amounts of protein were loaded for SDS-PAGE, and membranes were probed with anti-Casp-1, anti-Casp-11, anti-Gsdmd, anti-IL-1α, anti-IL-1β, anti-EF-Ts, and anti-Gapdh Abs, followed by enhanced chemiluminescence with secondary Abs conjugated to horseradish peroxidase.
Neutralization of endogenous IL-1α and IL-1β.
For in vivo neutralization of IL-1α and IL-1β, C57BL/6J WT mice were i.v. injected with 250 μg of anti-IL-1α (clone ALF-161; BioXCell), anti-IL-1β (clone B122; BioXCell), or an IgG isotype control (Armenian hamster IgG; BioXCell) Ab 24 h before the induction of mild rickettsiosis using 105 PFU of R. typhi, R. rickettsii, or R. montanensis.
Adoptive transfer of bone marrow-derived macrophages.
C57BL/6J WT mice were injected (i.v.) twice with PBS- or dichloromethylene biphosphate (Cl2MBP)-liposomes 72 and 48 h prior to macrophage transfer as described previously (43). Next, C57BL/6J WT mice were injected (i.v.) with WT, Il-1β−/−, or Il-1α−/− BMDMs (5 × 106 cells/mouse), followed by infection (24 h post-Mϕ transfer) with R. typhi, R. rickettsii, R. montanensis, or PBS (dose of 105 PFU).
Statistical analysis.
Endpoint studies of mice subjected to mild (105 PFU) and severe (106 PFU) rickettsiosis were analyzed by using Kaplan-Meir survival curves and the log-rank test (GraphPad Prism Software, version 8). The statistical significance was assessed using analysis of variance (ANOVA) with Tukey’s multiple-comparison posttest (GraphPad). Data are presented as the mean ± standard error of the mean (SEM), unless stated otherwise. The alpha level was set to 0.05.
ACKNOWLEDGMENTS
We gratefully acknowledge Amal Amer (The Ohio State University, OH, USA), Thirumala-Devi Kanneganti (St. Jude Children’s Research Hospital, TN, USA), Matthew Welch (University of California, Berkeley, CA, USA), and Ted Hackstadt (Rocky Mountain Laboratories, NIH, MT, USA) for generously providing us with essential biological specimens and reagents, including femurs from various knockout mice and rickettsial strains. We thank Magda Beier-Sexton for her administrative, technical, organizational, and editorial contributions, as well as Stefanie Vogel (University of Maryland School of Medicine, Baltimore, MD, USA) for critical discussions and editorial contributions to the manuscript.
This work was supported with funds from the NIAID/NIH grants (R01AI017828 and R01AI126853 to A.F.A.).
We declare no conflict of interest. The funding sources had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
O.H.V., M.S.R., and A.F.A. planned the research and analyzed and interpreted the data. O.H.V., J.C., H.G., N.R.D., and M.S.R. performed the experiments. A.F.A., O.H.V., L.D., and R.S. contributed to the overall project administration and supervision. O.H.V., M.S.R., and A.F.A. wrote the manuscript, and all authors participated in editing the manuscript.
Contributor Information
Oliver H. Voss, Email: ovoss@som.umaryland.edu.
Abdu F. Azad, Email: aazad@som.umaryland.edu.
Yasuko Rikihisa, Ohio State University.
REFERENCES
- 1.Ray K, Marteyn B, Sansonetti PJ, Tang CM. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 2.Personnic N, Bärlocher K, Finsel I, Hilbi H. 2016. Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol 24:450–462. doi: 10.1016/j.tim.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Sahni A, Fang R, Sahni SK, Walker DH. 2019. Pathogenesis of rickettsial diseases: pathogenic and immune mechanisms of an endotheliotropic infection. Annu Rev Pathol 14:127–152. doi: 10.1146/annurev-pathmechdis-012418-012800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehman SS, Noriea NF, Aistleitner K, Clark TR, Dooley CA, Nair V, Kaur SJ, Rahman MS, Gillespie JJ, Azad AF, Hackstadt T. 2018. The rickettsial ankyrin repeat protein 2 is a type IV secreted effector that associates with the endoplasmic reticulum. mBio 9:e00975-18. doi: 10.1128/mBio.00975-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahman MS, Ammerman NC, Sears KT, Ceraul SM, Azad AF. 2010. Functional characterization of a phospholipase A(2) homolog from Rickettsia typhi. J Bacteriol 192:3294–3303. doi: 10.1128/JB.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman MS, Gillespie JJ, Kaur SJ, Sears KT, Ceraul SM, Beier-Sexton M, Azad AF. 2013. Rickettsia typhi possesses phospholipase A2 enzymes that are involved in infection of host cells. PLoS Pathog 9:e1003399. doi: 10.1371/journal.ppat.1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rennoll-Bankert KE, Rahman MS, Gillespie JJ, Guillotte ML, Kaur SJ, Lehman SS, Beier-Sexton M, Azad AF. 2015. Which way in? The RalF Arf-GEF orchestrates Rickettsia host cell invasion. PLoS Pathog 11:e1005115. doi: 10.1371/journal.ppat.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rennoll-Bankert KE, Rahman MS, Guillotte ML, Lehman SS, Beier-Sexton M, Gillespie JJ, Azad AF. 2016. RalF-mediated activation of Arf6 controls Rickettsia typhi invasion by co-opting phosphoinositol metabolism. Infect Immun 84:3496–3506. doi: 10.1128/IAI.00638-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voss OH, Gillespie JJ, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF. 2020. Risk1, a phosphatidylinositol 3-kinase effector, promotes Rickettsia typhi intracellular survival. mBio 11:e00820-20. doi: 10.1128/mBio.00820-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q, Fan J, Billiar TR, Scott MJ. 2017. Inflammasome and autophagy regulation: a two-way street. Mol Med 23:188–195. doi: 10.2119/molmed.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Place DE, Kanneganti TD. 2018. Recent advances in inflammasome biology. Curr Opin Immunol 50:32–38. doi: 10.1016/j.coi.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue Y, Enosi Tuipulotu D, Tan WH, Kay C, Man SM. 2019. Emerging activators and regulators of inflammasomes and pyroptosis. Trends Immunol 40:1035–1052. doi: 10.1016/j.it.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Wiggins KA, Parry AJ, Cassidy LD, Humphry M, Webster SJ, Goodall JC, Narita M, Clarke MCH. 2019. IL‐1α cleavage by inflammatory caspases of the noncanonical inflammasome controls the senescence‐associated secretory phenotype. Aging Cell 18:e12946. doi: 10.1111/acel.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tapia VS, Daniels MJD, Palazón-Riquelme P, Dewhurst M, Luheshi NM, Rivers-Auty J, Green J, Redondo-Castro E, Kaldis P, Lopez-Castejon G, Brough D. 2019. The three cytokines IL-1β, IL-18, and IL-1α share related but distinct secretory routes. J Biol Chem 294:8325–8335. doi: 10.1074/jbc.RA119.008009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik A, Kanneganti T-D. 2018. Function and regulation of IL-1α in inflammatory diseases and cancer. Immunol Rev 281:124–137. doi: 10.1111/imr.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krakauer T. 2019. Inflammasomes, autophagy, and cell death: the trinity of innate host defense against intracellular bacteria. Mediators Inflamm 2019:2471215. doi: 10.1155/2019/2471215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell G, Isberg RR. 2017. Innate immunity to intracellular pathogens: balancing microbial elimination and inflammation. Cell Host Microbe 22:166–175. doi: 10.1016/j.chom.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seveau S, Turner J, Gavrilin MA, Torrelles JB, Hall-Stoodley L, Yount JS, Amer AO. 2018. Checks and balances between autophagy and inflammasomes during infection. J Mol Biol 430:174–192. doi: 10.1016/j.jmb.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storek KM, Monack DM. 2015. Bacterial recognition pathways that lead to inflammasome activation. Immunol Rev 265:112–129. doi: 10.1111/imr.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vladimer GI, Marty-Roix R, Ghosh S, Weng D, Lien E. 2013. Inflammasomes and host defenses against bacterial infections. Curr Opin Microbiol 16:23–31. doi: 10.1016/j.mib.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng D, Liwinski T, Elinav E. 2020. Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell Discov 6:36. doi: 10.1038/s41421-020-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahama M, Akira S, Saitoh T. 2018. Autophagy limits activation of the inflammasomes. Immunol Rev 281:62–73. doi: 10.1111/imr.12613. [DOI] [PubMed] [Google Scholar]
- 23.Bechelli J, Vergara L, Smalley C, Buzhdygan TP, Bender S, Zhang W, Liu Y, Popov VL, Wang J, Garg N, Hwang S, Walker DH, Fang R. 2019. Atg5 supports Rickettsia australis infection in macrophages in vitro and in vivo. Infect Immun 87:e00651-18. doi: 10.1128/IAI.00651-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke TP, Engström P, Chavez RA, Fonbuena JA, Vance RE, Welch MD. 2020. Inflammasome-mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nat Microbiol 5:688–696. doi: 10.1038/s41564-020-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smalley C, Bechelli J, Rockx-Brouwer D, Saito T, Azar SR, Ismail N, Walker DH, Fang R. 2016. Rickettsia australis activates inflammasome in human and murine macrophages. PLoS One 11:e0157231. doi: 10.1371/journal.pone.0157231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rumfield C, Hyseni I, McBride JW, Walker DH, Fang R. 2020. Activation of ASC inflammasome driven by Toll-like receptor 4 contributes to host immunity against rickettsial infection. Infect Immun 88:e00886-19. doi: 10.1128/IAI.00886-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engström P, Burke TP, Mitchell G, Ingabire N, Mark KG, Golovkine G, Iavarone AT, Rape M, Cox JS, Welch MD. 2019. Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence. Nat Microbiol 4:2538–2551. doi: 10.1038/s41564-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papp S, Moderzynski K, Rauch J, Heine L, Kuehl S, Richardt U, Mueller H, Fleischer B, Osterloh A. 2016. Liver necrosis and lethal systemic inflammation in a murine model of Rickettsia typhi infection: role of neutrophils, macrophages and NK cells. PLoS Negl Trop Dis 10:e0004935. doi: 10.1371/journal.pntd.0004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batista SJ, Still KM, Johanson D, Thompson JA, O’brien CA, Lukens JR, Harris TH. 2020. Gasdermin-D-dependent IL-1α release from microglia promotes protective immunity during chronic Toxoplasma gondii infection. Nat Commun 11:3687. doi: 10.1038/s41467-020-17491-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanneganti A, Malireddi RKS, Saavedra PHV, Vande WL, Van Gorp H, Kambara H, Tillman H, Vogel P, Luo HR, Xavier RJ, Chi H, Lamkanfi M. 2018. GSD MD is critical for autoinflammatory pathology in a mouse model of familial Mediterranean fever. J Exp Med 215:1519–1529. doi: 10.1084/jem.20172060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. 2016. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuchiya K, Hosojima S, Hara H, Kushiyama H, Mahib MR, Kinoshita T, Suda T. 2021. Gasdermin D mediates the maturation and release of IL-1α downstream of inflammasomes. Cell Rep 34:108887. doi: 10.1016/j.celrep.2021.108887. [DOI] [PubMed] [Google Scholar]
- 33.Fettelschoss A, Kistowska M, LeibundGut-Landmann S, Beer H-D, Johansen P, Senti G, Contassot E, Bachmann MF, French LE, Oxenius A, Kündig TM. 2011. Inflammasome activation and IL-1β target IL-1α for secretion as opposed to surface expression. Proc Natl Acad Sci USA 108:18055–18060. doi: 10.1073/pnas.1109176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moderzynski K, Papp S, Rauch J, Heine L, Kuehl S, Richardt U, Fleischer B, Osterloh A. 2016. CD4+ T cells are as protective as CD8+ T cells against Rickettsia typhi infection by activating macrophage bactericidal activity. PLoS Negl Trop Dis 10:e0005089. doi: 10.1371/journal.pntd.0005089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curto P, Simões I, Riley SP, Martinez JJ. 2016. Differences in intracellular fate of two spotted fever group Rickettsia in macrophage-like cells. Front Cell Infect Microbiol 6:80. doi: 10.3389/fcimb.2016.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guillotte ML, Gillespie JJ, Chandler CE, Rahman MS, Ernst RK, Azad AF. 2018. Rickettsia lipid A biosynthesis utilizes the late acyltransferase LpxJ for secondary fatty acid addition. J Bacteriol 200:e00334-18. doi: 10.1128/JB.00334-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillotte ML, Chandler CE, Verhoeve VI, Gillespie JJ, Driscoll TP, Rahman MS, Ernst RK, Azad AF. 2021. Lipid A structural divergence in Rickettsia pathogens. mSphere 6:e00184-21. doi: 10.1128/mSphere.00184-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Paolo NC, Shayakhmetov DM. 2016. Interleukin 1α and the inflammatory process. Nat Immunol 17:906–913. doi: 10.1038/ni.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters VA, Joesting JJ, Freund GG. 2013. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav Immun 32:1–8. doi: 10.1016/j.bbi.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi L, Song L, Maurer K, Dou Y, Patel VR, Su C, Leonard ME, Lu S, Hodge KM, Torres A, Chesi A, Grant SFA, Wells AD, Zhang Z, Petri MA, Sullivan KE. 2020. IL-1 transcriptional responses to lipopolysaccharides are regulated by a complex of RNA binding proteins. J Immunol 204:1334–1344. doi: 10.4049/jimmunol.1900650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Institutes of Health. 2002. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD. [Google Scholar]
- 42.Ammerman NC, Beier-Sexton M, Azad AF. 2008. Laboratory maintenance of Rickettsia rickettsii. Curr Protoc Microbiol Chapter 3:Unit-3A.5. doi: 10.1002/9780471729259.mc03a05s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voss OH, Murakami Y, Pena MY, Lee H-N, Tian L, Margulies DH, Street JM, Yuen PST, Qi C-F, Krzewski K, Coligan JE. 2016. Lipopolysaccharide-induced CD300b receptor binding to Toll-like receptor 4 alters signaling to drive cytokine responses that enhance septic shock. Immunity 44:1365–1378. doi: 10.1016/j.immuni.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voss OH, Arango D, Tossey JC, Villalona Calero MA, Doseff AI. 2021. Splicing reprogramming of TRAIL/DISC-components sensitizes lung cancer cells to TRAIL-mediated apoptosis. Cell Death Dis 12:287. doi: 10.1038/s41419-021-03567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Splenic data during infection of pathogenic and nonpathogenic Rickettsia species. (A) Representative images of spleens (at day 7) from C57BL/6J WT mice injected i.v. with R. typhi, R. rickettsii, R. montanensis, or PBS (dose of 106 PFU). (B) Spleen weight from injected animals was evaluated at day 7 day (n = 5). Error bars in panel B represent means ± SEM from five independent experiments. *, P ≤ 0.05; ***, P ≤ 0.005. Download FIG S1, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Splenic data after IL-1 signaling neutralization during infection of pathogenic and nonpathogenic Rickettsia species. (A) Representative images of spleens (day 7) from C57BL/6J WT mice injected i.v. with R. typhi, R. rickettsii, or R. montanensis (dose of 105 PFU), followed by administration of anti-IL-1β, anti-IL-1α, or IgG isotype control antibody (Ab) (250 μg Ab/mouse) (24 h post-Ab injection). (B) Spleen weight from injected animals was evaluated at day 7 (n = 5). Error bars in panel B represent means ± SEM from five independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. Download FIG S2, EPS file, 3.2 MB (3.2MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Splenic data after administration of recombinant IL-1α and IL-1β proteins during infection of pathogenic and nonpathogenic Rickettsia species. (A) Representative images of spleens (day 7) from C57BL/6J WT mice injected i.v. with rIL-1β or rIL-1α protein, followed by administration of R. typhi, R. rickettsii, R. montanensis, or PBS (dose of 106 PFU) at day 7. (B) Spleen weight from injected animals was evaluated at day 7 day (n = 5). Error bars in panel B represent means ± SEM from five independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. Download FIG S3, EPS file, 4.0 MB (4MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat inactivation of Rickettsia species resulted in a reduced IL-1α and IL-1β cytokine release by macrophages. (A and B) BMDMs from WT mice were infected with heat-inactivated or non-heat-treated R. typhi, R. rickettsii, or R. montanensis (MOI = 50) for 24 h. Culture supernatants were analyzed for production of IL-1β (A) and IL-1α (B) using Legendplex kits (BioLegend), followed by flow cytometry. (C) Bacterial burden in Rickettsia-infected BMDMs from WT mice was evaluated at 2 and 24 h postinfection by GltA RT-qPCR. Expression of the housekeeping gene Gapdh was used for normalization. Error bars in panels A to C represent means ± SEM from five independent experiments. NS, nonsignificant; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001. Download FIG S4, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gsdmd is involved in the release of IL-1α and IL-1β by macrophages upon Rickettsia infection. (A and B) BMDMs from WT and Gsdmd−/− mice were infected with R. typhi, R. rickettsii, or R. montanensis (MOI = 50) for 24 h. Culture supernatants were analyzed for production of IL-1β (A) and IL-1α (B) using Legendplex kits (BioLegend), followed by flow cytometry. (C) Bacterial burden in Rickettsia-infected BMDMs from WT and Gsdmd−/− mice was evaluated at 2 and 24 h postinfection by GltA RT-qPCR. Expression of the housekeeping gene Gapdh was used for normalization. Error bars in panels A to C represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. Download FIG S5, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Survival data after adoptive transfer of WT, Il-1β−/−, or Il-1α−/− macrophages and infection of pathogenic and nonpathogenic Rickettsia species into PBS-liposome-treated C57BL/6J WT mice. (A to D) PBS-treated C57BL/6J WT mice were injected i.v. with WT, Il-1β−/−, or Il-1α−/− BMDMs (5 × 106 cells/mouse), followed by infection (24 h post-Mϕ transfer) with R. typhi (B), R. rickettsii (C), R. montanensis (D), or PBS (dose of 105 PFU) (B to D; n = 12 for each treatment). Survival was monitored for 7 days. Download FIG S6, EPS file, 1.6 MB (1.6MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Splenic data after adoptive transfer of WT, Il-1β−/−, or Il-1α−/− macrophages and infection of pathogenic and nonpathogenic Rickettsia species into dichloromethylene biphosphate (Cl2MBP)-treated C57BL/6J WT mice. (A) Representative images of spleens (day 7) from Cl2MBP-treated C57BL/6J WT mice injected i.v. with WT, Il-1β−/−, or Il-1α−/− BMDMs (5 × 106 cells/mouse), followed by administration (24 h post-Mϕ transfer) of R. typhi, R. rickettsii, R. montanensis, or PBS (dose of 105 PFU). (B) Spleen weight from injected animals was evaluated at day 7 (n = 5). Error bars in panels B represent means ± SEM from five independent experiments. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01. Download FIG S7, EPS file, 3.8 MB (3.8MB, eps) .
Copyright © 2022 Voss et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.