Abstract
Continuous emergence of SARS-CoV-2 variants of concern (VOCs) is fueling the COVID-19 pandemic. Omicron (B.1.1.529) rapidly spread worldwide. The large number of mutations in its Spike raise concerns about a major antigenic drift that could significantly decrease vaccine efficacy and infection-induced immunity. A long interval between BNT162b2 mRNA doses elicits antibodies that efficiently recognize Spikes from different VOCs. Here, we evaluate the recognition of Omicron Spike by plasma from a cohort of SARS-CoV-2 naive and previously infected individuals who received their BNT162b2 mRNA vaccine 16 weeks apart. Omicron Spike is recognized less efficiently than D614G, Alpha, Beta, Gamma, and Delta Spikes. We compare with plasma activity from participants receiving a short (4 weeks) interval regimen. Plasma from individuals of the long-interval cohort recognize and neutralize better the Omicron Spike compared with those who received a short interval. Whether this difference confers any clinical benefit against Omicron remains unknown.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, spike glycoproteins, delayed mRNA vaccine regimen, variants of concern, humoral responses, neutralization, Omicron
Graphical abstract
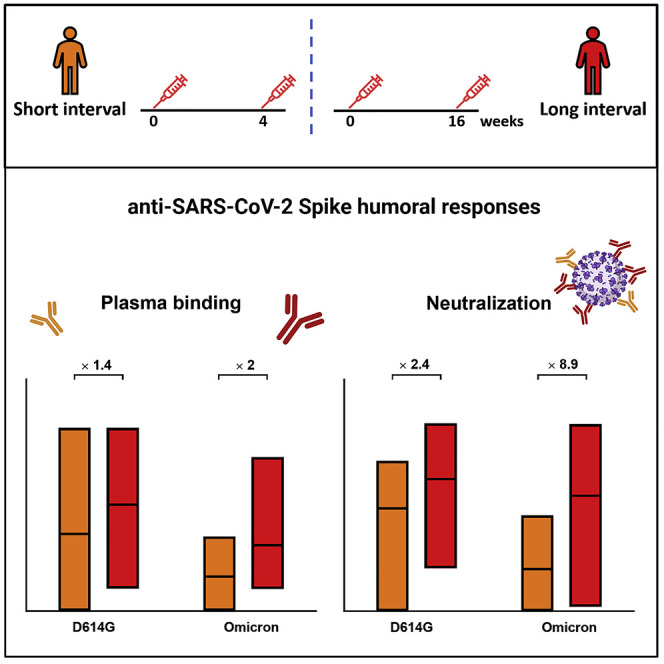
Chatterjee et al. report that extending the interval between doses in naive individuals leads to better recognition and neutralization of the SARS-CoV-2 Omicron Spike than in those receiving a 4-week interval. Vaccinated convalescent individuals present higher responses than naive vaccinated individuals
Introduction
SARS-CoV-2 variants are constantly evolving under immune selective pressure. Ongoing mutational events in the viral genome leads to the emergence of variants with unique properties, including increased transmission capabilities and resistance to antibodies elicited by both natural infection and vaccination. Based on transmission capabilities, virulence, and vaccine effectiveness, SARS-CoV-2 variants are classified as variants of concern (VOCs), variant of interest (VOIs), or variants under monitoring (VUMs) (WHO, 2021). In late 2020, the Alpha (B.1.1.7) variant emerged. The N501Y Spike mutation increased its affinity for the ACE2 receptor, leading to increased transmissibility (Davies et al., 2021; Prevost et al., 2021; Rambaut et al., 2020). The accumulation of E484K and K417N/T mutations along with N501Y in the receptor binding domain (RBD) led to the emergence of Beta (B.1.351) and Gamma (P.1) lineages, which rapidly spread worldwide (Amanat et al., 2021; ECDC, 2021; Tang et al., 2021). In April 2021, the Delta (B.1.617.2) variant emerged and quickly spread to most countries (Allen et al., 2022; Planas et al., 2021b), but is rapidly being replaced by Omicron (B.1.1.529). The World Health Organization designated Omicron as a VOC on November 26, 2021(WHO, 2021). Omicron accumulated more than 30 mutations in its Spike, raising concerns about a major antigenic drift that could significantly decrease vaccine efficacy.
Here we evaluated the recognition of the Omicron Spike by plasma from a cohort of SARS-CoV-2 naive and previously infected individuals who received the two BNT162b2 mRNA vaccine doses 16 weeks apart. We compared these responses with those elicited in individuals receiving a short dose interval regimen (4 weeks). Plasma from vaccinated previously infected individuals recognized more efficiently all tested Spikes (D614G, Alpha, Beta, Gamma, Delta, and Omicron) than those from naive vaccinated individuals. Omicron Spike was recognized less efficiently than D614G, Alpha, Beta, Gamma, and Delta Spikes. However, plasma from individuals receiving a long interval recognized and neutralized better the Omicron Spike compared with those who received a short interval.
Results
Recognition of Spike variants by plasma from vaccinated individuals
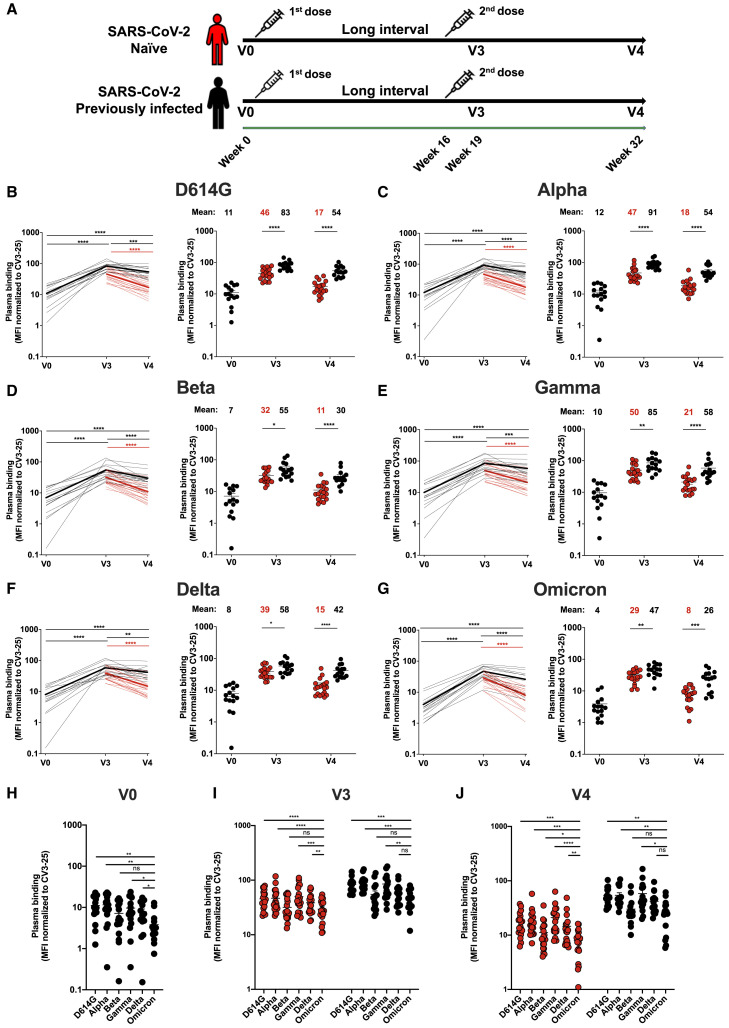
The antigenic profile of D614G, Alpha, Beta, Gamma, Delta, and Omicron Spikes was assessed with plasma collected 3 weeks (V3) and 4 months (V4) after the second dose of the BNT162b2 mRNA vaccine administered with a 16-week interval between doses (Figure 1 A) (Tauzin et al., 2022). Briefly, 293T cells were transfected with plasmids coding for full-length Spike variants. Two days post-transfection, cells were incubated with the indicated plasmas followed by flow cytometry analysis, as described (Anand et al., 2021; Beaudoin-Bussieres et al., 2020; Gasser et al., 2021; Prevost et al., 2020; Tauzin et al., 2021, 2022). Spike expression levels of VOCs were normalized to the signal obtained with the conformationally independent anti-S2 neutralizing CV3-25 antibody (Li et al., 2021; Prevost et al., 2021; Ullah et al., 2021) that efficiently recognized and neutralized all VOCs Spike, including Omicron (Figure S1). Using plasma from previously infected individuals or from naive double-vaccinated individuals, we observed a significant increase of recognition of all tested Spikes upon vaccination (Figures 1B–1G), in agreement with previous observations (Stamatatos et al., 2021; Tauzin et al., 2021, 2022). In all cases, the Omicron Spike was significantly less recognized than all other Spikes, with the exception of the Beta variant (Figures 1D, 1H–1J). At V3 and V4, levels of plasma binding against Omicron Spike in previously infected individuals were similar to those against Delta Spike in naive individuals (Figure 1 I and 1J). In agreement with previous observations (Tauzin et al., 2022), Spike recognition declined more rapidly in the naive group compared with previously infected individuals.
Figure 1.
Binding of vaccine-elicited antibodies to SARS-CoV-2 Spike variants
(A) SARS-CoV-2 vaccine cohort design.
(B–G) 293T cells were transfected with the indicated full-length Spike from different SARS-CoV-2 variants (D614G in B, Alpha in C, Beta in D, Gamma in E, Delta in F, and Omicron in G) and stained with the CV3-25 Ab or with plasma collected 3 weeks (V3) or 4 months (V4) after a second dose administered with a 16-week interval. Samples were analyzed by flow cytometry. The values represent the median fluorescence intensities (MFIs) normalized by CV3-25 Ab binding and presented as percentages of CV3-25 binding (B–G, left panels). Each curve represents the normalized MFIs obtained with the plasma of one donor at every time point. The mean of each group is represented by a bold line. In the right panels, plasma samples were grouped in different time points (V0, V3, and V4).
(H–J) Comparison of Spike recognition by plasma from naive and previously infected donors, represented by red and black points, respectively. Each symbol/points identifies one donor. Error bars indicate means ± SEM. For naive donors, n = 20 at V3 and V4. For previously infected donors vaccinated with two doses, n = 15 at V0 (H), V3 (I), and V4 (J). Statistical significance was tested using (B–G, left panels, (H, I, J) a Wilcoxon test, or (B–G, right panels) a Mann-Whitney test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, non-significant).
Impact of the interval between mRNA vaccine doses on Omicron Spike recognition and neutralization
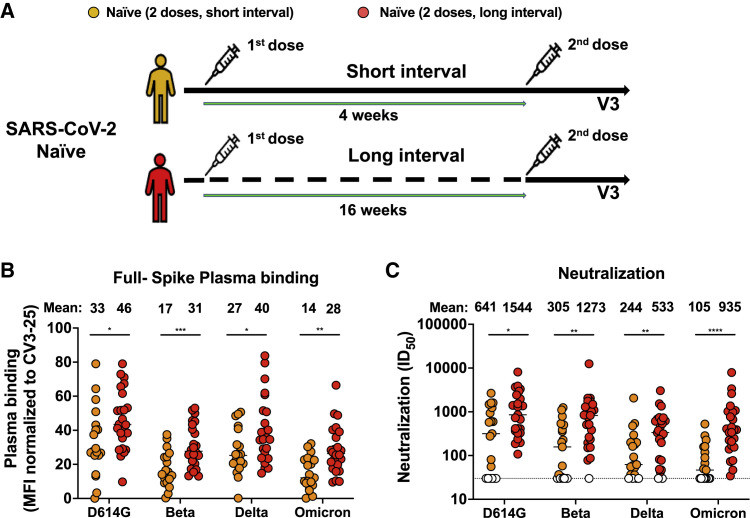
Recent reports suggested that vaccine regimens with a delayed boost elicit stronger humoral responses than the approved, short interval, regimen (Grunau et al., 2021; Tauzin et al., 2022). The long regimen interval has been associated with good vaccine efficacy against different VOCs (Skowronski et al., 2021). We therefore compared the capacity of plasma from naive vaccinated individuals who received the second dose with an interval of 16 weeks (median [range]: 111 days [76–120 days]) with those obtained from 19 SARS-CoV-2 naive donors who received their two doses 4 weeks apart (median [range]: 29 days [22–34 days]) (Table 1 and Figure 2 A). As shown in Figure 2B, plasma from naive individuals who received a 16-week interval between the two doses recognized significantly better all tested Spikes, including Omicron, than plasma from individuals who received a short interval between doses (4 weeks). The 16-week interval regimen elicited significantly better neutralization activity against pseudoviral particles bearing the D614G, Beta, Delta, and Omicron Spikes (Figure 2C). Strikingly, this increased neutralization was more pronounced for the Omicron Spike (8.9-fold increase) compared with the other emerging variant Spikes (D614G, Beta, and Delta) (2.2- to 4.2-fold increase). This suggests that the delayed boosting in naive individuals facilitates antibody maturation resulting in enhanced breadth able to provide detectable levels of recognition and neutralization against Omicron.
Table 1.
Characteristics of the vaccinated SARS-CoV-2 cohorts
| SARS-CoV-2 naive |
SARS-CoV-2 previously infected |
||
|---|---|---|---|
| Two doses Short interval (n = 19) |
Two doses Long interval (n = 25) |
Two doses Long interval (n = 15) |
|
| Age | 39 (20–74) | 50 (21–62) | 47 (29–65) |
| Sex | |||
| Male (n) | 12 | 11 | 10 |
| Female (n) | 7 | 14 | 5 |
| Days between symptom onset and V0a | N/A | N/A | 191 (85–234) |
| Days between symptom onset and the 1st dosea | N/A | N/A | 274 (166–321) |
| Days between the 1st and 2nd dosea | 29 (22–34) | 111 (76–120) | 110 (90–134) |
| Days between the 2nd dose and V3a | 22 (12–53) | 21 (14–34) | 22 (13–51) |
| Days between the 2nd dose and V4a | N/A | 112 (103–125) | 113 (90–127) |
Values displayed are medians, with ranges in parentheses.
Figure 2.
Omicron Spike recognition and neutralization with plasma from naive individuals who received a short versus a long mRNA vaccine dose interval
(A) SARS-CoV-2 vaccine cohort design.
(B) 293T cells were transfected with the full-length Spike from different SARS-CoV-2 variants (D614G, Beta, Delta, and Omicron) and stained with the CV3-25 Ab or with plasma from naive donors who received a short (4 weeks, yellow) or long (16 weeks, red) interval between doses collected 3 weeks after the second dose (V3) and analyzed by flow cytometry. The values represent the MFI normalized by CV3-25 Ab binding and presented as percentages of CV3-25 binding.
(C) Neutralizing activity was measured by incubating pseudoviruses bearing indicated SARS-CoV-2 Spikes (D614G, Beta, Delta, and Omicron), with serial dilutions of plasma for 1 h at 37°C before infecting 293T-ACE2 cells. Neutralization half-maximal inhibitory serum dilution (ID50) values were determined using a normalized non-linear regression using GraphPad Prism software. Undetectable measures are represented as white symbols, and limits of detection are plotted. Error bars indicate means ± SEM. For naive donors vaccinated with the short interval, n = 19. For naive donors vaccinated with the long interval, n = 25. Each symbol/points identifies one donor. Statistical significance was tested using a Mann-Whitney test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
Discussion
In the province of Québec, Canada, like in other jurisdictions worldwide, the prevalence of Omicron increased dramatically from the first case detected on November 23 to being the dominant variant less than 1 month later. Compared with the reference Wuhan-Hu-1 strain, the Omicron variant carries more than 50 non-synonymous mutations within its genome, more than 30 of which are located in the gene coding for the Spike glycoprotein. Several of these mutations affecting the RBD, the N-terminal domain (NTD), and the furin cleavage domain were observed in other VOCs (Viana et al., 2021), which is consistent with positive selection of favorable mutations. Previous in vitro studies already showed the association of some of these mutations with increased infectivity, ACE2 interaction (N501Y, P681H) (Gong et al., 2021; Saito et al., 2021), or immune evasion (K417N, N440K, G446S, S477N, E484A/K, Q493R) (Baum et al., 2020; Clark et al., 2021; Greaney et al., 2021a, 2021b, 2021c; Liu et al., 2020; Rappazzo et al., 2021; Starr et al., 2021; Weisblum et al., 2020). This unprecedented accumulation of Spike mutations raised concern about a major antigenic drift that could significantly decrease the efficacy of current vaccines (Andrews et al., 2021; Khoury et al., 2021; Schmidt et al., 2021b).
To get a better understanding of the antigenic profile, we compared the antigenicity of the Omicron Spike with those from D614G, Alpha, Beta, Gamma, and Delta VOCs. We used plasma from naive and previously infected individuals who received their two doses of the BNT162b2 mRNA vaccine 16 weeks apart. In agreement with previous observations, we found that previously infected vaccinated individuals recognized more efficiently all Spikes than naive individuals at the two timepoints analyzed (3 weeks and 4 months post second dose) (Stamatatos et al., 2021; Tauzin et al., 2021, 2022). Interestingly, we observed that recognition of all Spikes, including Omicron, decreased more rapidly in naive than previously infected individuals, as reported (Tauzin et al., 2022). The three antigenic exposures (infection +2 doses) of previously infected individuals compared with the two exposures in double-vaccinated naive individuals possibly explains their more sustained humoral response, suggesting that an additional exposition to the Spike antigen in the form of a third vaccine dose could elicit similar responses. Alternatively, if infection elicits a qualitatively broader humoral response linked to epitopes located outside the Spike glycoprotein, the effect of a third dose in naive individuals may remain qualitatively different. Independently of their infection history, all plasma recognized significantly less efficiently the Omicron Spike compared with Spikes from other VOCs (Figure 1). Of note, the sequence initially released for the Omicron Spike contained the Q493K substitution, but was then corrected to Q493R. Since initial studies on Omicron used the Q493K mutation, we verified whether the nature of the residue at 493 (either K or R) impacted plasma recognition or neutralization. We observed no significant differences among them (Figure S2). As indicated in the STAR methods section, all results generated in the current manuscript were done using the Q493R mutation.
Since recent studies have shown that recognition of full-length Spikes at the surface of transfected 293T cells strongly correlates with recognition of primary airway epithelial cells infected with authentic viruses as well as with antibody-dependent cellular cytotoxicity (Ding et al., 2022), these results suggest that Fc-mediated effector functions against Omicron could also be affected. Of note, low Spike recognition translated into increased Omicron neutralization resistance (Figure 2C). In agreement with previous observations (Stamatatos et al., 2021; Tauzin et al., 2021, 2022), plasma from vaccinated previously infected individuals recognized more efficiently Omicron and all other VOCs than vaccinated naive individuals (Figure 1). As naive double-vaccinated individuals have been well protected against the Delta variant, the observation of similar levels of plasma binding against Delta Spike in naive individuals and those against Omicron Spike in previously infected individuals may be important. This suggests that the benefits of hybrid immunity also apply to Omicron but this hypothesis will need confirmation through vaccine effectiveness studies.
Several reports (Cele et al., 2021; Garcia-Beltran et al., 2021; Planas et al., 2021a; Schmidt et al., 2021a; Zhang et al., 2021) have shown neutralization resistance using plasma from naive donors who received the approved regimen of the BNT162b2 mRNA vaccine (3- to 4-week vaccine interval). Strikingly, we observed that plasma from naive vaccinated donors who received their two doses according to the approved short 3- to 4-week interval, recognized and neutralized Omicron significantly less efficiently compared with the long 16-week interval. For all individuals, the level of Omicron Spike recognition remains lower than for the ancestral Spike, the antigen used in the current vaccines, and these levels decrease over time. Therefore, it will be important to determine in epidemiological studies if the vaccine interval advantage, as measured by these in vitro parameters, confers any clinical benefit against Omicron.
Limitations of the study
A limitation of our study is the relatively low number of individuals analyzed; however, we note that our results are in agreement with recent findings indicating that longer mRNA vaccine dosing intervals have improved immunogenicity (Grunau et al., 2021; Tauzin et al., 2022), which may have been associated with an optimized booster dose protection in Canada (Skowronski et al., 2021). Epidemiological studies will be required to establish the vaccine efficacy of the extended interval dosing against severe outcomes caused by Omicron.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LIVE-DEAD Fixable AquaVivid Cell Stain | Thermo Fischer Scientific | Cat# P34957 |
| CV3-25 | (Jennewein et al., 2021) | N/A |
| Alexa Fluor 647 AffiniPure Goat Anti-Human IgA + IgG + IgM (H+L) | Jackson ImmunoResearch | Cat # 109-605-064; RRID: AB_2337886 |
| Goat anti-Human IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat # A-21445; RRID: AB_2535862 |
| Biological samples | ||
| SARS-CoV-2 naïve donor blood samples | This paper | N/A |
| SARS-CoV-2 previously infected donor blood samples | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco's Modified Eagle's medium (DMEM) | Wisent | Cat# 319-005-CL |
| Penicillin/Streptomycin | Wisent | Cat# 450-201-EL |
| Fetal Bovine Serum (FBS) | VWR | Cat# 97068-085 |
| Phosphate Buffered Saline (PBS) | ThermoFischer Scientific | Cat# 10010023 |
| Puromycin Dihydrochloride | Millipore Sigma | Cat# P8833 |
| Passive lysis buffer | Promega | Cat # E1941 |
| D-Luciferin Potassium Salt | Prolume | Cat # 306 |
| Formaldehyde 37% | Thermo Fischer Scientific | Cat# F79-500 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat # D2650-5X5ML CAS: 67-68-5 |
| Experimental models: cell lines | ||
| HEK293T cells | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| 293T-ACE2 cells | (Prevost et al., 2020) | N/A |
| Recombinant DNA | ||
| pNL4.3 R-E− Luc | NIH AIDS reagent program | Cat# 3418 |
| pIRES2-EGFP | Clontech | Cat# 6029-1 |
| pCG1-SARS-CoV-2 D614G-Spike | (Beaudoin-Bussieres et al., 2020) | N/A |
| pCDNA3.1-SARS-CoV-2-B.1.1.7 Spike | (Tauzin et al., 2021) | N/A |
| pcDNA3.1-SARS-CoV-2-B.1.351 Spike | (Gong et al., 2021) | N/A |
| pCAGGS-SARS-CoV-2-B.1.617.2 Spike | (Gong et al., 2021) | N/A |
| pcDNA3.1-SARS-CoV-2-P.1 Spike | (Gong et al., 2021) | N/A |
| pCAGGS-SARS-CoV-2 B.1.1.529 Spike | This paper | N/A |
| Software and algorithms | ||
| Flow Jo v10.7.1 | Flow Jo | https://www.flowjo.com |
| GraphPad Prism v8.4.3 | GraphPad | https://www.graphpad.com |
| Microsoft Excel v16 | Microsoft Office | https://www.microsoft.com/en-ca/microsoft-365/excel |
| Others | ||
| BD LSRII Flow Cytometer | BD Biosciences | N/A |
| TriStar LB942 Microplate Reader | Berthold Technologies | N/A |
| Nanodrop Spectrophotometer ND-1000 | ThermoFisher Scientific | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrés Finzi (andres.finzi@umontreal.ca).
Materials availability
All unique reagents generated during this study are available from the lead contact without restriction.
Experimental model and subject details
Ethics statement
All work was conducted in accordance with the Declaration of Helsinki in terms of informed consent and approval by an appropriate institutional board. Blood samples were obtained from donors who consented to participate in this research project at CHUM (19.381) and from plasma donors who consented to participate in the Plasma Donor Biobank at Hema-Quebec (PLASCOV; REB-B-6-002-2021-003). Plasma was isolated by centrifugation, and samples stored at −80°C and in liquid nitrogen, respectively, until use.
Human subjects
The study was conducted in 25 SARS-CoV-2 naïve individuals (11 males and 14 females; age range: 21–62 years) vaccinated with a long interval, 19 SARS-CoV-2 naïve individuals (12 males and 7 females; age range: 20–74 years) vaccinated with a short interval and 15 SARS-CoV-2 previously-infected individuals (10 males and 5 females; age range: 29–65 years) vaccinated with a long interval. All this information is summarized in Table 1. No specific criteria such as number of patients (sample size), gender, clinical or demographic were used for inclusion, beyond PCR confirmed SARS-CoV-2 infection in adults.
Plasma and antibodies
Plasma from SARS-CoV-2 naïve and previously-infected donors were collected, heat-inactivated for 1 hour at 56°C and stored at −80°C until ready to use in subsequent experiments. The conformationally independent S2-specific monoclonal antibody CV3-25 (Gong et al., 2021; Jennewein et al., 2021; Li et al., 2021; Prevost et al., 2021; Ullah et al., 2021) was used as a positive control and to normalize Spike expression in our flow cytometry assays, as described (Gong et al., 2021; Tauzin et al., 2021, 2022). Alexa Fluor-647-conjugated goat anti-human Abs (Invitrogen) were used as secondary antibodies to detect plasma binding in flow cytometry experiments.
Cell lines
293T human embryonic kidney cells (obtained from ATCC) were maintained at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Wisent) containing 5% fetal bovine serum (FBS) (VWR) and 100 μg/mL of penicillin-streptomycin (Wisent). The 293T-ACE2 cell line was previously reported (Prevost et al., 2020).
Method details
Plasmids
The plasmids encoding the SARS-CoV-2 Spike variants; D614G, B.1.1.7, B.1.351, P.1 and B.1.617.2 were previously described (Beaudoin-Bussieres et al., 2020; Gong et al., 2021; Li et al., 2021; Tauzin et al., 2021, 2022). The plasmids encoding the B.1.1.529 Spike was generated by overlapping PCR using a codon-optimized wild-type SARS-CoV-2 Spike gene (GeneArt, ThermoFisher) that was synthesized (Biobasic) and cloned in pCAGGS as a template. The B.1.1.529 Spike coding sequence was derived from the sequence ID EPI_ISL_6640919. This sequence initially contained the Q493K substitution, as previously reported (Cameroni et al., 2021; Schmidt et al., 2021a; Shah and Woo, 2021). The ECDC (European Centre for Disease Prevention and Control) later informed that Omicron spike actually have an R mutation at position 493. We therefore generated and used an Omicron Spike bearing the Q493R mutation for the full manuscript (Figures 1 and 2 and Figure S1). Nevertheless, we compared whether the nature of the residue (either K or R) at this position impacted plasma recognition and/or neutralization activity; no significant differences were observed (Figure S2).
Cell surface staining and flow cytometry analysis
293T were transfected with full-length SARS-CoV-2 Spikes and a green fluorescent protein (GFP) expressor (pIRES2-eGFP; Clontech) using the calcium-phosphate method. Two days post-transfection, Spike-expressing 293T cells were stained with the CV3-25 Ab (5 μg/mL) as control or plasma from vaccinated individuals (1:250 dilution) for 45 min at 37°C. AlexaFluor-647-conjugated goat anti-human IgG (1/1000 dilution) were used as secondary Abs. The percentage of Spike-expressing cells (GFP + cells) was determined by gating the living cell population based on viability dye staining (Aqua Vivid, Invitrogen). Samples were acquired on a LSR II cytometer (BD Biosciences), and data analysis was performed using FlowJo v10.7.1 (Tree Star). The conformationally-independent anti-S2 antibody CV3-25 was used to normalize Spike expression, as reported (Gong et al., 2021; Li et al., 2021; Prevost et al., 2021; Ullah et al., 2021). CV3-25 was shown to be effective against all Spike variants (Gong et al., 2021; Li et al., 2021; Prevost et al., 2021; Ullah et al., 2021) and (Figure S1). The Median Fluorescence intensities (MFI) obtained with plasma were normalized to the MFI obtained with CV3-25 (Gong et al., 2021; Li et al., 2021; Prevost et al., 2021; Ullah et al., 2021) and presented as percentage of CV3-25 binding.
Virus neutralization assay
To produce SARS-CoV-2 pseudoviruses, 293T cells were transfected with the lentiviral vector pNL4.3 R-E− Luc (NIH AIDS Reagent Program) and a plasmid encoding for the indicated S glycoprotein (D614G, Alpha, Beta, Gamma, Delta or Omicron) at a ratio of 10:1. Two days post-transfection, cell supernatants were harvested and stored at −80°C until use. For the neutralization assay, 293T-ACE2 target cells were seeded at a density of 1×104 cells/well in 96-well luminometer-compatible tissue culture plates (PerkinElmer) 24h before infection. Pseudoviral particles were incubated with several plasma dilutions (1/50; 1/250; 1/1250; 1/6250; 1/31250) for 1h at 37°C and were then added to the target cells followed by incubation for 48h at 37°C. For CV3-25 neutralization, pseudoviral particles were incubated with increasing concentrations of CV3-25 (0.01, 0.0316, 0.1, 0.316, 1 and 3.16 μg/mL) for 1h at 37°C and were then added to the target cells followed by incubation for 48h at 37°C. Cells were lysed by the addition of 30 μL of passive lysis buffer (Promega) followed by one freeze-thaw cycle. An LB942 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15mM MgSO4, 15mM KH2PO4 [pH 7.8], 1mM ATP, and 1mM dithiothreitol) and 50 μL of 1mM d-luciferin potassium salt (Prolume). The neutralization half-maximal inhibitory dilution (ID50) represents the plasma dilution to inhibit 50% of the infection of 293T-ACE2 cells by pseudoviruses.
Quantification and statistical analysis
Statistical analysis
Symbols represent biologically independent samples from SARS-CoV-2 naïve or PI individuals. Lines connect data from the same donor. Statistics were analyzed using GraphPad Prism version 8.0.1 (GraphPad, San Diego, CA). Every dataset was tested for statistical normality and this information was used to apply the appropriate (parametric or nonparametric) statistical test. p values < 0.05 were considered significant; significance values are indicated as ∗P <0.05, ∗∗P<0.01, ∗∗∗P<0.001, ∗∗∗∗ P<0.0001, ns, non-significant.
Acknowledgments
The authors are grateful to the donors who participated in this study. The authors thank the CRCHUM BSL3 and Flow Cytometry Platforms for technical assistance. We thank Dr. Stefan Pöhlmann (Georg-August University, Germany) for the plasmid coding for SARS-CoV-2 Spike Wuhan-Hu-1 strain. We also thank Amélie Boivin and Yves Grégoire at Héma-Québec for helping to access the samples from the PLASCOV Biobank and all the plasma donors who participate in this biobank. This work was supported by le Ministère de l’Économie et de l’Innovation du Québec, Programme de soutien aux organismes de recherche et d’innovation to A.F. and by the Fondation du CHUM. This work was also supported by a Canadian Institutes of Health Research (CIHR) foundation grant #352417, by a CIHR operating Pandemic and Health Emergencies Research grant #177958, by a CIHR stream 1 and 2 for SARS-CoV-2 Variant Research to A.F., and by an Exceptional Fund COVID-19 from the Canada Foundation for Innovation (CFI) #41027 to A.F. and D.E.K. Work on variants presented was also supported by the Sentinelle COVID Quebec network led by the LSPQ in collaboration with Fonds de Recherche du Québec Santé (FRQS) to A.F. This work was also partially supported by a CIHR COVID-19 rapid response grant (OV3 170632) and CIHR stream 1 SARS-CoV-2 Variant Research to M.C. A.F. is the recipient of Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424. M.C. is a Tier II Canada Research Chair in Molecular Virology and Antiviral Therapeutics. V.M.L. is supported by a FRQS Junior 1 salary award. C.T. is the recipient of the Pfizer/Université de Montréal Chair on HIV translational research. D.E.K. is a FRQS Merit Research Scholar. G.B.B. is the recipient of a FRQS PhD fellowship, and J.P. is the recipient of a CIHR PhD fellowship. A.L. was supported by MITACS Accélération postdoctoral fellowships. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We declare no competing interests.
Author contributions
D.C., J.R., S.M., M.C., and A.F. conceived the study. D.C., A.T., J.R., M.C., and A.F. designed experimental approaches. D.C., A.T., L.M., S.Y.G., M.B., C.B., G.B.-B., Y.B., S.D., A.L., D.V., E.F., A.G., B.D., I.L., G.G., G.G.-L., H.M., J.Prévost, J.R., M.C., and A.F. performed, analyzed, and interpreted the experiments. J.Perreault, L.G., C.M., P.A., C.T., V.M.-L., D.E.K., and R.B. collected and provided clinical samples. G.D.S., C.T., V.M.-L., and D.E.K. provided scientific input related to VOCs and vaccine efficacy. D.C., J.R., and A.F. wrote the manuscript with inputs from others. Every author has read, edited, and approved the final manuscript.
Declaration of interests
A.F. has filed a provisional patent application on the anti-Spike monoclonal antibody CV3-25.
Published: February 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110429.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.
References
- Allen H., Vusirikala A., Flannagan J., Twohig K.A., Zaidi A., Chudasama D., Lamagni T., Groves N., Turner C., Rawlinson C., et al. Household transmission of COVID-19 cases associated with SARS-CoV-2 delta variant (B.1.617.2): national case-control study. Lancet Reg. Health Eur. 2022;12:100252. doi: 10.1016/j.lanepe.2021.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Thapa M., Lei T., Ahmed S.M.S., Adelsberg D.C., Carreño J.M., Strohmeier S., Schmitz A.J., Zafar S., Zhou J.Q., et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell. 2021;184:3936–3948.e10. doi: 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S.P., Prevost J., Nayrac M., Beaudoin-Bussieres G., Benlarbi M., Gasser R., Brassard N., Laumaea A., Gong S.Y., Bourassa C., et al. Longitudinal analysis of humoral immunity against SARS-CoV-2 Spike in convalescent individuals up to 8 months post-symptom onset. Cell Rep. Med. 2021;2:100290. doi: 10.1016/j.xcrm.2021.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., et al. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. Preprint at medRxiv. 2021 doi: 10.1101/2021.12.14.21267615. [DOI] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussieres G., Laumaea A., Anand S.P., Prevost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A., et al. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio. 2020;11:e02590-20. doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Saliba C., Bowen J.E., Rosen L.E., Culap K., Pinto D., De Marco A., Zepeda S.K., di Iulio J., Zatta F., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Preprint at bioRxiv. 2021 doi: 10.1101/2021.12.12.472269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khan K., Khoury D.S., Moyo-Gwete T., Tegally H., Scheepers C., Amoako D., Karim F., Bernstein M., et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. Preprint at medRxiv. 2021 doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617 e2618. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Adam D., Beaudoin-Bussières G., Tauzin A., Gong S.Y., Gasser R., Laumaea A., Anand S.P., Privé A., Bourassa C., et al. SARS-CoV-2 spike expression at the surface of infected primary human airway epithelial cells. Viruses. 2022;14:5. doi: 10.3390/v14010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC . 2021. Risk of spread of new SARS-CoV-2 variants of concern in the EU/EEA - first update.https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Preprint at medRxiv. 2021 doi: 10.1101/2021.12.14.21267755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R., Cloutier M., Prevost J., Fink C., Ducas E., Ding S., Dussault N., Landry P., Tremblay T., Laforce-Lavoie A., et al. Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Cell Rep. 2021;34:108790. doi: 10.1016/j.celrep.2021.108790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S.Y., Chatterjee D., Richard J., Prévost J., Tauzin A., Gasser R., Bo Y., Vézina D., Goyette G., Gendron-Lepage G., et al. Contribution of single mutations to selected SARS-CoV-2 emerging variants Spike antigenicity. Virology. 2021;563:134–145. doi: 10.1016/j.virol.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476 e466. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57 e49. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau B., Goldfarb D.M., Asamoah-Boaheng M., Golding L., Kirkham T.L., Demers P.A., Lavoie P.M. Immunogenicity of extended mRNA SARS-CoV-2 vaccine dosing intervals. JAMA. 2021;327:279–281. doi: 10.1001/jama.2021.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein M.F., MacCamy A.J., Akins N.R., Feng J., Homad L.J., Hurlburt N.K., Seydoux E., Wan Y.H., Stuart A.B., Edara V.V., et al. Isolation and characterization of cross-neutralizing coronavirus antibodies from COVID-19+ subjects. Cell Rep. 2021;36:109353. doi: 10.1016/j.celrep.2021.109353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Steain M., Triccas J.A., Sigal A., Davenport M.P., Cromer D. A meta-analysis of early results to predict vaccine efficacy against Omicron. Preprint at medRxiv. 2021 doi: 10.1101/2021.12.13.21267748. [DOI] [Google Scholar]
- Li W., Chen Y., Prevost J., Ullah I., Lu M., Gong S.Y., Tauzin A., Gasser R., Vezina D., Anand S.P., et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 2021;S2211-1247:01714–01719. doi: 10.1016/j.celrep.2021.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. Preprint at bioRxiv. 2021 doi: 10.1101/2021.12.14.472630. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Prevost J., Gasser R., Beaudoin-Bussieres G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S., et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost J., Richard J., Gasser R., Ding S., Fage C., Anand S.P., Adam D., Gupta Vergara N., Tauzin A., Benlarbi M., et al. Impact of temperature on the affinity of SARS-CoV-2 Spike glycoprotein for host ACE2. J. Biol. Chem. 2021;297:101151. doi: 10.1016/j.jbc.2021.101151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., et al. SARS-CoV-2 spike P681R mutation, a hallmark of the Delta variant, enhances viral fusogenicity and pathogenicity. Preprint at bioRxiv. 2021 doi: 10.1101/2021.06.17.448820. [DOI] [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., Silva J.D., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization properties of the SARS-CoV-2 Omicron variant. Preprint at medRxiv. 2021 doi: 10.1101/2021.12.12.21267646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Rutkowska M., Poston D., DaSilva J., Zhang F., Bednarski E., Cho A., Schaefer-Babajew D.J., Gaebler C., et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature. 2021;600:512–516. doi: 10.1038/s41586-021-04005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Woo H.G. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. Preprint at bioRxiv. 2021 doi: 10.1101/2021.12.04.471200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski D.M., Setayeshgar S., Febriani Y., Ouakki M., Zou M., Talbot D., Prystajecky N., Tyson J.R., Gilca R., Brousseau N., et al. Two-dose SARS-CoV-2 vaccine effectiveness with mixed schedules and extended dosing intervals: test-negative design studies from British Columbia and Quebec, Canada. Preprint at medRxiv. 2021 doi: 10.1101/2021.10.26.21265397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021:eabg9175. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Toovey O.T.R., Harvey K.N., Hui D.D.S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. 2021;82:e8–e10. doi: 10.1016/j.jinf.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Gong S.Y., Beaudoin-Bussieres G., Vezina D., Gasser R., Nault L., Marchitto L., Benlarbi M., Chatterjee D., Nayrac M., et al. Strong humoral immune responses against SARS-CoV-2 Spike after BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Host Microbe. 2022;30:97–109 e105. doi: 10.1016/j.chom.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussieres G., Brassard N., Laumaea A., Vezina D., Prevost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150 e1136. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I., Prevost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussieres G., Symmes K., Benlarbi M., Ding S., et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158 e2115. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Gaseitsiwe S., von Gottberg A.V., de Oliveira T. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Preprint at medRxiv. 2021 doi: 10.1101/2021.12.19.21268028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. Tracking SARS-CoV-2 Variants.https://www.who.int/en/activities/tracking-SARS-CoV-2-variants [Google Scholar]
- Zhang L., Li Q., Liang Z., Li T., Liu S., Cui Q., Nie J., Wu Q., Qu X., Huang W., et al. The significant immune escape of pseudotyped SARS-CoV-2 Variant Omicron. Emerg. Microbes Infect. 2021:1–11. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.