Abstract
Background
Viruses are the most abundant biological entities on the planet and drive biogeochemical cycling on a global scale. Our understanding of biogeography of soil viruses and their ecological functions lags significantly behind that of Bacteria and Fungi. Here, a viromic approach was used to investigate the distribution and ecological functions of viruses from 19 soils across China.
Results
Soil viral community were clustered more significantly by geographical location than type of soil (agricultural and natural). Three clusters of viral communities were identified from North, Southeast and Southwest regions; these clusters differentiated using taxonomic composition and were mainly driven by geographic location and climate factors. A total of 972 viral populations (vOTUs) were detected spanning 23 viral families from the 19 viromes. Phylogenetic analyses of the phoH gene showed a remarkable diversity and the distribution of viral phoH genes was more dependent on the environment. Notably, five proteins involved in phosphorus (P) metabolism-related nucleotide synthesis functions, including dUTPase, MazG, PhoH, Thymidylate synthase complementing protein (Thy1), and Ribonucleoside reductase (RNR), were mainly identified in agricultural soils.
Conclusions
The present work revealed that soil viral communities were distributed across China according to geographical location and climate factors. In addition, P metabolism genes encoded by these viruses probably drive the synthesis of nucleotides for their own genomes inside bacterial hosts, thereby affecting P cycling in the soil ecosystems.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-022-00401-9.
Keywords: Virus, Virome, Geographic location, PhoH, P metabolism, Nucleotide synthesis
Introduction
Viruses are the most abundant and diverse biological entities form and are major contributors to ecosystem functioning across all habitats [1]. Previous studies showed that viruses shape marine ecosystems by controlling the abundance and genomic diversity of their hosts through cell lysis [2–4] or lysogeny [5], and horizontal gene transfer [6–9]. Compared to around 1.01 × 1029 virus-like particles (VLPs) in marine environments, approximately 4.88 × 1030 VLPs were estimated to reside in global soils, accounting for 10% of the global viral abundance (4.80 × 1031) [1]. The potential roles of soil viruses in terrestrial ecosystem processes include impacting microbial mortality, biogeochemical cycling of soil elements, and food web dynamics [10]. Although soil viromes only contribute less than 1% of publicly available viral metagenomes [1], an increasing number of studies of viromes have focused on various soils, such as desert soil [11, 12], glacier soil [13], thawing permafrost soil [14], mangrove soil [9], mud volcanic soil [15], and Antarctic soil [16]. These studies revealed different patterns of soil viral community structure and largely uncharacterized viral assemblages. However, only a few studies have offered insight into how environmental factors influence viral communities. Soil pH was the main environmental driver of the viral community structure in agricultural soils [17]. Except soil pH, calcium content and site altitude were the main drivers of the Antarctic viral community structure [16].
In Chinese agricultural ecosystems, phosphorus (P) is an important biologically limiting nutrient that must be heavily supplemented for improving crop production [18]. Though lots of chemical P fertilizers have been applied to agricultural land, the P availability is still very low due to P slow diffusion and high fixation in soils [19]. Previous studies showed that P content in the marine ecosystem could affect the proportion of P allocated from hosts to viruses, as viruses have a higher proportion of P (C/N/P ≈ 20/6/1) [20] than Bacteria (69/16/1) [21, 22]. We considered the possibility that viruses in the soil ecosystem may also accelerate the uptake of soil P to synthesize their own genomes when P fertilizers were supplemented by the host cell. Thus, viral infection could cause the P present in the host bacteria to be disproportionately incorporated into the new phage particles, further resulting in P removal from soil biotic cycling and affecting plant and microbial P acquisition strategies [23]. However, it is not clear how viruses manipulate this process and whether this process is related to the P concentration or P fertilizer input in the soils.
Increasing evidence has shown that a certain number of putative auxiliary metabolic genes (AMGs) encoded by viruses are expressed during the infection cycle, and that AMG products reprogram host cell metabolism with direct impacts on biogeochemistry cycling [7, 24, 25]. In the genomes of globally abundant ocean viruses, more than two hundred viral-encoded AMGs have been identified [8], including carbon, nitrogen, sulfur, and P cycle related genes. Some viral AMGs, such as trzN [26], phoH [11], RNR [11], spoIIIE [27], carbon cycling related genes (CAZymes [9, 17], central C metabolism genes [14]), and oxidative phosphorylation related genes etc. [28] have been identified in soil ecosystems. Among them, the phoH gene encodes an ATP binding protein with undetermined function [29] and is presumed to belong to the Pho regulon and to regulate P uptake and metabolism under low-phosphate conditions [30]. It is known to be induced under phosphate stress in E. coli, while its expression is not upregulated during P starvation in marine cyanobacteria [31–33]. Despite the phoH gene is found widely distributed among both eubacteria and archaea [34, 35], our knowledge of their functions and potential mechanisms is still a mystery.
In this study, we aimed to investigate the distribution of viral communities and functions from 19 soil samples across China, and determine the main factors driving viral distribution and function. Furthermore, we explored whether the phoH gene and its homologs may play important roles in P cycling in soil ecosystems.
Materials and methods
Soil sampling and physicochemical properties
Between August 2015 and August 2016, a total of 19 soil samples were collected from ten provinces across China; these samples included ten agricultural soil samples and nine natural soil samples (Additional file 1: Fig. S1 and Additional file 2: Table S1). The agricultural soil samples, from five maize fields and five paddy fields, were located in seven provinces. The natural soil samples were also located in seven provinces and included forest, grassland, wetland, coastal, glacier, and mud volcanic soils (Additional file 2: Table S1). To study viral diversity and function in these soils, approximately 5 kg of each sample was collected and transported at 4°C back to the laboratory. At each site, a soil sample was collected from each of three separated 10 m × 10 m plots by pooling five upper 20-cm soil cores randomly taken from every plot. The three samples from each site were pooled and then processed as follows: 1 kg of soil was sieved to 1 mm for virus extraction, and 500 g of each soil sample was sieved to 2 mm and then stored at 4°C for physicochemical analyses.
A pH meter (Professional Meter PP-20, Sartorius, Germany) was used to measure soil pH and electrical conductivity (EC) at a ratio of 1:2.5 and 1:5 (soil to water, w/w), respectively. Organic matter (OM) was determined using the K2Cr2O7 oxidation method. Total nitrogen (TN) was measured using a Vario EL III analyzer (Elementar Analysensysteme GmbH, Hanau, Germany). Available P was determined using the Olsen method [36]. Available potassium (AK) was extracted with 0.5 M ammonium acetate and quantified using an atomic absorption spectrophotometer (ZEEnit700P, Analytik Jena AG, Jena, Germany). Mean annual temperature (MAT) and mean annual precipitation (MAP) data were from WorldClim Version2.
Virus extraction and purification
Viruses were extracted from the soil samples according to the method of Williamson et al. [37]. Briefly, 500 g of soil per sample was suspended in 1.5 L of glycine buffer (250 mM; pH = 8.5), shaken for 30 min, and centrifuged at 4000 g for 10 min at 4°C to precipitate soil particles. The supernatant was filtered sequentially through 1-mm, 0.45-µm, 0.20-µm tangential flow filters (GE Healthcare Life Sciences, Pittsburgh, PA, USA), and concentrated the filter liquid to less than 100 ml by 30-kDa tangential flow filters (GE Healthcare Life Sciences, Pittsburgh, PA, USA). The viruses in the filtrate were further concentrated using 30-kDa centrifugal ultrafiltration tubes (Merck Millipore Ltd., Tullagreen, Ireland) until the final sample volume was less than 1 ml. Finally, viral concentrates were treated with DNaseI (10 units DNaseI/100 μl) and incubated at 37°C for 1 h to remove free, non-encapsulated DNA. The presence of free and contaminating bacterial DNA was checked by PCR amplification of the 16S rRNA gene with primers 27F/1492R [38].
Viral DNA extraction and high-throughput sequencing
The Power Viral Environmental RNA/DNA Isolation kit (MO BIO Laboratories, Carlsbad, CA, USA) was used to extract total DNA. The REPLI-g Mini Kit (for multiple displacement amplification (MDA)) (Qiagen, Hilden, Germany) using Phi29 polymerase was applied to transfer ssDNA to dsDNA and obtain the concentration and quantity needed for high-throughput sequencing. For each sample, more than 1 ng of DNA was fragmented to approximately 400 bp and used as a template to create a metagenome library, which was constructed according to the TruSeq™ DNA Sample Prep Kit (Illumina, San Diego, CA, USA) protocol. The libraries were loaded onto flow cell channels for sequencing using an Illumina HiSeq2500 at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) to generate 300-bp paired-end reads.
Analysis of viromes
Data sets and assembly
The original raw reads of the 19 samples obtained from the Illumina HiSeq2500 were cleaned using Fastp software [39] for quality filtering and subsample the raw data. Firstly, adapter bases or poly [ATCG] bases (minlength = 10) in the 5′ or 3′ reads were removed. Secondly, those reads were deleted that meet any one of the following conditions: the number of N bases in the sequence exceeds 5 bp, the average sequence quality value QV < 20, sequence length < 18 bp, or sequence complexity < 30%. After quality control, each sample was independently assembled using metaSpades with default parameters [40], and contigs shorter than 10 kb were eliminated according to Minimum Information about an Uncultivated Virus Genome (MIUViG) [41]. A combination of VirSorter [42], VIBRANT [43] and DeepVirFinder [44] were used to detect viral contigs from each assembly. Based on the Discovery Environment 2.0 (https://de.cyverse.org), Virsorter was run in decontamination mode, and only categories 1, 2, 4 and 5 (higher confidence predictions) were retained, and combined phages in VIBRANT were considered viral. DeepVirFinder run according to its python script (https://github.com/jessieren/DeepVirFinder), and contigs with scores > 0.9 and p < 0.05 were considered viral [45]. All resulting viral contigs were combined and clustered at 98% identity with cd-hit-est software [46], resulting in 972 non-redundant genome fragments to create a viral Operational Taxonomic Units (vOTUs) database. Frap [1] was used to map quality-filtered reads from each sample to the vOTU database at 90% identity, with the genome size normalization option, to obtain the normalized vOTU table. Normalization was done by dividing the number of reads aligned database by the number of reads in the virome, then multiplying this by the mean genome length divided by the length of each viral contig. The number of viral reads was calculated by reads aligning to these vOTUs.
Viral taxonomy clusters and potential impact factors
An unsupervised random forest analysis was used to cluster the samples based on the normalized vOTU table and identify which environmental and/or geographical factors influenced viral community composition using the "randomForest" and "rfPermute" packages on the R platform [47]. Non-metric multi-dimensional scaling (NMDS) was used to analyze the random forest proximity matrix, to cluster the samples based on Ward distances, and to identify the subset of variables of importance for the random forest clustering. The effect of environmental factors and geographical coordinates on this dataset was tested using a supervised random forest permutational-based variable importance measures to identify the significant predictors of viral community composition.
Taxonomy annotation and comparison
Clean reads were classified using Kraken2 against the NCBI viral reference sequences (minikraken2_v1_8GB_201904) to identify viral reads [48]. The abundance of viral reads was computed by Bracken [49], which uses the taxonomy labels assigned by Kraken2 to estimate the number of reads present in each sample. UpSet analysis was further performed to visualize the interactive viral families among clusters by the "UpSetR" on the R platform [50].
Putative viral AMGs detection
DRAM-v [51] was used to identify putative AMGs in 972 vOTUs, and only results with viral_bitScore > 60 and viral_E-value < 1E−5 were retained. The contigs containing genes related to the P metabolism module were selected from the DRAM-v output, including genes encoding the predicted proteins dUTPase [52], MazG, PhoH, RNR, and Thy1 [53, 54]. Genomic maps of contigs encoding the phoH gene were generated with Easyfig [55].
Phylogenetic analysis of the phoH gene
Phylogenetic trees of the phoH gene amino acid sequences were reconstructed using MEGA-X software [56]. A total of 102 representative phoH gene amino acid sequences from viruses were collected (Additional file 2: Table S2), including 25 reference sequences from cultured phages, 15 from paddy water [57], 8 from sea water [7, 30], 9 form permafrost soil samples [58], 12 from other soil metagenomes [59], 25 reference sequences of soil viromes obtained from our previous work [17], and 8 sequences in this study. All selected amino acid sequences were aligned by ClustalW, and the gaps and ambiguously aligned positions were deleted. After alignment, a phylogenetic tree was constructed using the Jones–Taylor–Thornton (JTT) model and the maximum likelihood method, and support for tree structure was obtained using 1000 bootstraps. The output was visualized by Evolview v2 [60].
Data availability
Virome read data are available in the NCBI Short Read Archive (SRA) under BioProject ID PRJNA579576.
Results
Viral community structure
Soil samples from 10 provinces across China were used to generate 19 soil viromes, including 10 agricultural soil viromes and 9 natural soil viromes (Additional file 1: Fig. S1). A total of 186,503,518 reads (range: 4,782,132 to 15,945,343 per sample) passed quality control. Among them, 433,669 reads were identified as viral by Kraken 2, and most were still unclassified (Additional file 2: Table S3). A total of 972 de-replicated viral contigs (> 10 kb) were assembled and reserved for further analysis according to MIUViG [41]; the longest contig was 98,359 bp and the average contig length was 19,760 bp.
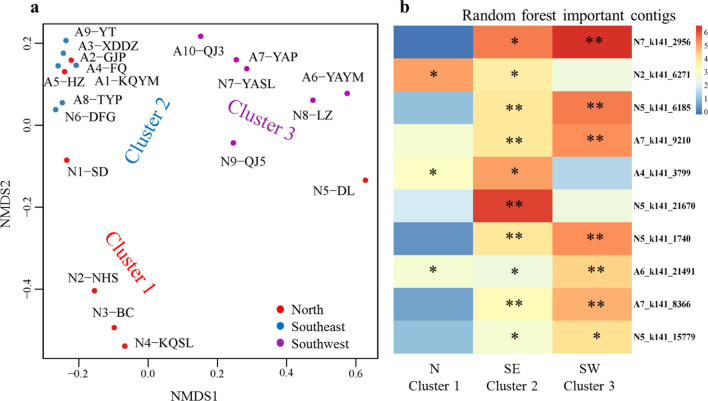
To identify similarities between soils, soil viromes were clustered using an unsupervised random forest analysis based on vOTU table (Additional file 2: Table S4). Three clusters of samples were identified (21.05% OOB (out-of-bag) estimate of error rate), and were related to the geographical distribution of the soil samples (Fig. 1a and Additional file 1: Fig. S1). Cluster 1 included four of seven North China samples, Cluster 2 included all six Southeast China samples, and Cluster 3 included all six samples from the Southwest of China (Fig. 1a and Additional file 1: Fig. S1). The top ten contigs with highest importance in differentiating clusters are shown in Fig. 1b and Additional file 1: Fig. S2.
Fig. 1.
Viral community structure. a Non-metric multidimensional scaling (NMDS) of viral community composition in 19 soil viromes obtained from an unsupervised random forest analysis followed by clustering using Ward distances (OOB estimate of error rate = 21.05%). Symbols are color-coded by site (red: North sites; blue: Southeast sites; purple: Southwest sites). b The heat map of contigs differentiating the geographical clusters in a random forest analysis supervised by geographical location and that there is a color gradient which represents the importance. P < 0.01, **; P < 0.05, *
To analyze and compare the viral community composition with respect to environmental factors, soil physical and chemical properties including pH, EC, OM, TN, AP, and AK, climate factors (MAT, Mean annual temperature, and MAP, mean annual precipitation (Additional file 2: Table S1)), and geographical coordinates were tested as potential predictors of viral frequencies in the vOTU table. The results indicated that only MAP, MAT, longitude, and latitude explained 12.78%, 8.2%, 21.18% and 22.08% of the variation in viral community composition, respectively. Soil physical and chemical properties didn’t show any relationship with viral community composition.
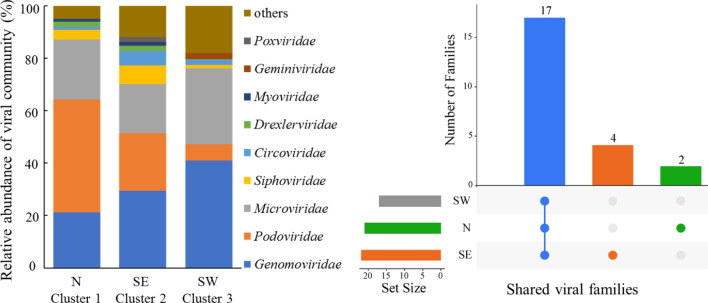
A total of 23 viral families (Fig. 2a, b and Additional file 2: Table S5) were identified from the 19 viromes by Kraken 2, including 15 families belonging to dsDNA viruses and eight families of ssDNA viruses. For ssDNA viruses, the Microviridae, Genomoviridae and Circoviridae families were widespread in all clusters. For dsDNA viruses, the Caudovirales (tailed viruses that infect Bacteria and Archaea) including Myoviridae, Siphoviridae, and Podoviridae were widespread in all clusters. Meanwhile, few numbers of giant viruses (Mimiviridae and Pandoraviridae) were distributed in all three clusters. In addition to these shared viruses, there were some specific viruses in Cluster 1 (North) and Cluster 2 (Southeast) (Fig. 2b). Such as, Anelloviridae and Hepadnaviridae were mainly present in Cluster 1 (North), and Bacilladnaviridae, Demerecviridae, Inoviridae, and Marseilleviridae only existed in Cluster 2 (Southeast) (Fig. 2b).
Fig. 2.
Viral taxonomy. a Relative abundances of the viral families among three NMDS clusters (top 10 are shown), and MDA was used in this study. b Shared viral families among three clusters displayed by UpSet analysis
P metabolism module
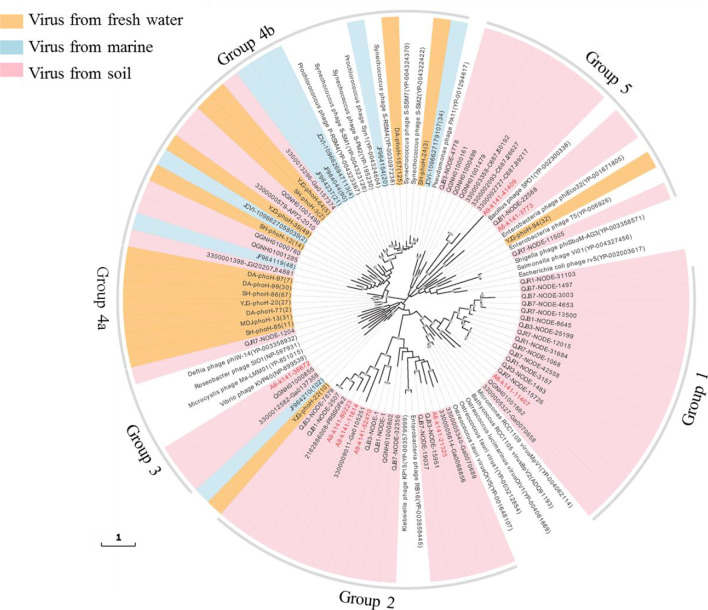
A phylogenetic tree of the phoH gene was built with 102 viral amino acid sequences from this study and others (Fig. 3). 15 representatives were collected from fresh water [57], eight representatives from sea water [7, 30], 25 reference sequences from cultured phages, and 54 phoH amino acid sequences from soil metagenomes [17, 58, 59] including eight representatives from agricultural maize fields in this study. All of the eight phoH amino acid sequences obtained from viromes of agricultural soils. Overall, the phylogenetic tree could be mainly divided into five groups. Group 1, 2, and 3 mainly contained viruses from soil samples, while group 4 and 5 contained viruses from different environments. Six phoH gene sequences in this study were grouped into Group 1, 2, and 3 with other global soil samples, and two were clustered into Group 5 with fresh water and other soil samples.
Fig. 3.
Phylogenetic tree of phoH gene sequences. Only representative sequences are displayed in the tree, and the number represents the number of original sequences. Sequences from fresh water, marine and soil are colored in lightyellow, blue and pink respectively. The red font in the figure represents the data from this study. The tree was bootstrapped with 1000 sub-replicates, and bootstrap scores > 50% are flagged with circles
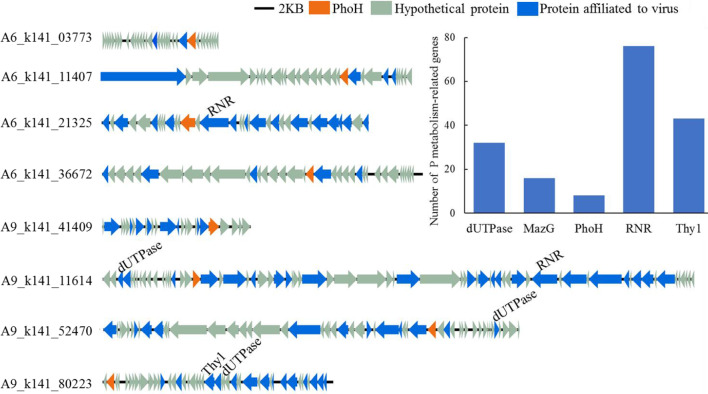
Putative AMGs were identified using DRAM-v [51]. Genes functionally related to phoH were further analyzed, which include five P metabolism-related nucleotide synthesis functions involving dUTPase, MazG, PhoH, Thy1, and RNR. A total of 175 viral ORFs belonging to the five P metabolism proteins were identified (Fig. 4), and they were mainly from agricultural soils (158 of 175 ORFs). Eight representative contigs (> 10 kb, all from maize fields) containing the phoH genes belonged to dsDNA viruses, and some accompanied genes encoding dUTPase, Thy1, or RNR (Fig. 4). In addition to these P metabolism proteins, these contigs encoded mostly hypothetical proteins.
Fig. 4.
Eight dsDNA viral contigs carrying AMGs predicted to be involved in phosphate metabolism. Arrows indicate ORFs and arrow color indicates predicted function. A histogram showing the number of genes in the Phosphate metabolism module, and the ordinate represents the number of viral ORFs
Discussion
Viruses play a vital role in the distribution of organisms and their contributions to global biogeochemical cycles [23]. However, our understanding of soil viruses, and the factors driving their distribution, lags far behind that of marine viruses. Further, the ways in which viruses participate in the biogeochemical cycling of soil elements have not been extensively investigated. This study provides evidence that geographic location and climate factors are key drivers of viral distribution in soils. Furthermore, the higher abundance of viral-encoded P metabolism genes in agricultural soils indicates that viruses have the potential roles of P cycling in these soil ecosystems.
The taxonomic distribution of soil viruses
The order Caudovirales, including Siphoviridae, Myoviridae and Podoviridae, was dominant in all of our soil samples, in agreement with previous studies [9, 11, 12, 16, 61]. In the Antarctic soil, Podoviridae presented at similar levels in all samples, whereas the abundances of Myoviridae and Siphoviridae were inversely correlated, as they may have direct competition for hosts in the same niche, and Siphoviruses are always present at higher abundances in neutral to alkaline pH soils [16]. However, our study showed different trends, with Myoviridae occurring in all samples with low abundance, whereas Siphoviridae and Podoviridae were mainly present in more acidic soils (Additional file 2: Table S5). More data is needed to find patterns, especially since so many viruses in the viromic data could not be classified and we used MDA.
Moreover, our soil viromes revealed diverse ssDNA viruses belonging to the Microviridae, Circoviridae, and Genomoviridae, (Fig. 2a). The broad presence of ssDNA viruses is likely due to the bias of MDA, which preferentially amplifies genomes of ssDNA viruses and thus leads to a quantitative bias [62–64]. Therefore, both ssDNA and dsDNA viruses were reported in a qualitative rather than quantitative way in this study. Meanwhile, the use of MDA leads to many short sequences. In this study, contigs less than 10 kb were ignored to avoid a misunderstanding of the soil virome. However, discovery of unknown function or partial viral genomes is still an important work.
Geographic location drives viral community composition and function
Viral community composition has been associated with a variety of environmental factors, such as host community composition, pH, soil depth and moisture, calcium content and site altitude [14, 16, 23, 58, 65]. According to the unsupervised random forests analysis, the viral communities and functions from 19 soil samples across China grouped into 3 clusters, which corresponded to geographical location well (Fig. 1a and Additional file 1: Fig. S1). A subsequent supervised random forest analysis showed that the main environmental driver of these clusters for viral community composition was MAP, MAT, longitude, and latitude. There have been few reports regarding location and climate factors and their effects on the distribution of viruses. Such as the altitude of Antarctic soils which probably linked to temperature could influence microbial metabolism and substantially impact viral communities and functions [16]. The temperature change along the latitude in this study may have similar effects, especially on viral community. All of the viruses differentiating these clusters were unclassified viruses. This highlighted the lack of knowledge and reference sequences for soil viruses.
Although phosphorus is an important factor of viral genome synthesis, the results do not imply any relationship between soil available P content and viral communities and functions. It is possible that our sampling time may be at different stages of phosphorus metabolism because of different fertilization time in each agricultural region. On the other hand, soil available P content may affect viral abundance more than viral community composition, and we will further focus on this point in the future.
Viruses may directly manipulate P cycling in soils
The phoH gene has been widely used as a signature gene for assessing viral phylogeny and diversity, and is encoded by various morphologically distinct viruses that infect a wide range of hosts, including autotrophic and heterotrophic Bacteria and Eukaryotes [30, 57]. A diversity of phoH genes have been found in viral communities inhabiting numerous environments, such as seawater [30], paddy water [57], and a Namib hypolith [11]. In these studies, phoH genes were distributed according to depth and location [30], biogeography [57], or were found to be entirely novel [11]. In this study, phylogenetic analyses showed that phoH sequences in groups 1, 2, and 3 (Fig. 3) were widely distributed in soils [57] from different sites of the world [17, 58, 59]. Group 4 and 5 contained viruses from different environments, including fresh water, sea water, and soil. The majority of the Namib hypolith phoH amino acid sequences clustered separately from other sequences and was omitted from our phylogenetic tree. These results support the inference that the distribution of viral phoH genes is more dependent on characteristics of the environment [66].
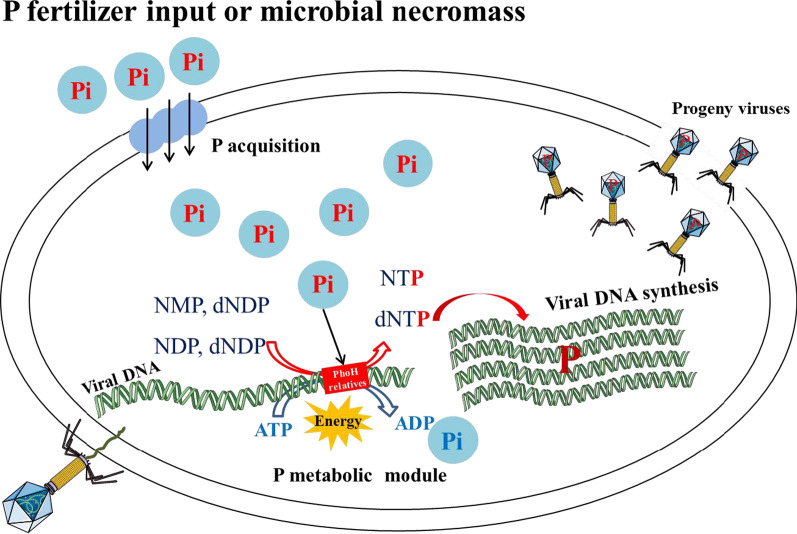
During the second Chinese soil survey [67], a database created from 2473 soil profiles was analyzed and showed relatively consistent C:P (136) and N:P (9.3) ratios, with a highly constrained C:N:P ratio of 134:9:1 for the surface soils from both of agricultural and natural soils [68]. This ratio indicates that the P content in Chinese soils is generally lower than that required by phages, which have a C:N:P ratio of 20:6:1 [20]. Due to P slow diffusion and high fixation in soils, plus the crops on the absorption of P for agricultural production [19], this means that P can be a major limiting factor for soil microbes, especially viruses. Based on this background, this P deficient environment may select for these viruses to regulate P uptake and metabolism through evolution of the phoH gene. It is interesting that all eight phoH gene sequences identified in this study were from viruses in agricultural soils. It is possible that agricultural soil is a rich environment in terms of dissolved organic matter, produced via photosynthesis, and nitrogen applied as fertilizer, but that these excesses of C and N result in P being limited. Once P fertilizer input, virus may prompt its host to quickly absorb inorganic P (Pi) and use PhoH to promote its own reproduction (Fig. 5).
Fig. 5.
Conceptual model of the P metabolism module of viruses
To better understand the metabolic potential of phoH genes, we searched for, but did not find, additional genes in the Pho regulon. However, it is interesting that four auxiliary metabolic potentials related to nucleotide synthesis, including dUTPase, MazG, Thy1, and RNR, were identified in association with phoH to act as a P metabolism module. Previous studies have demonstrated the presence of at least five proteins involved in P metabolism including PhoH, RNR, Thy1, endodeoxyribonuclease, and MazG pyrophosphatase in marine phage genomes [53, 54]. Similar modules were also found in two complete viral genomes from two agricultural soils in our previous data [17], including dUTPase, PhoH, RNR, and Thy1 (Additional file 1: Fig. S3). Here, five of the P metabolism genes were identified, especially in agricultural soils (Fig. 4). Among them, MazG is reported as a nucleoside triphosphate pyrophosphohydrolase, which can hydrolyze all eight of the canonical ribo- and deoxynucleoside triphosphates to their respective monophosphates and PP(i), with a preference for deoxynucleotides [69]. RNR, known as ribonucleoside diphosphate reductase, converts all four ribonucleotide diphosphates (rNDPs) to the respective deoxynucleoside diphosphates (dNDPs), which are then rapidly converted to dNTP [53, 70]. The dUTPase can catalyze dUTP to dUMP and release diphosphate, and provide a substrate (dUMP) for thymidylate synthase [52]. Thy1 can convert dUMP to dTMP depending on FAD, NADPH and 5,10-methylenetetrahydrofolate [71]. PhoH has been reported as a cytoplasmic protein with an ATP-binding activity and is predicted to be induced by P starvation [29]; however, its function remains unknown. Altogether, this information led us to hypothesize that PhoH can act as a nucleotide synthase, possibly binding and hydrolyzing ATP through its conserved nucleoside triphosphate hydrolase domain to obtain energy, and taking advantage of Pi from the agricultural soil (through the host cell) to catalyze the synthesis of nucleotides for the virus's own genome (conceptual model in Fig. 5). This model predicts the proliferation of a huge number of soil viruses playing an important role in depleting P from the soil ecosystem. Future work should focus on whether the concentration of Pi in soil is associated with the number of progeny produced by viruses, and also quantify the contribution of viruses to P loss from soil.
Conclusions
In summary, our analyses mainly explored viral community structure and function in soils across China. The results revealed that the distribution of viral communities was at least partly determined by geographical location and climate factors. Remarkably, AMGs related to P metabolism, including PhoH, RNR, Thy1, dUTPase and MazG, were mainly identified in viral genomes from agricultural soils, which suggested that viruses possibly take advantage of the Pi added to agricultural soils to synthesize their own genomes. As a consequence, these soil viruses have the potential to significantly contribute to P cycling in the soil ecosystem. Future investigations of the relationship between soil Pi content and viral ecology will reveal the specific mechanism of viral genome synthesis using soil-derived P and resulting depletion of soil P and provide more detailed insights into the contributions of viruses to the P cycle in soil ecosystems.
Supplementary Information
Additional file 1. Fig. S1. The distribution of the soil sampling sites. Fig. S2. Variable importance plot of contigs from random forest classification analysis based on geographical distribution. Fig. S3. Two complete viral genomes contain a P metabolism module.
Additional file 2. Table S1. Soil sampling information and physicochemical properties. Table S2. The amino acid sequences of phoH genes f.rom different environmental samples. Table S3. Overview of 19 virome data. Table S4. The vOTU table. Table S5. The abundance of viral reads identified by Kraken 2 (Family level).
Acknowledgements
We thank Dr. Taylor O'Connell, Mark Little, Nate Robinett, and Adam Barno at San Diego State University for their generous help with data analyses.
Abbreviations
- P
Phosphorus
- VLPs
Virus-like particles
- AMGs
Auxiliary metabolic genes
- vOTUs
Viral Operational Taxonomic Units
- NMDS
Non-metric multi-dimensional scaling
- NCBI
National Center for Biotechnology Information
- ORFs
Open reading frames
- RNR
Ribonucleoside reductase
- Thy1
Thymidylate synthase complementing protein
- JTT
Jones–Taylor–Thornton
- SRA
Short Read Archive
- MDA
Multiple displacement amplification
- Pi
Inorganic phosphorus
- N
North
- SE
Southeast
- SW
Southwest
- EC
Soil electrical conductivity
- OM
Soil organic matter
- TN
Soil total N content
- AP
Available phosphorus
- AK
Available potassium
- MAP
Mean annual precipitation
- MAT
Mean annual temperature
Authors' contributions
LLH, LMZ and JZH designed the research. LLH, DTY, LB and SD sampled the soils and conducted the laboratory analyses and the raw data collection. LLH, JZH, CS, AGC and FR performed the data processes and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Science Foundation of China (Grant Number: 41771289 and 41571248) and the China Scholarship Council.
Availability of data and materials
Virome read data are available in the NCBI Short Read Archive (SRA) under BioProject ID PRJNA579576.
Declarations
Ethics approval and consent to participate
Not applicable as there were no human, animal or pathogen subjects involved.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Li-Li Han, Email: llhan@rcees.ac.cn.
Dan-Ting Yu, Email: dtyu@fjnu.edu.cn.
References
- 1.Cobián Güemes AG, Youle M, Cantú VA, Felts B, Nulton J, Rohwer F. Viruses as winners in the game of life. Annu Rev Virol. 2016;3:197–214. doi: 10.1146/annurev-virology-100114-054952. [DOI] [PubMed] [Google Scholar]
- 2.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45(6):1320–1328. [Google Scholar]
- 3.Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4(6):739. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- 4.Thingstad TF, Våge S, Storesund JE, Sandaa R-A, Giske J. A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc Natl Acad Sci. 2014;111(21):7813–7818. doi: 10.1073/pnas.1400909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowles B, Silveira C, Bailey B, Barott K, Cantu V, Cobián-Güemes A, et al. Lytic to temperate switching of viral communities. Nature. 2016;531(7595):466. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 6.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459(7244):207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 7.Sharon I, Battchikova N, Aro E-M, Giglione C, Meinnel T, Glaser F, et al. Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 2011;5(7):1178. doi: 10.1038/ismej.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537(7622):689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 9.Jin M, Guo X, Zhang R, Qu W, Gao B, Zeng R. Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome. 2019;7(1):58. doi: 10.1186/s40168-019-0675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerson JB. Soil viruses: a new hope. MSystems. 2019;4(3):e00120–e219. doi: 10.1128/mSystems.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adriaenssens EM, Van Zyl L, De Maayer P, Rubagotti E, Rybicki E, Tuffin M, et al. Metagenomic analysis of the viral community in N amib D esert hypoliths. Environ Microbiol. 2015;17(2):480–495. doi: 10.1111/1462-2920.12528. [DOI] [PubMed] [Google Scholar]
- 12.Scola V, Ramond J-B, Frossard A, Zablocki O, Adriaenssens EM, Johnson RM, et al. Namib desert soil microbial community diversity, assembly, and function along a natural xeric gradient. Microb Ecol. 2018;75(1):193–203. doi: 10.1007/s00248-017-1009-8. [DOI] [PubMed] [Google Scholar]
- 13.Han L-L, Yu D-T, Zhang L-M, Wang J-T, He J-Z. Unique community structure of viruses in a glacier soil of the Tianshan Mountains, China. J Soils Sediments. 2017;17:852–860. [Google Scholar]
- 14.Trubl G, Jang HB, Roux S, Emerson JB, Solonenko N, Vik DR, et al. Soil viruses are underexplored players in ecosystem carbon processing. MSystems. 2018;3(5):e00076–e118. doi: 10.1128/mSystems.00076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu D-T, He J-Z, Zhang L-M, Han L-L. Viral metagenomics analysis and eight novel viral genomes identified from the Dushanzi mud volcanic soil in Xinjiang, China. J Soils Sediments. 2019;19(1):81–90. [Google Scholar]
- 16.Adriaenssens EM, Kramer R, Van Goethem MW, Makhalanyane TP, Hogg I, Cowan DA. Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome. 2017;5(1):83. doi: 10.1186/s40168-017-0301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi L, Yu DT, Du S, Zhang LM, Zhang LY, Wu CF, et al. Diversity and potential biogeochemical impacts of viruses in bulk and rhizosphere soils. Environ Microbiol. 2021;23(2):588–599. doi: 10.1111/1462-2920.15010. [DOI] [PubMed] [Google Scholar]
- 18.Kirkby EA, Johnston AEJ. Soil and fertilizer phosphorus in relation to crop nutrition. In: White PJ, Hammond JP, editors. The ecophysiology of plant–phosphorus interactions. Berlin: Springer; 2008. pp. 177–223. [Google Scholar]
- 19.Qiu J. Phosphate fertilizer warning for China. Nature News. 2010 doi: 10.1038/news.2010.498. [DOI] [Google Scholar]
- 20.Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz J. The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat Rev Microbiol. 2014;12(7):519. doi: 10.1038/nrmicro3289. [DOI] [PubMed] [Google Scholar]
- 21.Suttle CA. Marine viruses-major players in the global ecosystem. Nat Rev Microbiol. 2007;5(10):801. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 22.Sterner RW, Elser JJ. Ecological stoichiometry: the biology of elements from molecules to the biosphere. Princeton: Princeton University Press; 2002. [Google Scholar]
- 23.Kuzyakov Y, Mason-Jones K. Nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol Biochem. 2018;127:305–317. [Google Scholar]
- 24.Breitbart M, Thompson LR, Suttle CA, Sullivan MB. Exploring the vast diversity of marine viruses. Oceanography. 2007;20(2):135–139. [Google Scholar]
- 25.Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci U S. 2011;108(39):E757–E764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh D, Roy K, Williamson KE, White DC, Wommack KE, Sublette KL, et al. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl Environ Microbiol. 2008;74(2):495–502. doi: 10.1128/AEM.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Goethem MW, Swenson TL, Trubl G, Roux S, Northen TR. Characteristics of wetting-induced bacteriophage blooms in biological soil crust. MBio. 2019;10(6):e02287–e2319. doi: 10.1128/mBio.02287-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang X, Wagner RE, Zhuang J, DeBruyn JM, Wilhelm SW, Liu F, et al. Viral abundance and diversity vary with depth in a southeastern United States agricultural ultisol. Soil Biol Biochem. 2019;137:107546. [Google Scholar]
- 29.Kim S, Makino K, Amemura M, Shinagawa H, Nakata A. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J Bacteriol. 1993;175(5):1316–1324. doi: 10.1128/jb.175.5.1316-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldsmith DB, Crosti G, Dwivedi B, McDaniel LD, Varsani A, Suttle CA, et al. Development of phoH as a novel signature gene for assessing marine phage diversity. Appl Environ Microbiol. 2011;77(21):7730–7739. doi: 10.1128/AEM.05531-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Q, Chisholm SW. Marine viruses exploit their host's two-component regulatory system in response to resource limitation. Curr Biol. 2012;22(2):124–128. doi: 10.1016/j.cub.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 32.Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci. 2006;103(33):12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tetu SG, Brahamsha B, Johnson DA, Tai V, Phillippy K, Palenik B, et al. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J. 2009;3(7):835–849. doi: 10.1038/ismej.2009.31. [DOI] [PubMed] [Google Scholar]
- 34.Kazakov AE, Vassieva O, Gelfand MS, Osterman A, Overbeek R. Bioinformatics classification and functional analysis of PhoH homologs. In Silico Biol. 2003;3(1–2):3–15. [PubMed] [Google Scholar]
- 35.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3(5):e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen SR. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA Circ. 1954;939:1–19. [Google Scholar]
- 37.Williamson KE, Radosevich M, Wommack KE. Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol. 2005;71(6):3119–3125. doi: 10.1128/AEM.71.6.3119-3125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci U S. 1992;89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, et al. Minimum information about an uncultivated virus genome (MIUViG) Nat Biotechnol. 2019;37(1):29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kieft K, Zhou Z, Anantharaman K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8(1):1–23. doi: 10.1186/s40168-020-00867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren J, Song K, Deng C, Ahlgren NA, Fuhrman JA, Li Y, et al. Identifying viruses from metagenomic data using deep learning. Quantit Biol. 2020;8:1–14. doi: 10.1007/s40484-019-0187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ter Horst AM, Santos-Medellín C, Sorensen JW, Zinke LA, Wilson RM, Johnston ER, et al. Minnesota peat viromes reveal terrestrial and aquatic niche partitioning for local and global viral populations. bioRxiv. 2020. 10.1101/2020.12.15.422944.
- 46.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 47.Cutler A, Cutler DR, Stevens JR. Random forests. In: Zhang C, Ma Y, editors. Ensemble machine learning. Boston: Springer; 2012. pp. 157–175. [Google Scholar]
- 48.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):1–13. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. [Google Scholar]
- 50.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48(16):8883–8900. doi: 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J Gen Virol. 2002;83(10):2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- 53.Rohwer F, Segall A, Steward G, Seguritan V, Breitbart M, Wolven F, et al. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol Oceanogr. 2000;45(2):408–418. [Google Scholar]
- 54.Angly F, Youle M, Nosrat B, Srinagesh S, Rodriguez-Brito B, McNairnie P, et al. Genomic analysis of multiple Roseophage SIO1 strains. Environ Microbiol. 2009;11(11):2863–2873. doi: 10.1111/j.1462-2920.2009.02021.x. [DOI] [PubMed] [Google Scholar]
- 55.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Liu J, Yu Z, Jin J, Liu X, Wang G. Novel groups and unique distribution of phage phoH genes in paddy waters in northeast China. Sci Rep. 2016;6:38428. doi: 10.1038/srep38428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emerson JB, Roux S, Brum JR, Bolduc B, Woodcroft BJ, Jang HB, et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat Microbiol. 2018;3(8):870–880. doi: 10.1038/s41564-018-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graham EB, Paez-Espino D, Brislawn C, Neches RY, Hofmockel KS, Wu R, et al. Untapped viral diversity in global soil metagenomes. BioRxiv. 2019:583997.
- 60.He Z, Zhang H, Gao S, Lercher MJ, Chen W-H, Hu S. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44(W1):W236–W241. doi: 10.1093/nar/gkw370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zablocki O, van Zyl L, Adriaenssens EM, Rubagotti E, Tuffin M, Cary SC, et al. High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of antarctic soils. Appl Environ Microbiol. 2014;80(22):6888–6897. doi: 10.1128/AEM.01525-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reavy B, Swanson MM, Cock PJ, Dawson L, Freitag TE, Singh BK, et al. Distinct circular single-stranded DNA viruses exist in different soil types. Appl Environ Microbiol. 2015;81(12):3934–3945. doi: 10.1128/AEM.03878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parras-Moltó M, Rodríguez-Galet A, Suárez-Rodríguez P, López-Bueno A. Evaluation of bias induced by viral enrichment and random amplification protocols in metagenomic surveys of saliva DNA viruses. Microbiome. 2018;6(1):1–18. doi: 10.1186/s40168-018-0507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corinaldesi C, Tangherlini M, Dell’Anno A. From virus isolation to metagenome generation for investigating viral diversity in deep-sea sediments. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-08783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu Rev Virol. 2017;4(1):201–219. doi: 10.1146/annurev-virology-101416-041639. [DOI] [PubMed] [Google Scholar]
- 66.Li X, Sun Y, Wang X-Z, Liu J-J, Wang G-H. Research progress of new biomarker gene of phoH for bacteriophage genetic diversity. Biotechnol Bull. 2017;33(10):40–45. [Google Scholar]
- 67.Shi X, Yu D, Warner E, Pan X, Petersen G, Gong Z, et al. Soil database of 1: 1,000,000 digital soil survey and reference system of the Chinese genetic soil classification system. Soil Survey Horizons. 2004;45(4):129–136. [Google Scholar]
- 68.Tian H, Chen G, Zhang C, Melillo JM, Hall CA. Pattern and variation of C:N:P ratios in China’s soils: a synthesis of observational data. Biogeochemistry. 2010;98(1–3):139–151. [Google Scholar]
- 69.Zhang J, Inouye M. MazG, a nucleoside triphosphate pyrophosphohydrolase, interacts with Era, an essential GTPase in Escherichia coli. J Bacteriol. 2002;184(19):5323–5329. doi: 10.1128/JB.184.19.5323-5329.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stillman B. Deoxynucleoside triphosphate (dNTP) synthesis and destruction regulate the replication of both cell and virus genomes. Proc Natl Acad Sci. 2013;110(35):14120–14121. doi: 10.1073/pnas.1312901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogawa A, Sampei G-I, Kawai G. Crystal structure of the flavin-dependent thymidylate synthase Thy1 from Thermus thermophilus with an extra C-terminal domain. Acta Crystallogr Sect F Struct Biol Commun. 2019;75(6):450–454. doi: 10.1107/S2053230X19007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Fig. S1. The distribution of the soil sampling sites. Fig. S2. Variable importance plot of contigs from random forest classification analysis based on geographical distribution. Fig. S3. Two complete viral genomes contain a P metabolism module.
Additional file 2. Table S1. Soil sampling information and physicochemical properties. Table S2. The amino acid sequences of phoH genes f.rom different environmental samples. Table S3. Overview of 19 virome data. Table S4. The vOTU table. Table S5. The abundance of viral reads identified by Kraken 2 (Family level).
Data Availability Statement
Virome read data are available in the NCBI Short Read Archive (SRA) under BioProject ID PRJNA579576.
Virome read data are available in the NCBI Short Read Archive (SRA) under BioProject ID PRJNA579576.