Abstract
Staphylococcus aureus strains often carry in their genomes virulence genes that are not found in all strains and that may be carried on discrete genetic elements. Strains also differ in that they carry one of four classes of an accessory gene regulator (agr) locus, an operon that regulates virulence factor expression and that has been proposed to be a therapeutic target. To look at their distribution among hospital strains, we investigated 38 methicillin-sensitive S. aureus isolates, classifying the isolates by agr class and screening them for the presence and restriction fragment length polymorphisms (RFLPs) of 12 core and 14 accessory virulence genes. Twenty-three (61%) were agr class I, 10 (26%) were agr class II, and 5 (13%) were agr class III. None were agr class IV. The S. aureus strains had distinguishable RFLP profiles, although clusters of isolates with clearly related core gene profiles were found among our strains, including all five agr class III strains, two sets of six strains within agr class I, and six strains within agr class II. Within these clusters there was evidence of horizontal acquisition and/or loss of multiple accessory virulence genes. Furthermore, two isolates from the same patient were identical except for the presence of the sea gene, indicating that movement of mobile elements may occur in vivo. Several strong correlations with the carriage of virulence genes between strains were seen, including a positive correlation between tst and agr class III and negative correlations between tst and lukE-splB and between lukE-splB and seg-sei. This suggests that the core genome or the presence of accessory genetic elements within a strain may influence acquisition and loss of other elements encoding virulence genes.
Staphylococcus aureus is an important nosocomial pathogen causing a variety of clinical infections including septicemia, pneumonia, wound sepsis, septic arthritis, osteomyelitis, and postsurgical toxic shock syndrome (7, 56). It is also a significant cause of community-acquired disease. Genetic variation among S. aureus strains has been shown to be associated with pathogenic potential (5, 16). Variation occurs at the level of both core genes (present in >95% of isolates [31]) and accessory (variable) genes.
Many S. aureus accessory genes encode virulence factors such as enterotoxins A through I (sea to sei, respectively), toxic shock syndrome toxin 1 (tst), exfoliative toxins A and B (eta, etb), as well as leukocidins and surface-associated proteins, for example, collagen binding protein (cna) (39, 43). These genes are often carried on mobile genetic elements, such as phages and pathogenicity islands (SaPIs), which transfer horizontally between strains (sea, tst, eta) (4, 35, 57, 58) or which are suspected to do so (etb, seb) (24, 29). Transfer of tst can occur at an extremely high frequency (35) in vitro, although it is not known if this occurs in the clinical setting. There is also evidence that mobile accessory virulence genes are not distributed uniformly among strains. For example, seb and tst are never seen in the same strain (5), and it has been suggested that carriage of tst is correlated with agr class III strains and with phage group I strains (28, 30). Furthermore, some virulence genes can be carried on more than one element. For example, tst has been described on at least six related SaPIs that can be distinguished by Southern blotting (20, 34, 35).
Virulence gene expression in S. aureus is controlled by regulators such as agr (accessory gene regulator) (43), and four different mutually inhibitory classes have been described (26, 28). agr is a potential target for therapy, as the response can be modulated by synthetic peptides (37). However, these responses are agr class specific, and the distribution of agr types in clinical strains is poorly characterized.
The purpose of the present study was to investigate the distribution of genomic differences in clinical strains of S. aureus, particularly those that might have an effect on pathogenicity. We studied epidemiologically unrelated strains, collected from a 1,200-bed teaching hospital in southwest London, United Kingdom, chosen to reflect the spectrum of S. aureus-associated disease seen in our hospital. In order not to bias our collection with known clones of isolates, we studied only methicillin-sensitive S. aureus (MSSA) isolates. Our results confirm that hospital strains of MSSA comprise a heterogeneous population, although certain distinct lineages appear to be more prevalent. Within these lineages, horizontal movement of genetic elements encoding virulence factors is occurring, possibly in vivo. Furthermore, this horizontal movement is not random and may be restricted between certain genetic lineages.
MATERIALS AND METHODS
Isolates.
We characterized 38 strains of MSSA (Table 1). The strains were isolated by the Department of Medical Microbiology at St. George's Hospital and were chosen to reflect the spectrum of hospital S. aureus isolates. No isolates were known to be epidemiologically related, and the isolates comprised strains isolated from different patients on different wards and taken at different times. No strains were part of an outbreak, and none were determined to be from patients with cross infection by our Hospital Infection Control team. The isolates comprised 21 isolates from blood cultures, 9 isolates from sputum samples, 4 nasal isolates, 2 cerebrospinal fluid isolates, 3 lung isolates obtained postmortem from babies, 1 throat isolate obtained postmortem, 1 hip septic arthritis isolate, and 1 sternal osteomyelitis isolate. No isolates were collected from patients with scalded skin syndrome, food poisoning, or toxic shock syndrome. A second isolate from each of four patients was tested to determine the genetic stabilities of strains.
TABLE 1.
Distribution of agr class, core gene RFLPs, accessory gene carriage and RFLPs, 16S rRNA ribotypes and phage group among reference strains and MSSA isolatesa
| Cluster | Strain | Sourceb | agr classc | Core
genes
|
Accessory genes
|
16S rRNA ribotypee | Phage groupf | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rnaIII | sarA | ssp | spa | icaA | fnbA | efb | hlb | hlg | cna | cap5 | cap8 | lukE | splB | tst | sea | seb | sec | sed | seg | seh | sei | ||||||
| 8325-4 | Reference | I | 2.3 + 12 | 21 | 9 | 9.4 | 8 | 2.8 | 4 + 6 | 4 | 2.1 + 6 | — | 4 | — | 6 | 17 | — | — | — | — | — | — | — | — | I | I | |
| SA502A | Reference | II | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 | 4 + 7 | 3.5 + 7.7 | 2.1 + 6 | — | 4 | — | 3.8 | 9.4 | — | — | — | — | 4 | 8.2 | — | 8.2 | I | I | |
| RN8463 | Reference | III | 2.3 + 14 | 9.5 | 7 + 20 | 7.9 | 10 | 3.8 | 5.5 | 6 + 7.7 | 2.1 + 6 | 17 | — | 0.8 + 14 | — | — | 16 | 7.7 + 8.5 | — | — | — | 5.8 | — | 5.8 | II | I | |
| RN4850 | Reference | IV | 1 + 5.5 | 21 | 7 | 10 | 10 | 4.2 | 3.5 | 19 | 2.1 + 6 | 9.4 | — | 0.8 + 14 | 4.5 | 6 | — | — | — | — | — | 2.5 | — | 4 | V | II | |
| agr class III | PM2 | Sputum | III | 2.3 + 14 | 9.5 | 7 + 20 | 7.9 | 10 | 3.8 | 5.5 | 4.6 | 2.1 + 6 | 17 | — | 0.8 + 14 | — | — | 3 | — | — | 4 | — | — | — | — | II | I |
| PM47 | Blood culture | III | 2.3 + 14 | 9.5 | 7 + 20 | 7.9 | 10 | 3.8 | 5.5 | 9 + 19 | 2.1 + 6 | — | — | 0.8 + 14 | — | — | 16 | — | — | — | — | 5.8 | 14 | 5.8 | II | NT | |
| PM116 | Lung post mortem | III | 2.3 + 14 | 9.5 | 7 + 20 | 7.9 | 10 | 3.8 | 5.5 | 6 + 7.7 | 2.1 + 6 | 17 | — | 0.8 + 14 | — | — | 16 | 7.7 + 8.5 | — | — | — | 5.8 | — | 5.8 | II | I | |
| PM122 | Nose | III | 2.3 + 14 | 9.5 | 7 + 20 | 7.9 | 10 | 3.8 | 5.5 | 9 | 2.1 + 6 | 17 | — | 0.8 + 14 | — | — | 16 | — | — | — | — | 5.8 | — | 5.8 | II | I | |
| PM37 | Blood culture | III | 2.3 + 12 | 9.5 | 7 + 20 | 7.9 | 10 | 3.8 | 5.5 | 9 + 19 | 2.1 + 6 | — | — | 0.8 + 14 | — | — | 16 | — | — | — | — | 5.8 | 14 | 5.8 | II | I | |
| PM58 | Sputum | III | 2.3 + 12 | 9.5 | 7 + 20 | 7.9 | 10 | 3.8 | 5.5 | 4.6 | 2.1 + 6 | 17 | — | 0.8 + 14 | — | — | 6 | — | — | 4 | — | 5.8 | — | 5.8 | II | I | |
| SGH-1 | PM13 | Blood culture | I | 2.3 + 7.5 | 17 | 5 | 10 | 15 | 3.8 | 3.5 | 7.7 | 1.2 + 6 | 9.4 | — | 0.8 + 14 | — | — | — | — | — | — | — | 9.4 | — | 9.4 | VII | I |
| PM35 | Blood culture | I | 2.3 + 7.5 | 17 | 5 | 10 | 15 | 3.8 | 3.5 | 6 | 1.2 + 6 | 9.4 | — | 0.8 + 14 | — | — | — | — | — | 4 | — | 9.4 | — | 9.4 | VII | I | |
| PM52 | Blood culture | I | 2.3 + 7.5 | 17 | 5 | 10 | 15 | 3.8 | 3.5 | 6 | 1.2 + 6 | 9.4 | — | 0.8 + 14 | — | — | — | — | 5.5 | — | — | 9.4 | — | 9.4 | VII | I | |
| PM54 | Blood culture | I | 2.3 + 7.5 | 17 | 5 | 10 | 15 | 3.8 | 3.5 | 6 | 1.2 + 6 | 9.4 | — | 0.8 + 14 | — | — | — | — | 5.5 | — | — | 9.4 | — | 9.4 | VII | I | |
| PM83 | Blood culture | I | 2.3 + 7.5 | 17 | 5 | 10 | 15 | 3.8 | 3.5 | 6 | 1.2 + 6 | 9.4 | — | 0.8 + 14 | — | — | — | — | — | — | — | 9.4 | — | 9.4 | VII | I | |
| PM127 | Sputum | I | 2.3 + 7.5 | 17 | 5 | 10 | 15 | 3.8 | 3.5 | 6 | 1.2 + 6 | 9.4 | — | 0.8 + 14 | — | — | — | — | — | 4 | — | 9.4 | — | 9.4 | VII | I | |
| PM94 | Nose | I | 2.3 + 7.5 | 17 | 5 + 7 | 10 | 15 | 3.8 | 3.5 | 6 | 1.2 + 6 | 9.4 | — | 0.8 + 14 | 3.6 | 9.4 | — | — | — | 4 | — | 14 | — | 14 | VII | NT | |
| SGH-2 | PM20 | Blood culture | I | 2.3 + 12 | 21 | 9 | 9.4 | 8 | 2.8 | 4 + 6 | 4 + 7.3 + 7.7 | 2.1 + 6 | — | 4 | — | 6 | 17 | — | 2.5 + 6.7 | — | — | 4 | — | — | — | I | I |
| PM23 | Blood culture | I | 2.3 + 12 | 21 | 9 | 9.4 | 8 | 2.8 | 5.5 | 6 + 21 | 2.1 + 6 | — | 4 | — | 6 | 17 | — | — | — | — | — | — | — | — | I | I | |
| PM80 | Blood culture | I | 2.3 + 12 | 21 | 9 | 9.4 | 8 | 2.8 | 4 + 6 | 7 + 7.7 | 2.1 + 6 | — | 4 | — | 6 | 17 | — | 2.5 + 6.7 | — | — | 4 | — | — | — | I | I | |
| PM82 | Blood culture | I | 2.3 + 12 | 21 | 9 | 9.4 | 8 | 2.8 | 4 + 6 | 6 + 19 | 2.1 + 6 | — | 4 | — | 6 | 17 | — | 22 | — | — | — | — | —— | — | I | III | |
| PM118 | Sputum | I | 2.3 + 12 | 21 | 9 | 9.4 | 8 | 2.8 | 4 + 6 | 6 + 19 | 2.1 + 6 | — | 4 | — | 6 | 17 | — | — | — | — | — | — | — | — | I | I | |
| PM146 | Lung postmortem | I | 2.3 + 12 | 21 | 9 | 9.4 | 8 | 2.8 | 4 + 6 | 4 | 2.1 + 6 | — | 4 | — | 6 | 17 | — | — | — | — | — | — | — | — | I | NT | |
| SGH-3 | PM12 | Blood culture | II | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 2.8 + 3.8 | 4 + 6 | 3.5 + 4 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 9.4 | — | — | — | — | — | — | — | — | I | II |
| PM149 | Sputum | II | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 2.8 + 3.8 | 4 + 6 | 3.5 + 4 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 9.4 | — | — | — | — | — | — | — | — | I | II | |
| PM153 | Throat Swab | II | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 2.8 + 3.8 | 4 + 6 | 3.5 + 4 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 9.4 | — | — | — | — | — | — | — | — | I | II | |
| PM154 | Lung postmortem | II | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 2.8 + 3.8 | 4 + 6 | 3.5 + 4 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 9.4 | — | — | — | — | — | — | — | — | I | II | |
| PM19 | Blood culture | II | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 2.8 | 4 + 6 | 3.5 + 4 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 9.4 | — | — | — | — | — | — | — | — | I | II | |
| PM131 | Nose | II | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 2.8 | 4 + 6 | 3.5 + 4 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 9.4 | — | — | — | — | — | — | — | — | I | II | |
| PM148 | Sputum | II | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 2.8 | 4 + 6 | 3.5 + 4 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 9.4 | — | — | — | — | — | — | — | — | I | II | |
| PM45 | Blood culture | I | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 | 4 | 7 + 7.7 | 2.1 + 6 | 9.4 | — | 0.8 + 14 | 3 | 17 | — | 2.5 + 6.7 | — | — | — | — | — | — | I | NT | |
| PM46 | Blood culture | I | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 | 4 | 7 + 7.7 | 2.1 + 6 | 9.4 | — | 0.8 + 14 | 3 | 17 | — | 2.5 + 6.7 | — | — | — | — | — | — | I | NT | |
| PM99 | Sternal wound swab | I | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 | 4 | 3 + 19 | 2.1 + 6 | 9.4 | 4 | — | 3.8 | 17 | — | — | — | — | — | 8.2 | — | 8.2 | I | V | |
| PM126 | Sputum | I | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 | 4 | 19 | 2.1 + 6 | — | 4 | — | 3.6 | 17 | — | — | — | — | — | 8.2 | — | 8.2 | I | I | |
| PM50 | Blood culture | I | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 + 5 | 4 | 7 + 7.7 | 2.1 + 6 | 9.4 | — | 0.8 + 14 | 3 | 17 | — | 2.5 + 6.7 | — | — | — | — | — | — | I | III | |
| PM139 | Hip tissue | I | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 + 5 | 4 | 9 | 2.1 + 6 | 9.4 | — | — | 3 | 17 | — | — | — | — | — | — | — | — | VIII | III | |
| PM26 | Blood culture | I | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 10 | 3.8 | 7.7 + 19 | 2.1 + 6 | 9.4 | — | 0.8 + 14 | 3.8 | 8.9 | — | — | — | — | — | — | — | — | I | II | |
| PM48 | Blood culture | I | 2.3 + 12 | 21 | 7 | 9.4 | 8 | 3.8 | 4 | 7.7 + 12 | 2.1 + 6 | — | 4 | — | 4.5 | 9.4 | — | — | — | — | — | — | — | — | I | III | |
| PM63 | Sputum | I | 2.3 + 12 | 21 | 7 | 9.4 | 8 | 10 | 4 | 12 + 19 | 2.1 + 6 | 9.4 | 4 | — | 3.8 | 8.9 | — | — | — | — | — | — | — | — | I | II | |
| PM150 | Cerebrospinal fluid | II | 2.3 + 12 | 21 | 7 | 9.4 | 8 | 3.8 | 4 + 7 | 7.7 | 2.1 + 6 | — | 4 | — | 3.8 | 9.4 | — | — | 3 | — | 4 | 8.2 | — | 8.2 | I | III | |
| PM152 | Cerebrospinal fluid | II | 2.3 + 7.5 | 21 | 7 | 9.4 | 6 | 3.8 | 3.5 | 6 | 2.1 + 6 | 9.4 | — | 0.8 + 14 | 3.8 | 9.4 | — | 22 | 3 | — | — | — | — | — | IX | III | |
| PM49 | Blood culture | I | 2.3 + 7.5 | 21 | 7 | 6.6 | 8 | 3.8 | 5.5 | 7.7 + 9 | 2.1 + 6 | — | — | 0.8 + 14 | 3.8 | 15 | — | — | — | — | — | — | — | — | VI | NT | |
| PM16 | Blood culture | I | 2.3 + 7.5 | 21 | 6.5 | 6.6 | 10 | 3.8 | 0.8 + 3.5 | 3.5 + 7.7 | 2.1 + 6 | 6 | 4 | — | — | — | — | — | — | — | — | 8.2 | — | 8.2 | X | III | |
| PM10 | Sputum | I | 2.3 + 12 | 21 | 6.5 | 6.6 | 10 | 3.8 | 3.5 | 3.5 + 7.7 | 2.1 + 6 | 6 | 4 | — | — | — | — | — | — | — | — | 8.2 | — | 8.2 | III | NT | |
| PM1 | Nose | II | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 | 4 + 7 | 7 + 12 | 2.1 + 6 | — | 4 | — | 3.8 | 9.4 | — | 9.5 | — | — | — | 8.2 | — | 8.2 | I | I | |
| PM24 | Blood culture | II | 2.3 + 7.5 | 21 | 7 | 9.4 | 8 | 3.8 | 4 + 7 | 6 | 2.1 + 6 | — | 4 | — | 3.8 | 9.4 | — | — | — | — | — | 8.2 | — | 8.2 | I | I | |
The table summarizes the results of all experiments. The four reference strains are listed first, and then MSSA strains have been listed in cluster groups corresponding to those in Fig. 1. The following pairs of isolates were from the same patients: PM116 and PM122, PM52 and PM54, PM153 and PM154, and PM45 and PM46.
Source of each clinical isolate.
agr class was determined as described in Materials and Methods.
RFLP values are expressed as the size of ClaI fragment hybridizing to each core gene (rnaIII, sarA, ssp, spa, icaA, fnbA, efb, hlb, hlg) or accessory gene (cna, cap5, cap8, lukE, splB, tst, sea, seb, sec, sed, sed, seg, seh, sei), by Southern blotting. Numbers refer to the size of the ClaI fragment hybridizing to each probe in Southern blot (kb). When two bands hybridized to the same probe, both are listed separated by a plus sign; this occurs when there are two copies of the gene or there is a ClaI restriction site within the gene. Clusters (see Fig. 1) are indicated in the first column. —, absence of that gene.
Ribotypes were ascribed by us in this study.
Phage groups were assigned as described in Materials and Methods. NT, nontypeable by phage typing.
Reference strains used for comparison were 8324-5 (agr class I) (provided by S. Foster) and SA502A (class II), RN8463 (class III), and RN4850 (class IV) (all provided by R. P. Novick). The Public Health Laboratory Service (PHLS), Colindale, United Kingdom, provided enterotoxin-carrying strains American strain 578, 96/ST/31504, and 96/ST/31503 as templates for probe amplification. 8325-4 rnaIII::lacZ was provided by S. Foster for use in the characterization of the agr phenotype.
Selection of genes and primer design.
agr class-specific primers were designed for each of classes I, II, and III (Table 2). The primers specific for agr classes II and III spanned the variable region of the agrC gene, whereas the agr class I-specific primer spanned the agrD region. The regulator sarA was investigated, as was the agr response effector, rnaIII. Ten other core genes comprising those for hemolysins, surface binding proteins, and heat shock protein 70 (hla, hlb, hlg, icaA, ssp, spa, sbi, fnbA, efb, and hsp70, respectively) were studied, as were 14 accessory genes comprising those for enterotoxins A through I, toxic shock syndrome toxin 1, leukocidin E, a serine protease-like gene, collagen binding protein, and capsular types 5 and 8 genes (sea, seb, sec, sed, see, seg, seh, sei, tst, lukE, splB, cna, cap5, and cap8, respectively). The genes were chosen because they are known or potential virulence factors and are listed in Table 2. A 16SrRNA probe was used for ribotyping. sea and see primers were predicted to amplify PCR products with 78% nucleotide sequence homology with published sequences (GenBank accession numbers M18970 and M21319), but no other significant homologies (>40%) were predicted.
TABLE 2.
Gene probesa
| Probe | Gene product or function | 5′ primer sequence (5′ to 3′) | 3′ primer sequence (5′ to 3′) | Probe size (bp) | Source(s) of sequenceb | Templatec | Referenced |
|---|---|---|---|---|---|---|---|
| agr I | agrCDB of class I | 5′-CAC TTA TCA TCA AAG AGC C-3′ | 5′-CCA CTA ATT ATA GCT GG-3′ | 351 | X52543 | 8325-4 | 28 |
| agr II | agrC of class II | 5′-GTA GAG CCG TAT TGA TTC C-3′ | 5′-GTA TTT CAT CTC TTT AAG G-3′ | 464 | AF001782 | SA502A | 28 |
| agrIII | agrC of class III | 5′-CTG CAT TTA TTA GTG GAA TAC G-3′ | 5′-GTT TCA TTT CTT TAA GAG-3′ | 558 | EMRSA-16 | RN8463 | 28 |
| rnaIII | Mediator of the agr response | 5′-CAG AGA TGT GAT GGA AAA TAG TTG-3′ | 5′-AAT GAA GTA GAA CAG CAA CGC G-3′ | 824 | X52543 | 8325-4 | 43 |
| sarA | Staphylococcal accessory regulator A | 5′-GGC AAA TGT ATC GAG CAA GAT G-3′ | 5′-GCT TCA GTG ATT CAT TTA TTT ACT C-3′ | 398 | U46541 | 8325-4 | 10 |
| 16SrRNA | 16S rRNA | 5′-GTA GGT GGC AAG CGT TAT CC-3′ | 5′-CGC ACA TCA GCG TCA G-3′ | 228 | Reference 38 | 8325-4 | 45 |
| hsp70 | Heat shock protein 70 | 5′-GAC TTA GGT GGC GGT ACA TTT G-3′ | 5′-GAC AGC TTC TTG TAC TGC-3′ | 468 | D30690 | 8325-4 | 44 |
| ssp | Serine protease (V8) | 5′-GGA GGT TTT TAG ATG AAA GG-3′ | 5′-CGC CAT TGT CTG GAT TAT CAG G-3′ | 988 | Y00356 | 8325-4 | 8 |
| spa | Protein A, Immunoglobulin binding | 5′-GGT GTA GGT ATT GCA TCT G-3′ | 5′-CGA CGT CCA GCT AAT AAC G-3′ | 1303 | U54636 | 8325-4 | 36 |
| sbi | Immunoglobulin M and β2-glycoprotein I binding | 5′-CAC AGA GGA ACA ACG TAA CC-3′ | 5′-GAT TTA GCT AAG TAG CCG-3′ | 957 | AF027155 | 8325-4 | 59 |
| icaA | Intercellular adhesin | 5′-GTG GAT GAA TTA GAA GGC-3′ | 5′-TTA GCG TTG GGT ATT CCC-3′ | 1113 | AF086783 | 8325-4 | 15 |
| fnbA | Fibrinogen binding protein | 5′-CAC AAA CTG CAC AAC CAG C-3′ | 5′-GGA TTT GAT TCC TCA GAG GAC-3′ | 1522 | J04151 | 8325-4 | 51 |
| efb (fib) | Extracellular fibrinogen binding protein | 5′-AAC ATT AGC GGC AAT AGG-3′ | 5′-ATT CGC TCT TGT AAG ACC-3′ | 432 | X72014 | 8325-4 | 13 |
| cna | Collagen binding protein | 5′-AAG CAT TTG CAG CAC GAG-3′ | 5′-ATA TGA CCC ATA GCC TTG TGG-3′ | 740 | M81736 | RN8463 | 40 |
| cap5 | Capsule type 5 | 5′-ATG ACG ATG AGG ATA GCG-3′ | 5′-CTC GGA TAA CAC CTG TTG C-3′ | 881 | U81973 | 8325-4 | 33 |
| cap8 | Capsule type 8 | 5′-ATG ACG ATG AGG ATA GCG-3′ | 5′-CAC CTA ACA TAA GGC AAG-3′ | 1148 | U73374 | RN8463 | 49 |
| lukE | Leukocidin E | 5′-GAC TGA TTG CAC CTT TAG C-3′ | 5′-GCA ATT GAT GAG GCA ACT GAT G-3′ | 953 | Y13225 | 8325-4 | 21 |
| splB | Serine protease-like B gene | 5′-CCA TAT ACT GGT GTA GTT G-3′ | 5′-GGG TGT GGA TAA CCA ATC-3′ | 332 | U60589 | 8325-4 | 46 |
| hla | α-Hemolysin | 5′-CAC GTA TAG TCA GCT CAG-3′ | 5′-CTG AAG AAC GAT CTG TCC-3′ | 909 | X01645 | 8325-4 | 22 |
| hlb | β-hemolysin | 5′-AGT TGG TGC ACT TAC TGA C-3′ | 5′-TAC TAT AGG CTT TGA TTG GG-3′ | 920 | S72497 | 8325-4 | 42 |
| hlg | γ-Hemolysin leukocidin | 5′-CCA ATC CGT TAT TAG AAA ATG C-3′ | 5′-CCA TAG ACG TAG CAA CGG AT-3′ | 937 | X81586 | 8325-4 | 41 |
| tst | Toxic shock syndrome toxin 1 | 5′-ATC GTA AGC CCT TTG TTG-3′ | 5′-GTG GAT CCG TCA TTC ATT G-3′ | 578 | Reference 35, U93688 | RN8463 | 35 |
| sea | Enterotoxin A | 5′-CAT TGC CCT AAC GTT GAC-3′ | 5′-CGA AGG TTC TGT AGA AGT ATG G-3′ | 619 | M18970 | RN8463 | 3 |
| seb | Enterotoxin B | 5′-CTA AAC CAG ATG AGT TGC AC-3′ | 5′-CCA AAT AGT GAC GAG TTA GG-3′ | 489 | M11118 | PM52 | 29 |
| sec | Enterotoxin C | 5′-GTC TGT AGA TAA ATT TTT GGC AC-3′ | 5′-TAT GTC TAG TTC TTG AGC TG-3′ | 367 | X05815 | PM58 | 23 |
| sed | Enterotoxin D | 5′-GTG GTG AAA TAG ATA GGA CTG C-3′ | 5′-ATA TGA AGG TGC TCT GTG G-3′ | 385 | M28521 | SA502A | 2 |
| see | Enterotoxin E | 5′-GCT TTA AGC AAT CTT AGG C-3′ | 5′-CTA TCC ACA AGT TAA TTG GTA C-3′ | 531 | Reference 38 | American strain 578 | 14 |
| seg | Enterotoxin G | 5′-TCT TTA TAC GTC TCC ACC-3′ | 5′-GTC TAT TGT CGA TTG TTA CC-3′ | 326 | Reference 38 | 96/ST/31504 | 39 |
| seh | Enterotoxin H | 5′-AGA GTG ATG AAA TAA GTG G-3′ | 5′-TTT GAA TAC CAT CTA CCC-3′ | 270 | O'Neille | 96/ST/31503 | 53 |
| sei | Enterotoxin I | 5′-CAA CTC GAA TTT TCA ACA GGT AC-3′ | 5′-CAG GCA GTC CAT CTC CTG-3′ | 466 | Reference 38 | 96/ST/31504 | 39 |
| eta | Exfoliative toxin A | 5′-GTA GGA GCT AGT GCA TTT G-3′ | 5′-GCT CTC TAT CAA GAT GAG AC-3′ | 721 | M17357 | RN4850 | 32 |
| etb | Exfoliative toxin B | 5′-CCT TAC CTG TGA TTC CTT TTG-3′ | 5′-ATC AAC CGA ATA GAG TGA AC-3′ | 719 | M17348 | RN4850 | 24 |
| mecA | Methicillin resistance (PBP 2′) | 5′-GTG GAA TTG GCC AAT ACA GG-3′ | 5′-TGG ATA GCA GTA CCT GAG CC-3′ | 575 | Y14051 | PM59 | 48 |
All probes were prepared by PCR amplification (see Materials and Methods) with the primer pairs listed.
All primers were designed for this study by using the sequence from the GenBank accession number listed, unless indicated otherwise. The EMRSA-16 sequence is found at www.sanger.ac.uk/Projects/S_aureus.
The strain from which genomic DNA used as the template for PCR amplification was derived.
Reference to gene description or use in typing.
Designed by G. O'Neill, personal communication.
Southern blotting.
Probes were prepared by PCR amplification. Initial denaturation was for 3 min at 94°C, during which the Taq polymerase was added, followed by 32 cycles of denaturation at 94°C (30 s), annealing at 55°C (30 s), and extension at 72°C (45 to 90 s, determined at 1 min/kb). The probes were then gel purified on a QIAquick mini column (Qiagen), according to the manufacturer's instructions.
S. aureus genomic DNA was obtained after lysis of the cells with lysostaphin (25 μg/ml; Sigma) and extraction with Nucleobond Ax tips (Clontech). DNA was digested with ClaI (Promega) for 6 h at 37°C, electrophoresed on 0.7% (wt/vol) agarose gels, and transferred to nylon membranes for Southern blotting. The membranes were hybridized overnight with probes labeled with horseradish peroxidase (Amersham enhanced chemiluminescence), according to the manufacturer's instructions. The membranes were washed, and the signal was detected on X-ray film. As the signal fades after several hours, sequential probing of the same membrane without stripping was possible, and membranes were reused up to 10 times with no loss of signal. A restriction fragment length polymorphism (RFLP) value for each gene was ascribed by estimating visually the size (in kilobases) of the ClaI fragment hybridizing to the probe by comparison to a bacteriophage λ HindIII ladder. To confirm the robustness of the methods used, chromosomal DNA was extracted from a second subculture of the stored bank for selected isolates (isolates PM2, PM12, PM19, PM80, PM83, PM94, PM116, PM122, PM150, and PM152) and analyzed with a variety of accessory gene probes, as described above; identical results were obtained.
agr class designation.
Reaction with one of the agr-specific probes was used to assign each strain to a class. However, agr class IV strain RN4850 cross-reacted with the agr class I probe on Southern blotting, hybridizing to a ClaI band of 1.4 kb instead of the band of 2.3 kb typical of an agr class I strain. Strains of agr classes I and IV have a high degree of genetic homology (26). The agr class for strains of classes I through III was therefore ascribed by the presence of a 2.3-kb ClaI digestion product that reacted with the class-specific probe, whereas agr class IV strains were denoted by a 1.4-kb ClaI band that reacted with the agr class I-specific probe.
agr class designation was confirmed phenotypically as either class I or not class I by using an rnaIII::lacZ fusion reporter strain in an 8325-4 (agr class I) background (18). This strain was constructed as described by Chan and Foster (9). Test strains were grown for 6 h in brain heart infusion broth, and supernatant containing peptide was prepared as described by Ji et al. (28). The 8325-4 rnaIII::lacZ strain was grown in brain heart infusion broth to an optical density at 600 nm of 1, test supernatant was added, and the cultures were sampled at 2 and 3 h. The P3 promoter activity of the rnaIII::lacZ strain was measured by 4-methylumbelliferyl-β-d-galactopyranoside (MUG) assays (9). Supernatant from agr class I strains enhanced β-galactosidase activity by at least 30% over that for the control, as measured by the amount of luminescence of the MUG product; non-class I strains suppressed MUG production by at least 30% compared to that for the control.
Typing methods.
To confirm that strains were distinguishable, strains were kindly phage typed by the Staphylococcal Reference Laboratory (PHLS). Isolates were assigned to phage group I if they had any reaction, lytic or inhibitory (lack of growth), with phages of group I (phages 29, 52, 52A, 79, 80, 95); as phage group II if they reacted with phages of group II (phages 3A, 3C, 55, 71) and not with other groups; as phage group III if they reacted with phages of group III (phages 6, 42E, 47, 53, 54, 5, 75, 77, 83A, 84, 85, 88A, 90, 83C, 932) and not with phages of group I; and as phage group V if they reacted only with phages 94 and/or 96 (1).
Pulsed-field gel electrophoresis (PFGE) typing of selected strains was also kindly performed by the Staphylococcal Reference Laboratory. This method involves digestion of chromosomal DNA with SmaI and comparison of the size of the resultant bands (54).
Membrane arrays.
Since characterization of an individual strain by Southern blotting for each gene in turn is a lengthy procedure, we developed a rapid method of screening for agr class and the presence of several accessory virulence factors simultaneously using membrane arrays. Each Hybond-N nylon membrane (Amersham) was robotically spotted with 100 ng of DNA of each of the gene probes listed in Table 2 by using a MicroGrid (BioRobotics) instrument. The membrane was sitting on Whatman paper soaked in membrane denaturation buffer (0.5 M NaOH, 1.5 M NaCl). The DNA on the membrane was then neutralized (1.5 M NaCl, 1 M Tris [pH 7.4]) and UV fixed.
A total of 0.3 μg of genomic DNA from each test strain was extracted and digested with Sau3A for 6 h and cleaned on a QIAquick mini column (Qiagen), according to the manufacturer's instructions. DNA was labeled, hybridized, and detected with the ECL kit (Amersham). The resulting pattern of spots (positive or negative for each gene) was compared to the results from the Southern blots.
Statistics.
Fisher's exact test for approximate two-sided probability was used for comparisons of accessory gene distributions. Calculations used 38 strains and omitted the second strain of each pair. The Fisher's exact calculation for phage groups with agr groups used the statistics package Stata 5.0 (Stata Corp, College Station, Tex.).
BURST analysis (http://www.mlst.net/BURST/burst.htm) (16) was performed by using each of seven core gene locus RFLP values for genes rnaIII (including agr class), sarA, ssp, spa, icaA, fnbA, and hlg. Strains were included in a cluster if they had at least five of seven loci identical to at least one other strain (16).
RESULTS
agr class distribution.
By using the agr-specific probes to assign each strain to a class, 23 of 38 isolates (61%) were agr class I, 10 (26%) were agr class II, and 5 (13%) were agr class III (Table 1). No isolate was found to be agr class IV. We confirmed the results of our genotypic assays with a phenotypic assay because the original description of the agr classes was of mutual inhibition of agr function between strains by the agrD-encoded octapeptide. The results of the phenotypic assays ascribed isolates as agr class I or not class I. Our phenotypic results matched those of the genotypic assays, supporting our agr class assignations.
Diversity of S. aureus genes and evidence of clusters.
Of the 12 core genes present in all strains, only 3 were always found on the same-sized ClaI fragment. Probes for hla, sbi, and hsp70 always hybridized to fragments of 0.5 and 0.7 kb (double band), 6 kb, and 5 kb, respectively. In contrast, nine genes (rnaIII, sarA, ssp, spa, icaA, fnbA, efb, hlb, hlg) were found on variably sized ClaI fragments in some strains, and their distributions are shown in Table 1. Hybridization with a 16SrRNA probe (ribotyping) identified 10 different patterns of between three and six bands of between 2 and 10 kb. The distribution of accessory genes is also shown in Table 1. Of the accessory genes, see, eta, etb, and mecA were not carried by any isolate (data not shown).
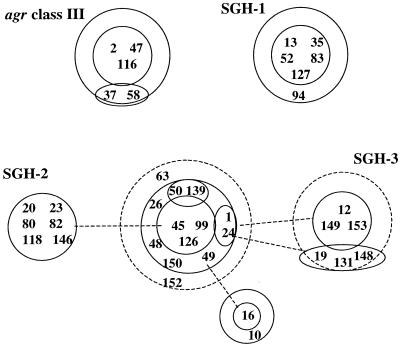
In Table 1 four clusters of isolates, each with unique and virtually identical core RFLP profiles and ribotypes, could be clearly differentiated, and these are illustrated by BURST analysis (Fig. 1). The first of these clusters included all five agr class III isolates. sarA, ssp, and spa were conserved on unique ClaI fragments in these isolates, and the cluster had a unique ribotype (Table 1). We have called the second cluster of six strains the SGH-1 (St. George's Hospital) cluster; it includes strains PM13, PM35, PM52, PM83, PM127, and PM94. The strains carried sarA, ssp, spa, icaA, and hlg on unique and identical ClaI fragments and had a unique ribotype. Another cluster of strains identified within agr class I, comprising strains PM20, PM23, PM80, PM82, PM118, and PM146 (SGH-2), were identical across all core genes. The fourth cluster of isolates identified (SGH-3) comprised strains PM12, PM149, PM153, PM19, PM131, and PM148. These isolates were agr class II and were identical across all core and accessory genes analyzed with the exception of fnbA. Surprisingly, phage typing did not group strains into clusters apart from cluster SGH-3.
FIG. 1.
BURST analysis of core genotypes of MSSA isolates. The relatedness of strains was determined by BURST analysis, based on core gene RFLP values (16). Numbers refer to the MSSA isolate numbers listed in Table 1. Core genotypes were defined by RFLP values for core genes rnaIII (including agr class), sarA, ssp, spa, icaA, fnbA, and hlg. Isolates in each central ring cluster have identical core genomes. Isolates in the first concentric ring have a core genome RFLP fingerprint that differs by one of the seven core gene RFLPs compared to those for isolates in the central ring. Isolates in the second concentric ring (dotted circle) differ by two core gene RFLPs from those for isolates in the central ring. Similarly, the solid lines connect rings of isolates that differ by a single RFLP, whereas dotted lines connect rings of isolates that differ by two RFLPs. At least four clusters of related strains can be identified and are called agr class III, SGH-1, SGH-2, and SGH-3 (Table 1).
Multiple accessory virulence genes may be mobile.
The clusters of S. aureus strains within our collection of hospital MSSA isolates are likely to represent conserved core lineages. However, within the conserved backgrounds of the first three clusters there appeared to be much variation in the carriage of accessory virulence genes. For example, four of the five agr class III strains carried seg and sei, three carried cna, and only two carried sec or seh. Notably, the ClaI fragments carrying cna, seg, and sei were unique to agr class III isolates. Furthermore, tst was carried on three different genetic elements (11, 34, 35) including SaP12, as judged by a tst ClaI fragment of 16 kb. None carried the SaPI1 int (integrase) gene (data not shown).
Of the six SGH-1 strains, one carried lukE and splB, three carried sec, and one carried seb. One isolate had a unique hlb RFLP profile, and the hlb gene is a known insertion site for toxin-carrying phages (12). Strains of SGH-2 varied in their carriage of sea and sed, with different ClaI fragments carrying sea within this group. Variation was seen not only in hlb but also in efb. SGH-3 strains carried few accessory genes, and no variation was seen in this cluster.
Because these results suggested that horizontal transfer of accessory virulence genes occurred frequently, we examined a second isolate of S. aureus from four of the patients to see if transfer might be occurring in the hospital setting (Table 1). One pair of isolates, taken from a baby who died with staphylococcal sepsis, appeared to be identical, except that the nasal strain (PM122) did not carry sea, whereas the lung isolate (PM116) taken 6 days later at postmortem examination was sea positive. The hlb band sizes in these strains also varied, and sea is known to be carried on a phage that inserts into the hlb gene, thus splitting the gene into two ClaI fragments. To confirm that the hlb gene had been disrupted, we amplified hlb by PCR with the primers listed in Table 2. We found that amplification was possible for PM122, which showed the expected band of 0.92 kb, but PM116 hlb could not be amplified. To further investigate these two strains, we compared their PFGE profiles, which showed no band differences, although their phage types differed by two strong lytic and seven inhibitory reactions. We concluded that an sea-carrying phage was inserted into hlb in one of two otherwise identical isolates from a patient.
A second pair of strains (PM153 and PM154) taken from the throat and lung of a baby at postmortem examination could not be differentiated by our RFLP or ribotyping methods but showed variable phage typing patterns, with three strong lytic differences. For the remaining two sets of pairs, neither our methods nor phage typing could differentiate the linked strains.
lukE and splB may be carried on the same genetic element.
We found that the gene encoding splB always co-occurred with lukE. Both were present in 68% of agr class I strains and 100% of agr class II isolates. The genomic sequence of strain 8325 (www.genome.ou.edu/staph) shows that in this strain lukE and splB are 12.6 kb apart. Between these two genes are two ClaI-cut sites, explaining why the RFLP band sizes are different. Downstream of lukE is lukD and then an open reading frame (ORF) with 44% amino acid homology to the Staphylococcus epidermidis epiB gene. epiB and the next six ORFs are predicted to be involved in lantibiotic synthesis (50), a known variable trait among S. aureus isolates. Farther downstream is a series of six serine protease-like genes in tandem, showing homology to splA through splF (GenBank accession number AF271715). The fact that lukE and splB are either present or entirely absent suggests that all these genes may be carried on the same accessory genetic element.
seg and sei are on the same genetic element.
seg and sei always co-occurred and were usually carried on the same-sized ClaI fragment, as expected (25, 27). They are 1.9 kb apart and within a cluster of seven enterotoxin-like genes (27). Downstream (2.2 kb) from seg is an ORF with 49% amino acid homology in the terminal 141 amino acids to a putative Escherichia coli φ27 phage protein (Shiga toxin 2e converting phage; GenBank accession number CAB93761). Downstream is a group of spl homologs similar to those downstream of lukE and splB.
Carriage of genetic elements is influenced by agr class and other genetic elements.
We confirmed the previously reported association between agr class III and tst (P < 0.00001). We were unable to demonstrate a correlation between tst carriage and phage group I strains (P = 0.21). As expected, seb was not seen with tst. We discovered a significant inverse correlation between lukE-splB and tst (P = 0.003), with lukE-splB being found only in tst-negative strains and, therefore, in agr class I and II strains (P = 0.003). We also found a significant negative correlation between lukE-splB and seg-sei, such that they were rarely found in the same strain (P = 0.0005). However, we did not demonstrate a significant correlation between seg-sei and tst (P = 0.28).
In contrast to Ryding et al. (47) and Booth et al. (6), we did not find a correlation between cna and cap8; however, we did find a negative correlation between cna and lukE (P = 0.003).
Membrane arrays.
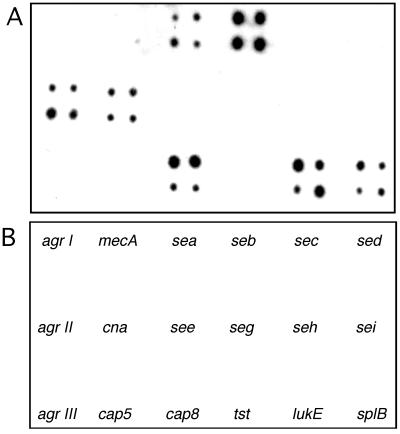
We developed membrane arrays as a rapid method for screening for the presence of several accessory virulence genes as well as agr class simultaneously. The patterns generated with membrane arrays correlated with the Southern blot results exactly, confirming their accuracy (Fig. 2). The entire collection could be characterized in 3 days.
FIG. 2.
Membrane array results for PM152. The array was manufactured as described in Materials and Methods. Each gene was spotted four times. (A) PM152; (B) order of genes spotted onto the membrane. agr I, II, and III refer to the agr class I-, II-, and III-specific probes listed in Table 2, respectively. The results illustrate the ability of membrane arrays to identify differences in accessory gene carriage in different strains. We have no explanation for the variable sizes of the spots on the arrays.
DISCUSSION
Our results demonstrate that hospital strains of MSSA are genetically diverse; however, within this diversity, some strains are clearly related and probably represent successful lineages. These conclusions are based upon the results generated by looking at core genes alone. Altered ClaI fragment sizes are presumably the result of a mutation in a nearby ClaI restriction site or a reorganization such as a deletion or an inversion. In addition, horizontal transfer of DNA between strains and recombinational replacement in the new host occurs in S. aureus (17, 19), which is probably mediated by generalized transducing phages. Therefore, we can predict that strains that differ in the ClaI fragments encoding one core gene are more closely related than strains that differ in the ClaI fragments encoding several core genes. By this approach, it is clear that at least four “clusters” of strains are present in our collection. Furthermore, two of these clusters are quite unrelated to the remaining strains, as they have multiple unique ClaI fragments encoding core genes. These lineages appear to be successful in our hospitals, which could be due to increased virulence. Our results are consistent with the findings of Day et al. (16), who were also able to group their S. aureus strains into related clusters and showed that conserved ancestral clones of isolates were more virulent.
The isolates of one of the successful lineages that we identified were all agr class III. However, van Leeuwen et al. (55) screened a collection of 55 MSSA isolates mostly taken from healthy nasal carriers and did not find any agr class III isolates. Our agr class III cluster may therefore be associated with virulence in hospitals, although since this cluster appears to be genetically distinct from the other isolates, it is not necessarily due to agr class per se. van Leeuwen et al. (55) also reported that only 15% of their strains were agr class II, while 26% of the strains in our collection were agr class II. Therefore, it is possible that agr class II strains are also more frequently associated with disease in hospitals, and this also needs to be investigated further.
Accessory virulence genes appear to be widely distributed in hospital strains, and all our strains carried at least one. Our results by RFLP analysis with ClaI also indicate that the same gene can be carried on a variety of elements; for example, sea can probably be carried on four distinct genetic elements.
More importantly, by focusing on three clusters of strains (SGH-1, SGH-2, and agr class III strains) we can see that the loss and/or acquisition of accessory genes tst, sea, seb, sec, sed, seg-sei, seh, cna, and lukE-splB is occurring in otherwise stable backgrounds. The data strongly suggest that vertical transmission of accessory genes to daughter cells does not account for all of this variation and that horizontal transfer of these genes between strains is occurring.
By looking at pairs of isolates taken from the same patient, we can predict that some of this variation may even be occurring in patients. In one pair of isolates, we have evidence that the sea phage either has been acquired by a nasal isolate that has gone on to cause infection in the lung or has been lost by an isolate residing in the nose. An alternative explanation is that the loss of the sea phage in the nasal isolate occurred during collection, storage, or laboratory growth. Any of these possibilities has important implications. First, if sea phage acquisition occurs in patients in vivo, we need to consider a possible source of this phage, what conditions promote phage acquisition, the frequency at which this occurs, and the effect of the phage on virulence. Alternatively, if phage can be lost in vivo, understanding the mechanism of loss may be beneficial for the treatment of infections. Finally, if phage is easily lost in the laboratory, we may need to reassess the reliability of published data on the frequency and distribution of accessory virulence genes in S. aureus populations. However, we found the sea gene to be stable after storage and subculture of PM116 in vitro (data not shown).
We examined a second isolate from only four patients and found variation in two, which suggests that horizontal transfer or loss of genetic elements may be frequent. The second pair of isolates showed an altered phage susceptibility profile, although we were unable to identify a difference using probes specific for selected accessory genes. Phage profiles are known to alter when a new phage or genetic element is integrated into the bacterial chromosome (4, 35), although the cause of this “phage immunity” is unknown. Furthermore, new S. aureus accessory virulence genes are continuously being described. Therefore, it seems likely that these two strains are closely related and that one has acquired or lost a phage or accessory genetic element, which may or may not carry virulence genes.
Increasing numbers of known and novel virulence genes have been shown to be encoded on mobile genetic elements. Our results suggest that lukE-splB, seg-sei, and seh may also be on genetic elements that can be acquired or lost. Some of these elements and putative elements were not evenly distributed between groups of strains. Vertical transfer could explain the nonrandom distribution of accessory genes; however, we believe that our evidence supports frequent horizontal transfer as well. If so, this suggests that acquisition of a mobile element may be restricted by host background (such as agr class) or the presence of other mobile elements. Mechanisms of restriction are under investigation in our laboratory.
The relatedness of diverse populations of S. aureus strains has been poorly characterized until recently due to a lack of appropriate methods. Epidemiological typing methods, such as phage typing and PFGE, are unsuitable for comparison of strains that are not epidemiologically linked because they rely on a comparison of patterns, while the causes of pattern variation are complex and poorly understood. Furthermore, our results and those of others (52) show that both methods are oversensitive to horizontal movement of mobile elements. Recently, the powerful technique of multilocus sequence typing, which involves the sequencing of seven core housekeeping genes and comparison of nucleotide differences, has been applied to invasive and carriage strains of S. aureus (16, 17). This approach also identified several clusters of related strains. However, since many known and putative virulence genes may be highly mobile, we believe that screening for these genes will enhance any such studies. Our rapid membrane array method is ideally suited for this and can easily be adapted to include more or different combinations of genes. It will also be appropriate for the screening of large numbers of clinically relevant isolates for their carriage of agr-class and genetic elements.
In conclusion, our results illustrate the genetic diversity of hospital MSSA strains and show that horizontal transfer of mobile genetic elements encoding virulence genes occurs frequently, probably in the hospital environment, and possibly in vivo. However, factors in the core genome of the recipient strain or other accessory genetic elements may restrict this transfer. These phenomena need to be investigated further, as they may have significant impact on the evolution and success of S. aureus strains in hospitals, as well as on the treatment of infections. This includes the potential for epidemic hospital-associated strains, including methicillin-resistant S. aureus strains, to acquire accessory virulence genes.
ACKNOWLEDGMENTS
This work was funded by Wellcome Trust Entry Level Fellowship 058051 to P.C.L.M.
We are grateful to H. Aucken and P. Kaufman, PHLS, for phage typing, PFGE typing, and provision of enterotoxin-positive strains. We thank P. D. Butcher, J. A. Mangan, and K. Laing for help with the membrane arrays and S. J. Foster and R. P. Novick for providing strains. We are grateful to M. Bland for advice on statistical analysis.
REFERENCES
- 1.Anderson E S, Williams R E O. Bacteriophage typing of the enteric pathogens and staphylococci and its use in epidemiology. J Clin Pathol. 1956;9:94–127. doi: 10.1136/jcp.9.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayles K W, Iandolo J J. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989;171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betley M J, Mekalanos J J. Nucleotide sequence of the type A staphylococcal enterotoxin gene. J Bacteriol. 1988;170:34–41. doi: 10.1128/jb.170.1.34-41.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betley M J, Mekalanos J J. Staphylococcal enterotoxin A is encoded by phage. Science. 1985;229:185–187. doi: 10.1126/science.3160112. [DOI] [PubMed] [Google Scholar]
- 5.Bohach G A, Fast D J, Nelson R D, Schlievert P M. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit Rev Microbiol. 1990;17:251–272. doi: 10.3109/10408419009105728. [DOI] [PubMed] [Google Scholar]
- 6.Booth M C, Pence L M, Mahasreshti P, Callegan M C, Gilmore M S. Clonal associations among Staphylococcus aureusisolates from various sites of infection. Infect Immun. 2001;69:345–352. doi: 10.1128/IAI.69.1.345-352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce J M. Epidemiology and prevention of nosocomial infections. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 309–329. [Google Scholar]
- 8.Carmona C, Gray G L. Nucleotide sequence of the serine protease gene of Staphylococcus aureus, strain V8. Nucleic Acids Res. 1987;15:6757. doi: 10.1093/nar/15.16.6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan P F, Foster S J. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus8325-4. Microbiology. 1998;144:2469–2479. doi: 10.1099/00221287-144-9-2469. [DOI] [PubMed] [Google Scholar]
- 10.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu M C, Kreiswirth B N, Pattee P A, Novick R P, Melish M E, James J F. Association of toxic shock toxin 1 determinant with a heterologous insertion at multiple loci in the Staphylococcus aureuschromosome. Infect Immun. 1988;56:2702–2708. doi: 10.1128/iai.56.10.2702-2708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman D C, Sullivan D J, Russell R J, Arbuthnott J P, Carey B F, Pomeroy H M. Staphylococcus aureusbacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J Gen Microbiol. 1989;135:1679–1697. doi: 10.1099/00221287-135-6-1679. [DOI] [PubMed] [Google Scholar]
- 13.Colque-Navarro P, Palma M, Soderquist B, Flock J I, Mollby R. Antibody responses in patients with staphylococcal septicemia against two Staphylococcus aureusfibrinogen binding proteins: clumping factor and an extracellular fibrinogen binding protein. Clin Diagn Lab Immunol. 2000;7:14–20. doi: 10.1128/cdli.7.1.14-20.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couch J L, Soltis M T, Betley M J. Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J Bacteriol. 1988;170:2954–2960. doi: 10.1128/jb.170.7.2954-2960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cramton S E, Gerke C, Schnell N F, Nichols W W, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureusand is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Day N P J, Moore C E, Enright M C, Berendt A R, Maynard Smith J, Murphy M F, Peacock S J, Spratt B G, Feil E J. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science. 2001;292:114–116. doi: 10.1126/science.1056495. [DOI] [PubMed] [Google Scholar]
- 17.Enright M C, Day N P, Davies C E, Peacock S J, Spratt B G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairhead H. Ph.D thesis. Sheffield, United Kingdom: University of Sheffield; 1998. [Google Scholar]
- 19.Feil E J, Enright M C, Spratt B G. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res Microbiol. 2000;151:465–469. doi: 10.1016/s0923-2508(00)00168-6. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald J R, Monday S R, Foster T J, Bohach G A, Hartigan P J, Meaney W J, Smyth C J. Characterization of a putative pathogenicity island from bovine Staphylococcus aureusencoding multiple superantigens. J Bacteriol. 2001;183:63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gravet A, Colin D A, Keller D, Girardot R, Monteil H, Prevost G, Giradot R. Characterization of a novel structural member, LukE-LukD, of the bicomponent staphylococcal leucotoxins family. FEBS Lett. 1998;436:202–208. doi: 10.1016/s0014-5793(98)01130-2. [DOI] [PubMed] [Google Scholar]
- 22.Gray G S, Kehoe M. Primary sequence of the alpha-toxin gene from Staphylococcus aureusWood 46. Infect Immun. 1984;46:615–618. doi: 10.1128/iai.46.2.615-618.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hovde C J, Hackett S P, Bohach G A. Nucleotide sequence of the staphylococcal enterotoxin C3 gene: sequence comparison of all three type C staphylococcal enterotoxins. Mol Gen Genet. 1990;220:329–333. doi: 10.1007/BF00260504. [DOI] [PubMed] [Google Scholar]
- 24.Jackson M P, Iandolo J J. Cloning and expression of the exfoliative toxin B gene from Staphylococcus aureus. J Bacteriol. 1986;166:574–580. doi: 10.1128/jb.166.2.574-580.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarraud S, Cozon G, Vandenesch F, Bes M, Etienne J, Lina G. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J Clin Microbiol. 1999;37:2446–2449. doi: 10.1128/jcm.37.8.2446-2449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarraud S, Lyon G J, Figueiredo A M, Gerard L, Vandenesch F, Etienne J, Muir T W, Novick R P. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182:6517–6522. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarraud S, Peyrat M A, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. egc, a highly prevalent operon of enterotoxin genes, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 28.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 29.Johns M B, Jr, Khan S A. Staphylococcal enterotoxin B gene is associated with a discrete genetic element. J Bacteriol. 1988;170:4033–4039. doi: 10.1128/jb.170.9.4033-4039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreiswirth B N, Lofdahl S, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 31.Lan R, Reeves P R. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
- 32.Lee C Y, Schmidt J J, Johnson-Winegar A D, Spero L, Iandolo J J. Sequence determination and comparison of the exfoliative toxin A and toxin B genes from Staphylococcus aureus. J Bacteriol. 1987;169:3904–3909. doi: 10.1128/jb.169.9.3904-3909.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J C, Xu S, Albus A, Livolsi P J. Genetic analysis of type 5 capsular polysaccharide expression by Staphylococcus aureus. J Bacteriol. 1994;176:4883–4889. doi: 10.1128/jb.176.16.4883-4889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindsay J A, Novick R P. Clinical isolates of Staphylococcus aureus encode tst on genetic elements related to S. aureus pathogenicity island-1 (SaPI1) In: Arbuthnott J, Furman B, editors. European conference on toxic shock syndrome. Proceedings of a symposium. London, United Kingdom: The Royal Society of Medicine Press Limited; 1998. pp. 137–139. [Google Scholar]
- 35.Lindsay J A, Ruzin A, Ross H F, Kurepina N, Novick R P. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 36.Lofdahl S, Guss B, Uhlen M, Philipson L, Lindberg M. Gene for staphylococcal protein A. Proc Natl Acad Sci USA. 1983;80:697–701. doi: 10.1073/pnas.80.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick R P, Muir T W. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureusresponsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monday S R, Bohach G A. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munson S H, Tremaine M T, Betley M J, Welch R A. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patti J M, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, Ryden C, Höök M. The Staphylococcus aureuscollagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun. 1994;62:152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prevost G, Cribier B, Couppie P, Petiau P, Supersac G, Finck-Barbancon V, Monteil H, Piemont Y. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureusATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun. 1995;63:4121–4129. doi: 10.1128/iai.63.10.4121-4129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Projan S J, Kornblum J, Kreiswirth B, Moghazeh S L, Eisner W, Novick R P. Nucleotide sequence: the beta-hemolysin gene of Staphylococcus aureus. Nucleic Acids Res. 1989;17:3305. doi: 10.1093/nar/17.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Projan S J, Novick R P. The molecular basis of pathogenicity. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 55–81. [Google Scholar]
- 44.Qoronfleh M W, Bortner C A, Schwartzberg P, Wilkinson B J. Enhanced levels of Staphylococcus aureusstress protein GroEL and DnaK homologs early in infection of human epithelial cells. Infect Immun. 1998;66:3024–3027. doi: 10.1128/iai.66.6.3024-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson J F, Aparicio P, Marples R R, Cookson B D. Ribotyping of Staphylococcus aureus: an assessment using well-defined strains. Epidemiol Infect. 1994;112:93–101. doi: 10.1017/s0950268800057459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rieneck K, Renneberg J, Diamant M, Gutschik E, Bendtzen K. Molecular cloning and expression of a novel Staphylococcus aureusantigen. Biochim Biophys Acta. 1997;1350:128–132. doi: 10.1016/s0167-4781(96)00216-3. [DOI] [PubMed] [Google Scholar]
- 47.Ryding U, Flock J L, Flock M, Soderquist B, Christensson B. Expression of collagen-binding protein and types 5 and 8 capsular polysaccharide in clinical isolates of Staphylococcus aureus. J Infect Dis. 1997;176:1096–1099. doi: 10.1086/516520. [DOI] [PubMed] [Google Scholar]
- 48.Ryffel C, Tesch W, Birch-Machin I, Reynolds P E, Barberis-Maino L, Kayser F H, Berger-Bächi B. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene. 1990;94:137–138. doi: 10.1016/0378-1119(90)90481-6. [DOI] [PubMed] [Google Scholar]
- 49.Sau S, Lee C Y. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J Bacteriol. 1996;178:2118–2126. doi: 10.1128/jb.178.7.2118-2126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnell N, Engelke G, Augustin J, Rosenstein R, Ungermann V, Götz F, Entian K D. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur J Biochem. 1992;204:57–68. doi: 10.1111/j.1432-1033.1992.tb16605.x. [DOI] [PubMed] [Google Scholar]
- 51.Signas C, Raucci G, Jonsson K, Lindgren P E, Anantharamaiah G M, Höök M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smeltzer M S, Hart M E, Iandolo J J. The effect of lysogeny on the genomic organization of Staphylococcus aureus. Gene. 1994;138:51–57. doi: 10.1016/0378-1119(94)90782-x. [DOI] [PubMed] [Google Scholar]
- 53.Su Y C, Wong A C. Identification and purification of a new staphylococcal enterotoxin, H. Appl Environ Microbiol. 1995;61:1438–1443. doi: 10.1128/aem.61.4.1438-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R, Jarvis W R, Kreiswirth B, Eisner W, Maslow J, McDougal L K, Miller J M, Mulligan M, Pfaller M A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Leeuwen W, van Nieuwenhuizen W, Gijzen C, Verbrugh H, van Belkum A. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrDgene, encoding a staphylococcal autoinducer peptide. J Bacteriol. 2000;182:5721–5729. doi: 10.1128/jb.182.20.5721-5729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent J L, Bihari D J, Suter P M, Bruining H A, White J, Nicolas-Chanoin M H, Wolff M, Spencer R C, Hemmer M. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee. JAMA. 1995;274:639–644. [PubMed] [Google Scholar]
- 57.Yamaguchi T, Hayashi T, Takami H, Nakasone K, Ohnishi M, Nakayama K, Yamada S, Komatsuzawa H, Sugai M. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol Microbiol. 2000;38:694–705. doi: 10.1046/j.1365-2958.2000.02169.x. [DOI] [PubMed] [Google Scholar]
- 58.Yoshizawa Y, Sakurada J, Sakurai S, Machida K, Kondo I, Masuda S. An exfoliative toxin A-converting phage isolated from Staphylococcus aureusstrain ZM. Microbiol Immunol. 2000;44:189–191. doi: 10.1111/j.1348-0421.2000.tb02481.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Jacobsson K, Vasi J, Lindberg M, Frykberg L. A second IgG-binding protein in Staphylococcus aureus. Microbiology. 1998;144:985–991. doi: 10.1099/00221287-144-4-985. [DOI] [PubMed] [Google Scholar]