Abstract
Family C G-protein-coupled receptors (GPCRs) operate as obligate dimers with extracellular domains that recognize small ligands, leading to G-protein activation on the transmembrane (TM) domains of these receptors by an unknown mechanism1. Here we show structures of homodimers of the family C metabotropic glutamate receptor 2 (mGlu2) in distinct functional states and in complex with heterotrimeric Gi. Upon activation of the extracellular domain, the two transmembrane domains undergo extensive rearrangement in relative orientation to establish an asymmetric TM6–TM6 interface that promotes conformational changes in the cytoplasmic domain of one protomer. Nucleotide-bound Gi can be observed pre-coupled to inactive mGlu2, but its transition to the nucleotide-free form seems to depend on establishing the active-state TM6–TM6 interface. In contrast to family A and B GPCRs, G-protein coupling does not involve the cytoplasmic opening of TM6 but is facilitated through the coordination of intracellular loops 2 and 3, as well as a critical contribution from the C terminus of the receptor. The findings highlight the synergy of global and local conformational transitions to facilitate a new mode of G-protein activation.
GPCRs share a common heptahelical transmembrane domain (7TM), but vary in the composition of their extracellular domains (ECD) and in their ligand-binding mechanisms2. Family A and B GPCRs share a similar G-protein-coupling mechanism3, whereby an outward displacement of TM6 accommodates binding of the α5 helix of Gα. Family C GPCRs are distinct in that they function as constitutive dimers and have large ECDs, at which ligand binding occurs distal to the 7TM. However, the lack of high-resolution structures of family C GPCRs in complex with G proteins has limited our understanding of the mechanisms that underlie receptor and G-protein activation.
The excitatory neurotransmitter l-glutamate (Glu) activates eight metabotropic glutamate receptors that regulate neuronal excitability and have vital roles in memory formation, pain management, addiction and anxiety4. Among them, mGlu2 has garnered attention as a drug target for the treatment of schizophrenia and depression5. The mGlu ECD includes a ligand-binding Venus flytrap domain (VFT) followed by a cysteine-rich domain (CRD) linker connected to the 7TM. Structural studies of isolated ECDs and a near full-length mGlu5 in the absence of G protein reveal that glutamate binding leads to the closure and rearrangement of the bilobed VFTs, bringing the CRDs in close proximity1,6,7. This conformational change results in the rearrangement of the transmembrane regions, in which the CRD has an essential role in transmitting structural changes from VFTs to the 7TM through its interactions with extracellular loop 2 (ECL2)1. In addition to orthosteric ligands at the ECD, negative (NAMs) or positive (PAMs) allosteric modulators of mGlu receptors are pursued as therapeutics8.
Here we show structures of the mGlu2 homodimer in the inactive state, in the Glu- and agonist-PAM (ago-PAM)-bound state, and in complex with the heterotrimeric protein Gi (active state). The structures reveal profound conformational transitions from an inactive to an active state, which demonstrate a new G-protein-coupling mechanism and provide a blueprint for receptor and G-protein activation by family C receptors.
Asymmetric 7TM dimer in inactive mGlu2
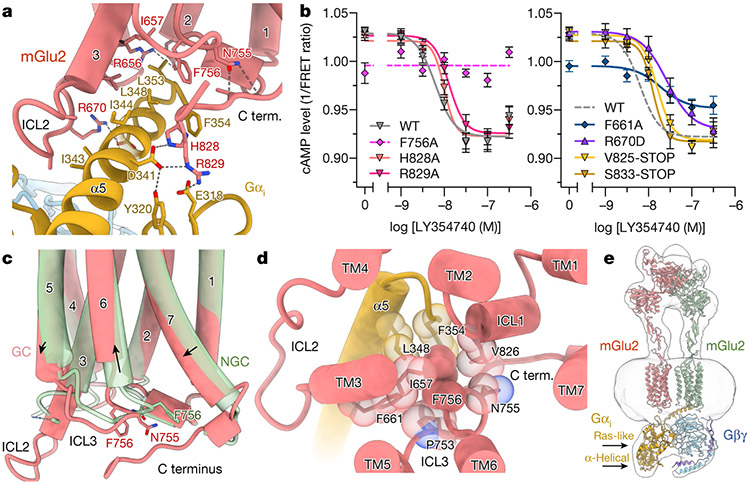
For structural studies, we separately expressed and purified full-length human mGlu2 and heterotrimeric Gi in insect cells (Extended Data Fig. 1). To stabilize the inactive-state mGlu2, we used the potent orthosteric antagonist LY3414959 and the NAM VU600196610. The cryo-electron microscopy (cryo-EM) map of inactive-state mGlu2 (Fig. 1a) revealed an open VFT (3.3 Å resolution at VFT–CRD) conformation with an inter-lobe angle of 44° on both protomers (measured between the Cα atoms of Gly451, Tyr144 and Arg2716) and well-separated CRDs, similar to the crystal structure of the ECD of antagonist-bound mGlu2 (PDB: 5KZQ)6 (Extended Data Figs. 2-4, Extended Data Table 1). The resolution of the inactive-state map was lowest in the transmembrane region (4.2 Å), but enabled unambiguous docking of the 7TMs and localization of the NAM in the 7TM of one protomer (Extended Data Fig. 4a, b). Although the overall architecture of mGlu2 resembles that of the mGlu5 apo structure1, differences in the VFT–CRD angles produce a distinct orientation of the 7TMs (Fig. 1a, Extended Data Fig. 5a-d). Whereas the 7TMs of apo mGlu5 are well separated, with TM5 helices being most proximal between protomers, the 7TMs of inactive mGlu2 come into proximity to form an asymmetric TM3–TM4 interface with the TM6 helices distal to the interaction plane (Extended Data Fig. 5e). In this interface, we also observe interactions between the cytoplasmic ends of TM4 and TM1. The dissimilarity in the disposition of the 7TM of mGlu2 from that of mGlu51 and the GABAB heterodimer11 indicates that there is a range of 7TM configurations across family C GPCRs, which are likely to have roles in signalling modulation (Extended Data Fig. 5e-h).
Fig. 1 ∣. Structures of mGlu2 alone and in complex with Gi.
a-c, Composite cryo-EM maps (top) and models (bottom) of mGlu2 in an inactive state in the presence of an orthosteric antagonist and NAM (a), in the Glu/ago-PAM-bound state (b) in the active-state coupled to nucleotide-free heterotrimeric Gi in the presence of Glu and ago-PAM (c). mGlu2, coral and green; NAM, not represented; antagonist, cyan; micelle, transparent; ago-PAM, light blue; glutamate, yellow; phospholipid, grey; Gαi, gold; Gβ, blue; Gγ, purple.
Mechanism of mGlu2 activation
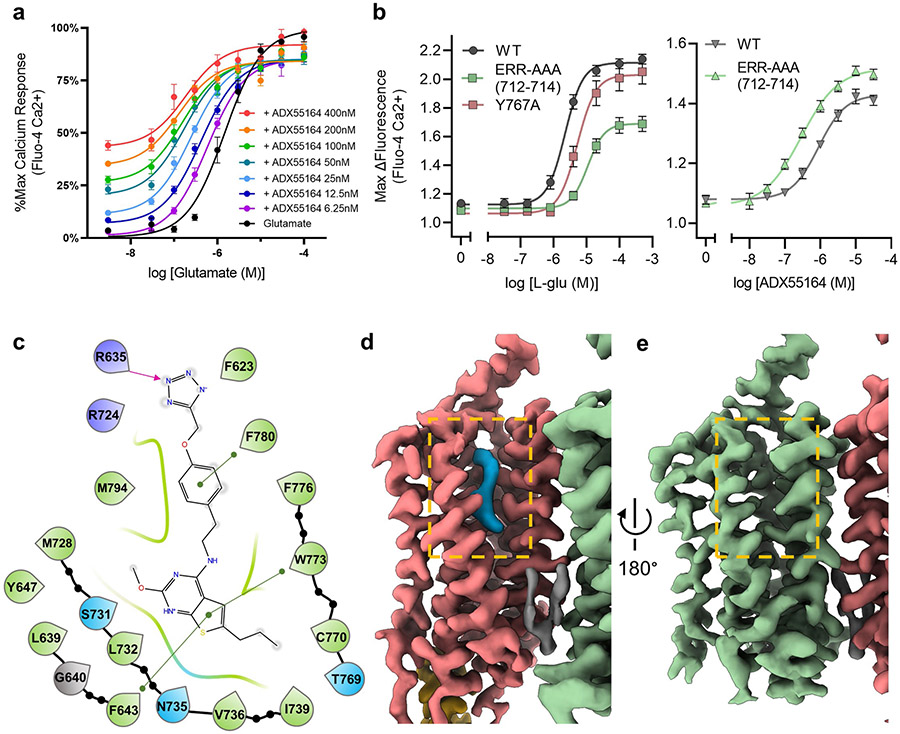
The cryo-EM maps of active-state conformations of mGlu2 alone and in complex with Gi (Fig. 1b, c) were obtained in the presence of the endogenous ligand glutamate and a new ago-PAM (ADX55164; Addex Therapeutics) that potentiates glutamate activation (Extended Data Fig. 6a). The antibody fragment scFv16 further stabilized the mGlu2–Gi complex. Owing to the elongated shape and segmental flexibility of the particle, the indicated resolutions of the global maps for mGlu2 and mGlu2–Gi were 3.3 ¥ and 3.6 Å, respectively; however, local refinements of the mGlu2–Gi complex enabled us to obtain maps in which the resolution was considerably improved (VFT–CRD, 2.7 Å; CRD–7TM, 3.2 Å; 7TM–Gαiβγ, 3.2 Å; Gβγ, 3.3 Å) (Extended Data Fig. 3). Both structures reveal a closed VFT conformation with an inter-lobe angle of 25° that drives the formation of a new ECD interface, similar to previously reported crystal structures of mGlu2 VFTs7. The closure of the VFT leads to the compaction of CRDs, resulting in a rearrangement of the 7TM bundles to form an asymmetric TM6–TM6 interface. The structure of mGlu2 bound to Glu and ago-PAM (Glu/ago-PAM-bound) and that of the G-protein-coupled state seem very similar, except for intracellular loop 2 (ICL2) and the C terminus, which are ordered only in the protomer coupled to Gi (Fig. 1b, c). In both structures, the ago-PAM resides within the 7TM of only one protomer at a site that overlaps the family A orthosteric site (Fig. 1b, c, Extended Data Fig. 6). Notably, Gi couples exclusively to the ago-PAM-bound 7TM, which also shows an opening through which the pocket is accessible to the plasma membrane (Extended Data Fig. 6d, e). By contrast, the corresponding pocket on the 7TM without PAM seems to be completely closed. We also observed a phospholipid density between the 7TMs in the mGlu2–Gi map. The PAM-binding pocket in our structures includes residues (Leu639 Phe643, Asn735, Trp773 and Phe776) that have been shown to affect the modulation of mGlu2 activity by several other mGlu2 PAMs12,13. We found that mutating residues Phe643, Tyr647, Asn735 and Trp773 abolished the potentiation of ADX55164 on glutamate-mediated mGlu2 signalling (Extended Data Fig. 7, Supplementary Table 1). Among them, Tyr647 coordinates a water molecule—similar to its role in NAM-stabilized mGlu514 (Extended Data Fig. 8a, b).
As well as the presence of ago-PAM, we observe additional differences between the G-protein-coupled (GC) and the non-G-protein-coupled (NGC) protomer in the complex. Superposition of the two protomers by alignment of the VFTs shows a lateral shift of around 12 Å between the 7TMs (measured between the Cα atoms of Thr768 on TM6) (Fig. 2a). This asymmetry in the relative 7TM configuration arises from differences in interaction networks between the ECLs and the CRD in each protomer (Fig. 2b), and also from asymmetry in the interaction between two TM6 helices, as also observed in our active-state structure of CaSR15. TM6 has been shown to be involved in the active-state interface of mGlu receptors, including mGlu216,17 and mGlu51, although the observed asymmetry has not been previously reported. The asymmetric interface spans the entire TM6, including Tyr767 from the GC subunit, tucked between Ile771 and Ile772 of the NGC subunit (Fig. 2c). Abrogation of this interaction leads to a reduction in glutamate-stimulated G-protein response as measured by Ca2+ mobilization using a chimeric Gq/o5 protein (Extended Data Fig. 6b).
Fig. 2 ∣. Active-state mGlu2 forms an asymmetric dimer.
a, The VFT of the NGC protomer of the mGlu2–Gi complex is aligned to the VFT of the GC protomer (glutamate, yellow). b, mGlu2 protomers in the same orientation. Ago-PAM (ADX55164; blue) binds only the GC protomer (left), and ECL2, ECL3 and TM6 are distinct between protomers. Differential interaction residues in each protomer are highlighted with key interactions shown (dashed lines). c, The asymmetric TM6–TM6 interface. d, The 7TM bundle of the mGlu2 NGC protomer is superposed to the 7TM bundle of the GC protomer. TM6 of the GC protomer is tilted towards the TM6 of the NGC protomer and shifted approximately half a helical pitch towards the extracellular side to form an asymmetric TM6–TM6 interface. e, Rearrangement of mGlu2 switch residues between GC and NGC subunits.
One notable difference between the GC and NGC subunits is the reorientation of Trp697 in TM4 and His723 on ECL2 (Fig. 2b). In the NGC protomer, Trp697 projects into the core of the 7TM bundle and interacts with ECL2 residue His723. By contrast, Trp697 in the GC protomer flips to face outside the 7TM bundle and is stabilized by Glu701. The positioning of Trp697 in the TM4 of the NGC protomer keeps TM5 closer to TM6, whereas its outwards conformation in the GC protomer allows for TM5 relaxation, which—together with Tyr781 reorientation in TM6—facilitates opening of the PAM-binding pocket. Intrasubunit CRD–ECL interactions also differ between protomers, leading to the closer proximity of ECL2 and ECL3 tips in the GC protomer compared to the NGC protomer. The ECL2 tip residues Glu712 and Arg714 point towards ECL3 in the GC protomer, but are not resolved in the NGC protomer (Fig. 2b). The triple-alanine mutation of mGlu2 ECL2 residues 712–714 (E712A/R713A/R714A, hereafter denoted the ERR-AAA mutant) decreases glutamate-stimulated G-protein activity (Extended Data Fig. 6b, Supplementary Tables 2, 3), which suggests a central role for this loop in coordinating structural transitions from the VFTs to the 7TMs as observed for mGlu51 and for GABAB11. The ago-PAM ADX55164 displayed increased potency and efficacy (Emax) in the ERR-AAA mutant; this supports the notion that the ECL2–ECL3 interaction between protomers is critical for allosteric coupling, and is consistent with previous observations that mGlu PAMs behave as agonists in the absence of ECDs18.
An increasing number of family A and B GPCR structures have revealed key switch residues, which undergo conformational changes that link agonist binding to intracellular G-protein activation. A well-characterized family A switch is the CWxP motif Trp residue, located in TM6, which changes its conformation upon agonist binding in some GPCRs19. As observed for the muscarinic acetylcholine 1 receptor, the rearrangement of the TM6 Trp side chain contributes to the opening of the cytoplasmic half of TM6 by bending at the proline residue of the motif, creating space for G-protein binding20,21 (Extended Data Fig. 8c, d). Although family C receptors do not possess a well-conserved ‘switch’ motif, most members retain a critical TM6 Trp residue. In addition, Phe776, which is situated one helical turn towards the extracellular side of Trp773, is also conserved in mGlu receptors. Key allosteric pocket residues, including Trp773 and Phe776, have been proposed to be involved in such switch mechanisms in mGlu receptors22-24. Superposition of 7TM between the GC and NGC subunits of the mGlu2–Gi structure shows conformational differences in TM5 and TM6 helices, with Trp773, Phe776 and Phe780 side chains seeming to act as switches (Fig. 2d, e). In the GC protomer, the same side chains rotate clockwise compared to their counterparts in the NGC subunit, resulting in an outwards movement of the extracellular part of TM6, which comes closer to the TM6 of the adjacent NGC protomer. This conformational change is also associated with an upwards shift of TM6 of the GC subunit towards the extracellular space. Notably, ago-PAM binding stabilizes the Trp773 conformation of the GC subunit, as its binding would not allow the conformation observed in the NGC subunit (Fig. 2e). Ago-PAM binding seems to be accompanied by a side-chain rearrangement of Leu732 on TM5—a residue that, along with Trp773, may form a dual toggle switch. Notably, these side-chain rearrangements, along with the displacement of TM6, result in an enlarged allosteric pocket in the GC protomer (around 800 Å3) relative to the NGC protomer (around 275 Å3).
G-protein-coupling mechanism in mGlu2
The hallmark of G-protein activation in family A and B GPCRs is the displacement of the cytosolic part of the TM6 helix to accommodate the insertion of the Gα α5 helix in a cleft between TM3, TM5 and TM6 (Extended Data Fig. 8e). The lack of cytosolic TM6 opening observed in our mGlu2–Gi structure underlines a fundamentally different binding mechanism for mGlu receptors and potentially for other family C GPCRs. In the mGlu2–Gi structure, the receptor dimer couples to only one G protein. This observation, together with evidence from previous studies on mGlu225 and other family C GPCRs26,27, confirms that homo- and heterodimers activate a single G protein. The G protein is well-ordered in a nucleotide-free state, with the Gα α5 helix not inserted in a cavity at the 7TM bundle but instead stabilized by a pocket formed between TM3, ICL2, ICL3 and the C terminus of mGlu2 (Fig. 3a, Extended Data Fig. 8f, g). The ICL2 of the GC protomer wraps one side of the C-terminal half of the Gi α5 helix through hydrophobic interactions and a polar interaction between ICL2 Arg670 and the α5 helix Asp350 (Fig. 3a). On the opposite side, receptor C-terminal residues His828 and Arg829 form a polar interaction network with the α5 helix Asp341, as well as with Tyr320 and Glu318 residues in the β6 strand and the α4–β6 loop, respectively, of the Gα Ras domain. His828, together with Phe756 on ICL3, forms hydrophobic interactions with the Gα C-terminal Phe354. We assessed these interactions by determining the effects of specific truncations or mutations of mGlu2 on the activation of Gi (inhibition of cytosolic cAMP) and of chimeric Gq/o5 (increase in cytosolic Ca2+). Truncation of the C-terminal tail of mGlu2—to produce a mutant denoted V825-STOP—significantly affected G-protein signalling (Fig. 3b, Extended Data Fig. 9, Supplementary Tables 2, 4). The addition of only eight residues of the C-terminal tail (denoted S833-STOP) partially recovered Gq/o5 activity, which suggests a role for interactions between further C-terminal residues and the Gα Ras domain, evident by unassigned densities in the cryo-EM map. Furthermore, single-point mutations of Arg670 and Arg829 decreased G-protein activation by mGlu2.
Fig. 3 ∣. G-protein coupling by mGlu2.
a, mGlu2 couples to Gi through an interface involving ICL2, ICL3, TM3 and the receptor C terminus (C term.). b, Truncation of the mGlu2 C terminus or mutation of critical residues of mGlu2 that are involved in the formation of the observed G-protein interface reduces Gi-mediated receptor activation by the agonist LY354740, as measured by inhibition of forskolin-stimulated cAMP formation. Data are mean ± s.e.m. from 9 independent experiments. WT, wild-type. c, Comparison of the cytoplasmic halves of the mGlu2 GC protomer and NGC protomer 7TM bundles shows reorganization of ICL3 along with ordering of ICL2 and the receptor C terminus upon G-protein binding. d, The conserved Phe756 establishes a hydrophobic core to pack residues from TM3, ICL2, the receptor C terminus and the α5 helix of Gαi. e, Cryo-EM map of the G-protein-coupled inactive-state mGlu2. Models of the intermediate state mGlu2 and GDP-bound Gαiβγ (PDB: 1GP2) with a closed α-helical domain are docked in the density map.
The critical elements for G-protein-coupling in the 7TMs adopt different conformations in the two protomers (Fig. 3c). The structure suggests that ICL3 orchestrates the ordering of ICL2, the C terminus and the cytoplasmic tip of TM3 to flank the Gα α5 helix. The upwards shift in TM6 of the GC subunit enables the rearrangement of ICL3 hydrophobic residues, including Phe756, which are now in a position to interact with ICL2, the C terminus of the receptor and the Gα α5 helix. Phe756, in particular, undergoes a 10 Å shift between protomers (Fig. 3c) and assumes a central position in organizing the hydrophobic packing between TM3, ICL2, ICL3, the α5 helix and the C terminus of the receptor (Fig. 3d). The interaction of Phe661, which is located at the beginning of ICL2, with Phe756 promotes ICL2 ordering and engagement of the G protein. Notably, mutation of Phe756 and Phe661 significantly impaired G-protein activation (Fig. 3b, d, Extended Data Fig. 9). Phe756 is well-conserved across family C GPCRs and has previously been shown to be essential for G-protein coupling27,28. Thus, ICL3 rearrangements in response to TM6 repositioning could be a central element for G-protein activation.
Intermediate activation states of mGlu2
During the classification of the mGlu2–Gi dataset, we identified two smaller particle populations with distinct ECD–7TM arrangements. Reconstruction of both classes at relatively lower resolution (4.3 Å and 4.5 Å) revealed that only one of the VFTs was closed in a rarely observed open–closed configuration28,29 (Extended Data Fig. 10a). In this conformation, the lower VFT lobes are separated by 30 Å—similar to the inactive state—thereby keeping the CRDs apart. The 7TMs assume the same asymmetric TM3–TM4 interface conformation as in the inactive state, with the two classes differing only in whether the closed VFT contributes TM3 or TM4 in this arrangement. These observations suggest that the closure of one VFT in mGlu2 is insufficient to establish an active-state 7TM interface.
From the same mGlu2–Gi dataset, we also isolated and reconstructed, at low resolution, a particle population with Gi bound to a receptor dimer displaying VFTs in the inactive state (Fig. 3e). In contrast to the mGlu2–Gi complex with active-state VFTs (Fig. 1c, Extended Data Fig. 10b), both Gα and Gβγ are located directly under both 7TMs, presumably because the 7TMs are not rotated to establish the TM6–TM6 interface. Notably, the envelope for G-protein in this map reveals density attributed to the Gα alpha-helical domain in the same position as in GDP-bound Gi; this indicates that the G protein is not engaged with the receptor in a way that would enable its nucleotide exchange, and thus represents a G-protein pre-coupled state. Although the low resolution precludes us from knowing whether pre-coupled Gi engages an ago-PAM-bound 7TM, this finding suggests that proper G-protein coupling and activation requires the formation of an active-state 7TM configuration in the homodimer. This notion is further supported by examining the superposition of the GC subunit–Gi complex structure with either protomer in the inactive-state mGlu2, which reveals steric clashes for the G protein (Extended Data Fig. 10c). Thus, the reorientation of 7TM could provide an additional mechanism for the regulation of G-protein coupling and activation.
Discussion
The cryo-EM structures of mGlu2 that we describe here collectively provide a dynamic view of activation and G-protein coupling by a prototypic family C GPCR homodimer (Fig. 4). In the absence of glutamate, or at concentrations that are insufficient to drive closure of both VFTs, the mGlu2 dimer remains in an inactive-state that involves an asymmetric TM3–TM4 interface. At higher glutamate levels, the closure of both VFTs enables rearrangement of the 7TMs, thereby establishing an asymmetric active-state TM6–TM6 interface similar to that of the active-state CaSR homodimer15. The upwards shift of TM6 in the GC protomer enables the restructuring of ICL3 to coordinate interactions of ICL2 and the C terminus with the Gα α5 helix. In contrast to family A and B GPCRs, which open up the 7TM bundle to accommodate the G protein, family C GPCRs may achieve this by creating a pseudo-cavity between ICL2 and the terminus of the receptor.
Fig. 4 ∣. mGlu2 conformational transitions upon activation.
Inactive-state mGlu2 adopts a conformation that has symmetric, open VFTs, distant CRDs and an asymmetric 7TM dimer with a TM3–TM4 interface. Glutamate binding closes one VFT to form a stable intermediate state in which the 7TM bundles remain in an inactive conformation. The binding of glutamate to both VFTs promotes an approximately 180° change in the relative configuration of the 7TM bundles to form an asymmetric TM6–TM6 interface. Ago-PAM (cyan) is observed bound only in one 7TM bundle. Upward movement of one TM6 in the TM6–TM6 interface enables the reorganization of cytoplasmic elements, the ordering of ICL2 and the engagement of the C terminus with the G protein.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Generation of heterotrimeric Gi protein
Heterotrimeric Gi protein was purified as described previously30. In brief, expression was achieved by the simultaneous infection of Trichoplusia ni Hi5 insect cells with two baculoviruses: one encoding the human Gαi1 subunit and a second one encoding the Gβ1 and Gγ2 subunits. A histidine tag was present at the N terminus of the Gβ1 gene. Cells were collected 48 h after infection. Upon hypotonic lysis, membranes were solubilized in the presence of 1% sodium cholate and 0.05% n-dodecyl-β-d-maltoside (DDM, Anatrace). A Ni-NTA affinity chromatography purification step, during which the detergent was exchanged to 0.05% DDM, was followed by cleavage of the N-terminal His-tag using human rhinovirus 3C protease (overnight at 4 °C). The protein was collected and was further purified by size exclusion chromatography on a Superdex 200 10/300 gel filtration column (GE) in 20 mM HEPES pH 7.5, 100 mM NaCl, 0.05% DDM, 5% glycerol, 1 mM MgCl2, 100 uM TCEP, 20 uM GDP and concentrated to 10 mg ml−1 for complexation with mGlu2.
Generation of scFv16
The secreted single-chain construct of Fab16 (scFv16) recognizes an epitope composed of the terminal part of the αN helix of Gαi1 and part of the Gβ1 subunit31, and was generated as previously described32. Purified scFv16 was concentrated to 13 mg ml−1 and flash-frozen in buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl and 15% glycerol for later use.
Expression constructs and purification of mGlu2
The native signal peptide of human mGlu2 (Uniprot Q14416) was replaced with an haemagglutinin (HA)-signal peptide (MKTIIALSY-IFCLVFA), a Flag-tag (DYKDDDD) followed by a triple Ala linker, all in front of the mature mGlu2 sequence from amino acid E19 in a manner as described previously for mGlu5 and GABAB1,11,33. The HA–Flag–-mGlu2 amino acid sequence was submitted to the signal peptide prediction server SignalP 4.1 server34, which returned the predicted cleavage site to be between the final Ala residue of the HA-signal peptide and the initial Flag-tag residue Asp. The C terminus of mGlu2 was not modified. For expression in insect cells, the HA–Flag–mGlu2 was subcloned into a pFastBac1 vector and baculovirus generated using the Bac-to-Bac system (Invitrogen). For expression in HEK293 cells, the HA–Flag–mGlu2 construct (termed wild-type receptor) was subcloned into pcDNA3.1(+) neo and used as a template for site-directed mutagenesis.
Spodoptera frugiperda (Sf9) insect cells (Expression Systems) were co-infected at a cell density of 2 × 106 cells per ml with HA–Flag–mGlu2 baculovirus. During expression, cells were exposed to l-glutamate and magnesium included in the ESF921 media (Expression Systems) and not treated with additional ligands or modulators. At 48 h post-infection cells were collected by centrifugation, washed once with phosphate-buffered saline containing protease inhibitors (leupeptin, soybean trypsin inhibitor, N-p-tosyl-l-phenylalanine chloromethyl ketone, tosyl-l-lysyl-chloromethane hydrochloride, phenylmethylsulfonyl fluoride, aprotinin, bestatin, pepstatin) and then rapidly frozen in liquid nitrogen.
Cell lysis was achieved by nitrogen decompression using a cell-disruption vessel (Parr Instruments) in buffer containing 40 mM Tris pH: 8.0, 150 mM NaCl, 0.1 mM MgCl2 and protease inhibitors. The lysate was centrifuged at 1,000g to remove nuclei and unbroken cells and the resulting supernatant was additionally centrifuged at 100,000g to isolate the membrane fraction. Membranes were resuspended by Dounce homogenization in buffer containing 20 mM HEPES pH 7.4, 500 mM NaCl and protease inhibitors.
Solubilization of membranes was initiated by the addition of 1% (w/v) detergent mix of lauryl maltose neopentyl glycol (LMNG) (Anatrace), cholesteryl hemisuccinate (CHS) (Anatrace) and sodium cholate (Sigma-Aldrich) at a 7:1:2 ratio in 20 mM HEPES pH 7.5, 500 mM NaCl. The solubilized membranes were then clarified by centrifugation at 100,000g and the supernatant loaded onto anti-DYKDDDDK G1 affinity resin (Genscript). The resin was washed with 20 mM HEPES pH 7.5, 150 mM NaCl (HBS150) with a 0.1% 8:1:1 detergent mix of LMNG, CHS and GDN. Protein was eluted using HBS150 supplemented with 10% glycerol, 0.1% of 8:1:1 detergent mix, and 0.1 mg ml−1 Flag peptide. Eluted protein was subsequently run on an Enrich SEC 650 column (Bio-Rad) in HBS150 with 0.002% of 8:1:1 detergent mix. Fractions containing dimeric mGlu2 were pooled and concentrated to 13–20 mg ml−1 for preparation of cryo-EM grids. Throughout purification buffers were supplemented with either 5 μM of VU6001966 and 5 μM LY341495 to stabilize inactive mGlu2, or with 5 μM ADX55164, 2 mM l-glutamate and 2 mM MgCl2 to stabilize the mGlu2 active state.
Complexation of mGlu2 and heterotrimeric Gi protein
For the formation of the mGlu2–Gi complex, purified Gi1 protein was pre-incubated with 0.1% 8:1:1 detergent mix at room temperature for 30 min. Following the pre-incubation of Gi1, mGlu2 was added at a ratio of 1:1.5 mGlu2 to Gi1, incubated for 2 h at room temperature before the addition of 0.5 μl apyrase (500 units per ml from New England BioLabs), and the reaction was allowed to proceed overnight at 4°C. The mGlu2–Gi complex was isolated from unbound components by size-exclusion chromatography in HBS150 buffer supplemented with 0.002% 8 LMNG: 1 CHS: 1 GDN detergent mix, and concentrated to 20 mg ml−1. ScFv16 was then added at a 1:2 ratio to mGlu2 and incubated for an additional 2 h at 4°C for preparation of cryo-EM grids.
[35S]GTPγS binding assays
mGlu2 stimulation of Gi nucleotide exchange was measured using a previously established [35S]GTPγS binding assay35. 200 nM of purified mGlu2, prepared in the presence of VU6001966 and LY341495 or l-glutamate and ADX55164, was incubated for 10 min at 25 °C with 100 nM Gi and 1 mM l-glutamate in a pre-incubation buffer (50 mM HEPES, pH 7.4, 1 mM DTT, 1 mM EDTA, 20 μM GDP, 3 μg ml−1 BSA). Reactions were performed in triplicate, and assays initiated by addition of an equal volume of [35S]GTPγS binding buffer (50 mM HEPES pH 7.4, 1 mM DTT, 1 mM EDTA, 20 μM GDP, 3 μg ml−1 BSA, 10 mM MgCl2, 50 mM NaCl, 2 μM GTPγS, [35S]GTPγS (~50,000 cpm pmol−1)). After a 10 min incubation, reactions were quenched with 20 mM Tris pH 7.7, 100 mM NaCl, 10 mM MgCl2, 1 mM GTP and 0.08 (m/v) deionized polyoxyethylene-10-lauryl-ether (C12E10), then filtered onto Protran BA85 nitrocellulose filters (GE Healthcare). Filters were washed with 20 mM Tris, pH 7.7, 100 mM NaCl and 2 mM MgCl2, dried and subjected to liquid scintillation counting.
Cryo-EM data acquisition and processing
A volume of 3.5 μl of around 15 mg ml−1 mGlu2 in the presence of VU6001966 and LY341495, around 14 mg ml−1 mGlu2 in the presence of l-glutamate and ADX55164, or around 20 mg ml−1 mGlu2/Gi heterotrimer in the presence of l-glutamate and ADX55164 supplemented with 0.05% (w/v) β-octyl glucoside detergent was applied under 100% humidity to UltrAuFoil gold grids (Quantifoil, Au300-R1.2/1.3, and R0.6/1) freshly glow-discharged at 10 mA for 45 s. The excess sample was blotted away for 1 s at 8 °C, and the grids were subsequently plunge-frozen using a Vitrobot Mark IV (Thermo Fisher Scientific). Frozen grids were imaged at cryogenic temperatures with a Titan Krios G2 (Thermo Fisher Scientific) transmission electron microscope with a post-column energy filter (20 mV slit width) operated at 300 kV on a Gatan K3 direct electron camera at a magnification of ×130,000 (0.8677 Å per pixel) for all the samples. Micrographs, dose-fractioned over 50 frames, were recorded with a dose rate of 1.32 e− Å−2 (0.05 s per frame) in super-resolution mode with a defocus range of 1.0–2.0 μm using SerialEM36. Beam-induced anisotropic image motion on the micrographs was corrected using MotionCor2 (v.1.2.6)37 with dose weighting. The contrast transfer function values of each micrograph were initially determined using Gctf (v.1.06)38 and further refined using Relion3.139. Particles were auto-picked using the reference-based picking algorithm in Relion. Auto-picked particle projections were transferred to CryoSPARC (v.2.15, v.3.0, v.3.1 and v.3.2)40 and subjected to several rounds of two-dimensional reference-free alignment to exclude damaged particles and contaminations. The initial models for both maps were obtained using a stochastic gradient descent algorithm in CryoSPARC. The particles in the well-defined 2D classes were further processed with several rounds of heterogeneous refinement to select distinct conformational states and remove particles populating poorly defined classes.
The inactive-state map of mGlu2 is symmetric at the VFTs but pseudosymmetric at the 7TMs embedded in the detergent micelle. Initial angular determination in 3D refinement algorithms in Relion and CryoSPARC failed to correctly identify the correct angular assignment for the pseudosymmetric particle. This problem was overcome by 7TM focused classification without angular and translational alignment following a C2 symmetry expansion of a globally refined map in Relion. Selected particle projections after classification were then transferred back to CryoSPARC for local refinement.
Heterogenous refinement of the mGlu2–Gi protein complex dataset yielded maps corresponding to active-state mGlu2 coupled to G protein, two intermediate states of mGlu2 and an inactive-state of mGlu2 coupled to a G protein. The particle subset for the G-protein-coupled inactive map was initially globally aligned in CryoSPARC and further locally aligned in Relion focusing only on the G protein using a recently implemented symmetry relaxation procedure41 along the C2 symmetry axis. The map was then additionally locally aligned using a mask covering the entire mGlu2–Gi protein complex.
Final global refinement of the inactive-state map, Glu/ago-PAM bound state map, and G-protein-coupled map was performed with the CryoSPARC non-uniform refinement algorithm and further locally refined using masks including only the VFTs with CRDs, CRDs with 7TMs, or 7TMs with a G-protein complex. Composite maps were generated with Chimera (v.1.17)42. Map sharpening was performed with CryoSPARC and deepEMhancer43.
Validation of cryo-EM maps and models was performed with Phenix (v.1.18.2-3874)44. UCSF Chimera (v.1.17)42 and ChimeraX (v.1.1)45 were used for map/model visualizations and figure preparation. Transfer of particles from CryoSPARC to Relion was performed by pyem (v.0.5)46.
Model building and refinement
Initial models of the VFTs were built by the rigid-body fitting of published mGlu2 ECD structures PDB: 5KZQ6 for the inactive-states and PDB: 5CNI7 for active-states for mGlu2 and heterotrimeric Gi protein complex structure PDB: 6N4B47 into the cryo-EM maps. Initial models of 7TMs were generated by a previously published mGlu51 cryo-EM structure based on homology modelling using SWISS-MODEL (November 2020)48. All other models were built by de novo modelling. Side chains and loop residues without sufficient EM density were stubbed or removed entirely.
The ligands were fit to cryo-EM maps using the GemSpot Maestro (2019-4) pipeline49 to identify the top five poses. The modelled pose was selected on the basis of the docking score and overall fit to the density.
The starting models were subjected to iterative rounds of manual refinement in Coot (0.9.3)50 and real-space refinement in Phenix44. For the inactive-state 7TM, the non-G-protein-coupled (NGC) protomer 7TM was rigid-body-fit to the map and further refined with Coot and Phenix. The final models were then visually inspected for general fit to the maps and the geometry was analysed using MolProbity (4.2)51 as part of the Phenix software suite. The final refinement statistics for both models are summarized in Extended Data Table 1. Allosteric pocket volume was calculated using Epock52. Ago-PAM binding schematic is generated by Schrödinger software53. All molecular graphics figures were prepared with UCSF ChimeraX45.
Pharmacological profiling of ADX55164 potentiation of mGlu2 by allosteric pocket mutagenesis using an intracellular calcium-mobilization assay
HEK293T cells were seeded in 6-well plates at a density of 0.8 × 106 cells in a 4 ml growth medium. The next day, the N-terminally Flag-tagged wild-type mGlu2 receptor or mutant receptor DNA carrying a single residue mutation were co-transfected with 0.1 μg chimeric Gαq/o5 subunit and 0.2 μg excitatory amino acid transporter 3 (EAAT3) expression vectors (both pcDNA3.1(+)neo) using jetPRIME (Polyplus) transfection agent, according to the manufacturer’s instructions. The amounts of mutant receptor DNA were adjusted beforehand (0.03–0.3 μg) to obtain comparable levels of cell-surface expression. 24 h after transfection, cells were seeded on poly-l-ornithine coated optical 384-well plates at a density of 20,000 cells per well in a medium lacking l-glutamate. 48 h after transfection, the cells were assayed. The cell plates were loaded with 40 μl per well of 3 μM of Fluo-4 (Thermo Fisher Scientific) and 3% of pluronic acid and incubated at 37 °C for 1 h. Plates were washed with DPBS and medium deprived with 35 μl per well of calcium buffer (1 mM CaCl2, 143 mM NaCl, 6 mM KCl, 1 mM MgSO4, 20 mM HEPES, 0.1% glucose and 0.125 mM sulfinpyrazone) for 5.5 h at 22 °C. To pharmacologically characterize the allosteric modulator ADX55164, the calcium response of mGlu2 was measured in a dose–response allosteric shift experiment, using a FLIPR Penta multi-well plate reader (Molecular Devices), with increasing concentrations of l-glutamate (ranging from 30 nM to 1 mM) in the absence and presence of the ago-PAM ADX55164 at different concentrations (ranging from 400 nM to 6.25 nM). Mutant receptors were tested with increasing concentrations of l-glutamate (ranging from 30 nM to 1 mM) in the absence and presence of ADX55164 at 200 nM. Using GraphPad Prism software (v.9.0.2), the difference in fluorescence measured was plotted against the log of the l-glutamate concentrations. The concentration–response curves were fitted to a four-parameter logistic curve by nonlinear regression using a 4 parameter logistic curve with GraphPad Prism. For each experiment, data were normalized to the maximum response of the wild-type receptor. A one-way ANOVA was performed to determine whether the potency (ΔpEC50) of l-glutamate with and without ADX55164 was significantly different from that of the wild-type. At the end of the assay, the surface expression level of each mGlu2 mutant was monitored in a buffer-treated well by live-cell staining of the N-terminal Flag-tag on the receptor with a monoclonal fluorescent antibody (anti-Flag M2 Cy3, Sigma-Aldrich A9594).
Cell-surface expression of wild-type and mutant mGlu2 for the PAM-binding site mutational assays were determined by flow cytometry, as previously described1, with the following modifications: a fraction of the HEK293T cells transfected for the calcium assay were re-seeded 24 h after transfection in a 6-well plate. On the day of the calcium experiment, living cells were stained with anti-Flag M2 Cy3 (Sigma-Aldrich A959) diluted 1:5,000 and surface expression was determined by flow cytometry using an Attune NxT FACS (Thermo Fisher Scientific). Living cells were gated using forward/sideward scatter and DAPI staining. Cell-surface expression levels for each mutant were determined by normalization of the total Cy3-fluorescences to the wild-type receptor total fluorescence.
Intracellular calcium mobilization assay for ECL, TM and G-protein interface mutants
HEK293 cells (ATCC CRL-1573) were transfected with the expression vector DNA encoding the N-terminally Flag-tagged wild-type mGlu2 and mutants along with a chimeric Gαq/o5 subunit and the EAAT3 essentially as described previously11. In brief, cells were brought into suspension in growth at a density of 0.20 million cells per ml. For each 1 ml of cell suspension transfected, a total of 1 μg DNA in 25 μl OptiMEM (Gibco 51985) was incubated with a mixture of 3 μl FuGene6 (Promega E2692) and 57 μl OptiMEM for 20 min at room temperature, before the addition of the DNA-FuGene6 mixture directly to the cells in suspension. The amount of DNA encoding the wild-type mGlu2 or mutant receptor ranged from 0.03 to 0.4 μg to obtain similar surface expression levels (absolute amounts are given in Supplementary Tables 2-4) and was supplemented with 0.2 μg of expression vector DNA encoding the chimeric Gαq/o5 subunit and 0.4 μg expression vector DNA encoding the EAAT3 in 25 μl OptiMEM (Gibco 51985). Empty vector DNA was added to give a total of 1 μg DNA transfected for each 1 ml of cells. 100 μl of transfected cell suspension was then seeded in poly-d-lysine coated 96-well plates at 20,000 cells per well (black clear-bottom plates for the intracellular calcium release assay and white clear-bottom plates for the receptor surface expression assay). The intracellular calcium mobilization assay was performed 48 h after transfection as described previously1. In brief, cells expressing the wild-type mGlu2 and mutants were loaded with Fluo-4 dye dissolved in HBSS buffer (HBSS Gibco 14025, 20 mM HEPES pH 7.5, 1 mM CaCl2, 1 mM MgCl2, 0.1% BSA and 2.5 mM probenecid) for 30 min and then stimulated with l-glutamate or PAM ADX55164 at different concentrations to determine the potency (half-maximal effective concentration; EC50) and maximal response (Emax) by nonlinear regression as described above.
Assay for determining receptor-mediated inhibition of forskolin-stimulated cAMP formation
HEK293 cells stably expressing an Epac1-based fluorescence resonance energy transfer (FRET) cAMP biosensor capable of reporting intracellular cAMP levels were transiently transfected as described previously54 with vector DNA encoding the N-terminally Flag-tagged wild-type mGlu2 and mutants as described above. The amount of DNA encoding the wild-type mGlu2 or mutant receptor ranged from 0.07 to 0.3 μg ml−1 of the transfected cell suspension to obtain similar surface expression levels (absolute DNA amounts given in Supplementary Tables 1-4) and was supplemented with empty vector DNA to give a total of 1 μg DNA transfected for each 1 ml of cells. 100 μl of transfected cell suspension was seeded in poly-d-lysine coated 96-well plates at 15,000 cells per well (black clear-bottom plates for the functional cAMP assay and white clear-bottom plates for the receptor cell surface expression assay). Assays were performed 48 h after transfection and seeding of cells.
Before initiating the kinetic measurement of cAMP levels, the growth medium in the 96-well plates was exchanged to 100 μl HBSS buffer for 1 h. Then HBSS buffer was replaced with fresh 50 μl HBSS buffer and incubated for 15 min before addition of forskolin to a final concentration of 3.3 μM (by addition of 50 μl HBSS buffer with 6.6 μM forskolin). The increase in intracellular cAMP levels was followed using the Epac1 biosensor by recording the change in FRET using an Envision plate reader (Perkin Elmer) equipped with filters for measuring the fluorescence of mCerulean fluorescent protein (474 nm) and mCitrine fluorescent protein (524 nm) upon mCerulean excitation (434 nm). After 14 min preincubation with forskolin, cells were stimulated with the mGlu2 agonist LY354740 diluted to different concentrations in 50 μl assay buffer containing 3.3 μM forskolin and the intracellular cAMP levels followed over 90 min (Extended Data Fig. 9). To determine the EC50 and Emax of LY354740 for the individual mutants, the FRET ratio at 60 min (or area under the curve 0–60 min as a supplementary readout) was plotted against the concentration of LY354740, and fitted by nonlinear regression as described above.
Cell surface ELISA assay of wild-type mGlu2 and mutants
Surface expression of wild-type mGlu2 and mutants used to assess ECD, 7TM and Gi protein interactions were determined to enable their comparison in the functional intracellular calcium mobilization and inhibition of forskolin-stimulated cAMP assays. The assay for estimation of surface-expressed receptors was performed as described previously11 except that only the HRP-conjugated antibody against Flag-epitope was used. In brief, on the day of the functional assay, cells expressing the Flag-tagged receptors growing in white clear-bottom plates were fixated with 4% paraformaldehyde in DPBS, blocked in 3% dry milk in DPBS, incubated with anti-Flag HRP-conjugated antibody in 3% dry milk in DPBS, washed 4 times in 3% dry milk in DPBS and 4 times in DPBS, and then finally added HRP substrate diluted in DPBS to emit luminescence upon conversion to its product by bound anti-Flag HRP-conjugated antibody. Luminescence levels were quantified using an Envision Plate reader (Perkin Elmer) and wild-type and mutant surface expression were reported relative to the signal from an identically transfected Flag-tagged wild-type condition.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Extended Data
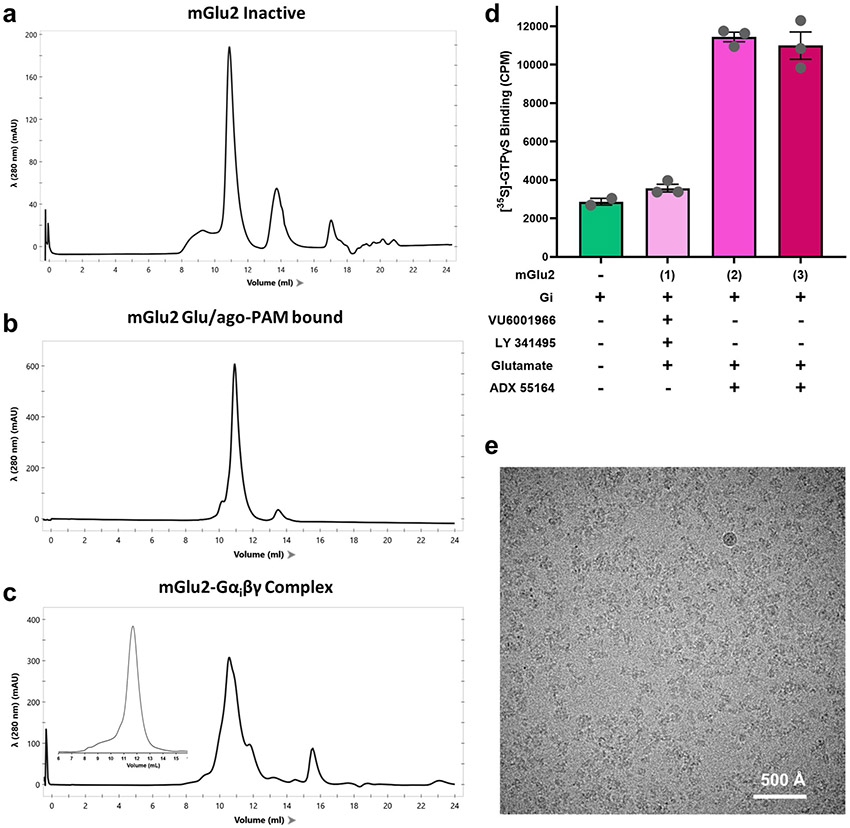
Extended Data Fig. 1 ∣. Preparation of cryo-EM samples.
a-c, Size-exclusion chromatography profiles of purified inactive-state mGlu2 (a), Glu/ago-PAM-bound state mGlu2 (b) and the mGlu2–Gi complex (c), repeated three times with similar results. Inset in c shows the size-exclusion profile of purified Gi heterotrimer. d, Gi protein nucleotide exchange stimulated by purified mGlu2 preparations in (1) inactive state, (2) Glu/ago-PAM-bound state and (3) active state preparation used for cryo-EM studies of the mGlu2–Gi complex, as determined in a GTPγS binding assay. mGlu2 purified in the presence of antagonist LY341495 and NAM VU6001966 did not produce a substantial increase in GαI GTPγS binding above the intrinsic binding of Gi alone. By contrast, mGlu2 purified in the presence of the agonist glutamate and ago-PAM ADX55164 produced a roughly 3.5-fold increase in Gi GTPγS binding. Data represent mean ± s.e.m. of reactions performed in triplicate. e, Representative cryo-EM micrograph of mGlu2–Gi–scFv complex from a single dataset with 45,371 micrographs.
Extended Data Fig. 2 ∣. Cryo-EM processing summary of mGlu2 in its inactive and Glu/ago-PAM-bound states.
a, Cryo-EM data processing workflow for the mGlu2 inactive state. b, Fourier shell correlation (FSC) curves for the mGlu2 inactive state cryo-EM maps of the ECD focused refinement and the global refinement. c, Angular distribution heat map of particles for reconstruction of the mGlu2 inactive state. d, Cryo-EM data processing workflow for the Glu/ago-PAM-bound state of mGlu2. e, FSC curves of the Glu/ago-PAM bound state of mGlu2 for the VFT focused refinement and the global refinement. f, Angular distribution heat map of particle projections in reconstruction of the Glu/ago-PAM-bound state of mGlu2. FSC curves and local refinement spectra were determined using CryoSPARC. Dashed lines represent the resolution at 0.143 FSC. All the processing steps were performed with Relion 3.1 (red) or CryoSPARC 3.1 (blue).
Extended Data Fig. 3 ∣. Cryo-EM processing summary for the mGlu2–Gi complex.
a, Cryo-EM data processing workflow for the mGlu2–Gi complex. b, FSC curves for the locally refined maps of the Gβγ, 7TM-Gαiβγ, CRD–7TM and VFT–CRD. Dashed lines represent the resolution at 0.143 FSC. c, Angular distribution heat map of particles for the mGlu2–Gi global reconstruction. FSC curves and local refinement spectra were calculated using CryoSPARC. All the processing steps were performed with Relion 3.1 (red) or CryoSPARC 3.1 (blue).
Extended Data Fig. 4 ∣. Agreement between cryo-EM map and model.
a, EM density and model for the 7TM of the mGlu2 inactive state. The 7TM model of mGlu2 from the mGlu2–Gi complex is rigid-body-docked to the mGlu2 inactive-state map (7TMA; green, 7TMB; coral, additional density inside the allosteric pocket of the 7TM; purple). b, Magnified view of a density inside the allosteric pocket of 7TMB that may correspond to negative allosteric modulator VU6001966. c, EM density, and model for the mGlu2–Gi complex; transmembrane helices of mGlu2 G-protein-coupled protomer (GC), transmembrane helices of non-G-protein-coupled protomer (NGC), ECL2, glutamate, ago-PAM ADX55164, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine as a representative phospholipid and α5 helix of Gαi. Densities were visualized with UCSF ChimeraX and zoned at 4 Å with a uniform threshold.
Extended Data Fig. 5 ∣. Comparison of structures across family C GCPRs.
The overall architecture of mGlu2 is similar to that of other family C GCPRs. The differences in the angle between VFT and CRD, and the angle between CRD and 7TMs leads to variations in the 7TM configuration. a-d, Models of single protomers of family C GPCRs are overlaid based on VFT superposition. Comparison of the inactive-state model of mGlu2 from this study with the mGlu5 apo state model (PDB: 6N52) (a), the mGlu5 active state model (PDB: 6N51) (b), the CaSR inactive state model (c) and the CaSR active state model (d, see ref. 15). e, Top-down view of mGlu2 7TM bundles shows that TM6 helices are distal to the 7TM interface in the inactive state (left) but form the active-state interface (right). f-h, The inactive state configuration of 7TMs of family C GPCRs are variable. Top-down view of the 7TMs of inactive-state (left) and active-state (right) family C GPCRs: mGlu5 (apo PDB: 6N52, active PDB: 6N51) (f), GABAB (inactive PDB: 6W2X, active PDB: 7C7Q) (g) and CaSR (h, ref. 15). The receptors display variable 7TM configuration.
Extended Data Fig. 6 ∣. Ago-PAM modulation of mGlu2.
a, The mGlu2 ago-PAM ADX55164, used to stabilize the active receptor conformation, potentiates the functional response of mGlu2 to l-glutamate. Receptor activation is measured by co-transfection of the mGlu2 with the neuronal glutamate transporter EAAT3 to remove extracellular glutamate and a chimeric Gq/o5 to enable intracellular calcium release as a readout. b, In a similar assay, mutation of the ECL2 tip (residues 712–714; ERR-AAA) and the Y767A mutation on the TM6–TM6 interface blunt glutamate-induced signalling (left) and compared to wild-type mGlu2, the ago-PAM ADX55164 has higher potency and higher Emax for the ERR-AAA mutant, consistent with a partial uncoupling of the ECD from the 7TM (right). c, Schematic of interactions between mGlu2 residues and ago-PAM ADX55164 bound within the 7TM core. Green, hydrophobic; blue, polar; purple, positively charged; magenta arrow, hydrogen bond; green line, π–π stacking; grey, glycine. d, e, Magnified views of the cryo-EM map of G-protein-coupled active-state mGlu2 GC (coral) (d) and NGC (green) (e) protomers show that ago-PAM (blue) binds only to the GC protomer, whereas the analogous pocket is not accessible by the membrane (pocket opening highlighted with orange box) on the NGC protomer. A phospholipid density between two 7TMs is shown in grey. The head group of the lipid molecule does not seem to interact with mGlu2 or Gi and the density might represent a lipid molecule with different head groups. All family C GPCR dimeric structures display elongated densities between two protomers, most likely corresponding to either cholesterol or phospholipids. Additionally, GABAB structures displayed a phospholipid molecule inside the 7TM, indicating the importance of lipid molecules in family C GPCR dimerization and activity. Data in a and b represent mean ± s.e.m. from four and five independent experiments measured in duplicate, respectively.
Extended Data Fig. 7 ∣. Ago-PAM binding-pose validation for mGlu2.
Residues within the binding pocket of ago-PAM ADX55164 were mutated to study their role in PAM activity. a, Mutant and wild-type receptor responses to increasing concentrations of glutamate in the absence (brown) or presence (green) of 200 nM ADX55164 were tested in an intracellular calcium release assay following co-transfection with the EAAT3 and Gq/o5. Responses were normalized to the maximum response of the wild-type receptor. The concentrations of glutamate are plotted on the x axis [log (M)]. The amount of receptor DNA transfected was increased up to tenfold to obtain mutant expression levels similar to that of the wild-type; however, for some mutants, such expression levels could not be reached. Data shown represent the mean ± s.e.m. from five independent experiments. b, Mutant and wild-type receptor surface expression levels were monitored by fluorescence microscopy using an N-terminal Flag-tag present on all constructs and an anti-Flag Cy3 antibody. Data in b represent images from three independent experiments. R720A, S731A and L732A mutants did not produce glutamate or ADX55164 responses and did not show surface expression in immunofluorescence studies (not shown). c, The change in pEC50 of mutant and wild-type receptors upon addition of 200 nM ADX55164 plotted from individual experiments along with individual data points for estimation of surface expression of mutants compared to the wild-type (100%) by flow cytometry. Statistics were derived from at least 4 independent experiments by one-way ANOVA and comparison of each mutant to the wild-type. A statistical difference from the wild-type is indicated by an asterisk (*). P values were corrected for multiple comparisons using Dunnett’s test and are provided in Supplementary Table 1.
Extended Data Fig. 8 ∣. Comparison of water coordination, 7TM activation and the G-protein interface across GPCRs.
a, EM density for the region of the observed water molecule coordinated inside the allosteric pocket of PAM-less 7TM of mGlu2 (labelled residues are conserved in mGlu5, and blue dashed lines represent hydrogen bonds between the water molecule and mGlu2). b, Comparison of the water molecule coordinated inside the allosteric pocket of mGlu5 (PDB: 4OO9, yellow; water, brown) and the PAM-less 7TM of mGlu2 (mGlu2–Gi structure, green; water, red). The homologous Tyr647 residue in mGlu5 (Tyr659) coordinates a water molecule in the NAM-stabilized mGlu5 allosteric pocket14,55,56 and has a role in ligand pharmacology by affecting allosteric modulator cooperativity23. The mGlu2–Gi map also reveals a density that corresponds to a water molecule within the PAM-less 7TM bundle, coordinated between residues Tyr647, Thr769, Ser801 and Gly802, similar to the water-molecule coordination in the allosteric pocket of NAM-stabilized mGlu5 structures14,55,56. The PAM-bound 7TM bundle shows a weaker density for this water molecule. Using JAWS simulations57, a statistical thermodynamics-based approach to determine water-molecule positioning, a water molecule was observed to be bound in the pocket containing the putative water site in both protomers. The conservation of these four residues and resolution of water molecules in mGlu2 and mGlu5 indicates the importance of water for mGlu ligand pharmacology. c, Superposition of M1 muscarinic receptor (M1R) inactive (PDB: 5CXV) and active (PDB: 6OIJ) states reveals TM6 movement (agonist: iperoxo, orange). d, Tryptophan toggle switch in M1R. e, f, Comparison of the overall Gi protein coupling arrangement on the cannabinoid receptor 1 (CB1), a representative family A GPCR (e) and mGlu2 (f). The non-G-protein-coupled protomer model is not shown for clarity. g, Magnified view of cryo-EM map of G-protein-coupled active-state mGlu2.
Extended Data Fig. 9 ∣. Functional analysis of mGlu2 mutants for Gi activation.
a, Truncation of the mGlu2 C terminus or mutation of critical residues of mGlu2 involved in the formation of the observed G-protein interface substantially decreases receptor activation by l-Glu as tested in an intracellular calcium release assay following co-transfection with the EAAT3 and Gq/o5. Data in a represent mean ± s.e.m. from five independent experiments. b, An Epac1-based FRET cAMP biosensor capable of reporting intracellular cAMP levels was used to investigate the effect of mutating critical residues of mGlu2 in the Gi-interacting interface. FRET between the fluorescent proteins decreases after cAMP binding to a fusion protein consisting of an Epac1-domain (residues 14–881) flanked by fluorescent proteins mCerulean and mCitrine. c, Representative kinetic traces of LY354740 (mGlu2 agonist)-induced inhibition of forskolin-stimulated cAMP formation through Gi activation by wild-type mGlu2 and mutants. The effect of the NAM MNI-137 and non-transfected cell traces are shown for comparison. Indicated are time points for mGlu2 ligand addition and for taking cAMP values for generation of the concentration-response curves (CRC) shown in Fig. 3b. Data in c represent mean ± s.d. from one representative experiment performed in triplicate.
Extended Data Fig. 10 ∣. Intermediate states of mGlu2–Gi activation.
a, Cryo-EM maps of mGlu2 in two distinct intermediate states with open-closed VFTs and inactive 7TM conformation. The protomer contributing TM3 to the interface (7TMA) displays an open VFT and the protomer contributing TM4 to the interface (7TMB) displays a closed VFT in one intermediate state (left), and the opposite in the other intermediate state (right). b, Reference-free cryo-EM 2D class average of the mGlu2–Gi protein complex shows heterotrimeric G-protein pre-coupling to mGlu2 in the inactive state (top), compared to an average of the nucleotide-free Gi coupled to mGlu2 (bottom). The α-helical domain of Gα is ordered in the pre-coupled state, indicating a GDP-bound G protein. This observation suggests that VFT activation and the approximately 180° rearrangement of the 7TMs to establish the TM6–TM6 interface would be necessary for G-protein activation. c, Cryo-EM model of G-protein-coupled active state model of mGlu2 and G protein overlaid to either of the 7TMs of mGlu2 in the inactive-state model. The αN helix of the Gα protein in both conformations would clash with the membrane, while Gβ would also clash with the adjacent protomer if the Gi is bound to TMB of the inactive state (clash represented by red stars).
Extended Data Table 1 ∣.
Cryo-EM data collection, refinement and validation statistics
| Inactive-State mGlu2 |
Glu + ago-PAM bound mGlu2 |
G-protein- coupled mGlu2 |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 130,000 | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 56 | 56 | 56 |
| Defocus range (μm) | −1.0~−2.0 | −1.0~−2.0 | −1.0~−2.0 |
| Pixel size (Å) | 0.8677 | 0.8677 | 0.8677 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 3,377,889 | 4,451,535 | 37,818,763 |
| Final particle images (no.) | 235,631 | 219,019 | 799,323 |
| Map resolution (Å) | 3.65 | 3.3 | 2.7-3.2 |
| FSC threshold | (0.143) | (0.143) | (0.143) |
| Refinement | |||
| Initial model used (PDB code) | 5KZQ, 6N52 | 5CNI, 6N51 | 5CNI, 6N51 |
| Model composition | |||
| Non-hydrogen atoms | 10609 | 10875 | 16040 |
| Protein residues | 1511 | 1525 | 2148 |
| Ligands | 2 | 3 | 3 |
| B factors (Å2) | |||
| Protein | 70.6 | 86.1 | 78.6 |
| Ligand | 70.6 | 86.1 | 78.6 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | 0.005 | 0.002 |
| Bond angles (°) | 0.882 | 0.937 | 0.502 |
| Validation | |||
| MolProbity score | 1.61 | 1.51 | 1.65 |
| Clashscore | 6.96 | 5.39 | 8.98 |
| Poor rotamers (%) | 0.00 | 0.00 | 0.00 |
| CaBLAM outliers (%) | 1.91 | 1.68 | 1.43 |
| EMRinger score | 2.80 | 2.63 | 2.76 |
| Ramachandran plot | |||
| Favored (%) | 96.57 | 96.54 | 97.03 |
| Allowed (%) | 3.43 | 3.46 | 2.97 |
| Outliers (%) | 0.06 | 0.00 | 0.00 |
Supplementary Material
Acknowledgements
We thank E. Montabana at the Stanford-SLAC cryo-EM facility for support with data collection and J.-P. Aubry of the FACS facility (University of Geneva) for assistance with the FACS experiments. We also thank G. Eskici for discussions and support with coding. This work was supported, in part, by T32GM089626 (J.G.M.), R01 NS092695 (G.S., B.K.K. and J.M.M.) and R01 NS028471 (B.K.K.). B.K.K. is a Chan Zuckerberg Biohub Investigator.
Footnotes
Competing interests B.K.K. is a co-founder of and consultant for ConfometRx, Inc.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-03680-3.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-03680-3.
Data availability
All data generated or analysed in this study are included in this article and the Supplementary Information. The cryo-EM density maps and corresponding coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB), respectively, under the following accession codes: EMD-23994 and 7MTQ (inactive-state mGlu2), EMD-23995 and 7MTR (Glu/ago-PAM-bound mGlu2), and EMD-23996 and 7MTS (mGlu2–Gi complex).
References
- 1.Koehl A et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attwood TK & Findlay JB Fingerprinting G-protein-coupled receptors. Protein Eng. 7, 195–203 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Weis WI & Kobilka BK The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem 87, 897–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niswender CM & Conn PJ Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol 50, 295–322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conn PJ, Lindsley CW & Jones CK Activation of metabotropic glutamate receptors as a novel approach for the treatment of schizophrenia. Trends Pharmacol. Sci 30, 25–31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell MD et al. Discovery of (1S,2R,3S,4S,5R,6R)-2-amino-3-[(3,4-difluorophenyl) sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid hydrochloride (LY3020371·HCl): a potent, metabotropic glutamate 2/3 receptor antagonist with antidepressant-like activity. J. Med. Chem 59, 10974–10993 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Monn JA et al. Synthesis and pharmacological characterization of C4-(thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid (LY2812223), a highly potent, functionally selective mGlu2 receptor agonist. J. Med. Chem 58, 7526–7548 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Galici R, Echemendia NG, Rodriguez AL & Conn PJ A selective allosteric potentiator of metabotropic glutamate (mGlu) 2 receptors has effects similar to an orthosteric mGlu2/3 receptor agonist in mouse models predictive of antipsychotic activity. J. Pharmacol. Exp. Ther 315, 1181–1187 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Kingston AE et al. LY341495 is a nanomolar potent and selective antagonist of group II metabotropic glutamate receptors. Neuropharmacology 37, 1–12 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Bollinger KA et al. Design and synthesis of mGlu2 NAMs with improved potency and CNS penetration based on a truncated picolinamide core. ACS Med. Chem. Lett 8, 919–924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papasergi-Scott MM et al. Structures of metabotropic GABAB receptor. Nature 584, 310–314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farinha A et al. Molecular determinants of positive allosteric modulation of the human metabotropic glutamate receptor 2. Br. J. Pharmacol 172, 2383–2396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundström L et al. Structural determinants of allosteric antagonism at metabotropic glutamate receptor 2: mechanistic studies with new potent negative allosteric modulators. Br. J. Pharmacol 164 (2b), 521–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christopher JA et al. Fragment and structure-based drug discovery for a class C GPCR: discovery of the mGlu5 negative allosteric modulator HTL14242 (3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J. Med. Chem 58, 6653–6664 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Gao Y et al. Asymmetric activation of the calcium sensing receptor homodimer. Nature 10.1038/s41586-021-03691-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue L et al. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol 11, 134–140 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Thibado JK et al. Differences in interactions between transmembrane domains tune the activation of metabotropic glutamate receptors. eLife 10, e67027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goudet C et al. Heptahelical domain of metabotropic glutamate receptor 5 behaves like rhodopsin-like receptors. Proc. Natl Acad. Sci. USA 101, 378–383 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trzaskowski B et al. Action of molecular switches in GPCRs - theoretical and experimental studies. Curr. Med. Chem 19, 1090–1109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda S, Qu Q, Robertson MJ, Skiniotis G & Kobilka BK Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thal DM et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature 531, 335–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez-Benito L et al. Molecular switches of allosteric modulation of the metabotropic glutamate 2 receptor. Structure 25, 1153–1162.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Fukuda J et al. Identification of a novel transmembrane domain involved in the negative modulation of mGluR1 using a newly discovered allosteric mGluR1 antagonist, 3-cyclohexyl-5-fluoro-6-methyl-7-(2-morpholin-4-ylethoxy)-4H-chromen-4-one. Neuropharmacology 57, 438–445 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Mühlemann A et al. Determination of key amino acids implicated in the actions of allosteric modulation by 3,3′-difluorobenzaldazine on rat mGlu5 receptors. Eur. J. Pharmacol 529, 95–104 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Liu J et al. Allosteric control of an asymmetric transduction in a G-protein-coupled receptor heterodimer. eLife 6, e26985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goudet C et al. Asymmetric functioning of dimeric metabotropic glutamate receptors disclosed by positive allosteric modulators. J. Biol. Chem 280, 24380–24385 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hlavackova V et al. Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J. 24, 499–509 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kniazeff J et al. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat. Struct. Mol. Biol 11, 706–713 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Ellaithy A, Gonzalez-Maeso J, Logothetis DA & Levitz J Structural and biophysical mechanisms of class C G-protein-coupled receptor function. Trends Biochem. Sci 45, 1049–1064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dror RO et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 348, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda S et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun 9, 3712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehl A et al. Structure of the μ-opioid receptor–Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen SG et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, von Heijne G & Nielsen H SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Stoveken HM, Hajduczok AG, Xu L & Tall GG Adhesion G-protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl Acad. Sci. USA 112, 6194–6199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Ilca SL et al. Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus. Nature 570, 252–256 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Sanchez-Garcia R et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Preprint at 10.1101/2021.01.12.426430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen EF et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daniel A, Eugene P & Cheng Y asarnow/pyem: UCSF pyem v0.5. 10.5281/zenodo.3576630 (2019). [DOI] [Google Scholar]
- 47.Krishna Kumar K et al. Structure of a signaling cannabinoid receptor 1–G protein complex. Cell 176, 448–458.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson MJ, van Zundert GCP, Borrelli K & Skiniotis G GemSpot: a pipeline for robust modeling of ligands into cryo-EM maps. Structure 28, 707–716.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams CJ et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurent B et al. Epock: rapid analysis of protein pocket dynamics. Bioinformatics 31, 1478–1480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friesner RA et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem 49, 6177–6196 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Vedel L, Bräuner-Osborne H & Mathiesen JM A cAMP biosensor-based high-throughput screening assay for identification of Gs-coupled GPCR ligands and phosphodiesterase inhibitors. J. Biomol. Screen 20, 849–857 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Christopher JA et al. Structure-based optimization strategies for G protein-coupled receptor (GPCR) allosteric modulators: a case study from analyses of new metabotropic glutamate receptor 5 (mGlu5) X-ray structures. J. Med. Chem 62, 207–222 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Doré AS et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511, 557–562 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Michel J, Tirado-Rives J & Jorgensen WL Prediction of the water content in protein binding sites. J. Phys. Chem. B 113, 13337–13346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed in this study are included in this article and the Supplementary Information. The cryo-EM density maps and corresponding coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB), respectively, under the following accession codes: EMD-23994 and 7MTQ (inactive-state mGlu2), EMD-23995 and 7MTR (Glu/ago-PAM-bound mGlu2), and EMD-23996 and 7MTS (mGlu2–Gi complex).