Abstract
Background
Human twin studies and other studies have indicated that chronic pain has heritability that ranges from 30% to 70%. We aimed to identify potential genetic variants that contribute to the susceptibility to chronic pain and efficacy of administered drugs. We conducted genome-wide association studies (GWASs) using whole-genome genotyping arrays with more than 700,000 markers in 191 chronic pain patients and a subgroup of 89 patients with postherpetic neuralgia (PHN) in addition to 282 healthy control subjects in several genetic models, followed by additional gene-based and gene-set analyses of the same phenotypes. We also performed a GWAS for the efficacy of drugs for the treatment of pain.
Results
Although none of the single-nucleotide polymorphisms (SNPs) were found to be genome-wide significantly associated with chronic pain (p ≥ 1.858 × 10−7), the GWAS of PHN patients revealed that the rs4773840 SNP within the ABCC4 gene region was significantly associated with PHN in the trend model (nominal p = 1.638 × 10−7). In the additional gene-based analysis, one gene, PRKCQ, was significantly associated with chronic pain in the trend model (adjusted p = 0.03722). In the gene-set analysis, several gene sets were significantly associated with chronic pain and PHN. No SNPs were significantly associated with the efficacy of any of types of drugs in any of the genetic models.
Conclusions
These results suggest that the PRKCQ gene and rs4773840 SNP within the ABCC4 gene region may be related to the susceptibility to chronic pain conditions and PHN, respectively.
Keywords: Genome-wide association study, single-nucleotide polymorphism, chronic pain, postherpetic neuralgia, gene-based/gene-set analysis
Introduction
An estimated 15–50% of the population experiences pain at any given time.1–3 Some pain is acute or subacute, but other forms of pain are chronic. 4 Chronic pain is a public health problem that affects the general population physically, psychologically, and socially. 5 Chronic pain is prevalent among the Japanese population, affecting 15.4–47% of individuals.5,6 The median prevalence of chronic pain was reported to be 26% among the adult population worldwide, ranging from 7% to 55%. 5 Chronic pain has been reported to be associated with health status, work productivity, impairments in daily activities, healthcare resource utilization, and economic burdens in Japan. 6 According to a recent report, people with chronic pain, particularly cancer-related pain, have a slightly higher risk of death. 7
Chronic pain conditions are complex traits with multiple etiologies. With regard to non-genetic and nonheritable factors, regression analyses have shown that chronic pain is associated with age, sex, unemployment, living status, exercise, 5 body mass index, fatigue, sleep, and mobility problems. 3 Human twin studies and other genetic studies have indicated that the heritability of chronic pain ranges from 30% to 70%. 8 Approximately 37%, 52–68%, and 35–58% of cases of neuropathic pain, low back pain, and neck pain, respectively, may be heritable.9,10 Previous genetic studies of candidate genes that are related to pain mechanisms found that human genetic variations were associated with various pain-related phenotypes.1,11,12 Pain-related genetic variations have also been identified for chronic pain conditions, such as the ADRB2,13,14 HTR2A, 15 SCN9A, 16 KCNS1, 17 CACNA2D3, 18 CACNG2, 19 COMT, 20 IL4, 14 and IL10 21 genes. Candidate genes for chronic postsurgical pain (CPSP) were systematically reviewed by Hoofwijk et al., 22 and candidate genes for neuropathic pain have been described in several previous reports.23–26 Chronic pain-related single-nucleotide polymorphisms (SNPs) have also been explored based on recent advances in high-density SNP arrays that can screen hundreds of thousands or millions of genetic markers throughout the human genome. For example, Jones et al. (2016) found that a SNP that was colocalized to the NGF gene, which encodes nerve growth factor, was associated with dysmenorrhea in a genome-wide association study (GWAS) of a cohort of females. 27 Peters et al. identified a common genetic variant on chromosome 5p15.2 that was associated with joint-specific chronic widespread pain (CWP) in a large-scale GWAS meta-analysis. 28 Genome-wide association studies have also been applied to investigate neuropathic pain. Several candidate loci were reported to be associated with pain conditions, including diabetic neuropathic pain.29–32
In the present study, we conducted GWASs of patients with chronic pain to identify potential genetic variants that contribute to the susceptibility to pain conditions and efficacy of several types of drugs that are used to treat pain. We also performed a GWAS to explore genetic factors that are associated with neuropathic pain, specifically postherpetic neuralgia (PHN).
Methods
Subjects with chronic pain and healthy subjects
We enrolled 194 adult patients who suffered from chronic pain who visited JR Tokyo General Hospital (Tokyo, Japan), Juntendo University Hospital (Tokyo, Japan), or Nihon University Itabashi Hospital (Tokyo, Japan) for the treatment of chronic pain and were apparently Japanese. Most of the patients were treated with analgesics before recruitment or were scheduled to be treated with analgesics at the time of recruitment in the study. We excluded patients with severe coexisting complications. The detailed demographic and clinical data of the subjects are provided in Table 1.
Table 1.
Demographic and clinical data of patient subjects.
| Demographic data | n | Minimum | Maximum | Mean | SD | Median | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender of all patients | |||||||||||
| Male | 89 | ||||||||||
| Female | 100 | ||||||||||
| Age (years) | 193 | 22 | 89 | 65.18 | 13.95 | 68.00 | |||||
| Weight (kg) | 182 | 34 | 98 | 57.32 | 12.21 | 57.00 | |||||
|
Status of patients |
Absence |
Presence |
Opioids |
Antidep-ressant |
Anticon-vulsant |
NSAIDs† |
GABA§ |
Ketamine |
Neuro-tropin |
Lidocaine |
Others |
| Nerve block | 132 | 25 | |||||||||
| Allodynia | 75 | 30 | |||||||||
| Administration of drugs | 50 | 66 | 99 | 25 | 58 | 7 | 5 | 18 | 4 | ||
|
Diagnosis (disease status) |
|
n |
Diagnosis (disease status) |
|
|
|
n |
||||
| Postherpetic neuralgia (PHN) | 92 | Spinal canal stenosis | 20 | ||||||||
| Lower back pain (LBP) | 13 | Postoperative pain | 12 | ||||||||
| Hernia of intervertebral disk | 8 | Neck pain | 8 | ||||||||
| Others | 46 | ||||||||||
†Non-steroidal anti-inflammatory drugs.
§Gamma-aminobutyric acid receptor modulators.
We also enrolled 282 healthy adult volunteers as controls who were disease-free, did not experience chronic pain, and who lived in or near the Kanto area in Japan. The detailed demographic data of the control subjects and their statistics are detailed in previous reports.33,34
The study protocol was approved by the Institutional Review Board of JR Tokyo General Hospital (Tokyo, Japan), Institutional Review Board of Juntendo University Hospital (Tokyo, Japan), Institutional Review Board of Nihon University Itabashi Hospital (Tokyo, Japan), and Institutional Review Board of Tokyo Metropolitan Institute of Medical Science (Tokyo, Japan). Written informed consent was obtained from all of the patients.
Patient characteristics and clinical data
In the patient subjects, we obtained data on surgical history, treatment history, pain status (e.g., presence/absence of nerve block and allodynia), drug treatments, and disease status (e.g., postherpetic neuralgia [PHN], spinal canal stenosis, lower back pain [LBP], etc.; Table 1). Some of the patients were affected by multiple diseases.
Various types of drugs were administered to the patients for the treatment of pain. In the present study, these drugs were divided into several groups for the analysis, including opioids (e.g., morphine and codeine), antidepressants (e.g., fluvoxamine and amitriptyline), anticonvulsants (e.g., gabapentin and pregabalin), nonsteroidal antiinflammatory drugs (NSAIDs; e.g., loxoprofen and diclofenac), γ-aminobutyric acid (GABA) receptor agonists that can be used as anticonvulsants or anxiolytics (e.g., clonazepam and diazepam), ketamine, neurotropin, lidocaine, and other drugs (e.g., Chinese herbal medicines and mexiletine). The detailed data on drug administration are provided in Table 1. Some patients received only one type of drug, whereas others received several types of drugs. Some of the drugs were effective for a number of patients, but others were not. Such drug administration and efficacy were comprehensively recorded for the statistical analyses.
Whole-genome genotyping and quality control
A total of 194 DNA samples from the patients were used for genotyping. Total genomic DNA was extracted from whole-blood samples using standard procedures. Whole-genome genotyping was performed using the Infinium assay II with an iScan system (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions, and two kinds of BeadChips were used for genotyping 153 and 41 patient samples, respectively: HumanOmni1-Quad v1.0 (total markers: 11,34,514) and HumanOmniExpress-12 v1.1 (total markers: 7,19,665). For genotyping 282 control samples, the HumanOmniExpressExome-8 v1.2 BeadChip (total markers: 9,64,193) was used. Other details for genotyping are described in the Supplementary Methods. The data for the whole-genome-genotyped samples were analyzed using GenomeStudio with the Genotyping module v3.3.7 (Illumina) to evaluate the quality of the results. In the data-cleaning process as detailed in the Supplementary Methods, three patient samples were excluded from further analyses, whereas no control samples were excluded based on this criterion. For the study of the effects of drugs in patients, 4,47,634 SNPs survived the entire filtration process and were used in the study. For the case-control study to compare genotypes between the patient and control subjects, more stringent criteria were used for filtration to remove spurious results, and 445,723 SNPs survived the entire filtration process and were used in the study. Furthermore, the TaqMan allelic discrimination assay (Life Technologies, Carlsbad, CA, USA) was performed to confirm the genotype data of the top 20 candidate SNPs if the data were suspected to be dubious.
Statistical analysis
A GWAS of patients with chronic pain was conducted to investigate associations between genetic variations and the susceptibility to chronic pain in all 191 patient subjects who passed the quality control criteria. A GWAS of a subgroup of 89 patients with PHN was also conducted because PHN was the most prevalent pain condition in our samples. A total of 282 control subjects were used in both of these analyses. Furthermore, another GWAS of only 191 patient subjects was also conducted to investigate the effects of drugs.
To explore associations between SNPs and disease status, Fisher’s exact tests were conducted in both analyses using both all patients and patients with PHN to compare genotype data between the patient and control subjects. To explore SNPs that were associated with the effects of drugs in patients, patient subjects were divided into two groups based on the effectiveness of five major kinds of drugs (i.e., opioids, antidepressants, anticonvulsants, NSAIDs, and GABA receptor agonists; Table 1), and Fisher’s exact tests were conducted to compare genotype data between the two groups. Trend, dominant, and recessive genetic models were used for all of the analyses because of insufficient knowledge of genetic factors that are associated with chronic pain, PHN, and the effectiveness of drugs that are used for the treatment of chronic pain. The association study included both female and male subjects for autosomal markers, although male genotypes were excluded from the analysis of X chromosome markers. All of the statistical analyses were performed using gPLINK v. 2.050, PLINK v. 1.07 (http://zzz.bwh.harvard.edu/plink/index.shtml; accessed July 15, 2018), 35 and Haploview v. 4.2. 36
For the correction of multiple testing in the GWAS, Bonferroni correction was used for the number of inferred Meff, defined in simpleM software,37–39 which is a multiple-testing correction method for genetic association studies that uses correlated SNPs. In our preliminary calculation, by substituting missing genotypes with homozygotes of minor or major alleles and heterozygotes, Meff was estimated to be 256,506–269,170. Therefore, statistical significance for the GWAS was defined as a corrected p < 0.05/269,170 = 1.858 × 10−7 in the present study.
To further understand the genetic backgrounds and molecular mechanisms that underlie complex traits, such as chronic pain and PHN, gene-based and gene-set approaches were adopted with Multi-marker Analysis of GenoMic Annotation (MAGMA) v1.06, 40 which is also available on the Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA GWAS) v1.3.3 platform, 41 as detailed in the Supplementary Methods. In the gene-set analysis, gene sets were defined using the Molecular Signatures Database (MSigDB) v6.1, 42 and a total of 10,654 gene sets (curated gene sets: 4737, GO terms: 5917) from MsigDB were tested.
Results
Identification of genetic polymorphisms associated with chronic pain and postherpetic neuralgia by GWAS
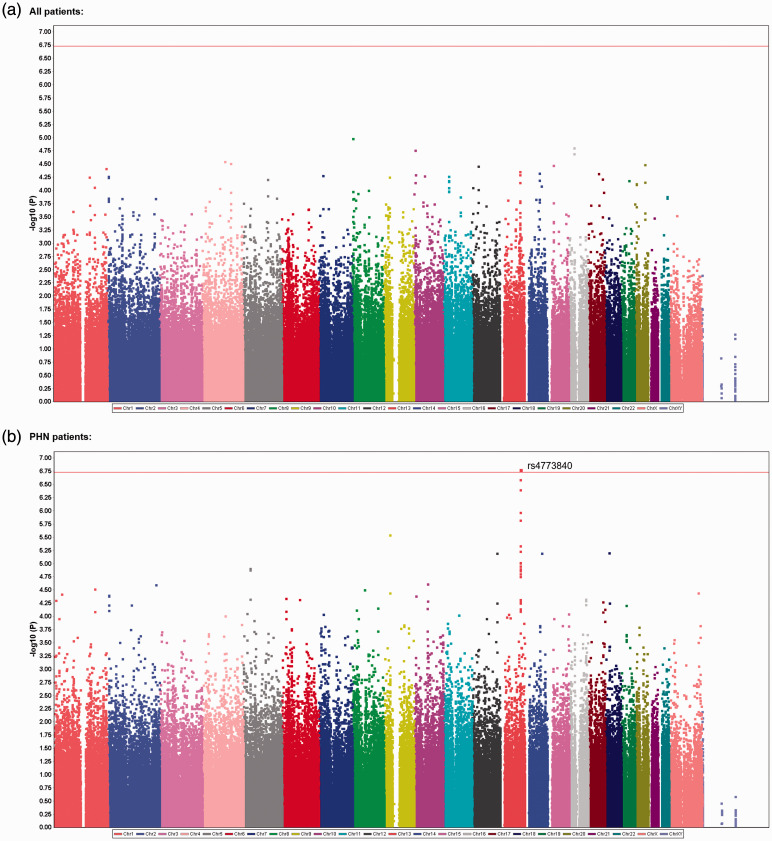
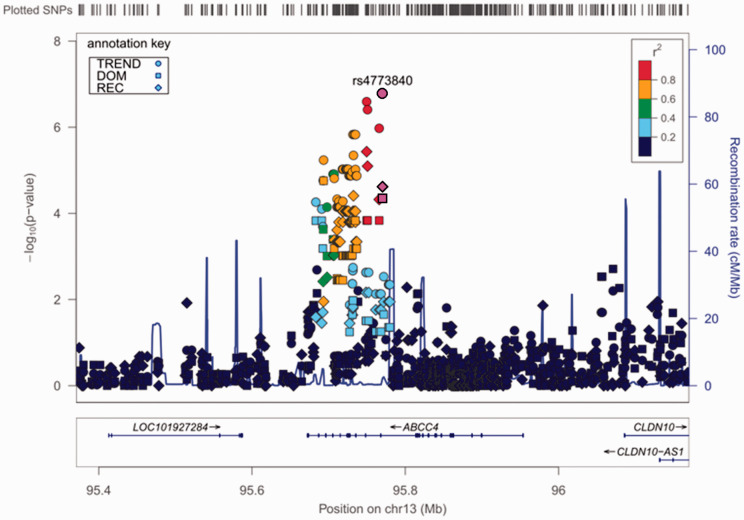
We comprehensively explored genetic variations that were associated with chronic pain conditions in a total of 191 patients who visited hospitals for treatment, and 282 adult healthy subjects were recruited as controls.33,34 In the GWAS of all patients, 4,45,723 SNPs that passed the quality control criteria were selected as candidate genetic polymorphisms in the trend, dominant, and recessive models. Among the highly ranked SNPs, genotype data for one SNP, rs6481467, was suspected to be dubious because of its cluster separation. After screening using the TaqMan allelic discrimination assay, the data were found to be erroneous for this SNP and thus were removed from the list of candidate SNPs. Table 2 shows the top 20 candidate SNPs in each genetic model after final quality control. However, none of the SNPs were genome-wide significantly associated with the phenotype (p ≥ 1.858 × 10−7; Table 2, Figure 1(a)). We then conducted another GWAS of the same SNPs by including only a subgroup of 89 patients with PHN. A significant association was found between the rs4773840 SNP that mapped to 13q32.1 and PHN in the trend model (nominal p = 1.638 × 10−7; Table 3, Figure 1(b)). The calculated log10 values (observed p value) for most of the analyzed SNPs were in accordance with or below the expected values based on the null hypothesis of a uniform distribution in the QQ plot (Supplementary Figures S1 and S2). The values for the rs4773840 SNP and other SNPs that ranked high in Table 3 were obviously above the expected values (Supplementary Figure S2). The gene that was located in this region of the rs4773840 SNP was ABCC4, which encodes adenosine triphosphate binding cassette subfamily C member 4. Most of the other SNPs in this gene region that ranked high in Table 3 were in relatively strong linkage disequilibrium (LD) with one another, and all of these SNPs were within the ABCC4 gene region (Figure 2). As shown in Table 3, an increment of the minor C allele carriage in the rs4773840 SNP was associated with a greater risk of PHN.
Table 2.
Top 20 candidate SNPs selected from GWAS for all patients.
| Model | Rank | CHR | SNP | Position | P | Related gene | Genotype (patients) |
Genotype (controls) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/B | B/B | A/A | A/B | B/B | |||||||
| Trend | 1 | 8 | rs10086452 | 3691292 | 0.00001026 | CSMD1 | 2 | 48 | 141 | 18 | 107 | 156 |
| Trend | 2 | 16 | rs12708686 | 25789460 | 0.00001532 | HS3ST4 | 5 | 70 | 116 | 28 | 133 | 121 |
| Trend | 3 | 10 | rs688391 | 6529658 | 0.00001721 | PRKCQ | 59 | 105 | 27 | 61 | 126 | 95 |
| Trend | 4 | 16 | rs9989408 | 25786610 | 0.0000198 | HS3ST4 | 8 | 76 | 107 | 34 | 140 | 108 |
| Trend | 5 | 4 | rs4141270 | 106242441 | 0.00002805 | 44 | 95 | 52 | 33 | 126 | 122 | |
| Trend | 6 | 4 | rs10518617 | 133841275 | 0.00003039 | 29 | 84 | 78 | 15 | 107 | 159 | |
| Trend | 7 | 20 | rs4811012 | 48294701 | 0.00003177 | 3 | 58 | 130 | 22 | 116 | 144 | |
| Trend | 8 | 15 | rs6493688 | 29560167 | 0.00003323 | 40 | 89 | 62 | 25 | 124 | 133 | |
| Trend | 9 | 12 | rs10844159 | 32288782 | 0.00003414 | BICD1 | 21 | 81 | 89 | 10 | 94 | 178 |
| Trend | 10 | 1 | rs10803183 | 242444561 | 0.00003789 | 5 | 58 | 128 | 6 | 36 | 240 | |
| Trend | 11 | 13 | rs4773840 | 94568426 | 0.00004323 | ABCC4 | 22 | 80 | 89 | 10 | 96 | 176 |
| Trend | 12 | 14 | rs11621135 | 70729362 | 0.00004646 | 10 | 65 | 115 | 2 | 66 | 214 | |
| Trend | 13 | 17 | rs2958927 | 50314685 | 0.00004719 | 29 | 81 | 77 | 17 | 104 | 161 | |
| Trend | 14 | 13 | rs1678353 | 94547567 | 0.00004959 | ABCC4 | 23 | 81 | 87 | 9 | 103 | 170 |
| Trend | 15 | 10 | rs4749828 | 9062151 | 0.00004966 | 15 | 80 | 95 | 6 | 89 | 187 | |
| Trend | 16 | 7 | rs12700309 | 21850980 | 0.00005138 | DNAH11 | 57 | 99 | 35 | 48 | 144 | 90 |
| Trend | 17 | 10 | rs17784350 | 50512270 | 0.00005223 | CHAT | 7 | 61 | 123 | 25 | 127 | 130 |
| Trend | 18 | 2 | rs2693818 | 6121959 | 0.0000536 | 31 | 80 | 79 | 59 | 166 | 57 | |
| Trend | 19 | 11 | rs6265 | 27636492 | 0.00005366 | BDNF-AS1,BDNF | 40 | 107 | 44 | 34 | 136 | 112 |
| Trend | 19 | 11 | rs11030104 | 27641093 | 0.00005366 | BDNF-AS1,BDNF | 40 | 107 | 44 | 34 | 136 | 112 |
| Dominant | 1 | 2 | rs2693818 | 6121959 | 0.0000009002 | 31 | 80 | 79 | 59 | 166 | 57 | |
| Dominant | 2 | 2 | rs6718476 | 6112647 | 0.0000009454 | 31 | 81 | 79 | 59 | 166 | 57 | |
| Dominant | 3 | 10 | rs688391 | 6529658 | 0.000001239 | PRKCQ | 59 | 105 | 27 | 61 | 126 | 95 |
| Dominant | 4 | 10 | rs604663 | 6544132 | 0.000002684 | PRKCQ | 52 | 110 | 29 | 57 | 128 | 97 |
| Dominant | 5 | 1 | rs10803183 | 242444561 | 0.000005297 | 5 | 58 | 128 | 6 | 36 | 240 | |
| Dominant | 6 | 11 | rs1488830 | 27593461 | 0.00003125 | BDNF-AS1 | 53 | 107 | 31 | 54 | 134 | 94 |
| Dominant | 7 | 18 | rs12964456 | 30023916 | 0.00003475 | NOL4 | 17 | 56 | 118 | 20 | 143 | 118 |
| Dominant | 8 | 4 | rs6531299 | 33872088 | 0.00003526 | 14 | 82 | 95 | 14 | 74 | 194 | |
| Dominant | 9 | 20 | rs6133220 | 551620 | 0.00003676 | 36 | 114 | 41 | 38 | 132 | 112 | |
| Dominant | 10 | 2 | rs941009 | 6058737 | 0.00003957 | 25 | 83 | 83 | 54 | 157 | 71 | |
| Dominant | 11 | 1 | rs6656194 | 164031638 | 0.00004554 | 33 | 98 | 60 | 29 | 111 | 142 | |
| Dominant | 12 | 7 | rs6461595 | 21724570 | 0.0000477 | DNAH11 | 41 | 111 | 39 | 53 | 122 | 107 |
| Dominant | 13 | 8 | rs2433150 | 6489560 | 0.00005107 | 5 | 40 | 146 | 13 | 104 | 165 | |
| Dominant | 14 | 13 | rs9532107 | 37187961 | 0.00005386 | TRPC4 | 14 | 67 | 110 | 33 | 140 | 109 |
| Dominant | 15 | 2 | rs10204095 | 57652544 | 0.0000553 | 5 | 37 | 148 | 10 | 101 | 166 | |
| Dominant | 16 | 4 | rs7670109 | 184691188 | 0.00005679 | 38 | 87 | 66 | 74 | 157 | 51 | |
| Dominant | 17 | 6 | rs13196989 | 184373 | 0.00005703 | 8 | 74 | 108 | 10 | 61 | 211 | |
| Dominant | 18 | 3 | rs7610425 | 150967983 | 0.00005804 | ANKUB1 | 9 | 90 | 92 | 11 | 82 | 189 |
| Dominant | 19 | 2 | rs12468070 | 6077432 | 0.00006067 | 25 | 84 | 82 | 56 | 155 | 71 | |
| Dominant | 20 | 14 | rs2167151 | 78933086 | 0.00006216 | NRXN3 | 17 | 84 | 90 | 14 | 82 | 186 |
| Recessive | 1 | 1 | rs4520412 | 15232554 | 0.0000008571 | KAZN | 25 | 110 | 56 | 92 | 115 | 75 |
| Recessive | 2 | 11 | rs1519480 | 27632288 | 0.000002159 | BDNF-AS1 | 0 | 61 | 130 | 25 | 101 | 156 |
| Recessive | 3 | 6 | rs3777799 | 133631276 | 0.000003063 | EYA4 | 22 | 54 | 111 | 4 | 93 | 185 |
| Recessive | 4 | 8 | rs12545634 | 26929236 | 0.00001289 | 39 | 77 | 75 | 19 | 121 | 142 | |
| Recessive | 5 | 2 | rs10205827 | 75356361 | 0.00002183 | 10 | 102 | 79 | 52 | 122 | 107 | |
| Recessive | 6 | 2 | rs10208470 | 75356624 | 0.00002186 | 10 | 102 | 79 | 52 | 122 | 108 | |
| Recessive | 7 | 7 | rs12538837 | 97522404 | 0.00004215 | 27 | 111 | 53 | 86 | 128 | 68 | |
| Recessive | 8 | 8 | rs10086635 | 26955860 | 0.00004484 | 48 | 76 | 67 | 30 | 142 | 110 | |
| Recessive | 9 | 4 | rs6826653 | 19736139 | 0.00004904 | 15 | 55 | 121 | 2 | 84 | 196 | |
| Recessive | 10 | 2 | rs9309489 | 75355228 | 0.00004915 | 11 | 101 | 79 | 52 | 122 | 108 | |
| Recessive | 11 | 10 | rs2026432 | 6547609 | 0.00004948 | PRKCQ | 25 | 106 | 60 | 81 | 130 | 71 |
| Recessive | 12 | 13 | rs9521844 | 110018508 | 0.00005096 | 0 | 61 | 130 | 19 | 94 | 169 | |
| Recessive | 13 | 9 | rs10959456 | 11002926 | 0.00005841 | 0 | 66 | 120 | 19 | 105 | 158 | |
| Recessive | 14 | 8 | rs9314506 | 3682052 | 0.00007367 | CSMD1 | 25 | 102 | 64 | 80 | 131 | 71 |
| Recessive | 15 | 13 | rs9555965 | 89459182 | 0.00007841 | 39 | 72 | 80 | 22 | 121 | 139 | |
| Recessive | 15 | 13 | rs9555966 | 89460007 | 0.00007841 | 39 | 72 | 80 | 22 | 121 | 139 | |
| Recessive | 17 | 6 | rs13203299 | 169184034 | 0.00008602 | 33 | 68 | 90 | 16 | 122 | 144 | |
| Recessive | 18 | 11 | rs12291063 | 27650677 | 0.00009339 | BDNF-AS1,BDNF | 0 | 53 | 138 | 18 | 92 | 172 |
| Recessive | 19 | 22 | rs7290832 | 25658787 | 0.00009952 | 38 | 81 | 72 | 21 | 149 | 112 | |
| Recessive | 20 | 9 | rs871095 | 138095067 | 0.0001101 | NACC2 | 41 | 93 | 57 | 24 | 145 | 113 |
Model, the genetic model in which candidate SNPs were selected by GWAS; CHR, chromosome number.Related gene, the nearest gene from the SNP site; A/A, homozygote for the minor allele in each SNP.A/B, heterozygote for the major allele in each SNP; B/B, homozygote for the major allele in each SNP.
Figure 1.
Manhattan plot of the GWAS results. (a) Plot of the analysis of all 191 patients with chronic pain in the trend model. (b) Plot of the analysis that including only patients with PHN. The red line indicates the threshold for a significant association.
Table 3.
Top 20 candidate SNPs selected from GWAS for patients with postherpetic neuralgia (PHN).
| Model | Rank | CHR | SNP | Position | P | Related gene | Genotype (patients) |
Genotype (controls) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/B | B/B | A/A | A/B | B/B | |||||||
| Trend | 1 | 13 | rs4773840 | 94568426 | 0.0000001638* | ABCC4 | 16 | 40 | 33 | 10 | 96 | 176 |
| Trend | 2 | 13 | rs1678353 | 94547567 | 0.000000255 | ABCC4 | 17 | 39 | 33 | 9 | 103 | 170 |
| Trend | 3 | 13 | rs1751057 | 94548737 | 0.0000003913 | ABCC4 | 17 | 39 | 33 | 10 | 102 | 170 |
| Trend | 4 | 13 | rs1678395 | 94563955 | 0.000001063 | ABCC4 | 16 | 40 | 33 | 11 | 101 | 170 |
| Trend | 5 | 13 | rs1678362 | 94529692 | 0.000001482 | ABCC4 | 16 | 41 | 32 | 12 | 103 | 167 |
| Trend | 5 | 13 | rs1751052 | 94531379 | 0.000001482 | ABCC4 | 16 | 41 | 32 | 12 | 103 | 167 |
| Trend | 5 | 13 | rs1189438 | 94532991 | 0.000001482 | ABCC4 | 16 | 41 | 32 | 12 | 103 | 167 |
| Trend | 8 | 9 | rs10114508 | 26892593 | 0.000002803 | 5 | 36 | 46 | 2 | 63 | 214 | |
| Trend | 9 | 13 | rs1729752 | 94530363 | 0.000004509 | ABCC4 | 18 | 39 | 32 | 14 | 108 | 160 |
| Trend | 10 | 13 | rs4148540 | 94491368 | 0.000005799 | ABCC4 | 13 | 45 | 31 | 16 | 94 | 172 |
| Trend | 10 | 13 | rs4148540 | 94491368 | 0.000005799 | ABCC4 | 4 | 42 | 43 | 66 | 136 | 80 |
| Trend | 12 | 18 | rs12458523 | 19074726 | 0.00000617 | CABLES1 | 11 | 43 | 35 | 14 | 82 | 186 |
| Trend | 13 | 14 | rs2167151 | 78933086 | 0.000006287 | NRXN3 | 6 | 44 | 39 | 6 | 80 | 196 |
| Trend | 14 | 12 | rs10851014 | 117614600 | 0.0000063 | 16 | 39 | 34 | 12 | 105 | 165 | |
| Trend | 15 | 13 | rs1678387 | 94515907 | 0.000009474 | ABCC4 | 16 | 39 | 34 | 12 | 105 | 165 |
| Trend | 15 | 13 | rs1678365 | 94516981 | 0.000009474 | ABCC4 | 16 | 39 | 34 | 12 | 105 | 165 |
| Trend | 15 | 13 | rs1189451 | 94520087 | 0.000009474 | ABCC4 | 16 | 39 | 34 | 12 | 105 | 165 |
| Trend | 15 | 13 | rs2619312 | 94521040 | 0.000009474 | ABCC4 | 16 | 39 | 34 | 12 | 105 | 165 |
| Trend | 15 | 13 | rs1751037 | 94521559 | 0.000009474 | ABCC4 | 16 | 39 | 34 | 12 | 105 | 165 |
| Trend | 15 | 13 | rs1189461 | 94521789 | 0.000009474 | ABCC4 | 16 | 39 | 34 | 12 | 105 | 165 |
| Trend | 15 | 13 | rs1189464 | 94523867 | 0.000009474 | ABCC4 | 16 | 39 | 34 | 12 | 105 | 165 |
| Dominant | 1 | 6 | rs4075048 | 19275975 | 0.00001134 | 0 | 4 | 85 | 4 | 65 | 213 | |
| Dominant | 2 | 14 | rs2167151 | 78933086 | 0.00001212 | NRXN3 | 11 | 43 | 35 | 14 | 82 | 186 |
| Dominant | 3 | 2 | rs6718476 | 6112647 | 0.00001274 | 11 | 38 | 40 | 59 | 166 | 57 | |
| Dominant | 3 | 2 | rs2693818 | 6121959 | 0.00001274 | 11 | 38 | 40 | 59 | 166 | 57 | |
| Dominant | 5 | 13 | rs4148540 | 94491368 | 0.00001754 | ABCC4 | 13 | 45 | 31 | 16 | 94 | 172 |
| Dominant | 6 | 6 | rs9368038 | 19298240 | 0.00001905 | 0 | 5 | 84 | 5 | 66 | 211 | |
| Dominant | 6 | 6 | rs9350106 | 19303045 | 0.00001905 | 0 | 5 | 84 | 5 | 66 | 211 | |
| Dominant | 8 | 12 | rs10851014 | 117614600 | 0.00002548 | 6 | 44 | 39 | 6 | 80 | 196 | |
| Dominant | 9 | 2 | rs4675047 | 226665422 | 0.00002799 | 3 | 24 | 62 | 29 | 125 | 119 | |
| Dominant | 10 | 1 | rs2176360 | 188083580 | 0.00002889 | 9 | 50 | 30 | 14 | 100 | 168 | |
| Dominant | 11 | 7 | rs4722067 | 21868091 | 0.00003014 | DNAH11 | 16 | 33 | 40 | 82 | 140 | 60 |
| Dominant | 12 | 16 | rs12596324 | 26039779 | 0.00003039 | HS3ST4 | 10 | 29 | 50 | 44 | 150 | 88 |
| Dominant | 13 | 6 | rs9358193 | 19281466 | 0.0000309 | 0 | 5 | 84 | 5 | 64 | 211 | |
| Dominant | 14 | 6 | rs648248 | 117187750 | 0.00003254 | FAM162B | 13 | 30 | 46 | 48 | 157 | 77 |
| Dominant | 15 | 9 | rs10114508 | 26892593 | 0.00003959 | 5 | 36 | 46 | 2 | 63 | 214 | |
| Dominant | 16 | 13 | rs4773840 | 94568426 | 0.00004453 | ABCC4 | 16 | 40 | 33 | 10 | 96 | 176 |
| Dominant | 17 | 8 | rs7822451 | 17266781 | 0.00004517 | MTMR7 | 5 | 29 | 55 | 35 | 143 | 104 |
| Dominant | 18 | 7 | rs10278297 | 135341940 | 0.00004885 | 15 | 33 | 41 | 49 | 168 | 65 | |
| Dominant | 19 | 1 | rs624912 | 236807876 | 0.00005329 | 7 | 21 | 61 | 32 | 126 | 123 | |
| Dominant | 20 | 8 | rs2658914 | 56511974 | 0.00005364 | XKR4 | 0 | 18 | 71 | 13 | 111 | 158 |
| Recessive | 1 | 13 | rs1678353 | 94547567 | 0.00000369 | ABCC4 | 17 | 39 | 33 | 9 | 103 | 170 |
| Recessive | 2 | 13 | rs1751057 | 94548737 | 0.000008018 | ABCC4 | 17 | 39 | 33 | 10 | 102 | 170 |
| Recessive | 3 | 18 | rs12458523 | 19074726 | 0.00001884 | CABLES1 | 4 | 42 | 43 | 66 | 136 | 80 |
| Recessive | 4 | 13 | rs4773840 | 94568426 | 0.00002414 | ABCC4 | 16 | 40 | 33 | 10 | 96 | 176 |
| Recessive | 5 | 12 | rs10849659 | 118331044 | 0.00002555 | CCDC60 | 19 | 28 | 42 | 15 | 132 | 135 |
| Recessive | 6 | 13 | rs1729752 | 94530363 | 0.00003901 | ABCC4 | 18 | 39 | 32 | 14 | 108 | 160 |
| Recessive | 7 | 2 | rs10208470 | 75356624 | 0.00004381 | 2 | 49 | 38 | 52 | 122 | 108 | |
| Recessive | 8 | 2 | rs10205827 | 75356361 | 0.00004401 | 2 | 49 | 38 | 52 | 122 | 107 | |
| Recessive | 9 | 12 | rs4465416 | 118338125 | 0.00004502 | CCDC60 | 19 | 28 | 42 | 16 | 131 | 135 |
| Recessive | 10 | 13 | rs9576139 | 36396944 | 0.00004547 | 0 | 37 | 52 | 36 | 108 | 138 | |
| Recessive | 11 | 13 | rs1678395 | 94563955 | 0.0000476 | ABCC4 | 16 | 40 | 33 | 11 | 101 | 170 |
| Recessive | 12 | 12 | rs4300442 | 118324515 | 0.00005037 | CCDC60 | 20 | 29 | 40 | 17 | 131 | 134 |
| Recessive | 13 | 2 | rs1015802 | 153792446 | 0.00005759 | 8 | 20 | 60 | 1 | 77 | 204 | |
| Recessive | 14 | 2 | rs11680628 | 153839089 | 0.00006176 | 8 | 20 | 61 | 1 | 75 | 206 | |
| Recessive | 15 | 2 | rs1439630 | 153839620 | 0.00006204 | 10 | 24 | 55 | 3 | 82 | 197 | |
| Recessive | 15 | 2 | rs7556698 | 153850240 | 0.00006204 | 10 | 24 | 55 | 3 | 81 | 198 | |
| Recessive | 17 | 9 | rs10981230 | 113851385 | 0.00007136 | MIR3134,SUSD1 | 35 | 38 | 16 | 50 | 152 | 80 |
| Recessive | 18 | 13 | rs9557470 | 100094751 | 0.00007228 | TMTC4 | 23 | 31 | 35 | 24 | 130 | 128 |
| Recessive | 19 | 1 | rs4129058 | 5310402 | 0.00007338 | 2 | 59 | 28 | 50 | 131 | 101 | |
| Recessive | 20 | 4 | rs7670109 | 184691188 | 0.00008034 | 35 | 37 | 17 | 51 | 157 | 74 | |
Model, the genetic model in which candidate SNPs were selected by GWAS; CHR, chromosome number.Related gene, the nearest gene from the SNP site; A/A, homozygote for the minor allele in each SNP.A/B, heterozygote for the major allele in each SNP; B/B, homozygote for the major allele in each SNP.*, Significant association after correction for multiple testing.
Figure 2.
Regional plot of a potent locus that was associated with PHN. The genomic region 400 kbp upstream and downstream of the rs4773840 SNP on chromosome 13 is illustrated. The results of the association analyses in each genetic model were plotted, with the information on annotated genes, estimated recombination rates, and the pairwise-calculated strength of linkage disequilibrium (LD; r2 values) with the rs4773840 SNP in this region.
Identification of genes and gene sets associated with chronic pain and postherpetic neuralgia by gene-based and gene-set analyses
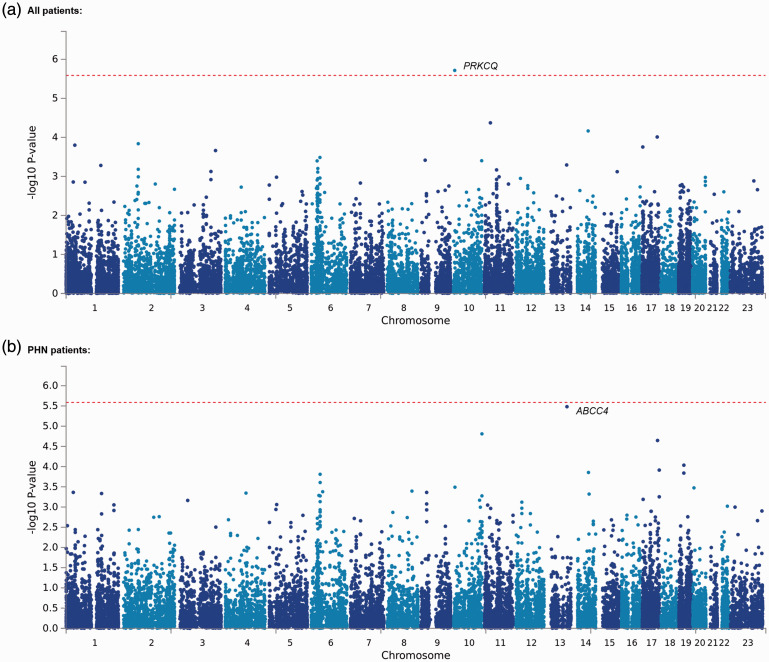
Considering the fact that the effects of individual markers tend to be too weak to be detected by comprehensive analyses, such as GWASs, that target only single polymorphisms, we conducted gene-based and gene-set analyses, which are statistical methods that are used to analyze multiple genetic markers simultaneously to determine their joint effect. In both analyses, we explored genes and gene sets that were associated with chronic pain conditions and PHN in a total of 191 patients, including 89 PHN patients and 282 control subjects, similarly to our GWAS by running MAGMA software, 40 which was available in the FUMA GWAS platform. 41 Consequently, the analyses of all patients included 4,45,723 SNPs of selected candidate genes and gene sets in the trend, dominant, and recessive models. Supplementary Tables S1 and S2 show the top 20 candidate genes that were identified in each genetic model in the gene-set analysis. The best candidate gene in the trend model that resulted from an analysis of all patients, PRKCQ, was significantly associated with the phenotype (adjusted p = 0.03722; Supplementary Table S1, Figure 3(a)). However, none of the genes were significantly associated with the phenotype in any of the genetic models that were used for the analysis of only PHN patients (Supplementary Table S2, Figure 3(b)). The association between PHN and the ABCC4 gene, for which the rs4773840 SNP was significantly associated with the phenotype, was only marginally significant in our gene-based analysis (adjusted p = 0.06364; Supplementary Table S2, Figure 3(b)). Tables 4 and 5 show the top 20 candidate gene sets that were identified in each genetic model in the gene-set analysis. As a result, the “go_fructose_metabolic_process” gene set was significantly associated with chronic pain in the recessive model (adjusted p = 0.003887; Table 4). Additionally, the “go_regeneration,” “go_reactive_oxygen_species_metabolic_process,” “go_arachidonic_acid_monooxygenase_activity,” and “go_translation_regulator_activity_nucleic_acid_binding” gene sets were significantly associated with PHN in the trend, dominant, and recessive models, respectively (adjusted p = 0.03587, 0.04548, 0.004380, and 0.01472, respectively; Table 5). The genes that were included in these gene sets are listed in Supplementary Table S3. The ABCC4 gene was not included in any of the gene sets; thus, the PRKCQ gene was included in the “go_regeneration” gene set (Supplementary Table S3). Among these genes, only three (PFKFB1, APOA4, and BCL2) were commonly included in two kinds of gene sets (Supplementary Table S3).
Figure 3.
Manhattan plot of the results of the gene-based analyses. (a) Plot of the analysis with all 191 patients with chronic pain in the trend model. (b) Plot of the analysis that included only patients with PHN. The dotted red line indicates the threshold for a significant association.
Table 4.
Top 20 candidate gene sets selected from gene-set analysis for all patients.
| Model | Rank | Gene set name | nGenes | Beta | SE | P | P a |
|---|---|---|---|---|---|---|---|
| Trend | 1 | go_transmembrane_receptor_protein_tyrosine_kinase_signaling_pathway | 490 | 0.14 | 0.0377 | 0.00010183 | 1 |
| Trend | 2 | go_morphogenesis_of_a_polarized_epithelium | 27 | 0.521 | 0.146 | 0.00018275 | 1 |
| Trend | 3 | chang_pou5f1_targets_up | 15 | 0.697 | 0.201 | 0.00027201 | 1 |
| Trend | 4 | pid_fanconi_pathway | 46 | 0.421 | 0.122 | 0.00028575 | 1 |
| Trend | 5 | go_oxidoreductase_activity_acting_on_paired_donors_with_incorporation_or_reduction_of_molecular_oxygen_reduced_flavin_or_flavoprotein_as_one_donor_and_incorporation_of_one_atom_of_oxygen | 24 | 0.613 | 0.184 | 0.00043085 | 1 |
| Trend | 6 | go_apical_protein_localization | 12 | 0.825 | 0.25 | 0.00048715 | 1 |
| Trend | 7 | delaserna_myod_targets_dn | 56 | 0.375 | 0.115 | 0.0005732 | 1 |
| Trend | 8 | go_execution_phase_of_apoptosis | 53 | 0.379 | 0.117 | 0.00059648 | 1 |
| Trend | 9 | go_atpase_activity_coupled | 299 | 0.149 | 0.0462 | 0.00064486 | 1 |
| Trend | 10 | liu_sox4_targets_dn | 299 | 0.152 | 0.0472 | 0.00066145 | 1 |
| Trend | 11 | firestein_ctnnb1_pathway | 32 | 0.475 | 0.149 | 0.00070207 | 1 |
| Trend | 12 | ning_chronic_obstructive_pulmonary_disease_dn | 117 | 0.23 | 0.0722 | 0.00072694 | 1 |
| Trend | 13 | mariadason_response_to_butyrate_curcumin_sulindac_tsa_1 | 9 | 1.11 | 0.349 | 0.00074306 | 1 |
| Trend | 14 | ross_aml_with_pml_rara_fusion | 72 | 0.316 | 0.1 | 0.00082081 | 1 |
| Trend | 15 | go_establishment_of_tissue_polarity | 17 | 0.57 | 0.181 | 0.00083939 | 1 |
| Trend | 16 | kondo_colon_cancer_hcp_with_h3k27me1 | 26 | 0.521 | 0.168 | 0.00098707 | 1 |
| Trend | 17 | go_enzyme_linked_receptor_protein_signaling_pathway | 675 | 0.1 | 0.0327 | 0.0010865 | 1 |
| Trend | 18 | go_atp_dependent_dna_helicase_activity | 33 | 0.411 | 0.135 | 0.0011325 | 1 |
| Trend | 19 | ikeda_mir30_targets_up | 115 | 0.232 | 0.0772 | 0.0013186 | 1 |
| Trend | 20 | go_gamma_tubulin_binding | 24 | 0.498 | 0.166 | 0.0013693 | 1 |
| Dominant | 1 | go_arachidonic_acid_monooxygenase_activity | 15 | 1.14 | 0.263 | 0.0000075774 | 0.08072962 |
| Dominant | 2 | go_oxidoreductase_activity_acting_on_paired_donors_with_incorporation_or_reduction_of_molecular_oxygen_reduced_flavin_or_flavoprotein_as_one_donor_and_incorporation_of_one_atom_of_oxygen | 24 | 0.75 | 0.188 | 0.000032941 | 0.350953414 |
| Dominant | 3 | pid_fanconi_pathway | 46 | 0.453 | 0.125 | 0.00013871 | 1 |
| Dominant | 4 | go_positive_regulation_of_receptor_recycling | 11 | 0.767 | 0.215 | 0.00018422 | 1 |
| Dominant | 5 | go_dna_double_strand_break_processing | 19 | 0.624 | 0.175 | 0.00018853 | 1 |
| Dominant | 6 | lenaour_dendritic_cell_maturation_up | 111 | 0.252 | 0.0754 | 0.00042167 | 1 |
| Dominant | 7 | kondo_colon_cancer_hcp_with_h3k27me1 | 26 | 0.574 | 0.172 | 0.00042761 | 1 |
| Dominant | 8 | go_apical_protein_localization | 12 | 0.842 | 0.255 | 0.00048821 | 1 |
| Dominant | 9 | reactome_xenobiotics | 15 | 0.874 | 0.266 | 0.00051506 | 1 |
| Dominant | 10 | delaserna_myod_targets_dn | 56 | 0.379 | 0.118 | 0.0006494 | 1 |
| Dominant | 11 | go_cytoplasmic_dynein_complex | 15 | 0.62 | 0.195 | 0.00072543 | 1 |
| Dominant | 12 | go_execution_phase_of_apoptosis | 53 | 0.373 | 0.12 | 0.0008959 | 1 |
| Dominant | 13 | go_cellular_response_to_exogenous_dsrna | 12 | 0.79 | 0.253 | 0.00091276 | 1 |
| Dominant | 14 | jechlinger_epithelial_to_mesenchymal_transition_up | 69 | 0.315 | 0.101 | 0.0009394 | 1 |
| Dominant | 15 | go_dna_metabolic_process | 728 | 0.0982 | 0.0317 | 0.00098413 | 1 |
| Dominant | 16 | taylor_methylated_in_acute_lymphoblastic_leukemia | 72 | 0.306 | 0.099 | 0.00099275 | 1 |
| Dominant | 17 | reactome_heparan_sulfate_heparin_hs_gag_metabolism | 52 | 0.385 | 0.126 | 0.0011419 | 1 |
| Dominant | 18 | go_dna_repair | 461 | 0.119 | 0.0391 | 0.0011488 | 1 |
| Dominant | 19 | go_poly_a_binding | 13 | 0.58 | 0.19 | 0.0011566 | 1 |
| Dominant | 20 | go_asymmetric_protein_localization | 19 | 0.594 | 0.195 | 0.0011843 | 1 |
| Recessive | 1 | go_fructose_metabolic_process | 14 | 1.24 | 0.25 | 0.00000036488 | 0.00388743152* |
| Recessive | 2 | kang_immortalized_by_tert_up | 86 | 0.349 | 0.0873 | 0.000032117 | 0.342174518 |
| Recessive | 3 | go_translation_factor_activity_rna_binding | 79 | 0.363 | 0.0956 | 0.000072849 | 0.776133246 |
| Recessive | 4 | go_regulation_of_hexokinase_activity | 11 | 0.886 | 0.238 | 0.00010025 | 1 |
| Recessive | 5 | haddad_t_lymphocyte_and_nk_progenitor_up | 75 | 0.344 | 0.0928 | 0.00010685 | 1 |
| Recessive | 6 | go_regulation_of_attachment_of_spindle_microtubules_to_kinetochore | 11 | 1.04 | 0.296 | 0.00022662 | 1 |
| Recessive | 7 | go_regulation_of_cell_projection_assembly | 148 | 0.247 | 0.0703 | 0.00022671 | 1 |
| Recessive | 8 | go_regulation_of_t_cell_tolerance_induction | 12 | 0.712 | 0.215 | 0.00045907 | 1 |
| Recessive | 9 | zwang_down_by_2nd_egf_pulse | 217 | 0.186 | 0.0564 | 0.00049442 | 1 |
| Recessive | 10 | go_regulation_of_membrane_lipid_metabolic_process | 13 | 0.782 | 0.238 | 0.00051065 | 1 |
| Recessive | 11 | kenny_ctnnb1_targets_up | 50 | 0.396 | 0.122 | 0.00056843 | 1 |
| Recessive | 12 | go_immunoglobulin_binding | 18 | 0.581 | 0.182 | 0.00068753 | 1 |
| Recessive | 13 | reactome_tca_cycle_and_respiratory_electron_transport | 115 | 0.26 | 0.0822 | 0.00076392 | 1 |
| Recessive | 14 | nielsen_synovial_sarcoma_dn | 19 | 0.791 | 0.25 | 0.00076547 | 1 |
| Recessive | 15 | doane_breast_cancer_esr1_dn | 48 | 0.376 | 0.119 | 0.00078823 | 1 |
| Recessive | 16 | go_dna_replication_dependent_nucleosome_organization | 31 | 0.839 | 0.267 | 0.0008361 | 1 |
| Recessive | 17 | go_t_cell_apoptotic_process | 15 | 0.651 | 0.207 | 0.00084519 | 1 |
| Recessive | 18 | go_lymphocyte_apoptotic_process | 18 | 0.605 | 0.193 | 0.0008619 | 1 |
| Recessive | 19 | go_regulation_of_pseudopodium_assembly | 13 | 0.735 | 0.237 | 0.00098328 | 1 |
| Recessive | 20 | lee_aging_cerebellum_dn | 80 | 0.292 | 0.0946 | 0.0010161 | 1 |
Model, the genetic model in which candidate gene sets were selected by analysis; nGenes, the number of genes in the data that are in the gene set; Beta, the regression coefficient of the gene set; SE, the standard error of the regression coefficient; P a , adjusted P-value for multiple testing; *, Significant association after the conservative Bonferroni correction.
Table 5.
Top 20 candidate gene sets selected from gene-set analysis for patients with postherpetic neuralgia (PHN).
| Model | Rank | Gene set name | nGenes | Beta | SE | P | Pa |
|---|---|---|---|---|---|---|---|
| Trend | 1 | go_regeneration | 153 | 0.308 | 0.0685 | 0.0000033672 | 0.0358741488* |
| Trend | 2 | go_reactive_oxygen_species_metabolic_process | 92 | 0.411 | 0.0922 | 0.0000042685 | 0.045476599* |
| Trend | 3 | go_organ_regeneration | 79 | 0.355 | 0.0966 | 0.00011875 | 1 |
| Trend | 4 | reactome_p2y_receptors | 12 | 1.03 | 0.282 | 0.0001333 | 1 |
| Trend | 5 | tuomisto_tumor_suppression_by_col13a1_up | 16 | 0.771 | 0.215 | 0.00016802 | 1 |
| Trend | 6 | go_regulation_of_mrna_3_end_processing | 27 | 0.494 | 0.141 | 0.00023174 | 1 |
| Trend | 7 | go_au_rich_element_binding | 21 | 0.655 | 0.192 | 0.00032672 | 1 |
| Trend | 8 | go_regulation_of_nuclear_transcribed_mrna_poly_a_tail_shortening | 11 | 0.765 | 0.226 | 0.00035697 | 1 |
| Trend | 9 | go_rna_destabilization | 16 | 0.601 | 0.178 | 0.00037747 | 1 |
| Trend | 10 | go_apical_protein_localization | 12 | 0.826 | 0.251 | 0.00050398 | 1 |
| Trend | 11 | murakami_uv_response_6hr_dn | 19 | 0.637 | 0.195 | 0.00053107 | 1 |
| Trend | 12 | go_superoxide_metabolic_process | 30 | 0.623 | 0.19 | 0.00053228 | 1 |
| Trend | 13 | go_negative_regulation_of_cellular_response_to_insulin_stimulus | 31 | 0.513 | 0.159 | 0.00060983 | 1 |
| Trend | 14 | go_execution_phase_of_apoptosis | 53 | 0.375 | 0.117 | 0.00068738 | 1 |
| Trend | 15 | hernandez_aberrant_mitosis_by_docetacel_4nm_up | 21 | 0.624 | 0.196 | 0.00072487 | 1 |
| Trend | 16 | go_regulation_of_mrna_polyadenylation | 10 | 0.629 | 0.198 | 0.00074015 | 1 |
| Trend | 17 | pid_nfat_tfpathway | 47 | 0.401 | 0.13 | 0.00099145 | 1 |
| Trend | 18 | go_regulation_of_transferase_activity | 920 | 0.0867 | 0.0283 | 0.0010735 | 1 |
| Trend | 19 | go_axon | 411 | 0.125 | 0.0413 | 0.0012388 | 1 |
| Trend | 20 | go_regulation_of_cellular_amide_metabolic_process | 344 | 0.136 | 0.0449 | 0.0012506 | 1 |
| Dominant | 1 | go_arachidonic_acid_monooxygenase_activity | 15 | 1.33 | 0.269 | 0.00000041113 | 0.00438017902* |
| Dominant | 2 | reactome_p2y_receptors | 12 | 1.23 | 0.294 | 0.000015196 | 0.161898184 |
| Dominant | 3 | go_regulation_of_mrna_polyadenylation | 10 | 0.791 | 0.206 | 0.000061699 | 0.657341146 |
| Dominant | 4 | go_regulation_of_mrna_3_end_processing | 27 | 0.551 | 0.147 | 0.000088695 | 0.94495653 |
| Dominant | 5 | go_long_chain_fatty_acid_metabolic_process | 87 | 0.342 | 0.0913 | 0.000090289 | 0.961939006 |
| Dominant | 6 | go_negative_regulation_of_binding | 127 | 0.273 | 0.074 | 0.00011268 | 1 |
| Dominant | 7 | go_neuron_apoptotic_process | 34 | 0.522 | 0.143 | 0.0001309 | 1 |
| Dominant | 8 | go_reactive_oxygen_species_metabolic_process | 92 | 0.347 | 0.0961 | 0.00015146 | 1 |
| Dominant | 9 | murakami_uv_response_6hr_dn | 19 | 0.724 | 0.203 | 0.00017847 | 1 |
| Dominant | 10 | graham_normal_quiescent_vs_normal_dividing_up | 64 | 0.433 | 0.122 | 0.00019318 | 1 |
| Dominant | 11 | go_regeneration | 153 | 0.252 | 0.0713 | 0.000204 | 1 |
| Dominant | 12 | reactome_signaling_by_notch4 | 12 | 0.933 | 0.264 | 0.00020967 | 1 |
| Dominant | 13 | tuomisto_tumor_suppression_by_col13a1_up | 16 | 0.772 | 0.224 | 0.00028268 | 1 |
| Dominant | 14 | go_arachidonic_acid_metabolic_process | 50 | 0.424 | 0.126 | 0.00036618 | 1 |
| Dominant | 15 | go_rna_destabilization | 16 | 0.626 | 0.186 | 0.00038011 | 1 |
| Dominant | 16 | 17 | 0.667 | 0.2 | 0.00041978 | 1 | |
| Dominant | 17 | go_negative_regulation_of_cellular_response_to_insulin_stimulus | 31 | 0.548 | 0.165 | 0.00044684 | 1 |
| Dominant | 18 | reactome_xenobiotics | 15 | 0.868 | 0.273 | 0.00073068 | 1 |
| Dominant | 19 | go_apical_protein_localization | 12 | 0.83 | 0.262 | 0.00075723 | 1 |
| Dominant | 20 | go_neuron_death | 46 | 0.4 | 0.127 | 0.00080182 | 1 |
| Recessive | 1 | go_translation_regulator_activity_nucleic_acid_binding | 17 | 1.06 | 0.226 | 0.0000013818 | 0.0147216972* |
| Recessive | 2 | galluzzi_permeabilize_mitochondria | 41 | 0.546 | 0.13 | 0.000014119 | 0.150423826 |
| Recessive | 3 | go_fructose_metabolic_process | 14 | 1.06 | 0.258 | 0.000020033 | 0.213431582 |
| Recessive | 4 | go_regulation_of_hexokinase_activity | 11 | 0.995 | 0.253 | 0.000043055 | 0.45870797 |
| Recessive | 5 | go_immunoglobulin_binding | 18 | 0.719 | 0.191 | 0.000084426 | 0.899474604 |
| Recessive | 6 | go_heat_shock_protein_binding | 88 | 0.329 | 0.0876 | 0.000088017 | 0.937733118 |
| Recessive | 7 | go_peptide_antigen_binding | 25 | 0.795 | 0.216 | 0.00011626 | 1 |
| Recessive | 8 | go_ikappab_kinase_complex | 11 | 1.02 | 0.282 | 0.0001527 | 1 |
| Recessive | 9 | mootha_glycolysis | 21 | 0.771 | 0.215 | 0.00016298 | 1 |
| Recessive | 10 | kang_immortalized_by_tert_up | 86 | 0.318 | 0.0922 | 0.00028655 | 1 |
| Recessive | 11 | bogni_treatment_related_myeloid_leukemia_up | 29 | 0.553 | 0.163 | 0.00033607 | 1 |
| Recessive | 12 | go_igg_binding | 7 | 0.947 | 0.281 | 0.00037687 | 1 |
| Recessive | 13 | ellwood_myc_targets_up | 13 | 0.839 | 0.249 | 0.00038154 | 1 |
| Recessive | 14 | dorsam_hoxa9_targets_up | 35 | 0.449 | 0.138 | 0.0005898 | 1 |
| Recessive | 15 | reactome_abortive_elongation_of_hiv1_transcript_in_the_absence_of_tat | 23 | 0.64 | 0.201 | 0.00073176 | 1 |
| Recessive | 16 | krieg_hypoxia_not_via_kdm3a | 716 | 0.109 | 0.0343 | 0.00073919 | 1 |
| Recessive | 17 | go_central_nervous_system_development | 841 | 0.0994 | 0.0316 | 0.00084853 | 1 |
| Recessive | 18 | shin_b_cell_lymphoma_cluster_9 | 19 | 0.659 | 0.212 | 0.0009452 | 1 |
| Recessive | 19 | go_regulation_of_protein_sumoylation | 21 | 0.596 | 0.192 | 0.00094559 | 1 |
| Recessive | 20 | holleman_daunorubicin_b_all_up | 10 | 1.16 | 0.374 | 0.00097434 | 1 |
Model, the genetic model in which candidate gene sets were selected by analysis; nGenes, the number of genes in the data that are in the gene set; Beta, the regression coefficient of the gene set; SE, the standard error of the regression coefficient; Pa, adjusted P-value for multiple testing.
*Significant association after the conservative Bonferroni correction.
Identification of genetic polymorphisms associated with the effects of drugs for the treatment of pain in patients
Various types of drugs were administered to the patients for the treatment of pain. Although some of these drugs were effective for some patients, others were not. We performed another GWAS of 191 patient subjects to explore SNPs that were associated with the efficacy of these drugs, which were divided into major five groups (opioids, antidepressants, anticonvulsants, NSAIDs, and GABA receptor agonists; Table 1). Supplementary Tables S4 to S8 show the top 20 candidates for these drugs in each genetic model. However, none of the SNPs were genome-wide significantly associated with the phenotypes (p ≥ 1.858 × 10−7; Supplementary Tables S4–S8). The best candidate SNPs with the lowest p values were rs7811258 SNP in the dominant model for opioids (nominal p = 1.655 × 10−6; Supplementary Table S4), rs10793705 SNP in the trend model for antidepressants (nominal p = 1.714 × 10−6; Supplementary Table S5), rs2300525 SNP in the dominant model for anticonvulsants (nominal p = 1.403 × 10−6; Supplementary Table S6), rs2195962 and rs12461406 SNPs in the dominant model for NSAIDs (nominal p = 3.573 × 10−6; Supplementary Table S7), and rs7094057 SNP in the trend model for GABA receptor agonists (nominal p = 3.311 × 10−6; Supplementary Table S8).
Discussion
To identify potential genetic variants that contribute to the susceptibility to chronic pain conditions and the effects of several types of drugs that are used to treat pain, we conducted an overall GWAS of patients with chronic pain and control subjects. We also explored genetic factors that are associated with PHN by performing another GWAS. The results suggested that carriers of the C-allele of the rs4773840 SNP within the ABCC4 gene region were more susceptible to PHN (Table 3), and several SNPs within or around the PRKCQ gene region jointly influenced the risk of developing chronic pain conditions. Furthermore, we found several gene sets that were possibly associated with these phenotypes. Meanwhile, we found no SNPs that were significantly associated with the efficacy of drugs for the treatment of pain. One of the reasons for this lack of an association might be related to the small sample size for each association analysis for each drug, which resulted in a lack of statistical power to detect positive associations. Indeed, the largest number of samples was only 99 in the analysis of anticonvulsant drugs among five major types of drugs (Table 1), whereas the total number of patients with chronic pain who were recruited in the study was 194, indicating that less than half of the patients were included in these analyses. Future studies with larger sample sizes will clarify which SNPs affect the efficacy of drugs to treat chronic pain.
Chronic pain is a common and heterogenous clinical condition. Previous studies have mostly explored genetic factors that are associated with chronic pain in a particular subset of patients, such as patients with CWP,13,15,28 CPSP, 22 chronic back pain, 43 and neuropathic pain, including diabetic neuropathic pain.23–25,29–32 The disease status of the patients in our samples was diverse, and the sample size for each disease status was fairly small (Table 1), thus hampering genetic association analyses of each patient subgroup, with the exception of patients with PHN. Therefore, the present study conducted analyses of overall patients with chronic pain and a subgroup of patients with PHN. Although the analysis of overall patients might present a risk that the genetic effects on each phenotype are obscured or not precisely detected, one could assume that some genetic factors that commonly affect chronic pain can be detected among all of the genetic factors. Postherpetic neuralgia is a neuropathic pain disorder that occurs most often in the elderly and is a major complication of herpes zoster, with spontaneous pain and stimulus-evoked pain, such as allodynia and hyperpathia.44–47 The genetic factors that contribute to PHN are poorly understood. Only a few studies have reported genetic variations that are associated with the susceptibility to PHN, including the human histocompatibility leukocyte antigen (HLA) locus, in which the HLA-A*3303, -B*4403, and -DRB1*1302 alleles have been shown to be associated with the risk of PHN.47–50 Although the present study did not investigate the HLA locus in detail because of an inability to precisely genotype HLA alleles using commercially available SNP arrays, we comprehensively explored genetic risk factors for PHN at the genome-wide level for the first time, which resulted in the identification of possibly associated SNPs, such as rs4773840 (Table 3).
The best candidate SNP with the lowest p value among the candidate SNPs for PHN was rs4773840, which is located in the intronic region of the ABCC4 gene on chromosome 13. The ABCC4 gene encodes the ABCC4 protein, which is a member of the MRP subfamily (MRP4) that is involved in multi-drug resistance and acts as an independent regulator of intracellular cyclic nucleotide levels and mediator of cyclic adenosine monophosphate (cAMP)-dependent signal transduction to the nucleus. 51 The mRNA of this gene was reported to be widely expressed in humans, with particularly high levels in the prostate, but it is barely detectable in the liver. 52 ABCC4 has been implicated in the transport of antiviral agents, anticancer drugs,53–55 and endogenous molecules, such as prostaglandins, steroids, bile acids, cyclic nucleotides, and folate.56–60 Indeed, ABCC4 is involved in the efflux of prostaglandin F2α, and the ABCC4 gene is reportedly upregulated in ovarian endometriosis tissue compared with normal endometrium tissue, 61 which would be a mechanism that underlies endometriosis, a chronic inflammatory disease that often involves severe pain or infertility.62,63 The disruption of cAMP and prostaglandin E2 transport by mrp4 deficiency in mice altered cAMP-mediated signaling and the nociceptive response. 64 These studies suggest that ABCC4 may be involved in some pain-related conditions in humans and mice. To date, many genetic variations within or around the ABCC4 gene have been identified and characterized in Japanese and other ethnically diverse populations.65,66 The functional impact of these variations, especially nonsynonymous polymorphisms, have been investigated in previous studies.67–71 In genetic association studies of disease status and symptoms, SNPs or copy number variations within or around the ABCC4 gene have been shown to be associated with airway inflammation in asthmatic individuals, 68 unfavorable clinical outcomes in children with acute lymphoblastic leukemia, 69 patients with esophageal squamous cell carcinoma, 72 patients with chemotherapy-induced peripheral neuropathy, 73 and measures of pain symptoms in patients with lung cancer and acute post-radiotherapy pain.74,75 However, none of these studies included the rs4773840 SNP or other SNPs that were in relatively strong LD with this SNP in our samples (r2 ≥ 0.8; Supplementary Figure S3). According to the Genotype-Tissue Expression (GTEx) portal (accessed July 10, 2019; Supplementary Methods), one of the SNPs that is in relatively strong LD with the rs4773840 SNP, rs2950957 (Supplementary Figure S3), significantly affects mRNA expression of the ABCC4 gene in the muscularis in the human esophagus. Single-nucleotide polymorphisms that are in relatively strong LD with the rs4773840 SNP include two synonymous SNPs in the coding region, rs1189466 and rs1678339 (Supplementary Figure S3), based on the Exome Aggregation Consortium (ExAC) Browser (accessed July 10, 2019; Supplementary Methods). When these SNPs were referred to SNPinfo Web Server and SNPnexus (accessed July 10, 2019; Supplementary Methods), they were predicted to affect splicing as exonic splicing enhancers or exonic splicing silencers, and the rs1678339 SNP was found to be within a putative transcription factor binding site in mice and humans. These results suggest that expression or splicing of the ABCC4 gene could be affected by the rs4773840 SNP and other SNPs that are in relatively strong LD with this SNP, which might be related to a mechanism that contributes to PHN.
In the gene-based analysis of all patients, the PRKCQ gene was significantly associated with the phenotype (Supplementary Table S1; Figure 3(a)). The PRKCQ gene encodes protein kinase Cθ (PKCθ), which is a family of serine- and threonine-specific protein kinases. The PRKCQ protein is a calcium-independent and phospholipid-dependent kinase that is important for T-cell activation and highly expressed in the thyroid and lymph nodes.76,77 Lidocaine, which is used as a local anesthetic, was shown to modulate inflammation in septic patients by decreasing chemokine-induced neutrophil arrest and transendothelial migration by inhibiting PKCθ activation. 78 The PKC inhibitor tamoxifen suppressed paclitaxel-, vincristine-, and bortezomib-induced cold and mechanical allodynia in mice, 79 although the specific role of PKCθ was not clearly revealed in this study. In genetic association studies of disease status and symptoms, SNPs within or around the PRKCQ gene were shown to be associated with type 1 diabetes 80 and Crohn’s disease,81,82 both of which may involve symptoms of neuropathy or pain as complications. Significant associations were found between Crohn’s disease and the nonsynonymous rs2236379 SNP.81,82 This SNP was found to be in relatively strong LD with the rs2026432 SNP in our samples according to the SNPinfo Web Server (r2 ≥ 0.8), which was among the top 20 candidate SNPs in the present study (Table 2). One of these SNPs may influence the susceptibility to both Crohn’s disease and chronic pain partly through the same mechanism, but future studies are required to confirm such a possibility. In the gene-set analysis, several significant associations were also found (Tables 4 and 5). Among the three genes that were commonly included in the two candidate gene sets (Supplementary Table S3), the BCL2 gene was reported to be upregulated in human cultured cells by capsaicin treatment, 83 which is known to affect inflammatory and pain pathways. However, the precise roles of the gene sets in chronic pain and PHN that were identified in the present study remain unknown and require further investigation.
A major limitation of this study would be the limited sample size. However, some of the previous GWAS have successfully identified SNPs significantly associated with the phenotypes examined in considerably small number of samples (i.e., approximately 200 or less samples).84,85 Moreover, stronger associations can be found in suitably stratified samples with homogenous property (i.g., diagnosis of PHN) than those in entire number of samples, even if such strong associations may be masked before stratification, as demonstrated in previous studies.86–89 Nevertheless, further studies will be warranted for replication of the results shown in the present study.
In conclusion, our GWASs identified several SNPs and genes associated with chronic pain and PHN, including the ABCC4 rs4773840 SNP and PRKCQ gene. The present findings require corroboration in future studies with larger sample sizes.
Supplemental Material
Supplemental material, sj-pdf-1-mpx-10.1177_1744806921999924 for Genome-wide association study identifies candidate loci associated with chronic pain and postherpetic neuralgia by Daisuke Nishizawa, Masako Iseki, Hideko Arita, Kazuo Hanaoka, Choku Yajima, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida and Kazutaka Ikeda in Molecular Pain
Acknowledgments
We thank Mr. Michael Arends for assistance with editing the manuscript. We are grateful to the volunteers for their participation in the study and anesthesiologists and surgeons for collecting the clinical data.
Footnotes
Author Contributions: DN, SK, MH, and KI conceived and designed the experiments. DN and JH performed the experiments. DN analyzed the data. DN and JH contributed reagents/materials/analysis tools. DN and KI wrote the paper. DN, AH, and KI collected DNA. MI, HA, KH, CY, JK, and SO collected clinical data and DNA.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Kazutaka Ikeda has received support from Asahi Kasei Pharma Corporation for a project that is unrelated to this research and speaker’s and consultant’s fees from MSD K.K., VistaGen Therapeutics, Inc., Atheneum Partners Otsuka Pharmaceutical Co. Ltd., Taisho Pharmaceutical Co. Ltd., Eisai, Daiichi-Sankyo, Inc., Sumitomo Dainippon Pharma, Japan Tobacco, Inc., and Nippon Chemiphar. The authors declare no other conflicts of interest.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI (no. 22790518, 233,903,772,479,054,426,293,347 JP16H06276 [AdAMS], 17H04324, 17K08970, 17K09052, 18K08829, 20K09259, and 20K07774), Ministry of Health, Labour, and Welfare (MHLW) of Japan (no. H26-Kakushintekigan-ippan-060), Japan Agency for Medical Research and Development (AMED; no. JP19ek0610011 and JP19dk0307071), Smoking Research Foundation (Tokyo, Japan), and Japan Research Foundation for Clinical Pharmacology (JRFCP). The funding agencies had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
ORCID iDs: Hideko Arita https://orcid.org/0000-0002-4151-5962
Kazutaka Ikeda https://orcid.org/0000-0001-8342-0278
Supplemental material: Supplemental material for this article is available online.
References
- 1.Young EE, Lariviere WR, Belfer I. Genetic basis of pain variability: recent advances. J Med Genet 2012; 49: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crook J, Rideout E, Browne G. The prevalence of pain complaints in a general population. Pain 1984; 18: 299–314. [DOI] [PubMed] [Google Scholar]
- 3.Jakobsson U. The epidemiology of chronic pain in a general population: results of a survey in Southern Sweden. Scand J Rheumatol 2010; 39: 421–429. [DOI] [PubMed] [Google Scholar]
- 4.Listed Na. Pain terms: a list with definitions and notes on usage. Recommended by the IASP subcommittee on taxonomy. Pain 1979; 6: 249. [PubMed] [Google Scholar]
- 5.Inoue S, Kobayashi F, Nishihara M, Arai YC, Ikemoto T, Kawai T, Inoue M, Hasegawa T, Ushida T. Chronic pain in the Japanese community–prevalence, characteristics and impact on quality of life. PLoS One 2015; 10: e0129262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takura T, Ushida T, Kanchiku T, Ebata N, Fujii K, DiBonaventura M, Taguchi T. The societal burden of chronic pain in Japan: an internet survey. J Orthop Sci 2015; 20: 750–760. [DOI] [PubMed] [Google Scholar]
- 7.Smith D, Wilkie R, Uthman O, Jordan JL, McBeth J. Chronic pain and mortality: a systematic review. PLoS One 2014; 9: e99048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke H, Katz J, Flor H, Rietschel M, Diehl SR, Seltzer Z. Genetics of chronic post-surgical pain: a crucial step toward personal pain medicine. Can J Anaesth 2015; 62: 294–303. [DOI] [PubMed] [Google Scholar]
- 9.Momi SK, Fabiane SM, Lachance G, Livshits G, Williams FM. Neuropathic pain as part of chronic widespread pain: environmental and genetic influences. Pain 2015; 156: 2100–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacGregor AJ, Andrew T, Sambrook PN, Spector TD. Structural, psychological, and genetic influences on low back and neck pain: a study of adult female twins. Arthritis Rheum 2004; 51: 160–167. [DOI] [PubMed] [Google Scholar]
- 11.Nishizawa D, Nagashima M, Satoh Y, Tagami M, Ikeda K. [Genetic polymorphisms and human sensitivity to pain and opioids]. Masui the Jpn J Anesthesiol 2009; 58: 1093–1101. [PubMed] [Google Scholar]
- 12.Kim DH, Schwartz CE. The genetics of pain: implications for evaluation and treatment of spinal disease. Spine J 2010; 10: 827–840. [DOI] [PubMed] [Google Scholar]
- 13.Hocking LJ, Smith BH, Jones GT, Reid DM, Strachan DP, Macfarlane GJ. Genetic variation in the beta2-adrenergic receptor but not catecholamine-O-methyltransferase predisposes to chronic pain: results from the 1958 British birth cohort study. Pain 2010; 149: 143–151. [DOI] [PubMed] [Google Scholar]
- 14.Sugaya K, Nishijima S, Yamada T, Miyazato M, Hatano T, Ogawa Y. Molecular analysis of adrenergic receptor genes and interleukin-4/interleukin-4 receptor genes in patients with interstitial cystitis. J Urol 2002; 168: 2668–2671. [DOI] [PubMed] [Google Scholar]
- 15.Nicholl BI, Holliday KL, Macfarlane GJ, Thomson W, Davies KA, O’Neill TW, Bartfai G, Boonen S, Casanueva FF, Finn JD, Forti G, Giwercman A, Huhtaniemi IT, Kula K, Punab M, Silman AJ, Vanderschueren D, Wu FC, McBeth J; European Male Ageing Study Group. Association of HTR2A polymorphisms with chronic widespread pain and the extent of musculoskeletal pain: results from two population-based cohorts. Arthritis Rheum 2011; 63: 810–818. [DOI] [PubMed] [Google Scholar]
- 16.Reimann F, Cox JJ, Belfer I, Diatchenko L, Zaykin DV, McHale DP, Drenth JP, Dai F, Wheeler J, Sanders F, Wood L, Wu TX, Karppinen J, Nikolajsen L, Mannikko M, Max MB, Kiselycznyk C, Poddar M, Te Morsche RH, Smith S, Gibson D, Kelempisioti A, Maixner W, Gribble FM, Woods CG. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc Natl Acad Sci U S A 2010; 107: 5148–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, Diatchenko L, Smith S, Cobos EJ, Zaykin D, Allchorne A, Gershon E, Livneh J, Shen PH, Nikolajsen L, Karppinen J, Mannikko M, Kelempisioti A, Goldman D, Maixner W, Geschwind DH, Max MB, Seltzer Z, Woolf CJ. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain 2010; 133: 2519–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, Diatchenko L, Gupta V, Xia CP, Amann S, Kreitz S, Heindl-Erdmann C, Wolz S, Ly CV, Arora S, Sarangi R, Dan D, Novatchkova M, Rosenzweig M, Gibson DG, Truong D, Schramek D, Zoranovic T, Cronin SJ, Angjeli B, Brune K, Dietzl G, Maixner W, Meixner A, Thomas W, Pospisilik JA, Alenius M, Kress M, Subramaniam S, Garrity PA, Bellen HJ, Woolf CJ, Penninger JM. A genome-wide drosophila screen for heat nociception identifies alpha2delta3 as an evolutionarily conserved pain gene. Cell 2010; 143: 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissenbaum J, Devor M, Seltzer Z, Gebauer M, Michaelis M, Tal M, Dorfman R, Abitbul-Yarkoni M, Lu Y, Elahipanah T, delCanho S, Minert A, Fried K, Persson A-K, Shpigler H, Shabo E, Yakir B, Pisanté A, Darvasi A. Susceptibility to chronic pain following nerve injury is genetically affected by CACNG2. Genome Res 2010; 20: 1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, Goldman D, Xu K, Shabalina SA, Shagin D, Max MB, Makarov SS, Maixner W. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet 2005; 14: 135–143. [DOI] [PubMed] [Google Scholar]
- 21.Shoskes DA, Albakri Q, Thomas K, Cook D. Cytokine polymorphisms in men with chronic prostatitis/chronic pelvic pain syndrome: association with diagnosis and treatment response. J Urol 2002; 168: 331–335. [PubMed] [Google Scholar]
- 22.Hoofwijk DM, van Reij RR, Rutten BP, Kenis G, Buhre WF, Joosten EA. Genetic polymorphisms and their association with the prevalence and severity of chronic postsurgical pain: a systematic review. Br J Anaesth 2016; 117: 708–719. [DOI] [PubMed] [Google Scholar]
- 23.Belfer I, Wu T, Kingman A, Krishnaraju RK, Goldman D, Max MB. Candidate gene studies of human pain mechanisms: methods for optimizing choice of polymorphisms and sample size. Anesthesiology 2004; 100: 1562–1572. [DOI] [PubMed] [Google Scholar]
- 24.Iseki M, Sato-Takeda M. [Implication of genetic polymorphism on neuropathic pain]. Masui Jpn J Anesthesiol 2009; 58: 1112–1121. [PubMed] [Google Scholar]
- 25.Belfer I, Dai F. Phenotyping and genotyping neuropathic pain. Curr Pain Headache Rep 2010; 14: 203–212. [DOI] [PubMed] [Google Scholar]
- 26.Veluchamy A, Hebert HL, Meng W, Palmer CNA, Smith BH. Systematic review and meta-analysis of genetic risk factors for neuropathic pain. Pain 2018; 159: 825–848. [DOI] [PubMed] [Google Scholar]
- 27.Jones AV, Hockley JR, Hyde C, Gorman D, Sredic-Rhodes A, Bilsland J, McMurray G, Furlotte NA, Hu Y, Hinds DA, Cox PJ, Scollen S. Genome-wide association analysis of pain severity in dysmenorrhea identifies association at chromosome 1p13.2, near the nerve growth factor locus. Pain 2016; 157: 2571–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters MJ, Broer L, Willemen HL, Eiriksdottir G, Hocking LJ, Holliday KL, Horan MA, Meulenbelt I, Neogi T, Popham M, Schmidt CO, Soni A, Valdes AM, Amin N, Dennison EM, Eijkelkamp N, Harris TB, Hart DJ, Hofman A, Huygen FJ, Jameson KA, Jones GT, Launer LJ, Kerkhof HJ, de Kruijf M, McBeth J, Kloppenburg M, Ollier WE, Oostra B, Payton A, Rivadeneira F, Smith BH, Smith AV, Stolk L, Teumer A, Thomson W, Uitterlinden AG, Wang K, van Wingerden SH, Arden NK, Cooper C, Felson D, Gudnason V, Macfarlane GJ, Pendleton N, Slagboom PE, Spector TD, Volzke H, Kavelaars A, van Duijn CM, Williams FM, van Meurs JB. Genome-wide association study meta-analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Ann Rheum Dis 2013; 72: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng W, Deshmukh HA, van Zuydam NR, Liu Y, Donnelly LA, Zhou K, Morris AD, Colhoun HM, Palmer CN, Smith BH, Wellcome Trust Case Control Consortium 2, Surrogate Markers for Micro- and Macro-Vascular Hard Endpoints for Innovative Diabetes Tools Study Group. A genome-wide association study suggests an association of Chr8p21.3 (GFRA2) with diabetic neuropathic pain. Eur J Pain 2015; 19: 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng W, Deshmukh HA, Donnelly LA, Torrance N, Colhoun HM, Palmer CN, Smith BA, Wellcome Trust Case Control Consortium 2, Surrogate Markers for Micro- and Macro-Vascular Hard Endpoints for Innovative Diabetes Tools Study Group. Genome-wide association study provides evidence of sex-specific involvement of Chr1p35.1 (ZSCAN20-TLR12P) and Chr8p23.1 (HMGB1P46) With diabetic neuropathic pain. EBioMedicine 2015; 2: 1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parisien M, Samoshkin A, Tansley SN, Piltonen MH, Martin LJ, El-Hachem N, Dagostino C, Allegri M, Mogil JS, Khoutorsky A, Diatchenko L. Genetic pathway analysis reveals a major role for extracellular matrix organization in inflammatory and neuropathic pain. Pain 2019; 160: 932–944. [DOI] [PubMed] [Google Scholar]
- 32.Gu Y, Qiu Z, Cheng N, Chen C, Hei Z, Li X. Identification of potential mechanism and hub genes for neuropathic pain by expression-based genome-wide association study. J Cell Biochem 2019; 120: 4912–4923. [DOI] [PubMed] [Google Scholar]
- 33.Nishizawa D, Fukuda K, Kasai S, Hasegawa J, Aoki Y, Nishi A, Saita N, Koukita Y, Nagashima M, Katoh R, Satoh Y, Tagami M, Higuchi S, Ujike H, Ozaki N, Inada T, Iwata N, Sora I, Iyo M, Kondo N, Won MJ, Naruse N, Uehara-Aoyama K, Itokawa M, Koga M, Arinami T, Kaneko Y, Hayashida M, Ikeda K. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Mol Psychiatry 2014; 19: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishizawa D, Fukuda KI, Kasai S, Ogai Y, Hasegawa J, Sato N, Yamada H, Tanioka F, Sugimura H, Hayashida M, Ikeda K. Association between KCNJ6 (GIRK2) gene polymorphism rs2835859 and post-operative analgesia, pain sensitivity, and nicotine dependence. J Pharmacol Sci 2014; 126: 253–263. [DOI] [PubMed] [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 37.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol 2008; 32: 361–369. [DOI] [PubMed] [Google Scholar]
- 38.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol 2010; 34: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao X. Multiple testing corrections for imputed SNPs. Genet Epidemiol 2011; 35: 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8: 1826–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suri P, Palmer MR, Tsepilov YA, Freidin MB, Boer CG, Yau MS, Evans DS, Gelemanovic A, Bartz TM, Nethander M, Arbeeva L, Karssen L, Neogi T, Campbell A, Mellstrom D, Ohlsson C, Marshall LM, Orwoll E, Uitterlinden A, Rotter JI, Lauc G, Psaty BM, Karlsson MK, Lane NE, Jarvik GP, Polasek O, Hochberg M, Jordan JM, Van Meurs JBJ, Jackson R, Nielson CM, Mitchell BD, Smith BH, Hayward C, Smith NL, Aulchenko YS, Williams FMK. Genome-wide meta-analysis of 158,000 individuals of European ancestry identifies three loci associated with chronic back pain. PLoS Genet 2018; 14: e1007601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dworkin RH, Portenoy RK. Pain and its persistence in herpes zoster. Pain 1996; 67: 241–251. [DOI] [PubMed] [Google Scholar]
- 45.Rowbotham MC, Fields HL. The relationship of pain, allodynia and thermal sensation in post-herpetic neuralgia. Brain 1996; 119: 347–354. [DOI] [PubMed] [Google Scholar]
- 46.Cluff RS, Rowbotham MC. Pain caused by herpes zoster infection. Neurol Clin 1998; 16: 813–832. [DOI] [PubMed] [Google Scholar]
- 47.Sato-Takeda M, Ihn H, Ohashi J, Tsuchiya N, Satake M, Arita H, Tamaki K, Hanaoka K, Tokunaga K, Yabe T. The human histocompatibility leukocyte antigen (HLA) haplotype is associated with the onset of postherpetic neuralgia after herpes zoster. Pain 2004; 110: 329–336. [DOI] [PubMed] [Google Scholar]
- 48.Ozawa A, Sasao Y, Iwashita K, Miyahara M, Sugai J, Iizuka M, Kawakubo Y, Ohkido M, Naruse T, Anzai T, Takashige N, Ando A, Inoko H. HLA-A33 and -B44 and susceptibility to postherpetic neuralgia (PHN). Tissue Antigens 1999; 53: 263–268. [DOI] [PubMed] [Google Scholar]
- 49.Sato M, Ohashi J, Tsuchiya N, Kashiwase K, Ishikawa Y, Arita H, Hanaoka K, Tokunaga K, Yabe T. Association of HLA-A*3303-B*4403-DRB1*1302 haplotype, but not of TNFA promoter and NKp30 polymorphism, with postherpetic neuralgia (PHN) in the Japanese population. Genes Immun 2002; 3: 477–481. [DOI] [PubMed] [Google Scholar]
- 50.Chung HY, Song EY, Yoon JA, Suh DH, Lee SC, Kim YC, Park MH. Association of human leukocyte antigen with postherpetic neuralgia in Koreans. Apmis 2016; 124: 865–871. [DOI] [PubMed] [Google Scholar]
- 51.Sassi Y, Lipskaia L, Vandecasteele G, Nikolaev VO, Hatem SN, Cohen Aubart F, Russel FG, Mougenot N, Vrignaud C, Lechat P, Lompre AM, Hulot JS. Multidrug resistance-associated protein 4 regulates cAMP-dependent signaling pathways and controls human and rat SMC proliferation. J Clin Invest 2008; 118: 2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee K, Belinsky MG, Bell DW, Testa JR, Kruh GD. Isolation of MOAT-B, a widely expressed multidrug resistance-associated protein/canalicular multispecific organic anion transporter-related transporter. Cancer Res 1998; 58: 2741–2747. [PubMed] [Google Scholar]
- 53.Schuetz JD, Connelly MC, Sun D, Paibir SG, Flynn PM, Srinivas RV, Kumar A, Fridland A. MRP4: a previously unidentified factor in resistance to nucleoside-based antiviral drugs. Nat Med 1999; 5: 1048–1051. [DOI] [PubMed] [Google Scholar]
- 54.Lee K, Klein-Szanto AJ, Kruh GD. Analysis of the MRP4 drug resistance profile in transfected NIH3T3 cells. J Natl Cancer Inst 2000; 92: 1934–1940. [DOI] [PubMed] [Google Scholar]
- 55.Leggas M, Adachi M, Scheffer GL, Sun D, Wielinga P, Du G, Mercer KE, Zhuang Y, Panetta JC, Johnston B, Scheper RJ, Stewart CF, Schuetz JD. Mrp4 confers resistance to topotecan and protects the brain from chemotherapy. Mol Cell Biol 2004; 24: 7612–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen ZS, Lee K, Walther S, Raftogianis RB, Kuwano M, Zeng H, Kruh GD. Analysis of methotrexate and folate transport by multidrug resistance protein 4 (ABCC4): MRP4 is a component of the methotrexate efflux system. Cancer Res 2002; 62: 3144–3150. [PubMed] [Google Scholar]
- 57.van Aubel RA, Smeets PH, Peters JG, Bindels RJ, Russel FG. The MRP4/ABCC4 gene encodes a novel apical organic anion transporter in human kidney proximal tubules: putative efflux pump for urinary cAMP and cGMP. J Am Soc Nephrol 2002; 13: 595–603. [DOI] [PubMed] [Google Scholar]
- 58.Reid G, Wielinga P, Zelcer N, van der Heijden I, Kuil A, de Haas M, Wijnholds J, Borst P. The human multidrug resistance protein MRP4 functions as a prostaglandin efflux transporter and is inhibited by nonsteroidal antiinflammatory drugs. Proc Natl Acad Sci U S A 2003; 100: 9244–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zelcer N, Reid G, Wielinga P, Kuil A, van der Heijden I, Schuetz JD, Borst P. Steroid and bile acid conjugates are substrates of human multidrug-resistance protein (MRP) 4 (ATP-binding cassette C4). Biochem J 2003; 371: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denk GU, Soroka CJ, Takeyama Y, Chen WS, Schuetz JD, Boyer JL. Multidrug resistance-associated protein 4 is up-regulated in liver but down-regulated in kidney in obstructive cholestasis in the rat. J Hepatol 2004; 40: 585–591. [DOI] [PubMed] [Google Scholar]
- 61.Sinreih M, Anko M, Kene NH, Kocbek V, Rizner TL. Expression of AKR1B1, AKR1C3 and other genes of prostaglandin F2alpha biosynthesis and action in ovarian endometriosis tissue and in model cell lines. Chem Biol Interact 2015; 234: 320–331. [DOI] [PubMed] [Google Scholar]
- 62.Guo SW, Wang Y. Sources of heterogeneities in estimating the prevalence of endometriosis in infertile and previously fertile women. Fertil Steril 2006; 86: 1584–1595. [DOI] [PubMed] [Google Scholar]
- 63.Rogers PA, D'Hooghe TM, Fazleabas A, Giudice LC, Montgomery GW, Petraglia F, Taylor RN. Defining future directions for endometriosis research: workshop report from the 2011 world congress of endometriosis in Montpellier, France. Reprod Sci 2013; 20: 483–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin ZP, Zhu YL, Johnson DR, Rice KP, Nottoli T, Hains BC, McGrath J, Waxman SG, Sartorelli AC. Disruption of cAMP and prostaglandin E2 transport by multidrug resistance protein 4 deficiency alters cAMP-mediated signaling and nociceptive response. Mol Pharmacol 2008; 73: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saito S, Iida A, Sekine A, Miura Y, Ogawa C, Kawauchi S, Higuchi S, Nakamura Y. Identification of 779 genetic variations in eight genes encoding members of the ATP-binding cassette, subfamily C (ABCC/MRP/CFTR. J Hum Genet 2002; 47: 147–171. [DOI] [PubMed] [Google Scholar]
- 66.Abla N, Chinn LW, Nakamura T, Liu L, Huang CC, Johns SJ, Kawamoto M, Stryke D, Taylor TR, Ferrin TE, Giacomini KM, Kroetz DL. The human multidrug resistance protein 4 (MRP4, ABCC4): functional analysis of a highly polymorphic gene. J Pharmacol Exp Ther 2008; 325: 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Banerjee M, Marensi V, Conseil G, Le XC, Cole SP, Leslie EM. Polymorphic variants of MRP4/ABCC4 differentially modulate the transport of methylated arsenic metabolites and physiological organic anions. Biochem Pharmacol 2016; 120: 72–82. [DOI] [PubMed] [Google Scholar]
- 68.Palikhe S, Uuganbayar U, Trinh HKT, Ban GY, Yang EM, Park HS, Kim SH. A role of the ABCC4 gene polymorphism in airway inflammation of asthmatics. Mediators Inflamm 2017; 2017: 3549375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mesrian Tanha H, Rahgozar S, Mojtabavi Naeini M. ABCC4 functional SNP in the 3' splice acceptor site of exon 8 (G912T) is associated with unfavorable clinical outcome in children with acute lymphoblastic leukemia. Cancer Chemother Pharmacol 2017; 80: 109–117. [DOI] [PubMed] [Google Scholar]
- 70.Tsukamoto M, Sato S, Satake K, Miyake M, Nakagawa H. Quantitative evaluation of drug resistance profile of cells expressing wild-type or genetic polymorphic variants of the human ABC transporter ABCC4. IJMS 2017; 18: 1435–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsukamoto M, Yamashita M, Nishi T, Nakagawa H. A human ABC transporter ABCC4 gene SNP (rs11568658, 559 G > T, G187W) reduces ABCC4-dependent drug resistance. Cells 2019; 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Y, Shi N, Lu H, Zhang J, Ma Y, Qiao Y, Mao Y, Jia K, Han L, Liu F, Li H, Lin Z, Li X, Zhao X. ABCC4 copy number variation is associated with susceptibility to esophageal squamous cell carcinoma. Carcinogenesis 2014; 35: 1941–1950. [DOI] [PubMed] [Google Scholar]
- 73.Johnson C, Pankratz VS, Velazquez AI, Aakre JA, Loprinzi CL, Staff NP, Windebank AJ, Yang P. Candidate pathway-based genetic association study of platinum and platinum-taxane related toxicity in a cohort of primary lung cancer patients. J Neurol Sci 2015; 349: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sloan JA, de Andrade M, Decker P, Wampfler J, Oswold C, Clark M, Yang P. Genetic variations and patient-reported quality of life among patients with lung cancer. J Clin Oncol 2012; 30: 1699–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee E, Takita C, Wright JL, Slifer SH, Martin ER, Urbanic JJ, Langefeld CD, Lesser GJ, Shaw EG, Hu JJ. Genome-wide enriched pathway analysis of acute post-radiotherapy pain in breast cancer patients: a prospective cohort study. Hum Genom 2019; 13: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun Z, Arendt CW, Ellmeier W, Schaeffer EM, Sunshine MJ, Gandhi L, Annes J, Petrzilka D, Kupfer A, Schwartzberg PL, Littman DR. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature 2000; 404: 402–407. [DOI] [PubMed] [Google Scholar]
- 77.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA-K, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson Å, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlén M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteom 2014; 13: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berger C, Rossaint J, Van Aken H, Westphal M, Hahnenkamp K, Zarbock A. Lidocaine reduces neutrophil recruitment by abolishing chemokine-induced arrest and transendothelial migration in septic patients. J Immunol 2014; 192: 367–376. [DOI] [PubMed] [Google Scholar]
- 79.Tsubaki M, Takeda T, Matsumoto M, Kato N, Yasuhara S, Koumoto YI, Imano M, Satou T, Nishida S. Tamoxifen suppresses paclitaxel-, vincristine-, and bortezomib-induced neuropathy via inhibition of the protein kinase C/extracellular signal-regulated kinase pathway. Tumour Biol 2018; 40: 1010428318808670. [DOI] [PubMed] [Google Scholar]
- 80.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, Warram JH, Todd JA. Analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet 2008; 40: 1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, Park YR, Raychaudhuri S, Pouget JG, Hubenthal M, Folseraas T, Wang Y, Esko T, Metspalu A, Westra HJ, Franke L, Pers TH, Weersma RK, Collij V, D'Amato M, Halfvarson J, Jensen AB, Lieb W, Degenhardt F, Forstner AJ, Hofmann A, Schreiber S, Mrowietz U, Juran BD, Lazaridis KN, Brunak S, Dale AM, Trembath RC, Weidinger S, Weichenthal M, Ellinghaus E, Elder JT, Barker JN, Andreassen OA, McGovern DP, Karlsen TH, Barrett JC, Parkes M, Brown MA, Franke A, International Genetics of Ankylosing Spondylitis Consortium (IGAS); International PSC Study Group (IPSCSG); Genetic Analysis of Psoriasis Consortium (GAPC); Psoriasis Association Genetics Extension (PAGE). Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016; 48: 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang YW, Xu XY, Zhang J, Yao X, Lu C, Chen CX, Yu CH, Sun J. Missense mutation in PRKCQ is associated with Crohn’s disease. J Dig Dis 2019; 20: 243–247. [DOI] [PubMed] [Google Scholar]
- 83.Paolillo N, Piccirilli S, Giardina E, Rispoli V, Colica C, Nistico S. Effects of paraquat and capsaicin on the expression of genes related to inflammatory, immune responses and cell death in immortalized human HaCat keratinocytes. Int J Immunopathol Pharmacol 2011; 24: 861–868. [DOI] [PubMed] [Google Scholar]
- 84.Anstee QM, Darlay R, Cockell S, Meroni M, Govaere O, Tiniakos D, Burt AD, Bedossa P, Palmer J, Liu YL, Aithal GP, Allison M, Yki-Jarvinen H, Vacca M, Dufour JF, Invernizzi P, Prati D, Ekstedt M, Kechagias S, Francque S, Petta S, Bugianesi E, Clement K, Ratziu V, Schattenberg JM, Valenti L, Day CP, Cordell HJ, Daly AK; EPoS Consortium Investigators. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol 2020; 73: 505–515. [DOI] [PubMed] [Google Scholar]
- 85.Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen C-Y, Choi KW, Coleman JRI, Dalvie S, Duncan LE, Gelernter J, Levey DF, Logue MW, Polimanti R, Provost AC, Ratanatharathorn A, Stein MB, Torres K, Aiello AE, Almli LM, Amstadter AB, Andersen SB, Andreassen OA, Arbisi PA, Ashley-Koch AE, Austin SB, Avdibegovic E, Babić D, Bækvad-Hansen M, Baker DG, Beckham JC, Bierut LJ, Bisson JI, Boks MP, Bolger EA, Børglum AD, Bradley B, Brashear M, Breen G, Bryant RA, Bustamante AC, Bybjerg-Grauholm J, Calabrese JR, Caldas-de-Almeida JM, Dale AM, Daly MJ, Daskalakis NP, Deckert J, Delahanty DL, Dennis MF, Disner SG, Domschke K, Dzubur-Kulenovic A, Erbes CR, Evans A, Farrer LA, Feeny NC, Flory JD, Forbes D, Franz CE, Galea S, Garrett ME, Gelaye B, Geuze E, Gillespie C, Uka AG, Gordon SD, Guffanti G, Hammamieh R, Harnal S, Hauser MA, Heath AC, Hemmings SMJ, Hougaard DM, Jakovljevic M, Jett M, Johnson EO, Jones I, Jovanovic T, Qin X-J, Junglen AG, Karstoft K-I, Kaufman ML, Kessler RC, Khan A, Kimbrel NA, King AP, Koen N, Kranzler HR, Kremen WS, Lawford BR, Lebois LAM, Lewis CE, Linnstaedt SD, Lori A, Lugonja B, Luykx JJ, Lyons MJ, Maples-Keller J, Marmar C, Martin AR, Martin NG, Maurer D, Mavissakalian MR, McFarlane A, McGlinchey RE, McLaughlin KA, McLean SA, McLeay S, Mehta D, Milberg WP, Miller MW, Morey RA, Morris CP, Mors O, Mortensen PB, Neale BM, Nelson EC, Nordentoft M, Norman SB, O'Donnell M, Orcutt HK, Panizzon MS, Peters ES, Peterson AL, Peverill M, Pietrzak RH, Polusny MA, Rice JP, Ripke S, Risbrough VB, Roberts AL, Rothbaum AO, Rothbaum BO, Roy-Byrne P, Ruggiero K, Rung A, Rutten BPF, Saccone NL, Sanchez SE, Schijven D, Seedat S, Seligowski AV, Seng JS, Sheerin CM, Silove D, Smith AK, Smoller JW, Sponheim SR, Stein DJ, Stevens JS, Sumner JA, Teicher MH, Thompson WK, Trapido E, Uddin M, Ursano RJ, van den Heuvel LL, Van Hooff M, Vermetten E, Vinkers CH, Voisey J, Wang Y, Wang Z, Werge T, Williams MA, Williamson DE, Winternitz S, Wolf C, Wolf EJ, Wolff JD, Yehuda R, Young RM, Young KA, Zhao H, Zoellner LA, Liberzon I, Ressler KJ, Haas M, Koenen KC. International Meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun 2019; 10: 4558–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Larsen MH, Albrechtsen A, Thorner LW, Werge T, Hansen T, Gether U, Haastrup E, Ullum H. Genome-wide association study of genetic variants in LPS-stimulated IL-6, IL-8, IL-10, IL-1ra and TNF-alpha cytokine response in a Danish Cohort. PLoS One 2013; 8: e66262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rimpela JM, Porsti IH, Jula A, Lehtimaki T, Niiranen TJ, Oikarinen L, Porthan K, Tikkakoski A, Virolainen J, Kontula KK, Hiltunen TP. Genome-wide association study of nocturnal blood pressure dipping in hypertensive patients. BMC Med Genet 2018; 19: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Igarashi M, Nogawa S, Kawafune K, Hachiya T, Takahashi S, Jia H, Saito K, Kato H. Identification of the 12q24 locus associated with fish intake frequency by genome-wide meta-analysis in Japanese populations. Genes Nutr 2019; 14: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang DW, Wang TM, Zhang JB, Li XZ, He YQ, Xiao R, Xue WQ, Zheng XH, Zhang PF, Zhang SD, Hu YZ, Shen GP, Chen M, Sun Y, Jia WH. Genome-wide association study identifies genetic susceptibility loci and pathways of radiation-induced acute oral mucositis. J Transl Med 2020; 18: 224–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-mpx-10.1177_1744806921999924 for Genome-wide association study identifies candidate loci associated with chronic pain and postherpetic neuralgia by Daisuke Nishizawa, Masako Iseki, Hideko Arita, Kazuo Hanaoka, Choku Yajima, Jitsu Kato, Setsuro Ogawa, Ayako Hiranuma, Shinya Kasai, Junko Hasegawa, Masakazu Hayashida and Kazutaka Ikeda in Molecular Pain