Abstract
Intercellular signals induce various cellular responses, including growth, proliferation, and differentiation, via the dynamic processes of signal transduction pathways. For cell fate decisions, ligand-binding induces the phosphorylation of ERBB receptors, which in turn activate downstream molecules. The ERBB family includes four subtypes, which diverged through two gene duplications from a common ancestor. Differences in the expression patterns of the subtypes have been reported between different organs in the human body. However, how these different expression properties influence the diverse phosphorylation levels of ERBB proteins is not well understood. Here we study the origin of the phosphorylation responses by experimental and mathematical analyses. The experimental measurements clarified that the phosphorylation levels heavily depend on the ERBB expression profiles. We developed a mathematical model consisting of the four subtypes as monomers, homodimers, and heterodimers and estimated the rate constants governing the phosphorylation responses from the experimental data. To understand the origin of the diversity, we analyzed the effects of the expression levels and reaction rates of the ERBB subtypes on the diversity. The difference in phosphorylation rates between ERBB subtypes showed a much greater contribution to the diversity than did the dimerization rates. This result implies that divergent evolution in phosphorylation reactions rather than in dimerization reactions after whole genome duplications was essential for increasing the diversity of the phosphorylation responses.
Significance
It is known that the expression patterns of a protein family are different between different organs in the human body. This difference is considered essential for the functional diversity between organs. However, the dynamical processes that translate expression patterns into function are not well understood. Here we study the origin of the diversity in ERBB phosphorylation patterns by combining experimental and mathematical methods. We confirmed experimentally that phosphorylation patterns depend on the ERBB expression profiles. Our mathematical model found that differences in phosphorylation rates make the greatest contribution to the diversity of phosphorylation patterns between ERBB subtypes.
Introduction
Signal transduction pathways govern the cellular response to an intercellular signal, thus affecting cell growth, proliferation, differentiation, and other fundamental physiological processes. In the ERBB-RAS-MAPK system (1), which is responsible for cell-fate decisions, more than 100 biomolecules are connected via chemical reactions or regulations of the reactions to constitute a complex network (2). In vertebrates other than teleostei, the ERBB family includes four subtypes, ERBB1, ERBB2, ERBB3, and ERBB4, which are all transmembrane proteins and receptor tyrosine kinases (3, 4, 5). ERBB1 is also called epidermal growth factor (EGF) receptor, coming from the name of its ligand (6).
ERBB proteins have more than 10 types of extracellular ligands (7) that are all small peptides produced by neighboring cells. Each ERBB paralog exhibits overlapping selectivity and specificity to a subset of ligand species that stimulates a given biological function. In general, ligand binding induces ERBB phosphorylation, which in turn activates the MAPK, PI3K-AKT, and other pathways in parallel (8). In other words, ERBB family proteins act as gatekeepers of signal transduction pathways. Ligand binding causes ERBB proteins to form homodimers or heterodimers (9,10). Phosphorylation of the protomers in the ERBB dimers takes place on tyrosine residues in the cytoplasmic domain (Fig. 1 a), triggering the activation of various cytoplasmic proteins in the signaling pathways (signal transduction), and finally altering the cell structure and function. The phosphorylation level of each ERBB protein depends on the combination of ERBB species in the dimer pair, and differences in the phosphorylation levels influence the downstream signaling. To understand the role of the ERBB network in signaling pathways totally, we need to understand the phosphorylation responses of systems consisting of multiple ERBB subtypes of diverse properties. It also is thought that different expression levels between ERBB subtypes should be responsible for the diversity of the phosphorylation responses. Additionally, it has been reported that different organs in the human body show different ERBB expression profiles (Fig. 1 c) (11, 12, 13). Further, the different expression patterns may be the origin of the diversity of the responses of ERBB signaling pathways between organs. In addition to the different expression profiles of ERBBs, the diversity of kinetic properties among the four subtypes are necessary for the diversity of phosphorylation responses. If the four subtypes have similar kinetic properties (binding, dimerization, and phosphorylation rates), then different ERBB expression profiles cannot induce large differences in dynamical behaviors. Therefore, the diversity of the phosphorylation responses should depend both on the expression profiles (dose-dependent diversity) and reaction rates (rate-dependent diversity) of ERBBs. As far as we know, the contributions of these two factors to the diversity of phosphorylation responses have not been examined thoroughly.
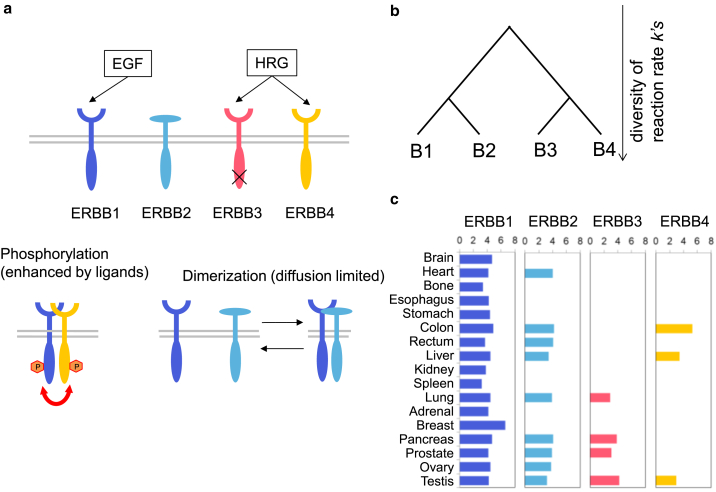
Figure 1.
(a) Summary of the three reactions (ligand binding, dimerization, phosphorylation) in the ERBB system. The specificity of the ligand binding is shown schematically. ERBB3 does not have kinase activity. (b) Schematic phylogenic tree of the ERBB family. (c) ERBB expression profiles from various human organs. Values are taken from ProteomicsDB (ERBB1: https://www.proteomicsdb.org/proteomicsdb/#protein/proteinDetails/51261/expression, ERBB2: https://www.proteomicsdb.org/proteomicsdb/#protein/proteinDetails/51718/expression, ERBB3: https://www.proteomicsdb.org/proteomicsdb/#protein/proteinDetails/53934/expression, ERBB4: https://www.proteomicsdb.org/proteomicsdb/#protein/proteinDetails/60520/expression).
The four members of ERBB are paralogs that diverged with two gene duplications from a common ancestor (Fig. 1 b) (14, 15, 16). The first divergence produced two members, the ancestor of ERBB1 and ERBB2 and that of ERBB3 and ERBB4. The present four members appeared in the second divergence. After each divergence, the ERBB family members are thought to have acquired different kinetic properties, including different rate constants for ligand binding, dimerization, and phosphorylation. For example, ERBB3 does not have kinase activity, probably because the activity was lost during evolution (17). However, it is difficult to determine all kinetic properties of these proteins under living cell conditions by experimental methods.
In this article, we study the origin of the diversity of the phosphorylation responses of ERBB systems consisting of the four subtypes as monomers, homodimers, and heterodimers by combining experimental measurements and mathematical analyses. In the experiments, EGF and hergulin (HRG) were chosen as the input signals of the ERBB system, since EGF binds to ERBB1 and HRG binds to ERBB3 and ERBB4 (18). We measured the phosphorylation responses to two ligands from three cell lines and found the phosphorylation levels highly depended on the ERBB expression profile. Mathematical modeling was used to estimate and identify differences in the reaction rate constants governing the phosphorylation responses between ERBB subtypes. From the mathematical analysis, we found that the diversity of the phosphorylation rates between ERBB subtypes has a much greater contribution to the diversity of the phosphorylation responses than the diversity of the dimerization rates does. This finding implies that divergent evolution in phosphorylation reactions rather than dimerization reactions after whole genome duplications was essential for increasing the diversity of the phosphorylation responses among subtypes.
Materials and methods
Time-lapse measurements
HeLa, A431, and MCF7 cells were cultured in DMEM (WAKO #044–29765) containing 10% FBS to high confluence in 35-mm dishes under 5% CO2 at 37°C. One day before the experiment, the cells were starved in DMEM without fetal bovine serum and phenol red. The cells were stimulated with 20 nM EGF (PEPROTECH) or 30 nM HRG (R&D Systems) and incubated for 5, 10, 15, 30, and 60 min at 25°C. At each time point, the cells were washed with PBS and lysed in 100 μL of 1× SDS sample buffer containing 1 mM Na3VO4 to avoid dephosphorylation during the sample preparation. The collected samples were boiled at 95°C for 30 min and then cooled on ice, and SDS-PAGE separation was executed in a 7.5%–10% polyacrylamide gel. The separated fractions were transferred to a membrane using a semidry blotting apparatus. After blocking the membrane with 2.5% skim milk in Tris-buffered saline with 0.1% Tween 20, the primary antibody reaction was carried out at 4°C overnight, in which anti-phospho-ERBB antibodies for pY1173 of ERBB1 (CST #4407), pY1221/1222 of ERBB2 (CST #2243), pY1289 of ERBB3 (CST #4791), and pY1284 of ERBB4 (CST #4757) were diluted to 1:300–1:500 and applied on the membrane. Then, incubation with a 1:1000 secondary antibody (anti-rabbit IgG) conjugated with horse radish peroxidase (horseradish peroxidase [HRP], CST #7074) was done at 25°C for 1 h. Chemical luminescence from the second antibody reacted with ECL prime reagent (GE Healthcare) was detected by a lumino image analyzer, ImageQuant LAS500 or LAS4000 (GE Healthcare).
ERBB expression profiles
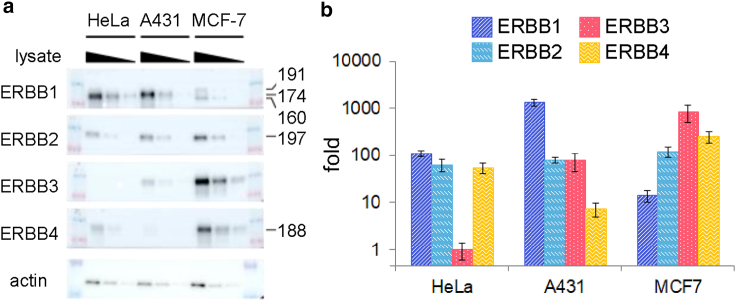
The relative expression levels of ERBB proteins in the cells were estimated from Western blotting. For quantitative comparisons, correction factors compensating for differences in the antibody titers were determined from the western blotting of ERBBs fused with GFP (ERBB-GFP), i.e., the expression of each type of ERBB-GFP was measured in the same extract of cells with anti-ERBB and anti-GFP (CST #2956) antibodies, and the titer of each anti-ERBB antibody was normalized against that of anti-GFP antibody (Fig. S1). Cross-talk between anti-ERBBs was observed as ERBB1 detected by anti-ERBB4 antibody; however, in most cases, the difference in the molecular weight between ERBBs allowed for the detection of specific staining. In MCF7 cells, the missed staining of ERBB4 (Fig. 3 a) might be because of an overlap with the minor upper band of the anti-ERBB1 staining. We excluded the overlapped band in the estimation to minimize the detection error, because the ERBB4 expression was relatively high; thus, the estimated lower limit value was used in the mathematical analysis described below.
Figure 3.
ERBB expression profiles in the three cell types. (a) Western blotting of ERBB proteins. The lysate of each cell line was diluted in three steps to confirm the linearity of the antibody staining. (b) Quantification of the ERBB expression level. Values were normalized to the ERBB3 expression in HeLa cells. Error bars, SE.
Overexpression experiments
A gene transfer plasmid that included the sequence of ERBB1, ERBB2, or ERBB3 was transfected to induce the overexpression of the given ERBB protein. The transfection was carried out with a combination of DNA and reagent product that maximized the expression level of transfected ERBB: for HeLa, A431, and MCF7 cells, this combination was a 10, 7.5, and 5 μg DNA/35-mm dish and the lipofection reagents Lipofectamine 3000, Lipofectamine LTX&PLUS, and Lipofectamine 2000 (Thermo Fisher Scientific), respectively. Two dishes of each transfectant were prepared and cultured for 1 day. One dish was used for Western blotting to measure the phosphorylation and expression levels of each ERBB, and the other was used to check the fraction of ERBB-overexpressing cells by fluorescence immunostaining. For the latter assay, cells were cultured on a coverslip in the dish for observation under a fluorescence microscope. In the Western blot analysis, only the cultures containing more than 90% of ERBB-overexpressing cells were used. To detect the total amount of ERBBs, anti-ERBB antibodies were used with dilutions of 1:500 for ERBB1 (SANTA CRUZ #sc-03), 1:300 for ERBB2 (CST #4290), 1:500 for ERBB3 (SANTA CRUZ #sc-285), and 1:300 for anti-ERBB4 (Santa Cruz B iotechnology #sc-283). The secondary antibodies were 1:2000-diluted HRP-linked anti-IgG antibodies (CST #7074 for rabbit and CST #7076 for mouse). ECL prime reagent (GE Healthcare) was used for acquiring the chemiluminescence signals. For the fluorescence immunostaining, cells without ligand stimulation were fixed using MeOH at −20°C for 2 min, washed with HBSS three times and left at 4°C overnight. After the blocking process with HBSS containing 2% BSA for 15 min, 1:300-diluted anti-ERBB1, anti-ERBB2, or anti-ERBB3 was used for the primary antibody incubation for 1 h at 25°C. Coverslips were washed three times with HBSS and then incubated with 1:1000 anti-IgG antibody conjugated with Alexa 488 (Thermo Fisher Scientific A-11034) for 1 h at 25°C The secondary antibody-labeled samples were observed under an inverted fluorescence microscope (Nikon Ti) with a 10× objective lens (Nikon PalnApo) and a filter set suitable for Alexa488. Image acquisition was carried out through a sCMOS camera (ORCA Flash 2.0, Hamamatsu) with NIS elements software (Nikon). The fluorescence intensity was measured for every cell, which was contoured manually using ImageJ software (NIH); the mean intensities are plotted in Fig. S2 b.
Quantification of Western blot staining
The intensities of the bands detected in the Western blotting were quantified using ImageJ software. Rectangular regions of interest were set in regions of the signal (band) and background; the latter was located far enough from the former so that there was no overlap. The signal intensity was defined as the difference in the average intensities of the two regions.
Experimental results
Phosphorylation of ERBB
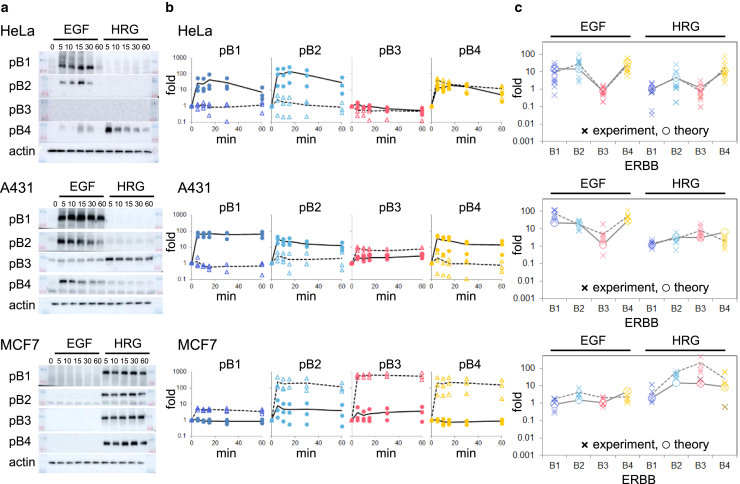
The phosphorylation of ERBB subtypes (ERBB1, ERBB2, ERBB3, and ERBB4) was detected semiquantitatively in A431, HeLa, and MCF7 cells stimulated with EGF or HRG (Fig. 2). There are multiple residues for tyrosine phosphorylation in each ERBB protein, and the phosphorylation levels have been shown to correlate between residues (19,20), allowing us to choose one representative residue for the phosphorylation assay. Additionally, A431, HeLa, and MCF7 cells are known to show different responses to the same ERBB ligands. We stimulated the cells with EGF or HRG with a saturation concentration of 17 nM (100 ng/mL) and 30 nM, respectively. At the present time, no intrinsic ligand is known for ERBB2 (Fig. S3). The initial response of the ligand-induced phosphorylation was quantified by western blotting at several time points within 60 min of the ligand application.
Figure 2.
Phosphorylation of ERBB induced by EGF or HRG ligands in HeLa, A431, and MCF7 cells. (a) Western blotting of ERBB1-B4 after ligand stimulation (numbers at the top indicate minutes). pB1, pB2, pB3, and pB4 denote phosphorylated ERBB1, ERBB2, ERBB3, and ERBB4, respectively. (b) The time course of the ERBB phosphorylation obtained from Western blotting. Fold changes compare the phosphorylation level with just before the ligand stimulation (time 0). Closed circles and open triangles indicate EGF and HRG stimulation, respectively. (c) The fold change of ERBB phosphorylation at 5 min. Crosses and open circles respectively represent the experimental results and global model fitting.
The time-course results indicated that the phosphorylation level changed dramatically within 5 min and then slowly changed the next 55 min. To focus on the initial responses directly induced by the ERBB-ligand interaction, we analyzed ERBB phosphorylation at 5 min, at which point feedback effects from downstream signaling are minimal (21). The three cell types showed distinct behaviors in their phosphorylation dynamics to the same ligand, and the two ligands induced distinct dynamics in the same cell type.
ERBB expression profile
The relative expression levels of ERBB1-B4 in HeLa, A431, and MCF7 cells were measured by Western blotting (Fig. 3 a). The densities of the observed bands for anti-ERBB antibodies were compensated for differences in the antibody titers (Fig. S1) to compare the expression levels (see Materials and methods). The estimated ERBB expression levels were normalized to the lowest expression level of ERBB (HeLa ERBB3) (Fig. 3 b). In the following calculation, we normalized the expression of each ERBB to the expression of ERBB3 in HeLa. We confirmed that the conclusions from the experiments were unaffected qualitatively by the choice of the normalization.
The observed tendency of the expression profiles was similar to previous studies: ERBB1 was abundantly expressed in A431 cells (22), and ERBB3 and ERBB4 were expressed more than ERBB1 in MCF7 cells (23).
ERBB overexpression profile
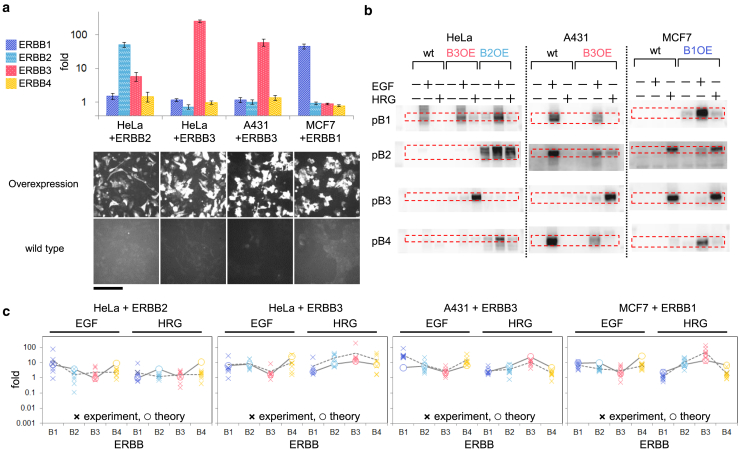
The large cell type-to-type differences in the expression levels among ERBB subtypes (Fig. 3) may relate to the diversity in their phosphorylation patterns (Fig. 2). To confirm this possibility, we examined the following four conditions: (1) ERBB2 overexpression in HeLa cells (HeLa + B2), (2) ERBB3 overexpression in HeLa (HeLa + B3), (3) ERBB3 overexpression in A431 cells (A431 + B3), and (4) ERBB1 overexpression in MCF7 cells (MCF7 + B1). These overexpressions were chosen based on our speculation that increasing the expression levels of initially lowly expressed genes induces large changes in the phosphorylation dynamics.
The overexpression levels of ERBB were measured by western blotting and found to be 51-, 260-, 61-, and 46-fold higher for HeLa + B2, HeLa + B3, A431 + B3, and MCF + B1, respectively, compared with the expression levels in parental cells (Fig. 4 a, upper). We confirmed that our measurements were not mixtures of cells with various expression levels by immunofluorescence (Fig. 4 a, lower, and Fig. S2), which showed that almost all cells (>90%) overexpressed the transfected ERBB. The phosphorylation of ERBBs in wild-type and ERBB-overexpressing cells was also measured by Western blotting (Fig. 4 b). The phosphorylation patterns of ERBBs were significantly affected by the overexpression. The overexpression levels and phosphorylation levels were quantified experimentally for the following mathematical analysis.
Figure 4.
ERBB phosphorylation in cells overexpressing ERBB. (a) ERBB expression levels (upper) and immunofluorescence images of overexpressed ERBB (lower) in the indicated cells. The locations of the cells are shown in Fig. S2a. The expression level of each ERBB was normalized to that in wild-type cells. Error bars, SE. Scale bar, 50 μm. (b) EGF or HRG induced ERBB phosphorylation in wild-type (wt) and ERBB-overexpressing (OE) cells. The red dotted rectangles indicate phosphorylated ERBB bands. (c) Fold change of the ERBB phosphorylation. Crosses and open circles respectively show experimental observations and results obtained from the global model fitting.
Mathematical analysis
Mathematical modeling
To clarify the effects of the expression profiles and kinetic properties on the phosphorylation levels, we constructed a mathematical model based on mass action differential equations of the chemical reactions (see (24, 25, 26, 27) for studies based on mass action kinetics). We labeled the ERBB subtypes as . The 4 subtypes can be phosphorylated, ligand-bound (for ), or both, thus giving 14 monomer states in total, which we write as . These monomers form dimers , such that there are 105 dimer states. In the early stage of the phosphorylation dynamics ( 5 min), there are three different classes of reactions: (1) ligand-binding and dissociation, (2) dimerization and dissociation, and (3) phosphorylation and dephosphorylation (Fig. 1 a). We do not consider the endocytosis or degradation of ERBBs in this early stage of the signal transduction at 25°C.
To decrease the number of variables and parameters in the model, we assumed that the ligand concentrations are negligible before the stimulation and that the ligand concentrations are sufficiently high and all ERBB proteins are ligand bound after the stimulation. We experimentally confirmed that phosphorylation levels after stimulation were saturated with the ligand concentrations used in our experiments (Fig. S3).
Thus, we can decrease the system size, since it is sufficient to consider either ligand-unbound states (before stimulation) or ligand-bound states (after stimulation), and we do not need to deal with their coexistence. More specifically, the concentrations were determined from the following equations:
| (1) |
| (2) |
Here, and are the concentrations of the monomers and dimers , respectively. The indices take only two states, {0 = unphosphorylated, 1 = phosphorylated}, and represents the other state, i.e., 1 and 0. and denote the phosphorylation/dephosphorylation rates of the monomers and dimers, respectively. For example, denotes the phosphorylation rate of ERBBi monomer, while denotes the dephosphorylation rate. We assume that ERBB3 receptors are incapable of phosphorylating themselves and other receptors have rates of zero; that is, and . Finally, and denote the dimerization rates and dissociation rates, respectively. An important point of our analysis is that we consider the steady-state Eqs. (1) and (2) both before and after ligand stimulation, and the effects of the stimulation are implemented using different values of the dimerization and dissociation rate constants and the dimer phosphorylation rate constants before and after the stimulation.
The model has four conserved quantities: the total amounts of ERBB1, ERBB2, ERBB3, and ERBB4. We measured these four quantities experimentally for each cell type. Given these conserved quantities and the reaction rate constants, Eqs. (1) and (2) determine the steady-state concentrations of all chemicals uniquely. To obtain a minimal model, we assumed that all cell types have the same reaction rate constants. As we show below, the diversity of the responses to a ligand stimulation among the cell types can be explained from the difference in the expression profiles of the ERBB receptors.
Parameter estimation
We estimated the parameter values of the reaction rate constants by performing a Markov Chain Monte Carlo (MCMC) simulation. For a given set of parameter values, we computed the phosphorylation levels of ERBB1, ERBB2, ERBB3, and ERBB4 under three external environments by numerically solving Eqs. (1) and (2): in the absence of ligands, in the presence of EGF, and in the presence of HRG. Then we compared the obtained phosphorylation levels with those measured experimentally with the energy function,
| (3) |
where the index = 1,2,3,4 labels the ERBB subtype, labels the three environmental conditions, and labels the seven cell types (HeLa, MCF7, A431, HeLa + B2, HeLa + B3, A431 + B3, and MCF7 + B1). Here, the four mutant types (HeLa + B2, HeLa + B3, A431 + B3, and MCF7 + B1) were also used in the parameter estimation to enlarge the data set for higher validity. and denote the respective experimentally and theoretically measured phosphorylation level of the ERBB receptor of cell under environment . Note that is the total concentration of phosphorylated ERBB molecules; therefore, both phosphorylated monomers and dimers contribute to .
In the MCMC, the parameter values were updated with a Metropolis method: every parameter value was randomly perturbed by , and then the energy in Eq. (3) was computed with the updated parameter set. If , the updated parameter set was accepted; if , the parameter set was accepted with probability with the inverse temperature . Through the MCMC, we constructed an ensemble of parameter sets that explains the experimental results with accuracy specified by . Note that, when perturbing the parameter values, maximum and minimum values were imposed; for the reaction rates, they were [10−5 s/nM, 1.0 s/nM], and for the multiplication factors owing to ligand stimulation, they were [1, 100]. The Mathematica code is available upon request.
Fig. 2C shows the experimentally measured and mathematically calculated fold changes in phosphorylation levels of ERBB receptors for two different ligands and three cultured cell types (HeLa, MCF7, and A431). The mathematical model explains the diversity of the phosphorylation levels of the ERBB receptors very well based on the difference in the receptor expression profiles. Fig. 4 C shows a comparison of phosphorylation profiles in mutant cell types (HeLa + B2, HeLa + B3, A431 + B3, and MCF7 + B1) between the experiments and mathematical calculations. The phosphorylation profiles by the mathematical model were consistent with the observed profiles of the ERBB overexpression mutants as well as the wild-type cells. We expect the estimated reaction rate constants to reflect the real phosphorylation dynamics.
The average estimated values of the reaction rate constants are shown in Table S1 and graphically represented in Fig. 5. In Fig. 5, the estimated reaction rates related to phosphorylation (Fig. 5 a) and dimerization (Fig. 5 b) are shown in three network styles, namely, the ratios of the phosphorylation rates to the dephosphorylation rates and the multipliers owing to EGF and HRG stimulation. The left multiplier networks in Fig. 5 characterize the effect of EGF, which binds to ERBB1. The right multiplier networks characterize the effect of HRG, which binds to ERBB3 and ERBBB4. The effect of ligand binding is highly variable among ERBB subtypes. We also show the distributions of these values in three panels in Fig. S4 a and b. Most of the estimated values show narrow distributions, which may allow us to discuss the estimated reaction rates based on the average values.
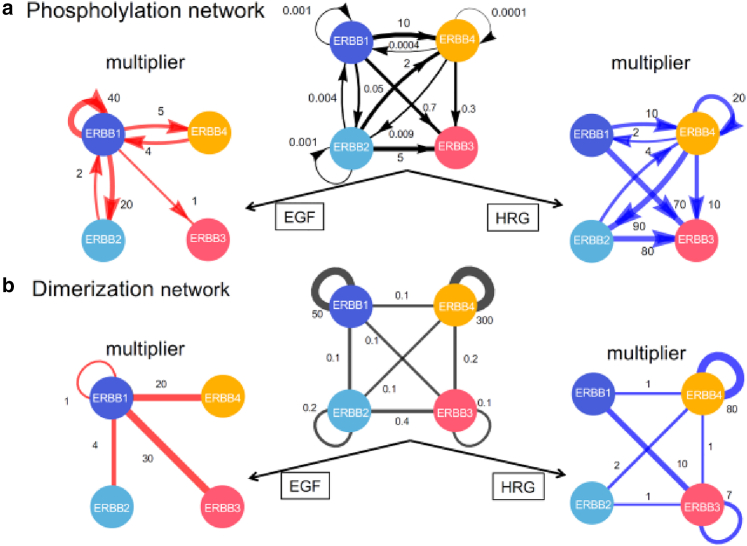
Figure 5.
Ligand effects on the phosphorylation and dimerization networks. (a) Phosphorylation networks. The black network in the middle shows effective phosphorylation rates on the edges, each of which is defined as the ratio of the dimer phosphorylation rate to the dephosphorylation rate. The left (red) and right (blue) networks represent multipliers on the effective rates owing to EGF and HRG stimulation, respectively. (b) Dimerization networks. The black network in the middle shows effective dimerization rates on the edges, each of which is defined as the ratio of the dimerization rate to the dissociation rate. The left (red) and right (blue) networks represent multipliers on the effective rates owing to EGF and HRG stimulation, respectively.
In the following, we discuss the characteristics of the estimated reaction rate constants under the three different conditions (namely, before ligand stimulation, after EGF stimulation, and after HRG stimulation) in view of the biological network response. First, we consider the phosphorylation and dimerization properties of the ERBB system before ligand stimulation (see the central networks in Fig. 5 a and b). Affinities between the ERBB protomers are strong, especially in B1 and B4 homodimers, even in the absence of any ligand. This prediction is consistent with previous experiments (9,10) that indicated the presence of ERBB preformed dimers (i.e., dimers before the ligand association). At basal states, the effective phosphorylation rates in homodimers, which are determined by the balance between spontaneous phosphorylation and dephosphorylation reactions, are much lower than 1. As a result, basal phosphorylation in the ERBB system is maintained at a low level despite the presence of preformed dimers.
Second, we examined the phosphorylation and dimerization responses of the ERBB system induced by EGF stimulation (see the left multiplier networks in Fig. 5 a and b). In Fig. 5 a, the increase of the autophosphorylation rate of ERBB1 by EGF was larger than the increase of the phosphorylation rates by ERBB2 or ERBB4. In Fig. 2, we observed enhanced phosphorylation levels of ERBB1 by EGF in both HeLa and A431. We can infer that the autophosphorylation of ERBB1 contributes to the enhanced phosphorylation level more than the phosphorylation rates in heterodimers. In Fig. 5 b, the dimerization rate of the ERBB1 homodimer was not increased by EGF stimulation. This observation suggests that a considerable number of preformed dimers contribute largely to the autophosphorylation of ERBB1 after the EGF signal.
Next, we investigated the responses induced by HRG stimulation (see the right multiplier networks in Fig. 5 a and b). In Fig. 2, we observed enhanced phosphorylation levels of ERBB4 by HRG in HeLa and MCF7. Fig. 5 a shows that the autophosphorylation rate of ERBB4 was increased compared with the phosphorylation rate by ERBB2 or ERBB4. In contrast, Fig. 5 b shows an increase in the dimerization rate of the ERBB4 homodimer by HRG. These results suggest that the autophosphorylation of ERBB4 contributes largely to the enhanced phosphorylation level in HeLa and MCF7 in a manner similar to the autophosphorylation of ERBB1 under EGF stimulation. In addition to the effect on autophosphorylation, the newly formed ERBB4 homodimers take another important role under HRG stimulation, which is in contrast with EGF stimulation, where ERBB1 preformed dimers take a dominant role. These predictions must be verified with future experiments, however.
Origin of the diversity of the phosphorylation responses
As we showed above, the ERBB phosphorylation responses strongly depend on the ERBB expression profiles. In addition to the different expression profiles of ERBBs, the diversity of the kinetic properties among the four subtypes is necessary for the diversity of the phosphorylation responses, because the same kinetic properties with different expression profiles cannot induce differences in phosphorylation behaviors. In the following, we analyze the diversity of the phosphorylation responses by distinguishing the diversity originated from the expression profiles, which we call dose-dependent diversity, and that from the reaction rates, which we call rate-dependent diversity.
Dose-dependent diversity
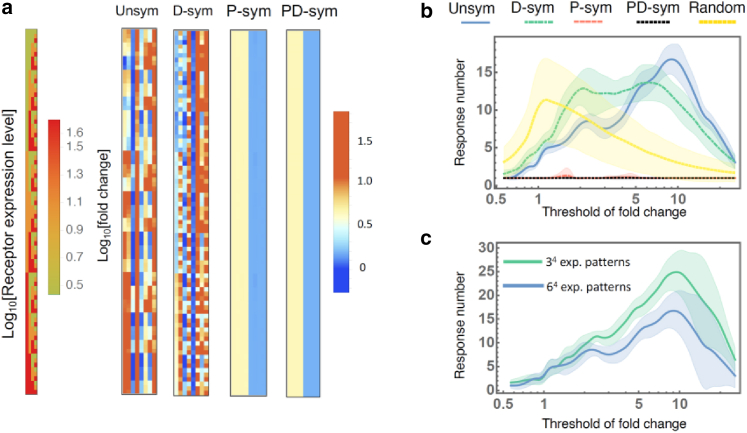
We examined the diversity of the phosphorylation responses by changing the ERBB expression patterns in the mathematical model. We prepared many expression profiles by setting three different expression levels (20, 100, 500) for each of the four subtypes, where all expression levels were normalized to ERBB3 in HeLa cells. (Discretizing the space of the expression levels into more than three levels does not change the results qualitatively.) For each of 81 hypothetical expression patterns (shown in the left-end column in Fig. 6 a), we calculated the phosphorylation levels before and after the ligand stimulation. The heatmap labeled "Unsym." (unsymmetrical) in Fig. 6 a shows the fold change in the phosphorylation levels of ERBB1-4 induced by EGF and HRG (columns) for each of the 81 ERBB expression patterns (rows). The pattern in the heatmap visualizes the diversity of the responses owing to differences in the expression profiles, namely, dose-dependent diversity.
Figure 6.
The diversity of phosphorylation patterns. (a) The receptor expression patterns (the leftmost column) and phosphorylation patterns obtained from the originally estimated parameters (unsymmetric kinetics; Unsym), symmetrized dimerization-associated parameters (D-sym), symmetrized phosphorylation-associated parameters (P-sym), and symmetrized phosphorylation- and dimerization-associated parameters (PD-sym). For the phosphorylation pattern, the averages of the fold changes over 100 sets of estimated parameters (generated by the MCMC method) are shown. (b) The response number against the threshold value used to discretize the fold change for Unsym, D-sym, P-sym, PD-sym, and random kinetics. (c) The number of response patterns for 34 expression levels, where each ERBB subtype expression level is taken from one of (20, 100, 500), and for 64 expression levels, where each ERBB subtype expression level is taken from one of (20, 43, 95, 210, 460, 1000). In (b) and (c), the solid curves represent the averages over 100 sets of estimated parameters, and the shaded regions indicate the standard deviations.
In the heatmap in Fig. 6 a, the responses of the four subtypes to two ligands (EGF and HRG) under 81 expression profiles are shown as continuous values. Each element of the 81 × 8 table is binarized depending on whether the element is higher or lower than a given threshold. The threshold of the phosphorylation level can be interpreted as the inability of the downstream molecules to detect phosphorylation (insensitivity) in the signal pathway. Then, each row (the response for each ERBB expression profile) is represented by an 8-digit binary number, e.g., 01011011, and we count the number of different eight-digit numbers within the 81 ERBB expression profiles. We call these numbers response numbers to quantify the diversity of the phosphorylation patterns caused by differences in the ERBB expressions. The response number is interpreted as the number of distinguishable phosphorylation patterns downstream of the ERBB system.
The response number depends on the threshold value used for the binarization. The curve labeled Unsym in Fig. 6 b shows the response number against the threshold value. To characterize the curve, we also calculated the response number using randomly sampled parameter values (Fig. 6 b). More specifically, we sampled random values from [0.00001,1] for reaction rates and from [1,100] for multipliers of the ligand stimulations. The response number curve for the randomized model had a peak around the threshold with a fold change equal to 1. Note that under a setting of a threshold close to 1 (high sensitivity), phosphorylation responses to the ligands and spontaneous fluctuations cannot be distinguished by the downstream pathway. In contrast, the response number curve obtained from the estimated parameter values (Fig. 6 b) has a single peak at the threshold of about 10. This result implies that the phosphorylation dynamics of ERBB in human cells realizes the highest diversity of responses at a threshold of fold change of 10, which seems to be sufficiently high to distinguish responses to ligands and spontaneous fluctuations.
The maximum value of the response number was less than 20, which is small compared to the eight-digit binary number (256 = 28) or the number of expression patterns . Note that even if we take a wider domain of the expression level space and discretize the domain using a greater number of points, the response number does not change drastically (Fig. 6 c). This result, together with Fig. 6 b, suggests that the upper bound of the diversity of the phosphorylation responses comes from the mathematical framework of the phosphorylation reaction (i.e., Eqs. (1) and (2)) rather than the choice of parameters.
Rate-dependent diversity
In this section, we discuss how differences in reaction rates (dimerization and phosphorylation) between different ERBB subtypes contribute to the diversity of the phosphorylation responses.
We first constructed symmetric models, where some reaction rate constants and the effects of the ligand stimulation were set to be equal among the different subtypes. More specifically, we considered three differently symmetrized models, D-symmetric, P-symmetric, and PD-symmetric, wherein particular subsets of reaction rate constants were set to be equal among the different subtypes (Table S1). In the D-symmetric model, the values of the reaction parameters related to dimerization (s and s, which are multipliers of the ligand stimulation) were set to be geometric averages of the four subtypes in the original unsymmetric model. Note that the symmetry of the multipliers for ligand binding implies uniformization of the ligand affinities between ERBBs. Similarly, in the P-symmetric model, the reaction rate constants related to phosphorylation (s and s) were set to be geometric averages of the original unsymmetric model. In the PD-symmetric model, four sets of parameters related to dimerization and phosphorylation (s, s, s and s) were set to be geometric averages. For each symmetric model, we calculated the phosphorylation dynamics and determined the steady states for all 81 expression profiles as done for the dose-dependent diversity analysis. We also calculated the response numbers for each symmetric model. From the decreases in the diversity in the symmetric models compared with the original unsymmetric model, we discuss the effects of the difference of the reaction rates to the diversity of the phosphorylation dynamics.
The heatmaps labeled D-sym, P-sym, and PD-sym in Fig. 6 a represent the diversities of the phosphorylation responses in each symmetric model. In the P-symmetric and PD-symmetric models, the color patterns in the heatmaps are much simpler than that in the unsymmetric and D-symmetric models.
In Fig. 6 b, the response numbers against the threshold in the symmetric models are plotted on the same chart with the unsymmetric model. The smaller response numbers in the symmetric models compared with the unsymmetric model show the effect of the variation in the reaction rate constants on the diversity of the phosphorylation responses (rate-dependent diversity). The response number of the D-symmetric model is comparable with that of the unsymmetric model. In contrast, in the P-symmetric and PD-symmetric models, the response numbers are significantly lower. This result implies that the diversity of the phosphorylation rate constants between the four ERBB subtypes makes a much larger contribution to the diversity of the phosphorylation responses than the dimerization parameters.
Discussion
Here we studied the phosphorylation responses of ERBB receptors, which initiate multiple signal transduction pathways, by observing multiple cells experimentally and mathematically. From our analysis, we found a diversity of phosphorylation responses depending on both the expression profiles and kinetic properties of the subtypes. Different expression profiles among organs in the human body have been reported (Fig. 1 c) (11, 12, 13), which may explain diverse organ-specific responses.
Using our mathematical analysis, we examined the origin of the diversity of the phosphorylation responses to intercellular signals by decomposing the responses into dose-dependent diversity and rate-dependent diversity responses. We first confirmed that differences in the ERBB expression profiles induce the diversity of the phosphorylation responses. We found that, under the parameter condition estimated from human cells, the maximum diversity of responses was realized when the sensitivity of the downstream molecules was sufficiently low (high in the threshold of the phosphorylation level) to distinguish spontaneous fluctuations from responses to ligands (Fig. 6 b). The maximum diversity of responses was smaller than the possible number of discrete phosphorylation levels (namely, 28). We expect that the number of different expression profiles of ERBB in human cells is also not large, since large differences in expression profiles do not always drive the large diversity of dynamic behaviors.
Second, we showed that different reaction rates between subtypes are necessary for the diversity of the phosphorylation responses. From the analysis, the difference in phosphorylation rate constants between ERBB subtypes was found to have a much larger contribution to the diversity of the phosphorylation responses than do the rate constants for dimerization. This finding implies that divergent evolution in phosphorylation reactions after gene duplications was essential to generating the diversity of phosphorylation responses among ERBB subtypes. The C-terminus phosphorylation domain, which is the substrate of the ERBB kinase domain, shows a much larger sequence difference than other domains. This difference may cause the difference in the phosphorylation rate constants among ERBB subtypes.
The estimated values of kinetic parameters of ERBB show the diversity in the phosphorylation and dimerization rates depends on the combination of subtypes in the pairs. The effects of ligand binding on the kinetic parameters were characterized in the four multiplier networks in Fig. 5. The differences of these networks between left (EGF) and right (HRG) are the origin of the different responses of the ERBB system to these ligands. From the multiplier networks, we predicted the reactions important for the observed changes in phosphorylation levels. For example, the increase in the phosphorylation level of ERBB1 induced by EGF stimulation (Fig. 2) is understood by the larger contribution of ERBB1 autophosphorylation than heterodimer phosphorylation. In addition, ERBB1 autophosphorylation is predicted to take place in ERBB1 preformed dimers rather than newly formed dimers under EGF stimulation. These predictions should be verified experimentally in the future.
We can compare the values of the kinetic parameters between ERBB subtypes and discuss their biological meaning. However, we should be careful when interpreting the absolute values of these parameters. It is difficult to determine all absolute values from experimental measurements. Actually, it is fundamentally impossible, because the values measured are relative to wild-type HeLa and therefore not absolute.
In this study, we used a simple mathematical model for the phosphorylation dynamics of ERBB and used it to analyze the steady states of the system. The aim of our analysis was to identify the origin of the diversity of the phosphorylation responses. To investigate transient behaviors or longer time dynamics, more biological factors need to be integrated. However, this integration would also make the analysis more difficult. The minimalistic approach used in the present study allowed us to characterize the effects of ligand binding and elucidate the origin of diversity of the phosphorylation responses with respect to expression levels and kinetic properties. Moreover, our approach gave results consistent with the diversity of the real system. The decomposition of dose-dependent diversity and rate-dependent diversity responses in the model are difficult to study experimentally. For the dose-dependent diversity analysis, we calculated the number of patterns in the phosphorylation responses under exhaustive changes in the ERBB expression profiles. As for the rate-dependent diversity analysis, we examined the phosphorylation responses under the symmetrization of subsets of reaction rate constants, where differences between ERBB subtypes created by evolutionary divergence are smoothed out. Again, such an analysis is not practical experimentally. Overall, by integrating experimental and theoretical methods, we obtained a deeper understanding of the behavior of the ERBB system based on extensive and evolutionary analyses.
Author contributions
T.O. conceived and carried out all of the mathematical analyses. H.M. carried out the experiments. Y.S. directed the experimental analysis and discussions. M.H. designed and carried out the experiments. A.M. organized the project. A.M., T.O., Y.S., and M.H. wrote the manuscript.
Acknowledgments
We thank H. Sato and A. Kanayama for experimental support. This research was supported by the CREST program (grant no. JPMJCR13W6, JPMJCR1922) of the Japan Science and Technology Agency (http://www.jst.go.jp/EN/index.html), the RIKEN iTHEMS Program, and the Joint Usage/Research Center Program of the Institute for Frontier Life and Medical Sciences, Kyoto University. A.M. was supported by MEXT Japan with Grants-in-Aid for Scientific Research (19H05670, 19H03196). Y.S. was supported by MEXT Japan with Grants-in-Aid for Scientific Research (19H05647) and J.S.T. with CREST (JPMJCR1912). M.H. was supported by MEXT Japan with Grants-in-Aid for Scientific Research(B) (18H01839) and Grant-in-Aid for Scientific Research on Innovative Areas (18H05414). All authors declare that there is no conflict of interest.
Editor: Pablo Iglesias.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.12.031.
Contributor Information
Michio Hiroshima, Email: m_hiroshima@riken.jp.
Atsushi Mochizuki, Email: mochi@infront.kyoto-u.ac.jp.
Supporting material
References
- 1.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oda K., Matsuoka Y., et al. Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005;1:2005.0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gschwind A., Fischer O.M., Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 4.Lemmon M.A., Schlessinger J., Ferguson K.M. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citri A., Yarden Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 6.Red Brewer M., Choi S.H., et al. Carpenter G. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarden Y., Sliwkowski M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 8.Scaltriti M., Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin. Cancer Res. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 9.Alroy I., Yarden Y. The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 10.Liu P., Sudhaharan T., et al. Wohland T. Investigation of the dimerization of proteins from the epidermal growth factor receptor family by single wavelength fluorescence cross-correlation spectroscopy. Biophys. J. 2007;93:684–698. doi: 10.1529/biophysj.106.102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samaras P., Schmidt T., et al. Wilhelm M. ProteomicsDB: a multi-omics and multi-organism resource for life science research. Nucleic Acids Res. 2020;48:D1153–D1163. doi: 10.1093/nar/gkz974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt T., Samaras P., et al. Wilhelm M. ProteomicsDB. Nucleic Acids Res. 2018;46:D1271–D1281. doi: 10.1093/nar/gkx1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelm M., Schlegl J., et al. Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 14.Stein R.A., Staros J.V. Insights into the evolution of the ErbB receptor family and their ligands from sequence analysis. BMC Evol. Biol. 2006;17:1–17. doi: 10.1186/1471-2148-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barberán S., Martín-durán J.M., Cebrià F. Evolution of the EGFR pathway in Metazoa and its diversification in the planarian Schmidtea mediterranea. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., He W., et al. Hu Y. Natural selection and functional diversification of the epidermal growth factor receptorEGFR family in vertebrates. Genomics. 2013;101:318–325. doi: 10.1016/j.ygeno.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Shi F., Telesco S.E., et al. Lemmon M.A. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. U S A. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olayioye M.A. New EMBO members’ review: the ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Lengerich B., Agnew C., et al. Jura N. EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc. Natl. Acad. Sci. U S A. 2017;114:E2836–E2845. doi: 10.1073/pnas.1617994114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagashima T., Shimodaira H., et al. Hatakeyama M. Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 2006;282:4045–4056. doi: 10.1074/jbc.M608653200. [DOI] [PubMed] [Google Scholar]
- 21.Nagashima T., Shimodaira H., et al. Hatakeyama M. Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 2007;282:4045–4056. doi: 10.1074/jbc.M608653200. [DOI] [PubMed] [Google Scholar]
- 22.Fabricant R.N., De Larco J.E., Todaro G.J. Nerve growth factor receptors on human melanoma cells in culture. Proc. Natl. Acad. Sci. U S A. 1977;74:565–569. doi: 10.1073/pnas.74.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong H., Kim J., et al. Kim A. Neuregulin-1 induces cancer stem cell characteristics in breast cancer cell lines. Oncol. Rep. 2014;32:1218–1224. doi: 10.3892/or.2014.3330. [DOI] [PubMed] [Google Scholar]
- 24.Shankaran H., Wiley H.S., Resat H. Modeling the effects of HER/ErbB1-3 coexpression on receptor dimerization and biological response. Biophys. J. 2006;90:3993–4009. doi: 10.1529/biophysj.105.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankaran H., Zhang Y., et al. Resat H. Quantifying the effects of co-expressing EGFR and HER2 on HER activation and trafficking. Biochem. Biophys. Res. Commun. 2008;371:220–224. doi: 10.1016/j.bbrc.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mac Gabhann F., Popel A.S. Dimerization of VEGF receptors and implications for signal transduction: a computational study. Biophys. Chem. 2007;128:125–139. doi: 10.1016/j.bpc.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kholodenko B.N., Demin O.V., et al. Hoek J.B. Quantification of short term signaling by the epidermal growth factor receptor. J. Biol. Chem. 1999;274:30169–30181. doi: 10.1074/jbc.274.42.30169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.