Abstract
SARS-CoV-2 variants of concern have emerged since the COVID-19 outburst, notably the lineages detected in the UK, South Africa, and Brazil. Their increased transmissibility and higher viral load put them in the spotlight. Much has been investigated on the ability of those new variants to evade antibody recognition. However, little attention has been given to pre-existing and induced SARS-CoV-2-specific CD8+ T cell responses by new lineages. In this work, we predicted SARS-CoV-2-specific CD8+ T cell epitopes from the main variants of concern and their potential to trigger or hinder CD8+ T cell response by using HLA binding and TCR reactivity in silico predictions. Also, we estimated the population's coverage for different lineages, which accounts for the ability to present a set of peptides based on the most frequent HLA alleles of a given population. We considered binding predictions to 110 ccClass I HLA alleles from 29 countries to investigate differences in the fraction of individuals expected to respond to a given epitope set from new and previous lineages. We observed a higher population coverage for the variant detected in the UK (B.1.1.7), and South Africa (B.1.351), as well as for the Brazilian P.1 lineage, but not P.2, compared to the reference lineage. Moreover, individual mutations such as Spike N501Y and Nucleocapsid D138Y were predicted to have an overall stronger affinity through HLA-I than the reference sequence while Spike E484K shows signs of evasion. In summary, we provided evidence for the existence of potentially immunogenic and conserved epitopes across new SARS-CoV-2 variants, but also mutant peptides exhibiting diminished or abolished HLA-I binding. It also highlights the augmented population coverage for three new lineages. Whether these changes imply more T cell reactivity or potential to evade from CD8+ T cell responses requires experimental verification.
Keywords: SARS-CoV-2, Lineages, Variants, CD8+ T cells, Class I HLA, Binding prediction
Abbreviations: aa, Amino acid; HLA, Human Leukocyte Antigen; IFN-γ, Interferon gamma; IL-2, Interleukin-2; MERS, Middle East respiratory syndrome; N, Nucleocapsid; nsSNVs, Non-synonymous single-nucleotide variants; RBD, Receptor-Binding Domain; REF, Reference lineage; S, Spike; SARS, Severe acute respiratory syndrome; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; SB, Strong Binder; TCR, T cell receptor; TNF-ɑ, Tumor necrosis factor alpha; VOI, Variant of interest; WB, Weak binder; COVID-19, Coronavirus disease of 2019
1. Introduction
Since the COVID-19 (Coronavirus disease of 2019) outburst, researchers are struggling in the entire world to better understand why the disease manifests in many different ways, generating a vast range of symptoms. During the last months, new SARS-CoV-2 lineages were detected in the United Kingdom, South Africa, and Brazil (CDC, 2020). Some of these new mutations are of global interest since they seem to increase SARS-CoV-2 transmission (Ozono et al., 2021; Volz et al., 2021). Most variants of concern have multiple mutations in the S and N gene (Hodcroft, 2021) which codes for immunodominant CD8+ T cell peptides (Grifoni et al., 2020; Schreibing et al., 2021; Tarke et al., 2021a). The emergence of these new lineages raised concerns about the reinfection of convalescent individuals as well as the effectiveness of available vaccines. Indeed, mRNA vaccine-elicited antibodies showed a reduced plasma neutralizing activity against new lineages (Wang et al., 2021). Consistently, Brazilian P.1 lineage seems to escape from neutralizing antibodies generated against previously circulating variants of SARS-CoV-2 as well as inactivated virus SARS-CoV-2 vaccine-elicited antibodies (de Souza et al., 2021).
Interestingly, although neutralizing antibody titers and memory B cell responses can be long-lived in some human coronavirus infections, many studies have evidenced the role of T cell response to SARS-CoV-2 (Braun et al., 2020; Sekine et al., 2020; Tarke et al., 2021a). SARS-CoV-2-specific CD8+ T cells from individuals with milder COVID-19 have higher perforin and granzyme activity than convalescent ones, showing an increased CD8+ T effector phenotype during the acute mild disease (Sekine et al., 2020). Of note, the cytotoxic activity is also increased in CD4+ T cells responding to SARS-CoV-2 (Meckiff et al., 2020).
Moreover, convalescent individuals showed naturally-occurring CD4+ and CD8+ memory T cells specific for Spike (S), Nucleocapsid (N), Membrane, and some non-structural ORF proteins leading to the production of single or multiple proinflammatory cytokines (IL-2, TNF-ɑ, and IFN-γ) (Breton et al., 2021; Grifoni et al., 2020; Peng et al., 2020; Sekine et al., 2020; Tarke et al., 2021a). These results highlight the importance of pre-existing and induced SARS-CoV-2-specific CD8+ and CD4+ T cell responses for immune protection in mild SARS-CoV-2 infection. Since then, studies suggesting the inclusion of specific peptides to elicit T cell response in vaccine design and in silico analysis of SARS-CoV-2 proteins have detected candidate epitopes specific for protective CD4+ or CD8+ T and B responses with low risks of allergy or autoimmunity (Jain et al., 2021; Safavi et al., 2020; Singh et al., 2020; Qamar et al., 2020). In fact, approved vaccines induce T cell response in addition to the production of neutralizing antibodies for immunization against SARS-CoV-2 (Corbett et al., 2020; Sahin et al., 2020; Vogel et al., 2021).
The quality and amplitude of adaptive immunity are highly dependent on the Human Leukocyte Antigen (HLA) complex-mediated presentation of epitopes to T cells. Previous works have already evidenced sets of SARS-CoV-2 peptides more likely to be presented through HLA molecules to CD8+ T cells (Barquera et al., 2020; Nguyen et al., 2020; Pretti et al., 2020). In a recent study, our group adopted a computational approach to map populational coverage for SARS-CoV-2-derived peptides based on ccClass I HLAs, which suggested a protective role of a higher S/N coverage ratio (Pretti et al., 2020). Since most of the new SARS-CoV-2 variants are located in the proteins S and N, we decided to investigate whether mutations in these regions could modify peptide presentation, antigenic coverage, and impact T cell responses.
2. Material and methods
2.1. SARS-CoV-2 lineages
BetaCoV/Wuhan/IPBCAMS-WH-01/2019 genome assembly (GenBank: MT019529.1) was downloaded and used as a wild-type reference (REF) sequence. Non-synonymous Single-Nucleotide Variants (nsSNVs) and deletions in S and N genes of four SARS-CoV-2 upcoming variants of interest (VOI) (20I/501Y·V1, 20H/501Y·V2, 20 J/501Y·V3, and 20B/S.484 K) were retrieved from the online database covariants.org (Hodcroft, 2021). A list of S and N mutations analyzed here can be found in Supplementary Table S1.
2.2. Binding and antigenicity predictions
Allele frequencies of 110 class I HLA-A and —B alleles from 39 countries (Supplementary Table S2, Fig. S2) were obtained from the repository Allelefrequencies.net (Pretti et al., 2020) considering the cumulative allele frequency of those alleles as close to 0.9 as possible for each country. Moreover, data were mostly retrieved from bone marrow registries, as they better represent the overall population. Binding predictions were performed as previously described (Pretti et al., 2020) and peptides classified as Strong (SB) and Weak Binders (WB) were used in the following analysis. In summary, netCTLpan1.1 (Stranzl et al., 2010) was used to obtain predictions for TAP transport and peptide cutting to accurately select the peptides more likely to be transported to the endoplasmic reticulum and cleaved. Those items of information were combined with HLA class I binding predictions performed by netMHCpan4.1 (Reynisson et al., 2020), which is more accurate since it has been trained with more HLA alleles. Importantly, residues from positions −10 to +10 in relation to the mutated amino acid (aa) were used to predict 8 to 11-mer binders. Only peptides that were distinct between each VOI and the REF were considered in the analysis. Peptide antigenicity was predicted for all supported (102 out of 110) HLA alleles among the 110 using PRIME (Schmidt et al., 2021). The prediction was carried out with default parameters by providing a list of all binders predicted by netCTLpan (Stranzl et al., 2010) and filtering for TAP and cleavage scores as previously described (Pretti et al., 2020). Peptides with scores equal to zero were excluded since they suggest no HLA presentation.
2.3. Search for conserved or convergent peptides in other viral strains
Unique VOI-derived peptides and their corresponding REF counterparts were searched against the entire set of viral proteins from the UniProtKB database release 2021_02 (UniProt Consortium, 2021). The dataset containing 2,559,061 entries was downloaded in csv format and only mutant peptides with an exact match to other viral proteins by using GNU grep were considered as VOI non-unique.
2.4. Population coverage
Predicted SB and WB class I peptide:HLA pairs derived from SARS-CoV-2 VOI and REF were used to calculate population coverage using the IEDB Population coverage software (Bui et al., 2006). The software was downloaded on 10/05/2021 and run locally with default parameters for binders exclusive to REF or VOI lineages. In this analysis, 29 countries were included (Austria, Brazil, Bulgaria, China, Croatia, Czech Republic, England, France, Germany, Indonesia, Israel, Italy, Japan, Malaysia, Mexico, Morocco, Oman, Poland, Portugal, Romania, Russia, Senegal, Spain, Thailand, Tunisia, Ireland Northern, South Africa, United States, and India).
2.5. Validation of predictions using in vitro data
The search of predicted binders in public databases was performed as follows. Data from T cell assays deposited in the Immune Epitope Database (IEDB) were retrieved on 18/05/2021 and filtered for linear epitopes, MHC-I restricted, from SARS-CoV-2, positive assays, and from Human hosts. Besides this, studies with less than 50 assays were excluded and those remaining were curated to ensure that peptides were tested isolatedly and not pooled together. In addition, only experiments using either ‘T cell CD8+’ or ‘PBMC’ that tested S or N-derived epitopes were selected. Epitopes identified by binding predictions were excluded from the final dataset.
2.6. Statistical and data analysis
The analysis was conducted on the R environment v4.0, using the R packages Biostrings v2.56 for peptide sequence manipulation and alignment, ggmsa v0.0.5 for visualization, and ggpubr v0.3.0 for statistical analysis. Wilcoxon test was used for mean comparisons unless stated otherwise. A p-value <0.05 was used as a threshold for mean comparisons except for the antigenicity score where a p < 0.01 was used instead.
3. Results
3.1. Binding profile of peptides derived from new SARS-CoV-2 lineages
Focused on evaluating the class I HLA binding profile of peptides derived from some of the SARS-CoV-2 variants, 26 nsSNVs and two deletions located within the genes coding for proteins S and N were obtained (Hodcroft, 2021). The mutations represent the genetic variability within these two proteins of four VOI circulating worldwide (Supplementary Table S1). We have originated a set of 8 to 11-mer peptides from 21-length aa sequences comprising each variant, within a window of 10 residues before and after the variant whenever possible (Fig. S1). The distribution of HLA frequencies among countries used in this work can be observed in Fig. S2 and Supplementary Table S2.
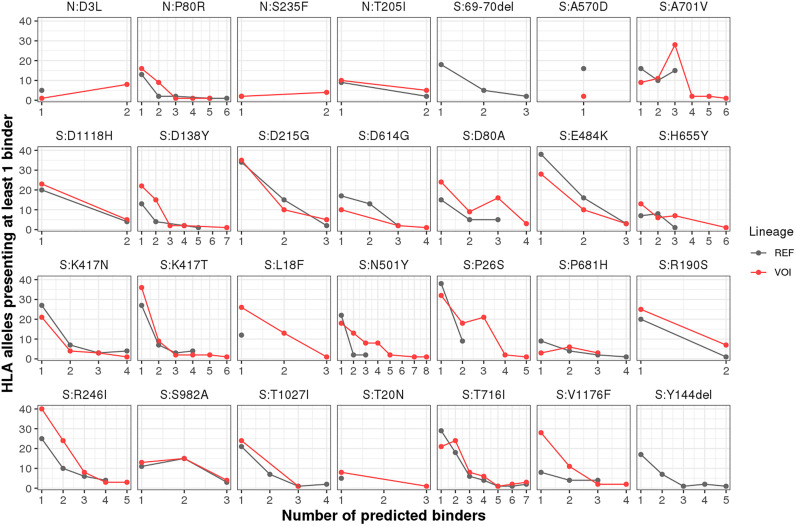
We obtained 352 unique peptides, comprising 8 to 11-mer predicted to be transported by TAP, cleaved by the proteasome, and bind to at least one of the 110 class I HLA-A and —B molecules. Of note, none of these viral peptides matched the human proteome. The peptides generated 2666 unique peptide:HLA combinations (Supplementary Table S3 and Table S4). Comparing the number of HLA-I alleles predicted to bind REF and VOI-derived peptides, we observed that peptides derived from the two deletions in S (69-70del, Y144del) lack the ability to bind to the HLA alleles set (Supplementary Table S3, Fig. 1). Importantly, six out of the 22 remaining nsSNVs in protein S generated fewer peptide:HLA pairs when compared to the REF sequence, and two of them (A570D and D614G) generated less than half the number of binders compared to the respective REF sequences. Notably, the nsSNV N501Y generated four times more binders than the REF sequence. All nsSNVs from protein N generated more binders compared to the REF sequences. The absence of peptide ligands derived from nsSNVs/deletions could contribute to the diminishing cellular immunity whilst a higher number of epitopes may favor T cytotoxic responses. Studying the relationship between antigen presentation and different mutations across VOI may shed light on population susceptibility to specific viral lineages.
Fig. 1.
Number of unique peptide:HLA-I pairs for REF and VOI-derived peptides. Binding predictions considering 110 class I HLA-A and —B alleles were performed on VOI and REF aa sequences using a combination of netMHCpan and netCTLpan predictors. Binder peptides were selected according to netMHCpan notation of strong (%Rank <0.5) and weak binder (%Rank <2.0) peptides; non-binder peptides were excluded.The number of HLA alleles presenting at least 1 binder is depicted for VOI (Red) and REF (Black) peptides. The nsSNV N:S235F did not generate any binder (Supplementary Table S2 and Supplementary Table S1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Number of unique peptide:HLA-I pairs for REF and VOI-derived peptides. Binding predictions considering 110 class I HLA-A and —B alleles were performed on VOI and REF aa sequences using a combination of netMHCpan and netCTLpan predictors. Binder peptides were selected according to netMHCpan notation of strong (%Rank <0.5) and weak binder (%Rank <2.0) peptides; non-binder peptides were excluded.The number of HLA alleles presenting at least 1 binder is depicted for VOI (Red) and REF (Black) peptides. The nsSNV N:S235F did not generate any binder (Supplementary Table S2 and Supplementary Table S1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Although we verified an increased number of potentially presented peptides derived from all mutations in protein N and the majority in protein S (16/24), lower affinities of variant peptides to the HLA-I molecule could counterbalance their elevated number. Therefore, we compared the HLA-affinities between VOI and REF-derived peptides (Fig. S3). A lower %Rank indicates peptides more likely to bind a given allele when compared with a set of natural binder peptides. No significant differences in affinities were found for N-derived peptides. In contrast, a significantly stronger binding (lower %Rank) was observed for peptides derived from the nsSNVs D138Y and N501Y. In contrast, peptides derived from E484K exhibited weaker binding compared to the respective REF-derived peptides. Of note, for the VOI 20I/501Y·V1, new mutations resulted in predicted epitopes with significantly stronger binding in protein S, while for epitopes derived from 20B/S.484 K, a significant decrease in binding was predicted when compared to the REF lineage (Fig. S3).
Recently, a handful of studies released information about the in vitro reactivity of SARS-CoV-2 epitope-specific T cell responses (Schulien et al., 2021; Tarke et al., 2021b; Tarke et al., 2021a). We then accessed data from the literature (Ferretti et al., 2020; Kared et al., 2020; Saini et al., 2021; Schulien et al., 2021; Tarke et al., 2021a) to check whether our binding predictions could be validated by in vitro results. The search identified 246 peptides derived from proteins S or N corresponding to 271 unique peptide:HLA combinations tested in vitro. Among those, 31 peptide:HLA pairs overlapped with our dataset which allowed us to compare our predictions with their reactivity detected by in vitro assays. Of the 31 peptide:HLA pairs, 23 of them were classified as ‘Positive’ by the in vitro assays, 3 as ‘Negative’, and 5 of them were either ‘Positive’ or ‘Negative’ depending on the study. Of note, the REF-derived peptides investigated by the published studies that induce T cell response fall into regions containing 18 distinct mutations in the VOIs (Supplementary Table S5). Importantly, the VOI-derived peptides could not be inspected in this analysis as they were not individually tested in vitro to this date, to the best of our knowledge (Tarke et al., 2021b).
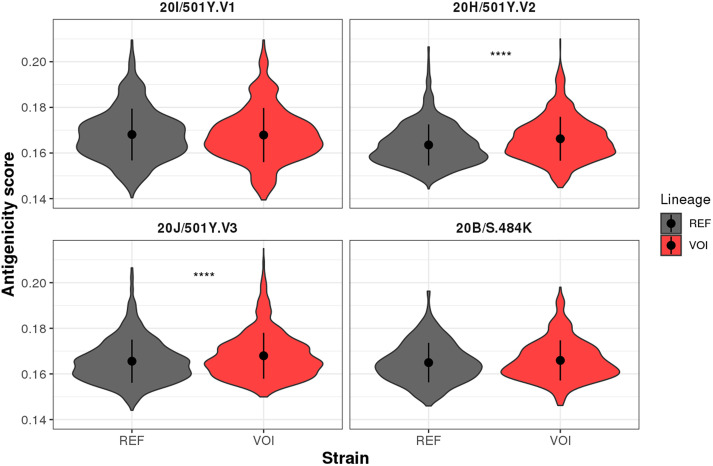
Since we were interested in investigating the antigenicity of SARS-CoV-2 variants, we compared T cell responses for REF and VOI-derived peptides using a predictor that considers TCR recognition of HLA-restricted epitopes (Schmidt et al., 2021). We obtained 7586 peptide:HLA combinations that potentially trigger the T cell receptor (Supplementary Table S4). We observed that the nsSNVs P80R, A701V, K417N, K417T, L18F, and R246I had greater antigenicity scores in regard to the REF sequence (Fig. S4). In contrast, nsSNVs T205I and Y144del showed lower antigenicity scores. Considering all nsSNVs associated with each VOI, an increased antigenicity was observed for 20H/501Y·V2 (p-value <0.0001) and 20 J/501Y·V3 (p-value <0.001) lineages (Fig. 2). We then associated those results with our previous binding predictions (Figure1, Fig. S3) considering only peptides with significant differences between REF and VOI lineages. We classified the significant results by mutation and highlighted the general interest in them either for their potential association with immune evasion or being used for vaccine development (Table 1). For instance, nsSNV 484 K is predicted to bind to fewer HLA alleles and weaker affinity than REF thus suggesting its potential role of fostering immune evasion. On the other hand, the majority of mutations were classified as potentially good targets for vaccine development since they are predicted to bind to more HLA alleles, with equal or stronger affinity, which suggests their ability to trigger a stronger TCR response compared to the REF peptide.
Fig. 2.
Overall antigenicity of SARS-CoV-2 VOI. Predictions were obtained using the list of 110 HLA alleles to the PRIME algorithm that calculated the likelihood of a peptide being presented through the HLA-I and triggering a T cell response. Non-zero scores for all peptide:HLA combinations are shown. All nsSNVs were aggregated according to the respective lineage they belong to. *** p < 0.001; ****p < 0.0001.
Table 1.
General interest of some VOI mutations. Characterization of mutant peptides in VOI lineages according to their presentation and antigenicity profiles in regard to REF peptides.
| Mutation | Number of HLA alleles | HLA binding | Immunogenicity | Associated VOI | Interest |
|---|---|---|---|---|---|
| S:E484K | Less | Weaker | Similar to the respective REF peptides | 20H/501Y.V2, 20 J/501Y·V3, 20B/S.484 K | Potentially associated with immune evasion |
| S:D138Y | More | Stronger | 20 J/501Y·V3 | Potentially good target for vaccine development | |
| S:N501Y | More | Stronger | 20I/501Y·V1, 20H/501Y.V2, 20 J/501Y·V3 | ||
| N:P80R, S:K417T | More | Similar | More than the respective REF peptides | 20 J/501Y·V3 | Potentially good target for vaccine development |
| S:A701V, S:R246I | More | Similar | 20H/501Y.V2 | ||
| S:L18F | More | Similar | 20H/501Y.V2, 20 J/501Y.V3 | ||
| S:T205I | More | Similar | Less than the respective REF peptides | 20H/501Y.V2 | Potentially good target for vaccine development |
| S:Y144del | Less | Similar | 20I/501Y·V1 | Potentially associated with immune evasion |
3.2. Occurrence of REF and VOI peptides in other viral strains
SARS-CoV-2 mutations could generate a gain of immunodominant peptides and hence, the potential for more CD8+ T cell reactivity. On the other hand, loss of immunodominant peptides may favor immune evasion. We investigated the presence of these newly acquired epitopes and their REF counterparts in other seasonal viral strains of Betacoronavirus according to the ICTV (King et al., 2018). We searched for identical peptides over the whole virus superkingdom. Epitopes have been divided into two categories regarding their Class-I HLA binding predictions, i.e. mutant peptides lacking HLA binding (Set A, Supplementary Table S6), and mutant peptides that acquired binding capacity (Set B, Supplementary Table S6), in regard to REF peptides. Overall we observed that there are more mutant peptides that are predicted to bind to Class-I HLAs (set B) than those predicted to lack binding affinities (set A). In addition, only two mutant peptides were also detected in other seasonal betacoronavirus. These two peptides were among those that potentially acquired binding capacities (Set B, Supplementary Table S6). This suggests that mutant epitopes are less likely to occur in other viruses, including Betacoronavirus. Given the protection provided by cross-reactive T-cell-mediated immunity, future studies are required to assess to what extent those mutant epitopes can confer better virus adaptability.
3.3. Most SARS-CoV-2 VOI show increased coverage for class I HLAs
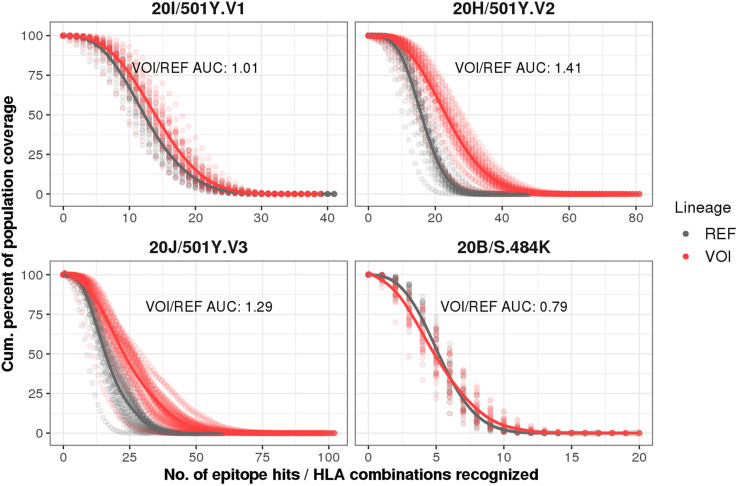
Despite the differences in binding profile and antigenicity, the HLA alleles presenting SARS-CoV-2 binder peptides are not necessarily equally distributed across different countries. Therefore, we investigated how different populations are predicted to be covered for the four VOI compared to the REF lineage using HLA frequency data from the 29 countries. In this specific analysis, we considered the allele frequencies of the class I HLA alleles in each analyzed population and the number of S and N-derived peptides (epitope hits) they are likely to present (Fig. 3 ). We observed a significant difference (p < 0.0001, Wilcoxon) when comparing the area under the curve of VOI and REF coverages for 20I/501Y·V1 (coverage ratio = 1.01), 20H/501Y·V2 (coverage ratio = 1.41), and 20 J/501Y·V3 lineages (coverage ratio = 1.29), but not for 20B/S.484 K (coverage ratio = 0.79, p = 0.35). Together, these findings indicate a remarkable increment in antigen coverage for three new lineages, increased HLA-I presentation for the majority of the analyzed nsSNVs as well as increased antigenicity scores for 20H/501Y·V2 and 20 J/501Y·V3. However, in vitro and in vivo approaches are needed to validate these findings.
Fig. 3.
Populational coverage for the four SARS-CoV-2 VOI compared to the REF sequence. Population coverage analyses were based on 110 HLA-A and —B alleles using allele frequency data from 29 countries. Each panel represents a different VOI (red) compared with the REF (gray). Tendency lines were drawn connecting the VOI and REF dots. The VOI/REF area under the curve (AUC) ratio is depicted. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The outburst of new SARS-CoV-2 variants capable of faster spreading than previous lineages is a major concern for health systems worldwide. Four of them bear nsSNV that confer more transmissibility than other circulating coronaviruses (Ozono et al., 2021; Volz et al., 2021), namely 20I/501Y·V1 (B.1.1.7 Lineage), which emerged in Britain and was detected in over 70 countries; 20H/501Y·V2 (B.1.351 Lineage), that was first detected in South Africa, and two that emerged in Brazil, 20J/501Y·V3 (P.1 Lineage) in Manaus and 20B/S.484K (P.2 Lineage), firstly identified in patients in Rio de Janeiro (Toovey et al., 2021; Hadfield et al., 2018). A case of reinfection was reported for P.2 lineage (Vasques Nonaka et al., 2021) and, to this date, it has already been detected in Europe, North and South America (Hadfield et al., 2018). Cytotoxic CD8+ T cells are key for immune protection against viral infections, and recent works have shown the importance of cross-reactive and induced SARS-CoV-2-specific CD8+ T cell responses to immune protection (Mateus et al., 2020; Schulien et al., 2021; Tarke et al., 2021a). Given the role of SARS-CoV-2-specific CD8+ T cells for COVID-19 resolution, we investigated the likelihood of four SARS-CoV-2 lineages to generate new epitopes capable to bind a set of cClass I HLA alleles representing countries from Europe, the Americas, Africa, and Asia to ultimately infer T cell-mediated immunity. Importantly, binding predictions use machine learning techniques and are more accurate for HLAs seen in the training set. It is worth mentioning that 55% of alleles investigated in our study were present in the tool's training set. Among the 49 absent alleles, 76% share the first HLA field with the other alleles, indicating the high reliability of our predictions. Despite the exhaustive curation of potential epitopes using strict criteria to enhance the accuracy of our analysis, such as the inclusion of TAP transport and proteasome processing predictions, data should not be taken as proof and in vitro testing to validate the findings is recommended.
Limited data exist on the magnitude and extension of CD8+ T cell responses associated with the new circulating SARS-CoV-2 variants. Mutations leading to new putative epitopes, or either the lack of conserved ones, should be investigated for their potential to either exacerbate or evade immune responses. We observed that mutations such as 69-70del in the gene coding for protein S reduce the number of HLA-I binders, which could lead to immune evasion. In fact, such deletion was previously associated with antibody escape (McCarthy et al., 2021). Despite no difference in affinities to HLAs, the D614G that has been related to increased transmissibility (Ozono et al., 2021; Volz et al., 2021) displayed half the number of potential binders than 614D, suggesting that lowering the number of potential binders would be permissive to infection due to lower immune recognition and response. The number of putative HLA ligands derived from REF or VOI sequences gives us an idea of whether there is material for T cell recognition since a higher number of presented peptides increases the probability of a robust and effective T cell response. On the other hand, fewer HLA ligands from immunodominant regions could be associated with immune evasion. A recent report performed in vitro assays and showed that other SARS-CoV-2 mutations have the ability to evade CD8+ T cell responses through reduced HLA-I binding (Agerer et al., 2021).
Another important aspect is whether the mutation-derived peptides are more or less immunogenic and likely to trigger the T cell response. For instance, E484K substitution has overall less affinity to class I HLA molecules than the respective REF peptides, but no difference in antigenicity scores. Although not mandatory to trigger a T cell response, tighter binding to HLA-I groove facilitates T cell response (Croft et al., 2019; Paul et al., 2013). Importantly, this nsSNV is located on the Receptor Binding Domain (RBD), the region responsible for the entry of the virus into the cell (Greaney et al., 2021). This finding is in line with other reports that identified evasion from antibodies associated with E484K (Greaney et al., 2021; Zhou et al., 2021) and with lineages bearing this mutation, especially the B.1.351 (Dejnirattisai et al., 2021). On the other hand, 501Y substitution has a stronger affinity to the HLA alleles than the REF peptides but, similarly to E484K, are able to escape from antibody recognition (Baum et al., 2020; Li et al., 2021). Accordingly, N501Y and D138Y substitutions showed increased HLA affinities and may be good targets to trigger CD8+ T cell responses. In addition, our previous work suggested that HLA presentation of N-derived peptides, rather than S-derived ones, was associated with high mortality rates (Pretti et al., 2020). Based on that, the fact that the B.1.1.7, B.1.351, and P.1 Lineage potentially present more N-derived epitopes than the original lineage could be a major concern to the protective response in convalescent and vaccinated individuals. In fact, all nsSNVs on the protein N, e.g. D3L, P80R, S235F, and T205I, generated more potential epitopes in comparison to the REF peptides. To date, there are no studies associating the nsSNVs in protein S such as D80A, P618H, T1027, and T716I with increased transmissibility or immune evasion, since most studies focused on nsSNVs located on the RBD. Overall, our data indicate that new mutant peptides in immunodominant regions exhibit altered HLA Class I binding and immunogenicity for CD8+ T cell response previously described by Schreibing and collaborators (Schreibing et al., 2021). While changes in this specific region leading to reduced or lack of affinities may culminate in the escape of the virus from the protective response of T cells, either in convalescent or vaccinated individuals, enhanced affinities could favor a more intense response.
When validating our results by comparing the predictions with publicly available in vitro data from the IEDB, we noticed that the overlap of peptide:HLA pairs was higher when considering the most frequent alleles. This is expected since in vitro testing does not usually consider less frequent HLAs. Although in silico analyses are able to estimate the binding affinities for less frequent alleles, they are usually less accurate. Moreover, since many epitopes known to trigger T cell responses by in vitro studies are lost in the new lineages, this could contribute to impair the recognition of SARS-CoV-2 by the host. In fact, a recent study indicated that SARS-CoV-2 mutations may help evade CD8+ T cell responses (Agerer et al., 2021). Unfortunately, we were able to check only the predictions associated with REF peptides, since there is no available data for individual peptides from the new lineages.
The overall susceptibility to new SARS-CoV-2 lineages is a major concern. We have used a populational coverage strategy to investigate whether the predicted ability of a broad population to recognize and present the viral epitopes is impacted by the new mutations in the VOIs. While the investigated countries exhibited higher coverage for the lineages B.1.351, P1, and B.1.1.7, P2 lineage had a trend to lower coverage. The absence of significant difference could be related to the fact that only a few variants are present in this lineage, which weakens the statistical power. Of note, Spain was the country less covered for all lineages except for P.2 when considering both REF and VOI coverages. Although more studies are still necessary, the data indicates less presentation of peptides from the nsSNV 484 K, and may suggest its ability to evade T cell response, helping to explain the high spreadability of new lineages bearing this mutation such as 20B/S.484 K lineage, which has been detected in other countries including England, Singapore, the USA, Norway, Argentina, Denmark, Ireland, and Canada (Toovey et al., 2021). Finally, besides investigating the populational T cell response to SARS-CoV-2 new lineages, the ability of the available vaccines to protect against those new lineages should be further explored (Le Bert et al., 2020; Mateus et al., 2020). On the other hand, being aware of the binding profile of mutant peptides may guide vaccine development efforts by ranking conserved epitopes of interest, thus reducing or eliminating the need to select new epitopes from emerging lineages.
5. Conclusion
Overall, our in silico analysis of antigenic coverage revealed higher HLA coverage associated with the B.1.351, P1, and B.1.1.7 lineages among human populations, suggesting that the CD8+ T response may be affected when compared to the wild SARS-CoV-2 lineage, probably enhancing the risk of T cell reactivity in naturally exposed individuals. In contrast, some individual mutant peptides were predicted to have either a weaker or lack of affinity through HLA—I, presenting potential signs of evasion from the immune system. Our results also may guide efforts to characterize and validate relevant peptides to trigger CD8+ T cell responses, and design new universal T cell-inducing vaccine candidates that minimize detrimental effects of viral diversification, at the same time inducing responses to a broad human population.
Authors contributions
MB, MP, and RG contributed to the design and conception of the study. MB and ASF contributed to important scientific discussions. MP, RG, and NMS performed all the analyses. MP and RG wrote the first draft of the manuscript. MB was responsible for the final approval of the submitted version. All the authors contributed to the article and approved the submitted version.
Availability of data and materials
The sequence of SARS-CoV-2 can be found at the GenBank with the ID: MT019529.1. The list of HLA-I alleles is available in a previous publication (Pretti et al., 2020). All software used in this work is free for academic use.
The following are the supplementary data related to this article.
List of SARS-CoV-2 nsSNV used in the analysis.
Distribution of HLA allelic frequency in the 39 analyzed countries.
Number of peptide:HLA-I pairs depicting Strong (SB) and Weak binders (WB).
All predicted peptide:HLA-I combinations.
Peptides matching in vitro tested peptides in the literature.
Occurrence of REF and VOI peptides among different virus strains*.
Global alignment of the 21-length aa sequence for each variant of four SARS-CoV-2 VOI to the REF sequence. Variants are surrounded by ten aa residues on each side, except by the nsSNV N:D3L, in which the variant falls at the beginning of the protein. Dashes (−) represent deletions (del) within the sequence.
Supplementary Fig. S2. Distribution of HLA allelic frequency in the 39 analyzed countries. Countries and HLAs were hierarchically clustered and distance was based on Pearson's correlation coefficient. Colour intensities represent the allelic frequency.
Supplementary Fig. S3. Binding affinities of VOI and respective REF-derived peptides. Binding affinities predictions (%Rank score) considered 110 class I HLA-A and —B alleles on 8 to 11-mer peptides derived from VOI (Red) and respective REF sequences (Gray). The lower the %Rank score the higher the likelihood of a peptide to bind a given HLA molecule. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Fig. S4. Predicted antigenicity of SARS-CoV-2 nsSNVs. For each nsSNV, unique peptide:HLA-I pairs were used to predict their overall TCR antigenicity. Only p values below 0.01 were considered significant. **p < 0.01; ***p < 0.001; ****p < 0.0001.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
The authors would like to thank the Bioinformatics Core Facility (INCA-RJ) for their support and the PhD fellowship for MP from the Brazilian National Cancer Institute (INCA-RJ).
References
- Agerer B., Koblischke M., Gudipati V., Montaño-Gutierrez L.F., Smyth M., Popa A., Genger J.-W., Endler L., Florian D.M., Mühlgrabner V., Graninger M., Aberle S.W., Husa A.-M., Shaw L.E., Lercher A., Gattinger P., Torralba-Gombau R., Trapin D., Penz T., Barreca D., Bergthaler A. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquera R., Collen E., Di D., Buhler S., Teixeira J., Llamas B., Nunes J.M., Sanchez-Mazas A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA. 2020;96:277–298. doi: 10.1111/tan.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Röhmel J., Voigt S., Schmitz J., Miltenyi S., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Breton G., Mendoza P., Hägglöf T., Oliveira T.Y., Schaefer-Babajew D., Gaebler C., Turroja M., Hurley A., Caskey M., Nussenzweig M.C. Persistent cellular immunity to SARS-CoV-2 infection. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H.-H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Emerging SARS-CoV-2 Variants | CDC. 2020. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html [WWW Document]. URL. (accessed 3.1.21)
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Graham B.S. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft N.P., Smith S.A., Pickering J., Sidney J., Peters B., Faridi P., Witney M.J., Sebastian P., Flesch I.E.A., Heading S.L., Sette A., La Gruta N.L., Purcell A.W., Tscharke D.C. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. U. S. A. 2019;116:3112–3117. doi: 10.1073/pnas.1815239116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza W.M., Amorim M.R., Sesti-Costa R., Coimbra L.D., Toledo-Teixeira D.A., Parise P.L., Barbosa P.P., Bispo-dos-Santos K., Mofatto L.S., Simeoni C.L., Brunetti N.S., Claro I.M., Duarte A.S.S., Coletti T.M., Zangirolami A.B., Costa-Lima C., Gomes A.B.S.P., Buscaratti L.I., Sales F.C., Costa V.A., Proenca-Modena J.L. Levels of SARS-CoV-2 lineage P.1 neutralization by antibodies elicited after natural infection and vaccination. SSRN J. 2021 doi: 10.2139/ssrn.3793486. [DOI] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., López-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Costa Clemens S.A., Naveca F.G., Nascimento V., Nascimento F., Fernandes da Costa C., Screaton G.R. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.055. 2939–2954.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., Whitton H.J., Virbasius A., Olivier K.J., Buckner L.R., Alistar A.T., Whitman E.D., Bertino S.A., Chattopadhyay S., MacBeath G. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53 doi: 10.1016/j.immuni.2020.10.006. 1095–1107.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. BioRxiv. 2021 doi: 10.1101/2020.12.31.425021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B. CoVariants: SARS-CoV-2 Mutations and Variants of Interest. 2021. https://covariants.org/ [WWW document]. URL.
- Jain N., Shankar U., Majee P., Kumar A. Scrutinizing the SARS-CoV-2 protein information for designing an effective vaccine encompassing both the T-cell and B-cell epitopes. Infect. Genet. Evol. 2021;87 doi: 10.1016/j.meegid.2020.104648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., Benner S.E., Patel E.U., Littlefield K., Laeyendecker O., Shoham S., Sullivan D., Casadevall A., Pekosz A., Nardin A., Fehlings M., Quinn T.C. CD8+ T cell responses in convalescent COVID-19 individuals target epitopes from the entire SARS-CoV-2 proteome and show kinetics of early differentiation. BioRxiv. 2020 doi: 10.1101/2020.10.08.330688. [DOI] [Google Scholar]
- King A.M.Q., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M.L., Rubino L., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Varsani A., Davison A.J. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2018) Arch. Virol. 2018;163:2601–2631. doi: 10.1007/s00705-018-3847-1. [DOI] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I.-C., Wang L.-F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G.-H., Tan Y.-J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., Zhao C., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Liu J., Liang H., Shi Y., Wang Y. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371.e9. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., Mallal S., Lammers M., Rubiro P., Quiambao L., Sutherland A., Yu E.D., da Silva Antunes R., Greenbaum J., Frazier A., Markmann A.J., Weiskopf D. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021 doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., Sette A., Ay F., Seumois G., Ottensmeier C.H., Vijayanand P. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell. 2020;183:1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., Thompson R.F. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94 doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Weiskopf D., Angelo M.A., Sidney J., Peters B., Sette A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 2013;191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D.I., Paesen G.C., Grimes J.M., Antson A.A., Bayfield O.W., Hawkins D.E.D.P., Ker D.-S., Dong T. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretti M.A.M., Galvani R.G., Vieira G.F., Bonomo A., Bonamino M.H., Boroni M. Class I HLA allele predicted restricted antigenic coverages for spike and nucleocapsid proteins are associated with deaths related to COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.565730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar Tahir Ul, Shahid F., Aslam Sadia, Ashfaq U.A., Aslam Sidra, Fatima I., Fareed M.M., Zohaib A., Chen L.-L. Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2. Infect. Dis. Poverty. 2020;9:132. doi: 10.1186/s40249-020-00752-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi A., Kefayat A., Mahdevar E., Abiri A., Ghahremani F. Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches. Vaccine. 2020;38:7612–7628. doi: 10.1016/j.vaccine.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., Pascal K., Maurus D., Brachtendorf S., Lörks V., Sikorski J., Koch P., Hilker R., Becker D., Eller A.-K., Grützner J., Türeci Ö. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. medRxiv. 2020 doi: 10.1101/2020.12.09.20245175. [DOI] [Google Scholar]
- Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Smith A.R., Magnin M., Racle J., Devlin J.R., Bobisse S., Cesbron J., Bonnet V., Carmona S.J., Huber F., Ciriello G., Speiser D.E., Bassani-Sternberg M., Coukos G., Baker B.M., Harari A., Gfeller D. Prediction of neo-epitope immunogenicity reveals TCR recognition determinants and provides insight into immunoediting. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreibing F., Hannani M., Ticconi F., Fewings E., Nagai J.S., Begemann M., Kuppe C., Kurth I., Kranz J., Frank D., Anslinger T.M., Ziegler P., Kraus T., Enczmann J., Balz V., Windhofer F., Balfanz P., Kurts C., Marx G., Marx N., Kramann R. Dissecting CD8+ T cell pathology of severe SARS-CoV-2 infection by single-cell epitope mapping. BioRxiv. 2021 doi: 10.1101/2021.03.03.432690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar Daul, Salvat Lago M., Decker A., Luxenburger H., Binder B., Bettinger D., Sogukpinar O., Rieg S., Panning M., Huzly D., Schwemmle M., Kochs G., Waller C.F., Neumann-Haefelin C. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Wullimann D.J., Kammann T., Emgård J., Parrot T., Folkesson E., Karolinska COVID-19 Study Group, Rooyackers O., Eriksson L.I., Henter J.-I., Sönnerborg A., Buggert M. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183 doi: 10.1016/j.cell.2020.08.017. 158–168.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Thakur M., Sharma L.K., Chandra K. Designing a multi-epitope peptide based vaccine against SARS-CoV-2. Sci. Rep. 2020;10:16219. doi: 10.1038/s41598-020-73371-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranzl T., Larsen M.V., Lundegaard C., Nielsen M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics. 2010;62:357–368. doi: 10.1007/s00251-010-0441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., Methot N., Phillips E., Mallal S., Frazier A., Rawlings S.A., Greenbaum J.A., Peters B., Smith D.M., Crotty S., Weiskopf D., Sette A. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., da Silva Antunes R., Frazier A., Rawlings S.A., Smith D.M., Peters B., Scheuermann R.H., Weiskopf D., Crotty S., Grifoni A., Sette A. Negligible impact of SARS-CoV-2 variants on CD4 + and CD8 + T cell reactivity in COVID-19 exposed donors and vaccinees. BioRxiv. 2021 doi: 10.1101/2021.02.27.433180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toovey O.T.R., Harvey K.N., Bird P.W., Tang J.W.-T.W.-T. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J. Inf. Secur. 2021 doi: 10.1016/j.jinf.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasques Nonaka C.K., Miranda Franco M., Gräf T., Almeida Mendes A.V., Santana de Aguiar R., Giovanetti M., Solano de Freitas Souza B. 2021. Genomic Evidence of a Sars-Cov-2 Reinfection Case With E484K Spike Mutation in Brazil. [DOI] [Google Scholar]
- Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., Loschko J., Maddur M.S., Ota-Setlik A., Tompkins K., Cole J., Lui B.G., Ziegenhals T., Plaschke A., Eisel D., Dany S.C., Sahin U. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184 doi: 10.1016/j.cell.2020.11.020. 64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G., Ramos V., Nussenzweig M.C. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Paesen G.C., Lopez-Camacho C., Slon-Campos J., Hallis B., Coombes N., Bewley K., Charlton S., Walter T.S., Skelly D., Screaton G.R. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.037. 2348–2361.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of SARS-CoV-2 nsSNV used in the analysis.
Distribution of HLA allelic frequency in the 39 analyzed countries.
Number of peptide:HLA-I pairs depicting Strong (SB) and Weak binders (WB).
All predicted peptide:HLA-I combinations.
Peptides matching in vitro tested peptides in the literature.
Occurrence of REF and VOI peptides among different virus strains*.
Global alignment of the 21-length aa sequence for each variant of four SARS-CoV-2 VOI to the REF sequence. Variants are surrounded by ten aa residues on each side, except by the nsSNV N:D3L, in which the variant falls at the beginning of the protein. Dashes (−) represent deletions (del) within the sequence.
Supplementary Fig. S2. Distribution of HLA allelic frequency in the 39 analyzed countries. Countries and HLAs were hierarchically clustered and distance was based on Pearson's correlation coefficient. Colour intensities represent the allelic frequency.
Supplementary Fig. S3. Binding affinities of VOI and respective REF-derived peptides. Binding affinities predictions (%Rank score) considered 110 class I HLA-A and —B alleles on 8 to 11-mer peptides derived from VOI (Red) and respective REF sequences (Gray). The lower the %Rank score the higher the likelihood of a peptide to bind a given HLA molecule. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Fig. S4. Predicted antigenicity of SARS-CoV-2 nsSNVs. For each nsSNV, unique peptide:HLA-I pairs were used to predict their overall TCR antigenicity. Only p values below 0.01 were considered significant. **p < 0.01; ***p < 0.001; ****p < 0.0001.
Data Availability Statement
The sequence of SARS-CoV-2 can be found at the GenBank with the ID: MT019529.1. The list of HLA-I alleles is available in a previous publication (Pretti et al., 2020). All software used in this work is free for academic use.
The following are the supplementary data related to this article.
List of SARS-CoV-2 nsSNV used in the analysis.
Distribution of HLA allelic frequency in the 39 analyzed countries.
Number of peptide:HLA-I pairs depicting Strong (SB) and Weak binders (WB).
All predicted peptide:HLA-I combinations.
Peptides matching in vitro tested peptides in the literature.
Occurrence of REF and VOI peptides among different virus strains*.
Global alignment of the 21-length aa sequence for each variant of four SARS-CoV-2 VOI to the REF sequence. Variants are surrounded by ten aa residues on each side, except by the nsSNV N:D3L, in which the variant falls at the beginning of the protein. Dashes (−) represent deletions (del) within the sequence.
Supplementary Fig. S2. Distribution of HLA allelic frequency in the 39 analyzed countries. Countries and HLAs were hierarchically clustered and distance was based on Pearson's correlation coefficient. Colour intensities represent the allelic frequency.
Supplementary Fig. S3. Binding affinities of VOI and respective REF-derived peptides. Binding affinities predictions (%Rank score) considered 110 class I HLA-A and —B alleles on 8 to 11-mer peptides derived from VOI (Red) and respective REF sequences (Gray). The lower the %Rank score the higher the likelihood of a peptide to bind a given HLA molecule. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Fig. S4. Predicted antigenicity of SARS-CoV-2 nsSNVs. For each nsSNV, unique peptide:HLA-I pairs were used to predict their overall TCR antigenicity. Only p values below 0.01 were considered significant. **p < 0.01; ***p < 0.001; ****p < 0.0001.