Abstract
Intrinsic molecular subtypes may explain marked variation between bladder cancer patients in prognosis and response to therapy. Complex testing algorithms and little attention to more prevalent, early‐stage (non‐muscle invasive) bladder cancers (NMIBCs) have hindered implementation of subtyping in clinical practice. Here, using a three‐antibody immunohistochemistry (IHC) algorithm, we identify the diagnostic and prognostic associations of well‐validated proteomic features of basal and luminal subtypes in NMIBC. By IHC, we divided 481 NMIBCs into basal (GATA3−/KRT5+) and luminal (GATA3+/KRT5 variable) subtypes. We further divided the luminal subtype into URO (p16 low), URO‐KRT5+ (KRT5+), and genomically unstable (GU) (p16 high) subtypes. Expression thresholds were confirmed using unsupervised hierarchical clustering. Subtypes were correlated with pathology and outcomes. All NMIBC cases clustered into the basal/squamous (basal) or one of the three luminal (URO, URO‐KRT5+, and GU) subtypes. Although uncommon in this NMIBC cohort, basal tumors (3%, n = 16) had dramatically higher grade (100%, n = 16, odds ratio [OR] = 13, relative risk = 3.25) and stage, and rapid progression to muscle invasion (median progression‐free survival = 35.4 months, p = 0.0001). URO, the most common subtype (46%, n = 220), showed rapid recurrence (median recurrence‐free survival [RFS] = 11.5 months, p = 0.039) compared to its GU counterpart (29%, n = 137, median RFS = 16.9 months), even in patients who received intravesical immunotherapy (p = 0.049). URO‐KRT5+ tumors (22%, n = 108) were typically low grade (66%, n = 71, OR = 3.7) and recurred slowly (median RFS = 38.7 months). Therefore, a simple immunohistochemical algorithm can identify clinically relevant molecular subtypes of NMIBC. In routine clinical practice, this three‐antibody algorithm may help clarify diagnostic dilemmas and optimize surveillance and treatment strategies for patients.
Keywords: bladder cancer, immunohistochemistry, GATA3, KRT5, p16
Introduction
Non‐muscle invasive bladder cancer (NMIBC) represents 75–80% of all bladder cancer diagnoses [1]. NMIBC presents challenges, including frequent recurrence which necessitates repeated cystoscopies, and a risk of progression to life‐threatening muscle‐invasive disease [1, 2]. NMIBCs include low‐ and high‐grade (stage pTa) noninvasive cancers, as well as superficially invasive (stage pT1) tumors. Their risks of recurrence and progression are variable [1, 2] and are currently assessed using clinical algorithms such as the American Urological Association (AUA) risk score [3] which inform the intensity of surveillance and treatment, including intravesical bacillus Calmette–Guérin (BCG) [1, 2, 3]. However, these algorithms do not capture the clinical variability seen in NMIBC, which may result from discrete biologic origins known as molecular subtypes. Here, we develop a streamlined approach to NMIBC subtyping and investigate its prognostic significance.
In bladder cancer, luminal and basal subtypes have been identified using mRNA profiles, predominantly from MIBC cases [4, 5, 6, 7, 8]. Reflecting distinct differentiation states and gene expression repertoires of basal stem cells or more differentiated luminal (intermediate and superficial) cells in benign urothelium, luminal and basal molecular subtypes may reflect distinct oncogenic pathways [9, 10, 11]. These subtypes have yielded a number of promising prognostic and predictive associations that may improve risk stratification beyond standard histologic assessment [4, 5, 6, 7, 12, 13]. A consensus subtyping scheme has been developed for MIBC [8], but not for NMIBC. Nevertheless, a basal subtype and two luminal subtypes, urothelial‐like (URO) and genomically unstable (GU), have been validated in both NMIBC and MIBC [4, 13, 14, 15, 16]. Both URO and GU express urothelial differentiation markers GATA3 and FOXA1 [17, 18], but inactivate cell cycle checkpoints through different mechanisms. URO cancers inactivate the cell cycle regulator RB1 indirectly through loss of CDKN2A, which encodes the p16INK4A cyclin‐dependent kinase inhibitor [19, 20, 21]. GU cancers inactivate RB1 directly through genomic loss and can have a worse prognosis than URO [13, 20, 22, 23]. GU cancers also demonstrate reduced p63 protein expression compared to URO. Conversely, basal/squamous cell carcinoma‐like (SCCL), the most lethal subtype, expresses basal urothelial stem cell markers (e.g. EGFR, KRT5, and KRT14) but not markers of urothelial differentiation (e.g. GATA3 and FOXA1) [10, 24].
While molecular subtyping could yield useful prognostic and predictive information, complex testing paradigms have prevented its clinical implementation. Motivated by the need to separate stromal signals from those of cancer cells, an immunohistochemistry (IHC)‐based Lund taxonomy was validated against mRNA subtypes for both MIBC and NMIBC [12, 14, 16, 25].
A mainstay of pathology practice, IHC‐based tests, should be easy to implement. However, Lund IHC subtyping uses up to 13 antibodies, posing a barrier to implementation [26]. We therefore identified antibodies routinely used in surgical pathology laboratories that represent cardinal features of luminal and basal subtypes. GATA3 and KRT5 staining distinguish luminal and basal subtypes [9, 27]. Expression of p16 further stratifies luminal subtypes into URO and GU [15, 26, 28]. Here, we validate the diagnostic and prognostic significance of molecular subtypes in an NMIBC cohort using this simplified three‐antibody algorithm.
Materials and methods
Cohort composition
Under Queen's University research ethics board approval, including a waiver of informed consent, NMIBC tumors (n = 481) representing 390 patients diagnosed between 2008 and 2016 were collected from Kingston Health Science Centre in Ontario, Canada. All Ta and T1 stage papillary tumors were included, following pathological review (DMB and LC) using the WHO 2004 grading system [29]. Samples were not required to be consecutive for a given patient. Pure carcinoma in situ (CIS) cases were excluded, as were cases with insufficient tissue. Clinical and pathological information is summarized in supplementary material, Tables S1 and S2. Subtype stability was assessed in a subset of metachronous samples from 49 patients (supplementary material, Table S3). In the absence of tumor location data for these metachronous samples, any NMIBC recurrence was considered regardless of anatomic location.
Clinical definitions
Progression‐free survival (PFS) was defined as time from NMIBC transurethral resection (TURBT) to confirmation of muscle‐invasive or metastatic disease. Recurrence‐free survival (RFS) was defined as time from TURBT to the next malignant TURBT diagnosis. Operative notes were reviewed to exclude re‐resections as recurrences. Adequate BCG treatment was defined as receipt of at least five induction BCG instillations in a previously BCG‐naïve patient.
Immunohistochemistry
Six tissue microarray (TMA) blocks were prepared from formalin‐fixed paraffin‐embedded TURBTs, of which 81% were sampled with two 1.0 mm cores, and the remainder with a single core. IHC assays were performed on a Bond III Autostainer (Leica Biosystems, Concord, Canada) using monoclonal antibodies listed in supplementary material, Table S4.
IHC analysis
Based on intensity (p16, GATA3) or localization (KRT5) of staining, tumor cell IHC scores were calculated as described previously [26, 28]. Percent IHC‐positive cells per core were quantified in deciles (0.1–0.9) using HALO software v1.94 (Indica Labs, Inc., Albuquerque, NM, USA) (supplementary material, Figure S1). Intensity was visually scored from 0 to 3. GATA3 was assessed as binary scores of 0 (no staining) or 1 (positive). As described [28], KRT5 was evaluated visually by proximity to the basal cell layer: 0, no staining; 1, staining confined to the basal cell layer; 2, mostly basal with some suprabasal staining; and 3, homogenous staining. In brief, tumor cell scores ranging from 0 to 1 were calculated as the product of the intensity or proximity score with the percentage score divided by the maximum score. Tumor cell scores for a given case were equal to the mean score of all cores. Detailed scoring methods are provided in supplementary material, Table S5.
Clustering and statistical analyses
All analyses were conducted using R version 3.5.3. Unsupervised hierarchical clustering was conducted using divisive methods and Euclidean distance measures with the base stats and cluster packages. Heatmaps were generated using gplots and RColorBrewer. Kaplan–Meier curves and Cox proportional hazards models were calculated using log‐rank statistics in the survminer and survival R packages. Forest plots were generated using ggplots. Follow‐up time for Kaplan–Meier curves ended when 10% of patients remained in each group [30]. Categorical clinical and pathological variables were tested using chi‐square tests or Fisher's exact test. Numerical pairwise or ANOVA testing was conducted using Mann–Whitney U or Kruskal–Wallis tests, respectively. Two‐sided P values of less than 0.05 were considered statistically significant.
Results
Overview of the three‐antibody algorithm
The three‐antibody algorithm (Figure 1A) is a two‐step process that identifies four subtypes of NMIBC. The first division uses GATA3 to separate basal (GATA3−) and luminal (GATA3+) subtypes as the ‘top‐level’ of classification universal to current bladder cancer subtyping schemes [4, 5, 6, 7, 8, 15, 16]. The basal subtype is verified with positive KRT5 staining. Division of the luminal subtypes uses KRT5, which is positive in the URO‐KRT5+ subtype, as well as p16, which is negative in the URO subtype and positive in the GU subtype.
Figure 1.
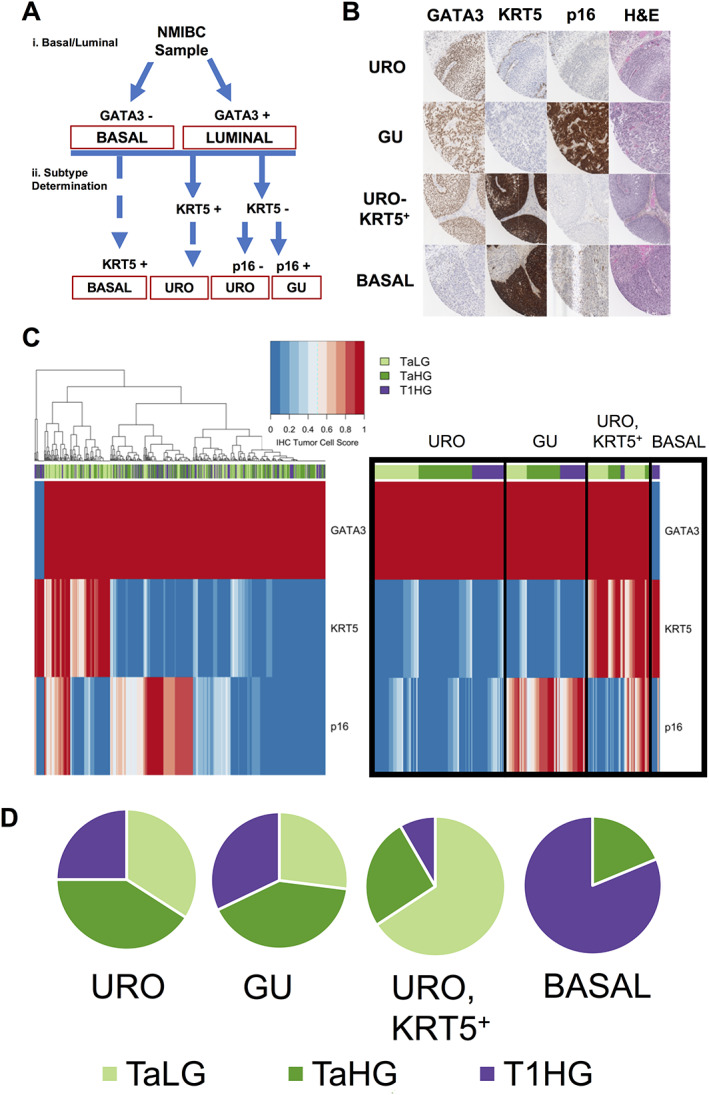
Subtyping algorithm and associations with stage and grade (A). Immunohistochemical algorithm for subtyping NMIBC samples. (i) GATA3 expression yields two categories (basal and luminal). (ii) Subtype is determined by KRT5 and p16 staining. KRT5‐positive staining classifies basal and URO (KRT5+) subtypes, whereas KRT5‐negative staining classifies the URO and GU subtypes. p16 staining defines the URO and GU subtypes based on negative and positive expression, respectively. (B) Representative IHC staining of NMIBC tumors using GATA3, KRT5, and p16 IHC. Images represent 0.5 mm from each 1.0 mm core. URO cases are strongly positive for GATA3 but negative for p16 expression, with KRT5 expression limited to the basal cell layer. GU cases are strongly positive for GATA3 and p16 expression and negative for KRT5 expression. URO (KRT5+) cases are strongly positive for GATA3 and KRT5 expression and negative for p16 expression. Basal cases are negative for GATA3 expression and highly positive for KRT5, with weak p16 expression. (C) Unsupervised hierarchical clustering of IHC scores identifies NMIBC subtypes from 481 tumor samples (left). Ordered heatmap (right) demonstrates stage and grade distributions across NMIBC subtypes. Stages are represented as: stage Ta, low‐grade samples (light green); stage Ta, high‐grade samples (green); and stage T1, high‐grade samples (purple). Basal tumors were classified as GATA3−, KRT5+. GU tumors were classified as GATA3+, KRT5−, p16+. URO subtype was classified as GATA3+, KRT5−, p16 weak or negative. Red represents high IHC tumor cell scores (high protein expression) and blue represents low IHC tumor cell scores (low protein expression). (D) Proportions of samples belonging to specific stages and grades, separated by subtype.
IHC staining patterns
GATA3
GATA3 is a transcriptional activator of luminal differentiation in urothelium and, along with additional transcription factors, can control the expression of luminal keratins [18]. Multiple studies have demonstrated that GATA3 can accurately classify luminal subtypes using IHC [9, 27]. GATA3 positivity was defined as necessary and sufficient to identify luminal cancers (Figure 1A). Homogenous strong nuclear GATA3 staining was found in 97% (n = 467) of NMIBC cancers [15, 28, 31] (Figure 1A, B). Only 3% (n = 16) were GATA3 negative and therefore defined as basal (see below).
KRT5
Using immunohistochemical detection of KRT5, we identified three patterns of basal cell differentiation. Basal tumors (n = 16, 3%) (Figure 1B) showed diffuse intense cytoplasmic staining in tumor cells (tumor cell score > 0.83) and were negative for GATA3 [15, 31]. As expected for the URO subtype [28], we observed stratified expression of KRT5 and KRT5 staining confined to the basal cell layer (n = 220, 46%). Surprisingly, a separate subset of GATA3‐positive tumors (n = 108, 23%) showed strong expression of KRT5 in an expanded suprabasal pattern (Figure 1B). As these were luminal tumors as defined by GATA3 expression, we called this novel subtype URO‐KRT5+. GU tumors (n = 137, 28%) were defined by intact p16 (see below) and demonstrated limited expression of KRT5.
p16 (CDKN2A)
Deletions of CDKN2A are frequent events in the URO subtype resulting in a lack of p16 protein expression [15, 31]. Conversely, p16 overexpression is characteristic of the GU subtype [15, 31]. We observed a range of intensities of p16 expression, with moderate and high expression belonging to the GU subtype (Figure 1B,C).
Clustering
Unsupervised hierarchical clustering of the IHC tumor cell scores identified four key clusters (Figure 1C) corresponding with the three previously identified subtypes [15, 26, 28, 31]: basal (GATA3−, KRT5+), genomically unstable (GATA3+, p16+, KRT5−), urothelial‐like (GATA3+, p16−, KRT5−), and the previously undescribed KRT5 positive urothelial‐like (GATA3+, KRT5+) subtype. For samples with two cores available for analysis, 86% demonstrated subtype agreement between the two cores.
Subtype switching across recurrences
A clinically useful subtyping scheme should be relatively stable across metachronous tumors. Using IHC, we classified multiple NMIBC recurrences in 49 patients (Figure 2 and supplementary material, Table S3). The majority were initially URO (n = 30), followed by GU (n = 13), URO‐KRT5+ (n = 3), and basal (n = 3). For most patients, recurrent tumors retained the same subtype as the initial tumor (n = 33/49, 67%). Similar to previous observations [31], BCG treatment had no discernible effect on subtype. The relative stability of subtypes across recurrent tumors indicates that this three‐antibody algorithm can provide clinically useful information at the time of diagnosis.
Figure 2.
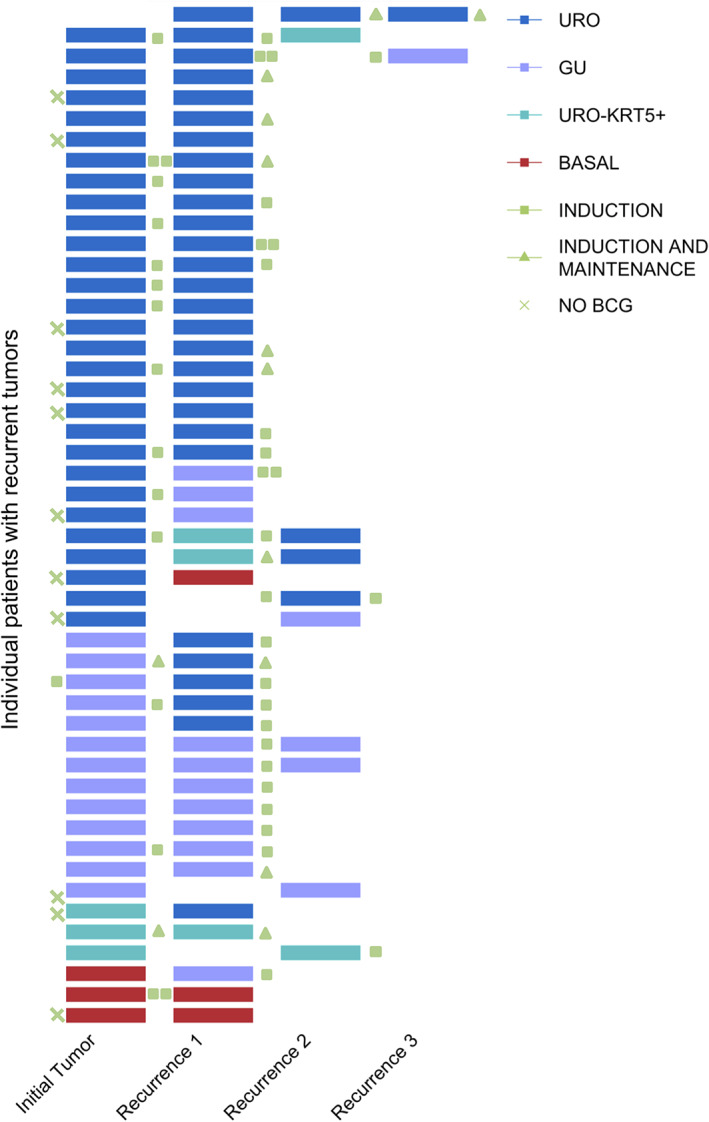
Recurrences across multiple patients (n = 49), with respect to molecular subtypes. Individual patients are represented by a single row, and each recurrence is represented by a colored bar which all occur within a 5‐year period. Majority of recurrences occurred within 1 year. Colors are representative of subtype: URO (blue), GU (purple), URO‐KRT5+ (turquoise), basal (red). Patients not treated with BCG are indicated by a green X. BCG treatment occurring between recurrences is categorized as induction (green square) or induction plus maintenance (green triangle).
Significant associations between subtype and clinical/pathological variables
Stage and grade
The basal subtype was dramatically enriched for high grade and stage, with 100% of tumors presenting as high grade, AUA high risk and over 80% of the subtype represented by invasive (stage pT1) tumors (Table 1, Figure 1D). Accordingly, the basal subtype showed a relative risk (RR) of 3.25 and an odds ratio (OR) of 13 for stage T1 disease (Table 1). Conversely, 66% of the URO‐KRT5+ subtype was pTa, low grade, and associated with low AUA risk scores, resulting in an RR and OR for low grade of 1.93 and 3.7, respectively (Table 1). Only 26 and 8% of the URO‐KRT5+ subtype were pTa high grade or pT1 high grade, respectively (Table 1, Figure 1D). In contrast, both the URO and GU subtypes showed relatively balanced frequencies across NMIBC grades and stages and across AUA risk scores (Table 1, Figure 1D). The basal and GU subtypes were significantly associated with concomitant CIS (p < 0.0001). In contrast, only 4% of URO‐KRT5+ cases presented with concomitant CIS (Table 1). Based on clinical and pathological features alone, basal tumors were most frequently associated with high‐risk features, followed by URO and GU, and finally, URO‐KRT5+ which was associated with lower‐risk features.
Table 1.
Clinical and histopathological characteristics of samples stratified by subtype.
| URO (n = 220) (%) | GU (n = 137) (%) | URO KRT5+ (n = 108) (%) | Basal (n = 16) (%) | P value | ||
|---|---|---|---|---|---|---|
| Stage and grade | Ta, low grade | 75 (34) | 37 (27) | 71 (66) | 0 (0) | <0.0001 |
| Ta, high grade | 90 (41) | 56 (41) | 28 (26) | 3 (19) | ||
| T1, high grade | 55 (25) | 44 (32) | 9 (8) | 13 (81) | ||
| Sex | Male | 182 | 108 | 86 | 10 | 0.24 |
| Female | 38 | 29 | 22 | 6 | ||
| CIS | Yes | 25 (11) | 32 (23) | 4 (4) | 5 (31) | <0.0001 |
| No | 195 (89) | 105 (76) | 104 (96) | 11 (69) | ||
| Variant histology | Squamous | 8 | 4 | 3 | 7 | 0.085 |
| Glandular | 0 | 3 | 0 | 0 | ||
| Micropapillary | 1 | 2 | 0 | 0 | ||
| AUA risk score | Low | 46 (21) | 28 (20.4) | 39 (36) | 0 (0) | <0.0001 |
| Intermediate | 42 (19) | 28 (20.4) | 36 (33) | 0 (0) | ||
| High | 132 (60) | 81 (59.2) | 33 (31) | 16 (100) | ||
| Time to recurrence post‐BCG | Early (<1 year) | 18 | 7 | 4 | 0 | 0.21 |
| Late (1–2 years) | 4 | 2 | 1 | 1 | ||
| No recurrence within 2 years | 36 | 41 | 16 | 5 | ||
| Median RFS | Days (months) | 349 (11.5) | 515 (16.9) | 1176 (38.7) | 276 (9.1) | 0.039* |
| Median PFS | Days (months) | NA | 3802 (124.9) | 4010 (131.8) | 1077 (35.4) | <0.00011* |
| OR for stage/grade relative to URO [95% CI] | Low grade | Reference | NA | 3.7 [2.28–6.02] | 0 | NA |
| High grade | 0.27 [0.17–0.44] | Infinite | ||||
| pTa | 3.67 [1.74–7.74] | 0.08 [0.02–0.28] | ||||
| pT1 | 0.27 [0.13–0.58] | 13 [3.6–47.3] | ||||
|
RR for stage and grade relative to URO [95% CI] |
Low grade | Reference | NA | 1.93 [1.53–2.42] | 0 | NA |
| High grade | 0.52 [0.39–0.69] | 1.51 [1.38–1.67] | ||||
| pTa | 1.22 [1.11–1.34) | 0.25 [0.09–0.7] | ||||
| pT1 | 0.33 [0.17–0.65] | 3.25 [2.34–4.5] |
NA, not applicable.
Log‐rank test.
Recurrence‐free survival
As high‐grade tumors present the highest risk for progression or recurrence events, we restricted further analyses to high‐grade samples. We observed significant associations between subtype and RFS (Figure 3A, p = 0.039). The basal subtype recurred the fastest, with a median RFS of 9.1 months (Table 1). The URO and GU subtypes followed, with a median RFS of 11.5 and 16.9 months, respectively. The URO‐KRT5+ subtype was the slowest to recur with a median RFS of 38.7 months, triple the RFS time of its URO counterpart (p = 0.011). We observed significant decreases in overall RFS between TaHG and T1HG tumors and conducted separate analyses for these tumor stages (Figure 3 and supplementary material, Figure S2). Stratification by stage and grade showed no overall significant differences between subtype and RFS for TaHG and T1HG tumors in Kaplan–Meier analysis (Figure 3C,E). However, due to the stage and grade distribution of basal and URO‐KRT5+ subtypes, there were limited basal TaHG and URO‐KRT5+ T1HG samples for analysis. Importantly, in pairwise and Kaplan–Meier analyses between the URO and URO‐KRT5+ subtype for TaHG tumors, we consistently observed that the URO‐KRT5+ subtype recurred significantly more slowly with a median RFS of 16.4 and 38.7 months, respectively (Figure 3C and supplementary material, Figure S2B,C, p = 0.047 and 0.046).
Figure 3.
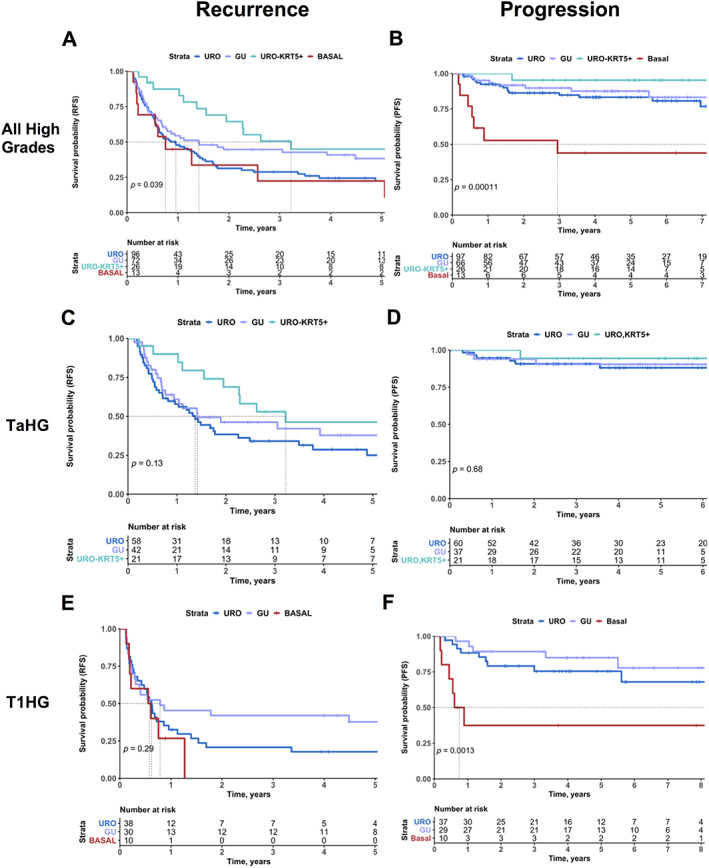
Prognostic associations of molecular subtypes in high‐grade NMIBC. RFS (A) and PFS (B) of high‐grade NMIBC patients, stratified by subtype. RFS (C) and PFS (D) of NMIBC patients with TaHG diagnoses, stratified by subtype. RFS (E) and PFS (F) of NMIBC patients with T1HG diagnoses, stratified by subtype. Progression is defined as progression to muscle invasion (greater than stage pT2). Colors represent subtype: URO (blue), GU (purple), URO‐KRT5+ (turquoise), and basal (red). P values listed according to log‐rank test.
Progression to muscle invasion
The basal subtype was significantly associated with more rapid progression to muscle invasion with a median PFS of 35.4 months (p = 0.00011) (Figure 3B, Table 1). High‐grade cases subtyped as URO‐KRT5+ and GU had a median PFS of 131.8 and 124.9 months, respectively (Figure 3B, Table 1). For the URO group, PFS was 79% at 7 years (Figure 3B). When all high‐grade cases were stratified by stage, there were no overall significant differences in PFS between subtypes for stage Ta tumors (Figure 3D). When stratified for stage pT1 tumors, URO and GU tumors demonstrated similar PFS (Figure 3F). However, stage pT1 high‐grade basal tumors showed the highest risk for progression to muscle invasion (Figure 3F, p = 0.0013). Subsequent multivariable analysis with AUA risk score and subtype indicated the basal subtype as a significant predictor for risk of progression (Figure 4).
Figure 4.
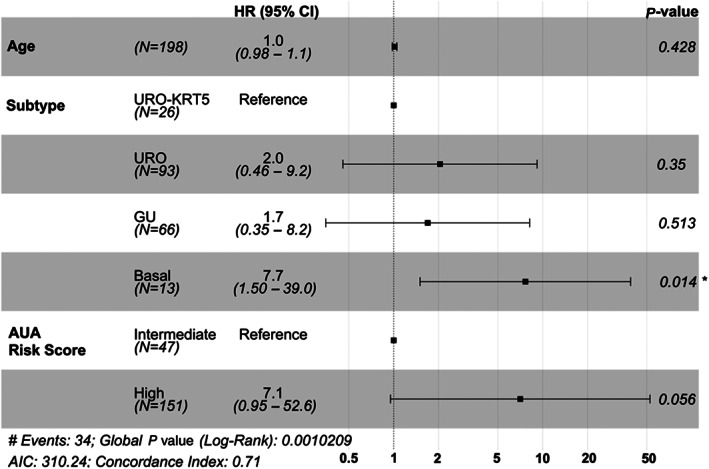
Forest plot for multivariable Cox regression analysis for PFS in high‐grade NMIBC tumors. P values listed according to log‐rank test, where * denotes p < 0.05.
Prognostic associations of URO and GU subtypes
Contrary to the findings in MIBC [12, 25], we observed that the GU subtype was associated with improved prognosis compared to URO. GU tumors exhibited a 5.5‐month improvement in RFS compared to URO (median RFS = 16.9 and 11.5, respectively) (Figure 3A). These RFS differences may reflect p16 status (Figure 1C), where URO tumors were defined by low p16 staining levels (<0.40), and GU tumor defined by intermediate and high p16 staining (>0.40). Indeed, tumors with low p16 recurred the fastest (median RFS = 13.5 months), and tumors with intermediate and high p16 expression (>0.40) recurred more slowly with a median RFS of 22.8 and 67.7 months, respectively (supplementary material, Figure S3).
As previous work in higher‐stage tumors reported that GU tumors were more likely than URO to progress [13], we confirmed GU subtyping using IHC assays for p63, which should be negative in approximately two‐thirds of GU tumors [28, 32] (supplementary material, Figure S4).
BCG response and subtype
We explored whether subtypes were associated with response to BCG. Both the URO and GU subtypes appeared to derive benefit from BCG, with fewer high‐grade recurrences after BCG treatment (Figure 5A, p < 0.0001). However, the GU subtype achieved the greatest RFS after adequate induction of BCG, experiencing fewer high‐grade recurrences than its URO counterpart (p = 0.033). As the URO‐KRT5+ subtype was significantly enriched for low‐grade tumors, many of these patients did not receive BCG, resulting in too few patients for analysis. Similarly, only six patients of the basal subtype received BCG and were excluded from the analysis. When categorized for time to recurrence, the GU subtype had significantly fewer high‐grade recurrences within 1 year post‐BCG and a greater proportion of patients with no recurrences within 2 years when compared to its URO counterpart (Figure 5B, p = 0.049).
Figure 5.
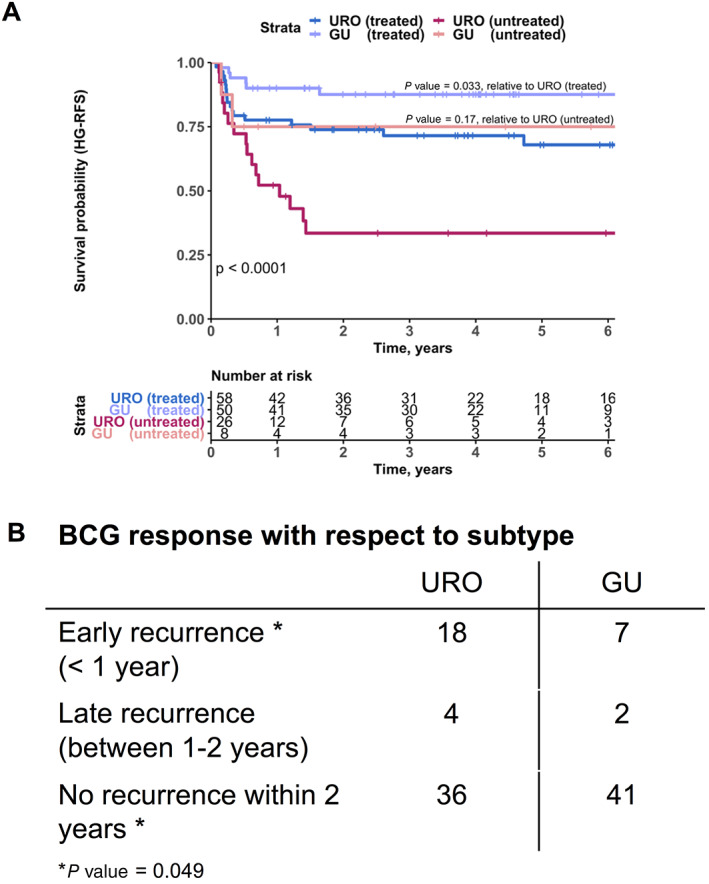
Subtype associations with BCG outcomes. (A) High‐grade RFS (HG‐RFS) for patients treated with adequate induction BCG versus untreated patients, stratified by subtype (URO versus GU). Colors represent patient treatment and subtype: treated URO (blue), untreated URO (magenta), treated GU (purple), and untreated GU (pink). P values are listed according to log‐rank test. Individual P values are listed for comparison of URO‐ and GU‐treated (p = 0.033) and ‐untreated (p = 0.17) patients, whereas comparison of all four groups demonstrated p < 0.0001. (B) BCG response with respect to subtype, observing the time to recurrence post‐BCG for URO and GU subtypes. P value is listed according to chi‐square test.
Discussion
This study describes a simple three‐antibody immunohistochemical algorithm that classifies NMIBC into four distinct subtypes. With a large body of evidence and investment in MIBC subtyping schemes, this study demonstrates a tractable way of validating these established classification schemes within the context of NMIBC. IHC‐based subtyping provides an important complement to RNA‐based approaches, which cannot feasibly be performed in every sample in a clinical laboratory. The antibodies used in this algorithm are routinely employed in surgical pathology [33, 34, 35, 36], and scoring can be performed digitally or visually [26] by qualitative assessment. The algorithm can therefore be readily implemented into the pathology workflow.
The validity of the four subtypes we observed is supported by unsupervised hierarchical clustering, which revealed three established clusters of NMIBC tumors (basal, URO, and GU) at expected frequencies, along with a novel cluster, URO‐KRT5+ [15, 26, 28, 31] (Figure 1C). Importantly, significant associations with progression and recurrence (Table 1, Figure 3A,B) indicate that identifying these subtypes can potentially provide important predictive and prognostic information and help guide clinical management if implemented into the pathology practice.
The novel URO‐KRT5+ subtype, constituting over 20% of NMIBC tumors, was enriched for low‐risk, low‐grade tumors that recur slower than any of the other subtypes. We also note that URO‐KRT5+ cases do not correspond to the RNA‐based subtype ‘UroB’, which occurs in more advanced stages and has increased basal marker expression and a poor outcome [15, 28]. Instead, URO‐KRT5+ most likely corresponds to UroA tumors, a luminal subtype (thus the URO designation) with expanded expression of KRT5. Recent work by Lindskrog et al transcriptomically characterized NMIBC and detected four mRNA classes [37]. Interestingly, class 3 tumors demonstrated positive immunohistochemical staining for GATA3 and KRT5, further supporting the presence of a luminal KRT5+ subtype [37]. Further characterization of URO‐KRT5+ tumors will be needed to determine whether they differ from their URO counterparts with regard to expression of other differentiation markers or genomic alterations. The URO‐KRT5+ subtype may represent a group of patients that requires less intense surveillance. If confirmed, identifying these tumors using this three‐antibody algorithm could reduce the economic burden, discomfort, and inconvenience of repeated cystoscopies.
In muscle‐invasive disease, the basal/SCCL subtype represents more aggressive, higher stage cancers [6, 8, 9, 12, 15, 38]. Here, we demonstrate similar aggressive features for the basal subtype in NMIBC, which is characterized by high stage, concomitant CIS, more rapid progression to muscle invasion, and faster recurrences. Although basal tumors are rare in NMIBC [13, 31, 38], when matched for stage and grade, these tumors presented statistically and clinically greater risk of progressing within 1 year when compared to the URO and GU subtypes (Figure 3F). The low frequency of this subtype observed in the current study confirms observations in previous NMIBC studies with similar proportions of basal tumors (4–10%) and associations with poor prognosis, identified by both RNA and IHC approaches [11, 13, 31, 38]. Despite their low frequency, identification of this aggressive subtype at diagnosis would provide valuable information for treatment intensification, such as closer surveillance and perhaps consideration for early cystectomy.
Universal to all subtyping schemes in NMIBC and MIBC is the broad (top‐level) classification into two categories, luminal and basal. Studies suggest that top‐level classification into luminal and basal subtypes is sufficient to identify prognostically different groups in NMIBC [22, 39, 40]. However, both the Lund taxonomy and others identify biological and prognostic differences between luminal subtypes, identifying genomically unstable and urothelial‐like subtypes [13, 14]. While the URO and GU subtypes are evenly distributed for grade and stage, we observed that the URO subtype recurred approximately 5.5 months earlier than the GU subtype. We observed that these differences in RFS were associated with p16 protein expression (low, intermediate, and high) (supplementary material, Figure S3). The division of URO and GU subtypes is supported by previous reports that a subset of early bladder cancers are defined by deep deletion or loss of chromosome 9p, its CDKN2A locus, and the cognate protein, p16 [4, 15, 25, 41, 42, 43]. GU tumors are an exception, with intact CDKN2A and p16 protein expression [15, 25]. Further supporting this ‘GU’ label, profiling of stage pT1 cohorts has indicated the presence of a ‘genomically unstable’ subtype, linking expression of p16 and E2F3 with a greater proportion of copy number gains and rearranged genomes. The results reported here suggest that the ‘GU’ label can be extended to encompass noninvasive NMIBC. Future work could investigate whether these p16‐positive cases are themselves genomically unstable or precursors of those that will become unstable if they progress.
The URO and GU subtypes are further defined by their rate of recurrence after BCG. In MIBC, the GU subtype is associated with poor prognosis along with a higher mutational burden and greater immune infiltration, resulting in increased responses to immune checkpoint inhibitors [44]. In NMIBC cohorts, Patschan et al demonstrated increased CD3+ infiltration in GU and basal/SCCL tumors compared with URO tumors [13]. Additional research in NMIBC has demonstrated that CD3D, an immune response gene involved in T‐cell signaling, is associated with poor prognosis when expressed at low levels [45]. Therefore, a lack of immune infiltration and adequate immune signaling in the URO subtype may explain the resulting poor RFS post‐BCG treatment. Furthermore, Meeks et al demonstrated that a significantly higher mutational burden was associated with response to BCG in a subset of non‐progressing tumors that also have intact CDKN2A [46]. The current work would suggest that GU tumors match these biological and clinical characteristics, with intact CDKN2A (p16) expression that results in better RFS post‐BCG treatment. In contrast to MIBC studies [15, 25, 38], this may explain why we do not observe a poor prognosis from the GU group, as they experience fewer recurrences and progression events, particularly after BCG treatment. Alternatively, it is possible that the GU subtype becomes more aggressive in higher stage (pT1 and MIBC) cohorts due to enrichment of additional molecular alterations [12, 13]. The comprehensive study published by Hedegaard et al does not observe differences in BCG response with respect to molecular classes; however, only 18% (n = 88/467) of their cohort was given BCG compared to 46% (n = 180/390) in this cohort [43]. To our knowledge, this is the first report detailing a significant relationship between the GU subtype and improved RFS after BCG. This supports that top‐level classification into one luminal subtype is insufficient to identify the prognostic differences.
An important addition to this study was the ability to observe the stability of molecular subtypes across recurrent NMIBC. Sjödahl et al observed that true molecular subtype switches were a rare event, and that URO tumors frequently remained URO while GU tumors typically had previous URO or GU tumors [31]. Similarly, we observed that subtypes are relatively stable over time. Future work in larger cohorts could investigate whether the frequency of true subtype switches was correlated with the time to each patient's recurrence. Furthermore, we also observed no significant associations between BCG treatment and changes in subtype (Figure 2). However, this is confounded by the fact that patients with GU tumors have fewer recurrences post‐BCG compared to URO, and thus there are fewer recurrences in which to track subtype changes. The parallels between these studies further suggest that this IHC subtyping method validates the biological and clinical characteristics of the Lund subtypes identified by both IHC and mRNA profiling methods.
The accuracy of grading and staging of NMIBC by histomorphology alone can be affected by limited inter‐ and intra‐observer reproducibility [47, 48, 49, 50]. Furthermore, spatial heterogeneity between TMA cores and whole sections is expected, and rules will need to be developed to incorporate scoring into routine clinical practice. However, matching TMA cores from samples demonstrated 86% agreement on subtype, and similar to observations from Sjödahl et al., the majority of these subtype differences (54%) were between URO and GU, resulting from borderline p16 threshold changes [31]. Given that these subtypes remain consistent across recurrences and between cores of the same sample, it seems likely that subtype heterogeneity is limited and that the three‐antibody assay can be successfully applied to whole slides.
Overall, we demonstrate that this IHC‐based algorithm has important clinical associations and potential utility in research and in clinical applications. Although challenging because it requires large cohorts to observe modest effect sizes, this IHC algorithm could be employed in future biomarker studies to improve risk stratification in NMIBC. In the meantime, especially those where grade or stage is uncertain, these IHC assays could be easily implemented in a pathology workflow to provide additional information on the likelihood that a cancer is low grade or high grade, or whether it is minimally invasive or noninvasive. In these instances, the basal staining pattern would indicate that a sample is very unlikely to be low grade and is 2.34–4.5 times more likely to be invasive than noninvasive (stage pT1). The URO‐KRT5 subtype, in contrast, might indicate the opposite, as it is 1.53–2.42 times more likely to be low grade than high grade and fewer than 10% are invasive. Furthermore, these IHC assays may provide clinicians with additional information regarding risk of recurrence and progression. More specifically, we observe that relative to the URO‐KRT5 subtype, the basal subtype recurs almost 2.5 years earlier and progresses 8 years earlier. Even when stratified for stage and grade (e.g. TaHG tumors), we continued to observe significant differences in RFS between the URO and URO‐KRT5+ subtypes (Figure 3C and supplementary material, Figure S2C). Additionally, we observed improved RFS post‐BCG treatment for the GU subtype relative to its URO counterpart. These observations further support the potential value of identifying URO and GU as two separate luminal categories, which is currently not a universal component of subtyping models. Future work validating these observations in multicenter cohorts will be crucial for evaluating its overall clinical utility.
Author contributions statement
CLJ and DMB conceived and drafted the manuscript. CLJ created all figures and tables. CLJ and CH selected cases and constructed the cohort and associated TMAs. LC, KYMR and DMB performed pathological review. KV, JJ and VFB contributed to clinical data collection. DRS contributed to clinical data interpretation. GS contributed to conception of IHC‐based profiling methods. RJG contributed to the bioinformatic analyses. All authors contributed to manuscript editing.
Supporting information
Figure S1. Example staining patterns for GATA3 scores, KRT5 proximity scores, and p16 intensity scores
Figure S2. Time to recurrence stratified by stage and subtype
Figure S3. Recurrence‐free survival for URO and GU samples, stratified by p16 protein expression cutoffs determined by unsupervised hierarchical clustering
Figure S4. Clustering and survival analysis indicate that p63 was unnecessary to further classify URO and GU subtypes
Table S1. Characteristics of all patients in the cohort
Table S2. Characteristics of all samples (n = 481) representing the total cohort of 390 NMIBC patients
Table S3. Sample characteristics of metachronous samples from 49 high‐grade NMIBC patients
Table S4. Antibodies used for immunohistochemical staining
Table S5. Scoring and assessment methods that classify each subtype
Acknowledgements
We are grateful for the assistance of Lee Boudreau in TMA construction and Shakeel Virk for slide scanning. We also thank Winnie Fu for assistance with slide retrieval. This study was conducted with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario (DMB). Additional support (30%) was provided by the Cancer Research Society and Bladder Cancer Canada through the Operating Grant Funding Program (DMB). Fellowship support was provided by Queen's University through the R.J. Wilson Fellowship and R. Samuel McLaughlin Fellowship (CLJ).
No conflicts of interest were declared.
References
- 1. Babjuk M, Böhle A, Burger M, et al. EAU guidelines on non‐muscle‐invasive urothelial carcinoma of the bladder: update 2016. Eur Urol 2017; 71: 447–461. [DOI] [PubMed] [Google Scholar]
- 2. Cambier S, Sylvester RJ, Collette L, et al. EORTC nomograms and risk groups for predicting recurrence, progression, and disease‐specific and overall survival in non–muscle‐invasive stage Ta–T1 urothelial bladder cancer patients treated with 1–3 years of maintenance bacillus Calmette‐Guérin. Eur Urol 2016; 69: 60–69. [DOI] [PubMed] [Google Scholar]
- 3. Chang SS, Boorjian SA, Chou R, et al. Diagnosis and treatment of non‐muscle invasive bladder cancer: AUA/SUO guideline. J Urol 2016; 196: 1021–1029. [DOI] [PubMed] [Google Scholar]
- 4. Sjödahl G, Lauss M, Lövgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 2012; 18: 3377–3386. [DOI] [PubMed] [Google Scholar]
- 5. Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high‐grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 2014; 111: 3110–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle‐invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014; 25: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robertson AG, Kim J, Al‐Ahmadie H, et al. Comprehensive molecular characterization of muscle‐invasive bladder cancer. Cell 2017; 171: 540–556.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamoun A, de Reyniès A, Allory Y, et al. A consensus molecular classification of muscle‐invasive bladder cancer. Eur Urol 2020; 77: 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dadhania V, Zhang M, Zhang L, et al. Meta‐analysis of the luminal and basal subtypes of bladder cancer and the identification of signature immunohistochemical markers for clinical use. EBioMedicine 2016; 12: 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He X, Marchionni L, Hansel DE, et al. Differentiation of a highly tumorigenic basal cell compartment in urothelial carcinoma. Stem Cells 2009; 27: 1487–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Volkmer J‐P, Sahoo D, Chin RK, et al. Three differentiation states risk‐stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci U S A 2012; 109: 2078–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seiler R, Ashab HAD, Erho N, et al. Impact of molecular subtypes in muscle‐invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol 2017; 72: 544–554. [DOI] [PubMed] [Google Scholar]
- 13. Patschan O, Sjödahl G, Chebil G, et al. A molecular pathologic framework for risk stratification of stage T1 urothelial carcinoma. Eur Urol 2015; 68: 824–832. [DOI] [PubMed] [Google Scholar]
- 14. Robertson AG, Groeneveld CS, Jordan B, et al. Identification of differential tumor subtypes of T1 bladder cancer. Eur Urol 2020; 78: 533–537. [DOI] [PubMed] [Google Scholar]
- 15. Sjödahl G, Eriksson P, Liedberg F, et al. Molecular classification of urothelial carcinoma: global mRNA classification versus tumour‐cell phenotype classification. J Pathol 2017; 242: 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sjödahl G, Jackson CL, Bartlett JM, et al. Molecular profiling in muscle invasive bladder cancer: more than the sum of its parts. J Pathol 2019; 247: 563–573. [DOI] [PubMed] [Google Scholar]
- 17. Warrick JI, Walter V, Yamashita H, et al. FOXA1, GATA3 and PPARɣ cooperate to drive luminal subtype in bladder cancer: a molecular analysis of established human cell lines. Sci Rep 2016; 6: 38531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fishwick C, Higgins J, Percival‐Alwyn L, et al. Heterarchy of transcription factors driving basal and luminal cell phenotypes in human urothelium. Cell Death Differ 2017; 24: 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chin L, Pomerantz J, DePinho RA. The INK4a/ARF tumor suppressor: one gene—two products—two pathways. Trends Biochem Sci 1998; 23: 291–296. [DOI] [PubMed] [Google Scholar]
- 20. Shariat SF, Tokunaga H, Zhou J, et al. p53, p21, pRB, and p16 expression predict clinical outcome in cystectomy with bladder cancer. J Clin Oncol 2004; 22: 1014–1024. [DOI] [PubMed] [Google Scholar]
- 21. Gan X, Lin X, He R, et al. Prognostic and clinicopathological significance of downregulated p16 expression in patients with bladder cancer: a systematic review and meta‐analysis. Dis Markers 2016; 2016: 5259602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Breyer J, Wirtz RM, Otto W, et al. In stage pT1 non‐muscle‐invasive bladder cancer (NMIBC), high KRT20 and low KRT5 mRNA expression identify the luminal subtype and predict recurrence and survival. Virchows Arch 2017; 470: 267–274. [DOI] [PubMed] [Google Scholar]
- 23. Krüger S, Mahnken A, Kausch I, et al. P16 immunoreactivity is an independent predictor of tumor progression in minimally invasive urothelial bladder carcinoma. Eur Urol 2005; 47: 463–467. [DOI] [PubMed] [Google Scholar]
- 24. Chan KS, Espinosa I, Chao M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor‐initiating cells. Proc Natl Acad Sci U S A 2009; 106: 14016–14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marzouka N, Eriksson P, Rovira C, et al. A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort. Sci Rep 2018; 8: 3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sjödahl G. Molecular subtype profiling of urothelial carcinoma using a subtype‐specific immunohistochemistry panel. In: Urothelial Carcinoma: Methods and Protocols, Schulz WA, Hoffmann MJ, Niegisch G (Eds). Springer: New York, 2018; 53–64. [DOI] [PubMed] [Google Scholar]
- 27. Guo CC, Bondaruk J, Yao H, et al. Assessment of luminal and basal phenotypes in bladder cancer. Sci Rep 2020; 10: 9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bernardo C, Eriksson P, Marzouka N, et al. Molecular pathology of the luminal class of urothelial tumors. J Pathol 2019; 249: 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eble JN (Ed.). Pathology and genetics of tumours of the urinary system and male genital organs. Editorial and Consensus Conference in Lyon, France, 14–18 December, 2002. Reprint. IARC Press: Lyon, 2006.
- 30. Pocock SJ, Clayton TC, Altman DG. Survival plots of time‐to‐event outcomes in clinical trials: good practice and pitfalls. Lancet 2002; 359: 1686–1689. [DOI] [PubMed] [Google Scholar]
- 31. Sjödahl G, Eriksson P, Patschan O, et al. Molecular changes during progression from nonmuscle invasive to advanced urothelial carcinoma. Int J Cancer 2020; 146: 2636–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sjödahl G, Lövgren K, Lauss M, et al. Toward a molecular pathologic classification of urothelial carcinoma. Am J Pathol 2013; 183: 681–691. [DOI] [PubMed] [Google Scholar]
- 33. Chang A, Amin A, Gabrielson E, et al. Utility of GATA3 immunohistochemistry in differentiating urothelial carcinoma from prostate adenocarcinoma and squamous cell carcinomas of the uterine cervix, anus, and lung. Am J Surg Pathol 2012; 36: 1472–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Darragh TM, Colgan TJ, Cox JT, et al. The lower anogenital squamous terminology standardization project for HPV‐associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Arch Pathol Lab Med 2012; 136: 1266–1297. [DOI] [PubMed] [Google Scholar]
- 35. Miettinen M, McCue PA, Sarlomo‐Rikala M, et al. GATA3: a multispecific but potentially useful marker in surgical pathology: a systematic analysis of 2500 epithelial and nonepithelial tumors. Am J Surg Pathol 2014; 38: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu PG, Weiss LM. Expression of cytokeratin 5/6 in epithelial neoplasms: an immunohistochemical study of 509 cases. Mod Pathol 2002; 15: 6–10. [DOI] [PubMed] [Google Scholar]
- 37. Lindskrog SV, Prip F, Lamy P, et al. An integrated multi‐omics analysis identifies prognostic molecular subtypes of non‐muscle‐invasive bladder cancer. Nat Commun 2021; 12: 2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tan TZ, Rouanne M, Tan KT, et al. Molecular subtypes of urothelial bladder cancer: results from a meta‐cohort analysis of 2411 tumors. Eur Urol 2019; 75: 423–432. [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez Pena MDC, Chaux A, Eich M‐L, et al. Immunohistochemical assessment of basal and luminal markers in non‐muscle invasive urothelial carcinoma of bladder. Virchows Arch 2019; 475: 349–356. [DOI] [PubMed] [Google Scholar]
- 40. Rebola J, Aguiar P, Blanca A, et al. Predicting outcomes in non‐muscle invasive (Ta/T1) bladder cancer: the role of molecular grade based on luminal/basal phenotype. Virchows Arch 2019; 475: 445–455. [DOI] [PubMed] [Google Scholar]
- 41. Lindgren D, Sjödahl G, Lauss M, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One 2012; 7: e38863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pietzak EJ, Bagrodia A, Cha EK, et al. Next‐generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol 2017; 72: 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive transcriptional analysis of early‐stage urothelial carcinoma. Cancer Cell 2016; 30: 27–42. [DOI] [PubMed] [Google Scholar]
- 44. Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD‐L1 blockade by contributing to exclusion of T cells. Nature 2018; 554: 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen X, Jiang F, Jia C, et al. Comprehensive gene expression analysis in NMIBC using RNA‐seq reveals new therapy strategies. Front Oncol 2019; 9: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meeks JJ, Carneiro BA, Pai SG, et al. Genomic characterization of high‐risk non‐muscle invasive bladder cancer. Oncotarget 2016; 7: 75176–75184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Soukup V, Čapoun O, Cohen D, et al. Prognostic performance and reproducibility of the 1973 and 2004/2016 World Health Organization grading classification systems in non–muscle‐invasive bladder cancer: a European Association of Urology non‐muscle invasive bladder cancer guidelines panel systematic review. Eur Urol 2017; 72: 801–813. [DOI] [PubMed] [Google Scholar]
- 48. May M, Brookman‐Amissah S, Roigas J, et al. Prognostic accuracy of individual uropathologists in noninvasive urinary bladder carcinoma: a multicentre study comparing the 1973 and 2004 World Health Organisation classifications. Eur Urol 2010; 57: 850–858. [DOI] [PubMed] [Google Scholar]
- 49. Bol MG, Baak JP, Buhr‐Wildhagen S, et al. Reproducibility and prognostic variability of grade and lamina propria invasion in stages Ta, T1 urothelial carcinoma of the bladder. J Urol 2003; 169: 1291–1294. [DOI] [PubMed] [Google Scholar]
- 50. Bosschieter J, Hentschel A, Savci‐Heijink CD, et al. Reproducibility and prognostic performance of the 1973 and 2004 World Health Organization classifications for grade in non–muscle‐invasive bladder cancer: a multicenter study in 328 bladder tumors. Clin Genitourin Cancer 2018; 16: e985–e992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Example staining patterns for GATA3 scores, KRT5 proximity scores, and p16 intensity scores
Figure S2. Time to recurrence stratified by stage and subtype
Figure S3. Recurrence‐free survival for URO and GU samples, stratified by p16 protein expression cutoffs determined by unsupervised hierarchical clustering
Figure S4. Clustering and survival analysis indicate that p63 was unnecessary to further classify URO and GU subtypes
Table S1. Characteristics of all patients in the cohort
Table S2. Characteristics of all samples (n = 481) representing the total cohort of 390 NMIBC patients
Table S3. Sample characteristics of metachronous samples from 49 high‐grade NMIBC patients
Table S4. Antibodies used for immunohistochemical staining
Table S5. Scoring and assessment methods that classify each subtype


