Abstract
Both SARS-CoV-2 infections and COVID-19 vaccines elicit memory T cell responses. Here, we report the development of 2 pools of experimentally defined SARS-CoV-2 T cell epitopes that, in combination with spike, were used to discriminate 4 groups of subjects with different SARS-CoV-2 infection and COVID-19 vaccine status. The overall T cell-based classification accuracy was 89.2% and 88.5% in the experimental and validation cohorts. This scheme was applicable to different mRNA vaccines and different lengths of time post infection/post vaccination and yielded increased accuracy when compared to serological readouts. T cell responses from breakthrough infections were also studied and effectively segregated from vaccine responses, with a combined performance of 86.6% across all 239 subjects from the 5 groups. We anticipate that a T cell-based immunodiagnostic scheme to classify subjects based on their vaccination and natural infection history will be an important tool for longitudinal monitoring of vaccinations and for establishing SARS-CoV-2 correlates of protection.
Keywords: SARS-CoV-2, T cells, epitope, viruses, COVID-19, vaccination, breakthrough infection, immunodiagnostic tool
Graphical abstract
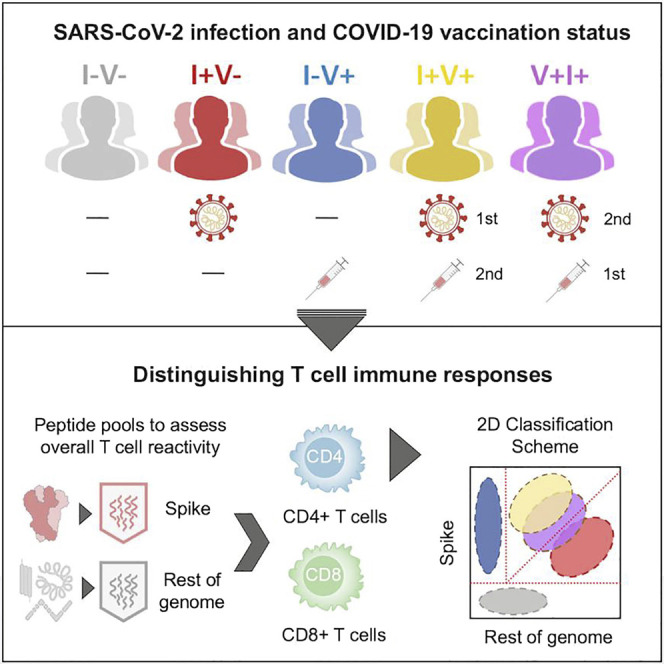
Yu et al. developed an assay using epitope pools to effectively discriminate T cell responses of subjects based on their SARS-CoV-2 infection and COVID-19 vaccination history. This T cell-based classification scheme could potentially be used as an immunodiagnostic tool for longitudinal monitoring of vaccination responses and for establishing correlates of protection.
Introduction
Immune memory against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is associated with cellular and humoral adaptive immunity (Painter et al., 2021; Rydyznski Moderbacher et al., 2020; Sette and Crotty, 2021). Progress has been made in defining the correlation of protection based on neutralizing antibody titers (Khoury et al., 2021), but a full mechanistic understanding of protection from symptomatic and severe disease might require comprehensive characterization of both antibody responses and effector and memory B and T cell responses (Feng et al., 2021; Koup et al., 2021; Krammer, 2021a, b).
Broad measurement of T cell responses is hindered by the lack of immunodiagnostics tools with effective predictive power able to discriminate pre-existing immunity, vaccination, and infection (Ogbe et al., 2021; Peeling and Olliaro, 2021; Sekine et al., 2020; Vandenberg et al., 2021). While SARS-CoV-2 T cell responses are detected in nearly all individuals who have recovered from symptomatic coronavirus disease 19 (COVID-19) (Grifoni et al., 2020b; Le Bert et al., 2020; Tarke et al., 2021a), they can also be found in 20%–50% of unexposed individuals (Mateus et al., 2020; Sette and Crotty, 2020; Tarke et al., 2021a). However, recent evidence suggests that SARS-CoV-2 infection generates a largely novel repertoire of T cells, with over 80% of the epitopes not recognized in unexposed donors (Mateus et al., 2020; Tarke et al., 2021a). In addition, mRNA or viral vector vaccines boost the spike (S) protein-specific immune responses in both unexposed and convalescent individuals without affecting the responses to non-S SARS-CoV-2 components (Bertoletti et al., 2021; Lozano-Ojalvo et al., 2021; Mateus et al., 2021). Further complexity is associated with evaluating responses in subjects previously infected and subsequently vaccinated and, conversely, previously vaccinated and subsequently infected (breakthrough infections) (Goel et al., 2021; Lucas et al., 2021; Niessl et al., 2021; Rovida et al., 2021).
We have shown that SARS-CoV-2-specific T cells can be detected and quantitated using peptide pools in various T cell assays (da Silva Antunes et al., 2021; Dan et al., 2021; Grifoni et al., 2020a; Mateus et al., 2020; Tarke et al., 2021b), which have proven useful to derive information about the kinetics and magnitude of SARS-CoV-2-specific T cell responses in both COVID-19 infection and vaccination (Dan et al., 2021; Mateus et al., 2021). Subsequent studies detailed the repertoire of epitope specificities recognized in a cohort of COVID-19 convalescent subjects (Tarke et al., 2021a). More recently, a meta-analysis of experimental curated data from the Immune Epitope Database revealed a large repertoire of over 1,400 epitopes defined in 25 different studies (Grifoni et al., 2021). Here, we used this information to develop SARS-CoV-2-specific peptide pools optimized for broader epitope repertoire and wider human leukocyte antigen (HLA) coverage for both CD4+ and CD8+ T cell responses. Accordingly, 2 pools of experimentally (E) defined epitopes derived from the non-S remainder (R) of the SARS-CoV-2 proteome (CD4RE and CD8RE) were established.
Several platforms and strategies have been developed to assess T cell responses in both vaccinated or infected individuals, using different readouts and technologies, such as cytokine release assays (ELISPOT or ELISA) (Krishna et al., 2021; Kruse et al., 2021; Martínez-Gallo et al., 2021; Murugesan et al., 2022; Tan et al., 2021; Tormo et al., 2022) or flow cytometry-based assays (blast transformation or intracellular cytokine staining [ICS]) (Lind Enoksson et al., 2021; Zelba et al., 2021). These assays mainly rely on the characterization of responses to the S or nucleocapsid (N) antigens and therefore do not address the entire SARS-CoV-2 proteome and the remarkable breadth of T cell responses generated against this pathogen (Grifoni et al., 2021).
In this study, we developed an immunodiagnostic T cell assay using a pool of overlapping peptides spanning the entire S protein in combination with E defined non-S pools to classify subjects based on their vaccination and infection history. This tool showed high predictive power to discriminate responses based on distinctive COVID-19 immune profiles, including hybrid immunity from breakthrough infections. Using a validation cohort, we demonstrated the clinical applicability of this tool for assessing immune responses in diverse individuals, including those who received different vaccine platforms and at different lengths of time post vaccination and infection.
Results
Cohorts associated with known infection and vaccination history
239 participants were enrolled in the study and classified into 5 groups based on known vaccination and infection history: 50 non-infected, non-vaccinated (I−V−); 50 infected and non-vaccinated (I+V−); 66 infected and then vaccinated (I+V+); 50 non-infected and vaccinated (I−V+); and 23 vaccinated and then infected (V+I+). An overview of the characteristics from all the participants is provided in Table 1 . For the I+V−, I+V+, and V+I+ groups, SARS-CoV-2 infection was determined by PCR-based testing during the acute phase of infection or verified by serological detection of antibodies against the SARS-CoV-2 S protein receptor binding domain (RBD) region at the time of blood donation.
Table 1.
Description of donor cohort characteristics and demographics
| Cohort name | I−V− | I+V− | I−V+ | I+V+ | V+I+ | |
|---|---|---|---|---|---|---|
| number of donors | 50 | 50 | 66 | 50 | 23 | |
| gender (M/F) | (26, 24) | (21, 29) | (28, 38) | (23, 27) | (7, 16) | |
| median age (years) | 25 (17–64) | 42 (19–67) | 40 (21–74) | 38 (21–73) | 30 (22–68) | |
| race | white (n [%]) | 32 (64%) | 37 (74%) | 38 (58%) | 39 (78%) | 15 (65%) |
| Hispanic/Latino (n [%]) | 8 (16%) | 7 (14%) | 11 (17%) | 6 (12%) | 5 (22%) | |
| Asian (n [%]) | 8 (16%) | 4 (8%) | 16 (24%) | 3 (6%) | 3 (13%) | |
| Black (n [%]) | 2 (4%) | 2 (4%) | 2 (3%) | 2 (4%) | 0 (0%) | |
| sample collection date | 2013–2019 | 2020–2021 | 2021 | 2021 | 2021 | |
| COVID-19 vaccination status | none | none | vaccinated | vaccinated | vaccinated | |
| Pfizer (n [%]) | – | – | 30 (45%) | 25 (50%) | 15 (65%) | |
| Moderna (n [%]) | – | – | 36 (55%) | 25 (50%) | 8 (35%) | |
| days from second dose of vaccination | – | – | 16 (13–190) | 32 (7–188) | 163(55–271) | |
| SARS-CoV-2 status | Ab(−) | Ab(+) or PCR(+) | Ab(+) and PCR(−) | Ab(+) or PCR(+) | PCR(+) | |
| SARS-CoV-2 PCR (n [%]) | positive | 0 (0%) | 47 (94%) | 0 (0%) | 45 (90%) | 23 (100%) |
| unknown | – | 3 (6%) | – | 5 (10%) | – | |
| spike (S) antibody response (n [%]) | median | 3.0 | 191.8 | 4,157.0 | 4,654.0 | 8,783.0 |
| range | 3.0–23.9 | 3.0–7,326.0 | 410.6–3,2033.0 | 159.2–2,5876.0 | 2,165.0–3,5319.0 | |
| nucleocapsid (N) antibody response (n [%]) | median | 4.9 | 177.9 | 16.3 | 73.2 | 241.5 |
| range | 3.0–339.8 | 3.0–11755.0 | 3.0–109.0 | 3.0–1873.0 | 72.6–5044.0 | |
| post-symptom onset (days) | median | – | 119 | – | 354 | 32 |
| range | – | 20–308 | – | 57–508 | 18–93 | |
| symptoms (n [%]) | asymptomatic | – | 0 (0%) | – | 0 (0%) | 0 (0%) |
| mild | – | 44 (88%) | – | 45 (90%) | 23 (100%) | |
| moderate | – | 3 (6%) | – | 3 (6%) | 0 (0%) | |
| severe | – | 3 (6%) | – | 2 (4%) | 0 (0%) | |
Summary of donor characteristics: non-infected, non-vaccinated, I−V−; infected and non-vaccinated, I+V−; infected and then vaccinated, I+V+; non-infected and vaccinated, I−V+; and vaccinated and then infected, V+I+.
The study primarily consisted of subjects recruited in San Diego, California (see STAR Methods for more details). Among individuals with history of COVID-19 disease, the majority were symptomatic mild disease cases, owing to the nature of the study recruitment design. Specifically, 44 donors (88%) for I+V−, 45 donors (90%) for I+V+, and 23 donors (100%) for V+I+ had mild symptoms; 3 donors (6%) of I+V− and I+V+ groups had moderate symptoms; and 3 (6%) and 2 donors (4%) from the I+V− and I+V+ groups, respectively, had severe symptoms. The median days of blood collection post-symptom onset (PSO) were 119 (20–308), 354 (57–508), and 32 (18–93) for I+V−, I+V+, and V+I+ groups, respectively. For the I−V+, I+V+, and V+I+ groups, the vaccinated subjects received 2 doses of mRNA vaccines BNT162b2 (Pfizer/BioNTech) or mRNA-1273 (Moderna) as verified by vaccination records and positive plasma SARS-CoV-2 S protein RBD immunoglobin G (IgG) titers. Similar distribution of Pfizer- or Moderna-administered vaccines (45%–55%) were present in vaccinated subjects from either the I−V+ or I+V+ group, while in the V+I+ group, 15 (65%) subjects had received the BNT162b2 vaccine and 8 (35%) the mRNA-1273 vaccine.
The median days of blood collection post second dose of vaccination (PVD) were 16 (13–190), 32 (7–188), and 163 (55–271) for I−V+, I+V+, and V+I+ groups, respectively. All of the I−V− subjects were collected before the attributed pandemic period (2013–2019) and confirmed seronegative with undetectable SARS-CoV-2 S protein RBD IgG titers. In all cohorts, the median ages were relatively young (25 [17–64], 42 [19–67], 40 [21–74], 38 [21–73], 30 [22–68] for I−V−, I+V−, I−V+, I+V+, and V+I+ groups, respectively) with the female gender well represented and different ethnicities represented. In our study, participants were further divided in an exploratory cohort (120 donors; Table S1), an independent validation cohort (96 donors; Table S2), and a third cohort of breakthrough infections (V+I+; 23 donors; Table 1).
Differential SARS-CoV-2 CD4+ T cell responses in unexposed, convalescent, and vaccinated subjects
To detect SARS-CoV-2 T cell reactivity, we previously routinely utilized a pool of overlapping peptides spanning the entire S sequence (253 peptides) and a pool of predicted HLA class II binders from the R of the genome (CD4R; 221 peptides) (Grifoni et al., 2020b) (Tables S3 and S4). Here, to further optimize detection of non-S reactivity, we designed epitope pools based on E defined epitopes, from the non-S sequences of the SARS-CoV-2 proteome. The CD4RE and CD8RE megapools (MPs) consisted of 284 and 621 peptides, respectively (Tables S3 and S4). A pool of epitopes derived from an unrelated ubiquitous pathogen Epstein-Barr virus (EBV) (Carrasco Pro et al., 2015) was used as a specificity control (Table S3).
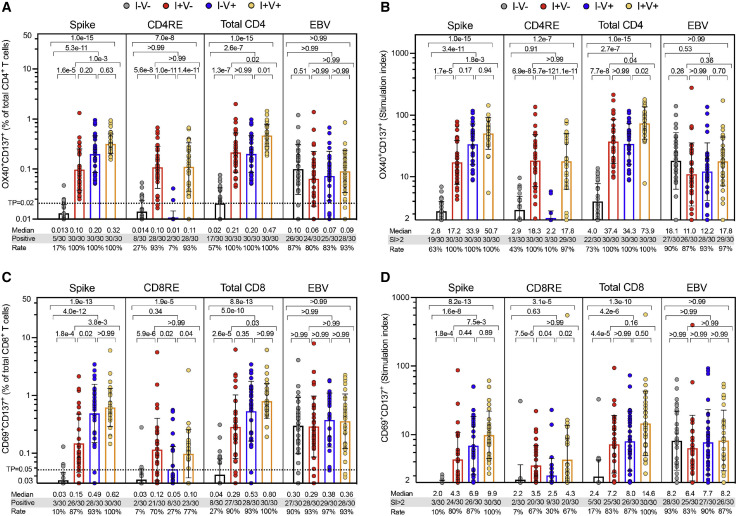
T cell reactivity was assessed by the activation-induced marker (AIM) assays (da Silva Antunes et al., 2021), and data were represented as either absolute magnitude or stimulation index (SI). As shown in Figure 1 A, SARS-CoV-2-specific CD4+ T cell responses were detected in all convalescent and/or vaccinated individuals and approximately 50% of non-infected, non-vaccinated individuals. Similar results were observed when responses were plotted as SI (Figure 1B). Unexposed subjects were associated with significantly lower reactivity as compared to all the other groups (p values ranging 1.3e−7 to 1.0e−15), and convalescent and vaccinated (I+V+) subjects exhibited higher responses than convalescent (I+V−) subjects (p = 0.02 and p = 0.04 for absolute magnitude and SI, respectively) or vaccinated (I−V+) subjects (p = 0.01 and p = 0.02 for absolute magnitude and SI, respectively) (Figures 1A and 1B). Importantly, CD4RE responses were able to differentiate convalescent subjects (I+V− or I+V+) from unexposed and vaccinated (I−V+) subjects with p values ranging 5.6e−8 to 5.7e−12 and vaccinated (I−V+) from infected and vaccinated (I+V+) subjects (p = 1.4e−11 and p = 1.1e−11 for absolute magnitude and SI, respectively) (Figures 1A and 1B). As expected, no statistically significant difference in EBV reactivity was observed when the 4 groups were compared (Figures 1A and 1B).
Figure 1.
SARS-CoV-2-specific CD4+ and CD8+ T cell responses in the study groups
SARS-CoV-2-specific CD4+ and CD8+ T cell responses were measured as percentages of AIM+ (OX40+CD137+) CD4+ T cells (A and B) or AIM+ (CD69+CD137+) CD8+ T cells (C and D) after stimulation of PBMCs with peptides pools encompassing spike-only (Spike) MPs or the experimentally defined CD4RE and CD8RE MPs representing all the proteome without spike. EVB MP was used as a control. Graphs show individual response of spike, CD4RE, or CD8RE and the combination of both (total CD4+ or total CD8+) plotted as background subtracted (A and C) or as SI (B and D) against DMSO negative control. Geometric mean with standard deviation (SD) for the 4 different groups is shown. Kruskal-Wallis test adjusted with Dunn’s test for multiple comparisons was performed, and p values < 0.05 considered statistically significant. I−V−, unexposed and unvaccinated (n = 30); I+V−, infected and non-vaccinated (n = 30); I+V+, infected and then vaccinated (n = 30); I−V+, non-infected and vaccinated (n = 30). Threshold of positivity (TP) is indicated. Median response and the number or percentage of positive responding donors for each group is shown.
Differential SARS-CoV-2 CD8+ T cell and IFNγ FluoroSpot responses in unexposed, convalescent, and vaccinated subjects
SARS-CoV-2-specific CD8+ T cell responses were also broadly detected among all the cohorts studied. CD8+ T cell responses were detected in 90%–100% of the convalescent and/or vaccinated individuals and approximately in 25% of non-infected, non-vaccinated individuals (Figure 1C). Similar responses were observed when plotted as SI (Figure 1D). As observed for CD4+ T cell responses, CD8+ T cell responses of unexposed subjects (I−V−) were discriminated from all the other groups (p values ranging 2.6e−5 to 8.8e−13), and I+V+ infected/vaccinated subjects exhibited higher responses than I+V− convalescent (p = 0.03 and p = 0.16 for absolute magnitude and SI, respectively). Identical results were observed parsing S and CD8RE responses separately, with both able to differentiate convalescent (I+V−) from unexposed and vaccinated (I−V+) subjects (p values ranging 0.02 to 5.9e−6). Importantly, CD8RE responses were able to differentiate vaccinated from infected/vaccinated (I+V+) subjects (p = 0.04 and p = 0.02 for absolute magnitude and SI, respectively) (Figures 1C and 1D). When the 4 groups were compared, no statistically significant difference in EBV reactivity was observed (Figures 1C and 1D).
In parallel, an IFNγ FluoroSpot assay was also employed to evaluate the CD4+ and CD8+ T cell responses using a threshold of 20 IFNγ spot-forming cells (SFCs) per million peripheral blood mononuclear cells (PBMCs). Responses were detected in many infected or vaccinated individuals, and similar results were observed for S, CD4RE, or CD8RE when considering both the absolute magnitude or SI, albeit with predictably lower sensitivity and specificity than AIM assay (Figures S1A and S1B).
Improved performance of the CD4RE pool based on experimentally defined epitopes
Results from both AIM and IFNγ FluoroSpot assays demonstrated that the newly developed CD4RE pool had both improved sensitivity and specificity, compared to the previously used CD4R pool of predicted epitopes (Figures S1C and S1D). In more detail, higher positive CD4+ T cell responses in I+V− (28/30 [93%] versus 26/30 [87%]; p = 2.0e−4) and I+V+ (28/30 [93%] versus 23/30 [77%]; p = 5.0e−6) and lower non-specific response in I−V− (8/30 [27%] versus 14/30 [47%]; p = 0.037) and I−V+ (2/30 [7%] versus 4/30 [13%]; p = 0.031) were detected using CD4RE when compared to CD4R in the AIM assay (Figure S1C). Similar results were shown by IFNγ FluoroSpot assay, albeit with lower sensitivity compared to AIM (Figure S1D). These results demonstrate that the use of E defined, as opposed to predicted, epitopes provides higher signal in SARS-CoV-2-exposed subjects while lowering responses from non-exposed subjects. The fact that E defined epitopes yield better results is consistent with mass spectrometry studies showing the divergence of predicted from HLA-eluted SARS-CoV-2 immunopeptidome (Knierman et al., 2020; Pan et al., 2021; Weingarten-Gabbay et al., 2021).
Classification of subjects with different exposure history based on S and CD4RE reactivity
We reasoned that unexposed (I−V−) subjects would be unreactive to E defined SARS-CoV-2 peptide pools, while uninfected vaccinated (I−V+) subjects should react only to the S pool. We further reasoned that infected (I+V−) subjects should recognize both S and CD4RE, but infected and vaccinated (I+V+) subjects would have a higher relative S reactivity than infected only (I+V−), as is often the case with hybrid immunity (Crotty, 2021), due to exposure to S twice, once during infection and the other during vaccination.
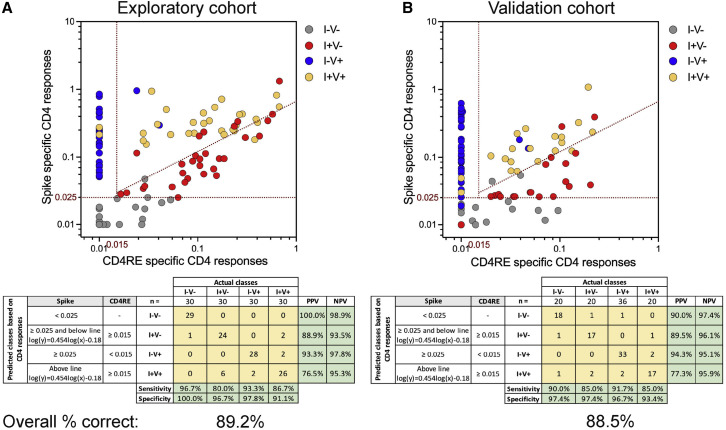
As shown in Figure 2 A, S- and CD4RE-specific CD4+ T cell responses derived from the AIM assay were arranged in a two-dimensional plot. Each dot represents a single subject from a total of 120 donors (30 for each of the 4 groups; Table S1). Optimal cutoffs were established to discriminate the 4 groups, and the positive predictive value (PPV), negative predictive value (NPV), sensitivity, and specificity were calculated for each individual group.
Figure 2.
COVID-19 clinical classification scheme using SARS-CoV-2-specific CD4+ T cell responses
CD4+ T cell responses to spike and CD4RE MPs were measured as percentage of AIM+ (OX40+CD137+) CD4+ T cells and plotted in 2 dimensions as absolute magnitude in order to discriminate the 4 study groups with known COVID-19 status of infection and/or vaccination in 2 independent cohorts: (A) exploratory cohort (n = 120) and (B) validation cohort (n = 96). I−V−, unexposed and unvaccinated (n = 30 and n = 20); I+V−, infected and non-vaccinated (n = 30 and n = 20); I+V+, infected and then vaccinated (n = 30 and n = 20); I−V+, non-infected and vaccinated (n = 30 and n = 36). Red dotted lines indicate specific cutoffs. Table inserts depict the diagnostic exam results in 4×4 matrix. Sensitivity, specificity, PPV, NPV, and overall percentage of subjects classified correctly is shown.
Subjects with S responses lower than 0.025% were classified predictively as unexposed (I−V−) (Figure 2A). 29 out of 29 subjects with responses matching this criterion were correctly classified (100% of PPV), while nearly all the actual I−V− subjects (29 out of 30) were found to be associated with responses below the threshold, corresponding to a sensitivity of 96.7% (Figure 2A [gray circles]). Subjects with S responses greater than 0.025% and CD4RE responses lower than 0.015% were classified predictively as I−V+. 28 out of 30 subjects with responses matching this threshold were correctly classified (93.3% of PPV), and 28 out of the 30 I−V+ subjects detected within this threshold (93.3% of sensitivity) (Figure 2A [blue circles]).
Lastly, subjects with S and CD4RE responses above 0.025% and 0.015%, respectively, and above or below a diagonal line (log(y) = 0.454log(x)−0.18) were classified as I+V+ or I+V−, respectively. 24 out of 27 subjects with responses matching the lower compartment (I+V−) were correctly classified (88.9% of PPV), while 24 out of the 30 I+V− subjects were found to be associated with this threshold (80% of sensitivity) (Figure 2A [red circles]). Conversely, the majority of subjects (26 out of 34) with responses matching the upper compartment (I+V+) were correctly classified (76.5% of PPV), while 26 out of the 30 I+V+ subjects studied were found to be associated with this threshold, corresponding to a sensitivity of 86.7% (Figure 2A [yellow circles]). Further statistical examinations to assess the robustness of the classification scheme as a potential diagnostic test were performed, specifically assessments of specificity and negative predictive value (NPV). High specificity and NPV were observed for each individual group with a range of 91.1%–100% and 93.5%–98.9%, respectively (Figure 2A). In summary, good PPV, NPV, sensitivity, and specificity values were observed across all the groups with an overall classification accuracy of 89.2%.
Validation of the classifier in an independent cohort
To confirm the accuracy of this classification scheme, we assessed CD4+ T cell responses in an independent validation cohort of 96 donors (20 each for I−V−, I+V−, I+V+, and 36 for I−V+; Table S2). As shown in Figure 2B, using the same cutoffs as described above for S and CD4RE responses, similar PPV, NPV, sensitivity, and specificity to the experimental cohort was observed across all the groups in the validation cohort with an overall classification accuracy of 88.5%. To further validate the robustness of this classification scheme, the same data (Figure 2) were plotted as a function of the SI (Figures S2A and S2B). Strikingly, these results paralleled the observations using the absolute magnitude, with a similar overall classification accuracy (86.7% and 85.4% for the exploratory and validation cohorts, respectively).
Applying the same classification scheme using either absolute magnitude or SI for IFNγ responses yielded an overall classification accuracy of 72.5% and 60.0%, respectively (Figures S2C and S2D). A lower accuracy was observed when CD8+ T cell responses from AIM assay were analyzed, as compared to CD4+ T cell responses (data not shown). Overall, these results demonstrate the feasibility of an integrated classification scheme in assessing CD4+ T cell responses as a clinical immunodiagnostic tool. Importantly, it also displays the potential to discriminate previously undetected infection, including in vaccinated individuals.
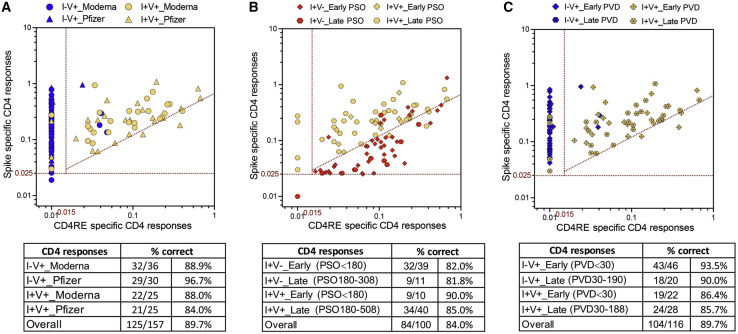
The classification scheme is applicable to different vaccine platforms and different lengths of time post infection/post vaccination
To gain further insights into the applicability of the classification scheme, we sought to further test and validate this tool across vaccine platforms and longer time points PSO or post vaccination. First, we looked at the response classification as a function of whether vaccinated subjects received BNT162b2 or mRNA-1273 vaccines. As shown in Figure 3 A, the overall classification accuracy when using the different mRNA vaccines was 89.7%. Specifically, both vaccines showed similar magnitude for both total CD4+ and CD8+ T cell responses in the I−V+ or I+V+ groups (Figures S3A and S3B). The accuracy of the classification scheme for the different types of vaccines in the combined I−V+ or I+V+ groups was almost identical (88.5% and 90.9% for the mRNA-1273 and BNT162b2 vaccines, respectively) (Table 2 ).
Figure 3.
COVID-19 clinical classification scheme is applicable to different mRNA vaccines and different lengths of time post infection/post vaccination
CD4+ T cell responses to spike and CD4RE MPs were measured as percentage of AIM+ (OX40+CD137+) CD4+ T cells and plotted in 2 dimensions as absolute magnitude in order to discriminated between (A) different types of mRNA vaccines (Moderna versus Pfzier) among vaccinated groups (I−V+ and I+V+), (B) different lengths of time post infection among infected groups (I+V− and I+V+), and (C) different lengths of time post vaccination among vaccinated groups (I−V+ and I+V+). Early infection, PSO ≤ 180; late infection, PSO > 180; early post vaccination, PVD ≤ 30; late post vaccination, PVD > 30. I−V+, non-infected and vaccinated (n = 66); I+V−, infected and non-vaccinated (n = 50); I+V+, infected and then vaccinated (n = 50). Red dotted lines indicate specific cutoffs. Table inserts depict the overall percentage of subjects classified correctly.
Table 2.
Summary of the percentage correct and applicability of classification scheme
| Variable | Group | AIM assaya (% correct) |
|---|---|---|
| type of vaccine | mRNA-1273 | 88.5 |
| BNT162b2 | 90.9 | |
| days PSO | early | 83.7 |
| late | 84.3 | |
| days post vaccination | early | 91.2 |
| late | 87.5 |
CD4+ T cell responses for S and CD4RE pools
Next, we looked at the response classification as a function of the length of time PSO. The overall classification accuracy was of 84.0% (Figure 3B). No differences were observed in the magnitude of both total CD4+ and CD8+ T cell responses between early (≤180 days) and late (>180 days) time points from PSO in either the I+V− or the I+V+ groups (Figures S3C and S3D). CD4+ T cell reactivity associated with different time from PSO was also plotted as a continuous variable (Figure S4A). The accuracy of the classification scheme when considering the different PSO time points was 82.0% and 81.8% in the I+V− group and 90.0% and 85.0% in the I+V+ group for the early and late time points, respectively (Figure 3B).
We also looked at the responses as a function of the length of time from the second dose of vaccination. The overall classification accuracy was of 89.7% (Figure 3C). No differences were observed in the magnitude of both total CD4+ or CD8+ T cell responses between early (≤30 days) or late (>30 days) time points from the last dose of vaccination in either the I−V+ or the I+V+ groups (Figures S3E and S3F). CD4+ T cell reactivity associated with different post-vaccination dates was also plotted as a continuous variable (Figure S4B). The accuracy of the classification scheme when considering the different vaccine time points was 93.5% and 90.0% in the I−V+ group and 86.4% and 85.7% in the I+V+ group for the early and late time points, respectively (Figure 3C).
Lastly, as an alternative to the T cell classification scheme, we classified subjects based on S RBD and N antibody responses. An overall classification accuracy of 69% (Figure S5A) was observed when previously described standard clinical cutoffs were employed (Dan et al., 2021; Grifoni et al., 2020b; Tarke et al., 2021a). The attempt to classify infected individuals at late PSO time points resulted in even lower accuracies (Figure S5B), consistent with reports that N positivity is relatively short lived (Dan et al., 2021; Ibarrondo et al., 2020; Ortega et al., 2021). We next examined the possibility that this low classification accuracy might be reflective of suboptimal thresholds. By setting more stringent cutoffs based on the optimal classification of the exploratory cohort, we achieved an overall classification accuracy of 84.2% (Figure S5C). However, when the same classification scheme was applied to the validation cohort, the overall accuracy decreased to 52.1% (Figure S5D), indicating that the previous value was likely a result of data overfitting. Overall, the use of antibody responses failed to yield a useful classification scheme, unlike the classification scheme using CD4+ T cell responses, which proved to be a robust tool that can accurately classify subjects regardless of the days post infection/post vaccination or type of vaccine administered (Table 2).
CD4+ T cell reactivity of subjects associated with breakthrough infections
Breakthrough infections are defined as cases of previously COVID-19 vaccinated individuals associated with positive SARS-CoV-2 PCR tests (Bergwerk et al., 2021; Kustin et al., 2021; Mizrahi et al., 2021). Studies of antibody or T cell responses associated with breakthrough infection are scarce (Collier et al., 2021; Rovida et al., 2021). Breakthrough infection might be associated with increased immune responses as a result of the re-exposure (hybrid immunity) (Collier et al., 2021). In other cases, subjects experiencing breakthrough infections might be associated with general weaker immune responsiveness or decrease of vaccine effectiveness (Klompas, 2021; Mizrahi et al., 2021).
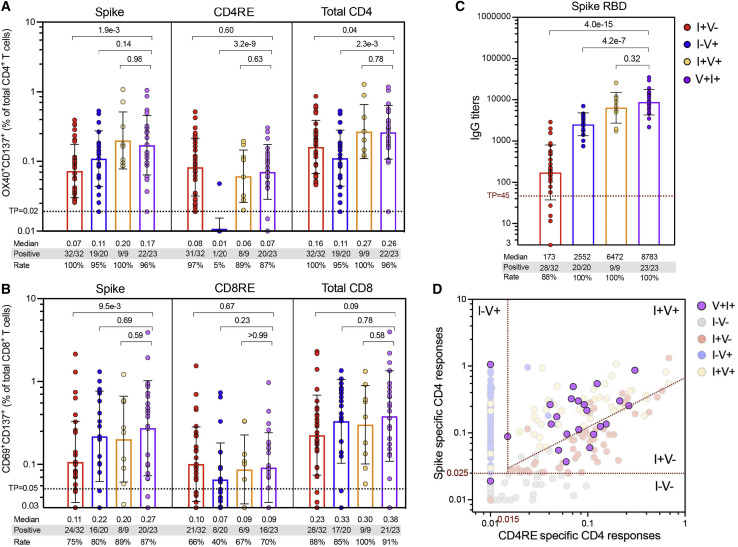
Here, we assessed S and CD4RE T cell responses in a group (n = 23) of breakthrough infected individuals (V+I+). Responses were compared to the vaccinated (I−V+), infected (I+V−), or infected and then vaccinated (I+V+) groups matching the V+I+ intervals of vaccination and infection (55−271 and 18−93 days, respectively). As shown in Figure 4 A, CD4+ T cell responses from V+I+ subjects were associated with significant higher levels compared to I+V− (p = 0.04) and I−V+ (p = 2.3e−3) subjects and similar magnitude as the I+V+ subjects. CD8+ T cell responses had comparable levels across all the groups (Figure 4B). Similar to CD4+ T cell responses, S RBD IgG titers from V+I+ subjects were equivalent to I+V+ subjects and significantly higher than I+V− (p = 4.2e−7) and I−V+ (p = 4.0e−15) subjects (Figure 4C). Thus, at the population level, breakthrough infections are associated with CD4+ T cell and S IgG responses that resemble hybrid immunity.
Figure 4.
SARS-CoV-2 T cell and antibody response in breakthrough infection cases: comparison to other study groups
(A and B) SARS-CoV-2-specific T cell responses were measured as percentage of (A) AIM+ (OX40+CD137+) CD4+ T cells or (B) AIM+ (CD69+CD137+) CD8+ T cells after stimulation of PBMCs with spike and CD4RE or CD8RE peptide pools.
(C) Comparison of anti-spike RBD IgG titers in the plasma of the different study groups. For both T cell and antibody determinations, only donors matching the V+I+ intervals of vaccination and infection (55–271 and 18–93 days, respectively) were plotted.
(A-C) Graph bars show geometric mean with SD. Threshold of positivity (TP), median response, and the number or percentage of positive responding donors for each group is indicated. Kruskal-Wallis test adjusted with Dunn’s test for multiple comparisons was performed, and p values < 0.05 considered statistically significant.
(D) V+I+ CD4+ T cell responses plotted using the two-dimensional classification scheme with the specific cutoffs attributed to the different study groups (red dotted lines). I−V−, unexposed and unvaccinated (n = 50); I+V−, infected and non-vaccinated (n = 50); I+V+, infected and then vaccinated (n = 50); I−V+, non-infected and vaccinated (n = 66); V+I+, vaccinated and then infected (n = 23).
The classification scheme captures heterogeneity in breakthrough infections
At the level of the T cell response classification scheme, individuals who had COVID-19 were effectively segregated from non-infected groups (unexposed and vaccinated) (Figure 4D). We further expected that the V+I+ breakthrough infections would be classified in the same manner of I+V+ hybrid immunity samples. Approximately two thirds (15/23) of subjects were identified by the same thresholds associated with responses from the I+V+ group (“high responders”), while the remaining third were classified similarly to I+V− subjects (“low responders”). No obvious difference in terms of age, PSO, PVD, disease severity, or length of infection from vaccination was detected between these donors and the high responders sub-group of 15 donors (Figure S6; Table 1).
In summary, while T cell responses following breakthrough infections (V+I+) are effectively segregated from the responses of uninfected donors (vaccinated or not) and follow the same pattern of responses of individuals vaccinated following natural infection (I+V+), in the majority of the cases, the classification scheme revealed heterogeneity in the CD4+ T cell responses of breakthrough donors.
Validation of the classification scheme with whole study cohort
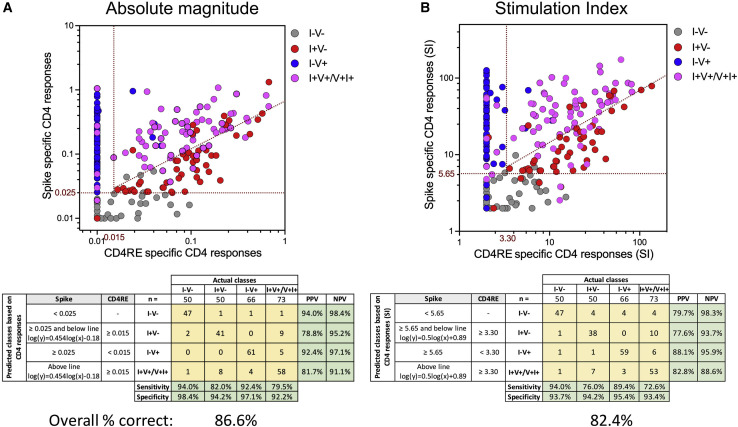
Finally, we summarized the overall accuracy of the classification scheme across the 5 cohorts used in this study, including breakthrough infections. For this purpose, we clustered individuals that had been infected and vaccinated, irrespective of the event that occurred first, into a single group, i.e., I+V/V+I+ (Figure 5 ). When the 239 subjects with distinct COVID-19 status of infection and/or vaccination were combined, the classification scheme achieved a high overall accuracy, either as function of absolute magnitude (86.6%) or SI (82.4%). Also, high specificity and NPV were retained for each individual group with a range of 92.2%–98.4% and 88.6%–98.4%, respectively. These results further illustrate the highly predictive power of this classification scheme and its broad clinical applicability.
Figure 5.
Overall COVID-19 clinical classification scheme
(A and B) CD4+ T cell responses to spike and CD4RE MPs were measured as percentage of AIM+ (OX40+CD137+) CD4+ T cells and plotted in two dimensions as (A) SFCs per million PBMCs or (B) stimulation index (SI), in order to discriminate the 5 study groups with known COVID-19 status of infection and/or vaccination. I−V−, unexposed and unvaccinated (n = 50); I+V−, infected and non-vaccinated (n = 50); I−V+, non-infected and vaccinated (n = 66); I+V+/V+I+, infected and then vaccinated (I+V+, n = 50) merged with vaccinated and then infected (V+I+, n = 23). Red dotted lines indicate specific cutoffs. Table inserts depict the diagnostic exam results in 4×4 matrix. Sensitivity, specificity, PPV, NPV of all the subjects that participated in this study (n = 239), and overall percentage classified correctly is shown.
Discussion
There is a need to understand the roles of SARS-CoV-2 T cell responses as potential correlates of disease outcome and/or correlates of vaccine protection from infection or severe disease. Herein, we present the results of T cell quantitation based on the determination of relative activity directed against S and the rest of the genome by the use of optimized pools of E defined epitopes (CD4RE and CD8RE). We report successful classification of subjects with different COVID-19 vaccination or natural infection history in the 85%–90% range of accuracy. We further show that the strategy is applicable to characterizing immune responses in a group of infected vaccinees (i.e., breakthrough infections).
Although previous reports studied responses to SARS-CoV-2 in either unexposed, COVID-19-infected, or vaccinated individuals (da Silva Antunes et al., 2021; Dan et al., 2021; Goel et al., 2021; Grifoni et al., 2020b; Le Bert et al., 2020; Mateus et al., 2021), this is the first demonstration, to the best of our knowledge, that a simple assay strategy can classify T cell responses measured simultaneously in 5 different groups of known COVID-19 status of infection and/or vaccination. The improved sensitivity and specificity resulted from the concept of considering the relative magnitude of responses against the S and “rest of the genome” components, which overcomes issues related to the fact that magnitude of responses may wane over time, and also from the inclusion of E defined epitopes, which we show are associated with improved signal and selectivity as compared to previously utilized predicted epitopes.
We suggest that the combined use of overlapping S and CD4RE pools can be used to detect differential and relative reactivity to different SARS-CoV-2 antigens and therefore classify individuals based on SARS-CoV-2 infection and COVID-19 vaccine status. More importantly, this approach allows to identify bona fide exposition to SARS-CoV-2, even in individuals that have been vaccinated, thus effectively distinguishing COVID-19 vaccine and infection history. This is of importance, as current COVID-19 diagnostic practices rely heavily on subjectively reported history, clinical records, and lab modalities with imperfect performance, leading to limited reliability. For example, in longitudinal vaccination studies, it will be important to monitor whether subjects enrolled in the studies might have been associated with asymptomatic infection (Kustin et al., 2021; Mizrahi et al., 2021; Pouwels et al., 2021) or even associated with abortive seronegative infections (Swadling et al., 2021). Also, diagnosis of past COVID-19 infections based on T cell reactivity could be an element considered in the context of booster vaccinations. Monitoring the differential T cell reactivity associated with vaccination versus infection might provide important information in terms of correlating T cell immunity with protection from infection and disease in a setting where an increasingly high fraction of the general population might have been associated with both vaccination and infection. Continued monitoring of vaccine- versus infection-induced T cell responses might be of interest in light of the ongoing controversy over whether vaccinations protect against long COVID (Massey et al., 2021a, 2021b; Taquet et al., 2021) or provide improved protection for immunocompromised vulnerable subjects. Distinguishing T cell responses induced by vaccination versus infection might be also of interest in the context of individual COVID-19 certifications (e.g., “health passes”) and further characterizing individuals that might have been exposed but have not tested positive or had false negative results for COVID-19 using a molecular or antigen diagnostic test.
Our study builds on the well-known fact that infected individuals mount a T cell response against multiple SARS-CoV-2 antigens and that individuals vaccinated with mRNA vaccines are mounting only a T cell response to S. A detailed classification of T cells response in different categories of vaccinated/infected individuals has not been described and compared as in the current study. Indeed, the use of our developed pools, spanning all the antigens from SARS-CoV-2, allowed for detection of SARS-CoV-2 responses with increased sensitivity and specificity compared to other studies performing T cell assays using only S or other SARS-CoV-2 antigens (Krishna et al., 2021; Kruse et al., 2021; Martínez-Gallo et al., 2021; Murugesan et al., 2020; Tan et al., 2021; Zelba et al., 2021).
We also show that similar results were observed when relative versus absolute determinations were employed to measure T cell responses (i.e., using SI or absolute magnitude), which allows for a more generalized use of the classification tool in different flow-cytometer platforms. The robustness of the T cell-based classification scheme was validated in an independent cohort exhibiting identical performances and was applicable to different types of mRNA vaccines, even when considering extended periods of time elapsed from infection and/or vaccination. T cell responses might differ according to the vaccine platform. Also, despite the wide range of time interval following the second vaccine dose between groups, and even when considering extended periods of time elapsed from infection and/or vaccination, the classification scheme performance remained unchanged.
The strength of the approach is further demonstrated by the fact that T cell responses act as a better classifier than antibody responses, consistent with the notion that antibody responses to N protein are short lived (Dan et al., 2021; Ibarrondo et al., 2020; Ortega et al., 2021). Also, while applicable to data generated by FluoroSpot cytokine assays, despite the lower intrinsic sensitivity of this assay, we anticipate that this assay strategy will be broadly applicable to other readouts, such as ICS (Cohen et al., 2021; Mateus et al., 2021) and whole blood in an interferon-gamma release assay (Murugesan et al., 2020; Petrone et al., 2021; Tan et al., 2021).
T cell responses from breakthrough infections were also evaluated, and high levels of CD4+ and CD8+ T cell reactivity were observed. Elevated T cell responsiveness was paralleled by high levels of S RBD IgG. Interestingly, these responses were of similar magnitude as responses from a group of individuals infected and then vaccinated (I+V+ in our study) whose features are commonly associated with hybrid immunity (Crotty, 2021). Notably, breakthrough infections were also associated with higher CD4+ T cell and S RBD IgG responses compared to only-infected or only-vaccinated subjects. These results suggest that T and B cell reactivity associated with breakthrough infections is increased as a result of re-exposure. However, the classification tool system also revealed significant heterogeneity in responses in some subjects, possibly linking some breakthrough infections to lower adaptive responses. A more detailed analysis at T cell epitope level could better define whether differences in T cell responses occur in this category of “low responders” when compared to only-vaccinated and only-infected individuals.
Limitations of the study
Although our findings were validated in several different cohorts, further validation in larger and more ethnically diverse populations and with different HLA backgrounds is warranted. Further studies using cohorts of asymptomatic subjects associated with PCR positive tests and lack of clinical symptoms are required to address the performance of a T cell-based immunodiagnostic scheme in the identification of asymptomatic infections. Further additional studies will also have to address the performance of the classification scheme in assessing responses associated with variants of concern such as delta and omicro, and responses observed after 3 vaccine administrations. The study of additional cohorts representative of different vaccine platforms (e.g., Ad26.COV2.S, ChAdOx1 nCoV-19, or CoronaVac) will be important to generalize these findings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD3 (AF532) (UCHT1) | LifeTech | Cat#: 58-0038-42; RRID: AB_11218675 |
| anti-CD4 (BV605) (RPA-T4) | BD Biosciences | Cat#: 562658; RRID: AB_2737935 |
| anti-CD8 (BUV496) (RPA-T8) | BD Biosciences | Cat#: 612942; RRID: AB_2563505 |
| anti-CD14 (V500) (M5E2) | BD Biosciences | Cat#: 561391; RRID: AB_10611856 |
| anti-CD19 (V500) (HIB19) | BD Biosciences | Cat#: 561121; RRID: AB_10562391 |
| anti-CD137 (APC) (4B4-1) | Biolegend | Cat#: 309810; RRID: AB_830672 |
| anti-CD134 (PE-Cy7) (Ber-ACT35) | Biolegend | Cat#: 350012; RRID: AB_10901161 |
| anti-CD69 (PE) (FN50) | BD Biosciences | Cat#: 555531; RRID: AB_2737680 |
| anti-CD45RA (BV421) (HI100) | Biolegend | Cat#: 304130; RRID: AB_10965547 |
| anti-CCR7 (FITC) (G043H7) | Biolegend | Cat#: 353216; RRID: AB_10916386 |
| Live/Dead Viability (eF506/Aqua) | Invitrogen | Cat#: 65-0866-18; RRID: N/A |
| Biological samples | ||
| Human blood samples | La Jolla Institute | https://www.lji.org |
| Human blood samples | UC San Diego Health | https://health.ucsd.edu/Pages/default.aspx |
| Human blood samples | BioIVT | https://bioivt.com/ |
| Chemicals, peptides, and recombinant proteins | ||
| Spike megapool | Grifoni et al., 2020b | https://doi.org/10.17632/s6gpnrmxg2.1 |
| CD4RE megapool | Grifoni et al., 2021 | https://doi.org/10.17632/s6gpnrmxg2.1 |
| CD8RE megapool | Grifoni et al., 2021 | https://doi.org/10.17632/s6gpnrmxg2.1 |
| CD4R megapool | Grifoni et al., 2020b | https://www.cell.com/cell/pdf/S0092-86742030610-3.pdf |
| EBV megapool | Carrasco Pro et al., 2015 | https://www.hindawi.com/journals/jir/2015/763461/ |
| SARS-CoV-2 Receptor Binding Domain (RBD) protein | La Jolla Institute | https://www.lji.org |
| SARS-CoV-2 nucleocapsid (N) protein | Genscript | https://www.genscript.com |
| Software and algorithms | ||
| GraphPad Prism Version 9 | GraphPad Software | https://www.graphpad.com |
| Microsoft Excel Version 16.16.27 | Microsoft | https://www.microsoft.com/en-us/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact: Dr. Ricardo da Silva Antunes (rantunes@lji.org).
Materials availability
Epitope pools used in this study will be made available to the scientific community upon request, and following execution of a material transfer agreement (MTA), by contacting A.S. (alex@lji.org) and R.d.S.A (rantunes@lji.org). Likewise, biomaterials archived from this study may be shared for further research with MTA.
Method details
Human subjects and PBMC isolation
The Institutional Review Boards of the University of California, San Diego (UCSD; 200236X) and the La Jolla Institute for Immunology (LJI; VD-214) approved the protocols used for blood collection for all the subjects who donated at all sites. The vast majority of the blood donations were collected through the UC San Diego Health Clinic and at the La Jolla Institute for Immunology (LJI). Additional samples were obtained from contract research organizations (CRO) under the same LJI IRB approval. All samples with the exception of the I-V- study group were collected during COVID-19 pandemic from 2020-2021. Pre-pandemic blood donations of the I-V- group were performed from 2013-2019. Each participant provided informed consent and was assigned a study identification number with clinical information recorded. Subjects who had a medical history and/or symptoms consistent with COVID-19, but lacked positive PCR-based testing for SARS-CoV-2 and subsequently had negative laboratory-based serologic testing for SARS-CoV-2, were then excluded; i.e., all COVID-19 cases in this study were confirmed cases by SARS-CoV-2 PCR or SARS-CoV-2 serodiagnostics, or both. Adults of all races, ethnicities, ages, and genders were eligible to participate, but the association of gender on the results of the study was not explicitly measured. Study exclusion criteria included lack of willingness to participate, lack of ability to provide informed consent, or a medical contraindication to blood donation (e.g., severe anemia). In all cases, PBMCs were isolated from whole blood by density gradient centrifugation according to manufacturer instructions (Ficoll-Hypaque, Amersham Biosciences, Uppsala, Sweden). Cells were cryopreserved in liquid nitrogen suspended in FBS containing 10% (vol/vol) DMSO (Sigma-Aldrich). Plasma was obtained by centrifugation (400 g for 15 min at 4°C) of whole blood and collection of the upper layer, prior to PBMC isolation and cryopreserved at −80°C.
Design and production of new SARS-CoV-2 epitope pools
To study T cell responses against SARS-CoV-2, we used a megapool (MP) of 15-mer peptides overlapping by 10 spanning the entire spike protein sequence (253 peptides; Table S3) as previously described (Grifoni et al., 2020b). For the rest of the SARS-CoV-2 proteome, and in order to design epitope pools with increased HLA coverage and broadly recognized by demographically and geographically diverse populations, experimental defined epitopes from non-spike (R) region of SARS-CoV-2 were selected based on our recent meta-analysis (Grifoni et al., 2021). Briefly, peptides were synthetized and pooled to include both dominant (recognized in 3 or more donors/studies) and subdominant epitopes. To improve specificity, overly short or long ligands which could cause “false positive” signals (Paul et al., 2018), were excluded and only peptides of sizes ranging 15-20 and 9-10 amino acids, respectively in CD4RE and CD8RE pools were included, resulting in the generation of CD4RE and CD8RE MPs with 284 and 621 peptides, respectively (Table S3). Epitopes were further classified in dominant and subdominant based on the frequency of individual responses as previously described (Grifoni et al., 2021). In addition, detailed information of the MPs composition with peptide sequences, length, ORFs of origin, and HLA coverages are specified in Table S4. Alternatively, a MP for the remainder genome consisting of dominant HLA class II predicted CD4+ T cell epitopes (221 peptides), as previously described (Grifoni et al., 2020b) was also used as control (Table S3). In addition, an EBV pool of previously reported experimental class I and class II epitopes (Carrasco Pro et al., 2015) with 301 peptides was used as positive control. All peptides were synthesized by TC peptide lab (San Diego, CA), pooled and resuspended at a final concentration of 1 mg/mL in DMSO.
SARS-CoV-2 RBD spike and nucleocapsid ELISAs
The SARS-CoV-2 ELISAs have been described in detail previously (Dan et al., 2021). Briefly, 96-well half-area plates (ThermoFisher 3690) were coated with 1 ug/mL of antigen and incubated at 4°C overnight. Antigens included recombinant SARS-CoV-2 RBD protein obtained from the Saphire laboratory at LJI or recombinant nucleocapsid protein (GenScript Z03488). The next day, plates were blocked with 3% milk in phosphate-buffered saline (PBS) containing 0.05% Tween-20 for 1.5 h at room temperature. Plasma was heat inactivated at 56°C for 30 to 60 min. Plasma was diluted in 1% milk containing 0.05% Tween-20 in PBS starting at a 1:3 dilution followed by serial dilutions by three and incubated for 1.5 h at room temperature. Plates were washed five times with 0.05% PBS-Tween-20. Secondary antibodies were diluted in 1% milk containing 0.05% Tween-20 in PBS. Anti-human IgG peroxidase antibody produced in goat (Sigma A6029) was used at a 1:5,000 dilution. Subsequently, plates were read on Spectramax Plate Reader at 450 nm, and data analysis was performed using SoftMax Pro. End-point titers were plotted for each sample, using background-subtracted data. Negative and positive controls were used to standardize each assay and normalize across experiments. Limit of detection (LOD) was defined as 1:3 of IgG. Spike RBD IgG or nucleocapsid IgG thresholds of positivity (TP) for SARS-CoV-2 infected or COVID-19 vaccinated individuals were established based on uninfected and unvaccinated subjects (I-V-).
Activation induced cell marker (AIM) assay
The AIM assay was performed as previously described (Mateus et al., 2020). Cryopreserved PBMCs were thawed by diluting the cells in 10 mL complete RPMI 1640 with 5% human AB serum (Gemini Bioproducts) in the presence of benzonase [20 mL/10ml]. Cells were cultured for 20 to 24 h in the presence of SARS-CoV-2 specific and EBV pools (1ug/mL) in 96-wells U bottom plates with 1x106 PBMC per well. An equimolar amount of DMSO was added as a negative control and phytohemagglutinin (PHA, Roche (San Diego, CA) 1 mg/mL) was used as the positive control. The cells were stained with CD3 AF532, CD4 BV605, CD8 BUV496, and Live/Dead Aqua. Activation was measured by the following markers: CD137 APC, OX40 PE-Cy7, and CD69 PE. The detailed information of the antibodies used are summarized in Table S5. All samples were acquired on a ZE5 cell analyzer (Biorad laboratories, Hercules, CA) and analyzed with FlowJo software (Tree Star, Ashland, OR).
CD4+ and CD8+ T cells responses were calculated as percent of total CD4+ (OX40+CD137+) or CD8+ (CD69+CD137+) T cells. The background was removed from the data by subtracting the wells stimulated with DMSO. The Stimulation Index (SI) was calculated by dividing the counts of AIM+ cells after SARS-CoV-2 pools stimulation with the ones in the negative control. A positive response was defined as SI32 and AIM+ response above the threshold of positivity after background subtraction. The limit of detection (0.01% and 0.03 for CD4+ and CD8+ T cells, respectively) was calculated based on 2 times 95% CI of geomean of negative control (DMSO), and the threshold of positivity (0.02% for CD4+ and 0.05% for CD8+ T cells) was calculated based on 2 times standard deviation of background signals according to previous published studies (Dan et al., 2021; Mateus et al., 2020). The gating strategy utilized is shown in Figure S7, as well as reactive CD4+ and CD8+ T cell responses to SARS-CoV-2, EBV and PHA positive control from a representative donor.
IFNγ FluoroSpot assay
The FluoroSpot assay was performed as previously described (Tarke et al., 2021a). PBMCs derived from 80 subjects from 4 clinical cohorts (20 each for I-V-, I+V-, I-V+, and I+V+ cohorts) were stimulated in triplicate at a single density of 2x105 cells/well. The cells were stimulated with the different MPs analyzed (1ug/mL), PHA (10mg/mL), and DMSO (0.1%) in 96-well plates previously coated with anti-cytokine antibodies for IFNγ, (mAbs 1-D1K; Mabtech, Stockholm, Sweden) at a concentration of 10ug/mL. After 20-24 h of incubation at 37°C, 5% CO2, cells were discarded and FluoroSpot plates were washed and further incubated for 2 h with cytokine antibodies (mAbs 7-B6-1-BAM; Mabtech, Stockholm, Sweden). Subsequently, plates were washed again with PBS/0.05% Tween20 and incubated for 1 h with fluorophore-conjugated antibodies (Anti-BAM-490). Computer-assisted image analysis was performed by counting fluorescent spots using an AID iSPOT FluoroSpot reader (AIS-diagnostika, Germany). Each megapool was considered positive compared to the background based on the following three criteria: 20 or more IFNγ spot forming cells (SFC) per 106 PBMC after background subtraction (Threshold defined as 2 times standard deviation of background signals), a stimulation index (SI) greater than 2, and statistically different from the background (p < 0.05) in either a Poisson or t test as previously described (Oseroff et al., 2005).
Statistical analysis
Experimental data were analyzed by GraphPad Prism Version 9 (La Jolla, CA) and Microsoft Excel Version 16.16.27 (Microsoft, Redmond, WA). The statistical details of the experiments are provided in the respective figure legends. Data were analyzed by Wilcoxon test (two-tailed) to compare between two paired groups, and Kruskal-Wallis test adjusted with Dunn’s test for multiple comparisons to compare between multiple groups. Data were plotted as geometric mean with geometric SD p values < 0.05 (after adjustment if indicated) were considered statistically significant. For the classification scheme, statistical determinations and metrics were executed as previously described (Trevethan, 2017). Briefly, for each individual group the following calculations were performed: 1) positive predictive value (PPV) = (True Positives)/(True Positives+False Positives); 2) negative predictive value (NPV) = (True Negatives)/(True Negatives+False Negatives); 3) sensitivity = (True Positives)/(True Positives+False Negatives); and 4) specificity = (True Negatives)/(True Negatives+False Positives).
Study approval
This study was approved by the Human Subjects Protection Program of the UC San Diego Health under IRB approved protocols (UCSD; 200236X), or under IRB approval (LJI; VD-214) at the La Jolla Institute for Immunology. All donors were able to provide informed consent, or had a legal guardian or representative able to do so. Each participant provided informed consent and was assigned a study identification number with clinical information recorded.
Acknowledgments
We wish to acknowledge all subjects for their participation and for donating their blood and time for this study. We are grateful to the La Jolla Institute for Immunology clinical core’s relentless efforts and for the support of Sanguine, BioIVT, and iSpecimen in obtaining blood samples. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award number U19 AI142742 and contract numbers 75N93019C00065 and 5N9301900066. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, E.D.Y., R.d.S.A., and A. Sette; methodology, E.D.Y., J.M.D., E.W., E.G., A. Sutherland, A.T., B.G., J.C., R.I.G., J.M., and A.G.; formal analysis, E.D.Y., J.M.D., R.d.S.A, and A. Sette; investigation, E.D.Y., S.C., R.d.S.A., and A. Sette; support of investigation, S.I.R., S.A.R., D.M.S., G.F., and A.F.; funding acquisition, D.W., S.C., and A. Sette; writing, E.D.Y., S.C., R.d.S.A., and A. Sette; supervision, E.D.Y., R.d.S.A., S.C., and A. Sette.
Declaration of interests
A. Sette is a consultant for Gritstone Bio, Flow Pharma, Arcturus Therapeutics, ImmunoScape, CellCarta, Avalia, Moderna, Fortress, and Repertoire. S.C. is a consultant for Avalia. La Jolla Institute for Immunology (LJI) has filed for patent protection for various aspects of SARS-CoV-2 epitope pools design. All other authors declare no conflict of interest.
Published: February 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.02.003.
Supplemental information
Data and code availability
-
•
The datasets generated and analyzed in this study will be shared by the lead contact upon reasonable request. Additional supplemental items are available from Mendeley Data at https://doi.org/10.17632/s6gpnrmxg2.1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoletti A., Le Bert N., Qui M., Tan A.T. SARS-CoV-2-specific T cells in infection and vaccination. Cell. Mol. Immunol. 2021;18:2307–2312. doi: 10.1038/s41423-021-00743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Pro S., Sidney J., Paul S., Lindestam Arlehamn C., Weiskopf D., Peters B., Sette A. Automatic Generation of Validated Specific Epitope Sets. J. Immunol. Res. 2015;2015:763461. doi: 10.1155/2015/763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen K.W., Linderman S.L., Moodie Z., Czartoski J., Lai L., Mantus G., Norwood C., Nyhoff L.E., Edara V.V., Floyd K., et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med. 2021;2:100354. doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier A.Y., Brown C.M., McMahan K., Yu J., Liu J., Jacob-Dolan C., Chandrashekar A., Tierney D., Ansel J.L., Rowe M., et al. Immune Responses in Fully Vaccinated Individuals Following Breakthrough Infection with the SARS-CoV-2 Delta Variant in Provincetown, Massachusetts. medRxiv. 2021 doi: 10.1101/2021.10.18.21265113. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Hybrid immunity. Science. 2021;372:1392–1393. [Google Scholar]
- da Silva Antunes R., Pallikkuth S., Williams E., Dawen Yu E., Mateus J., Quiambao L., Wang E., Rawlings S.A., Stadlbauer D., Jiang K., et al. Differential T-Cell Reactivity to Endemic Coronaviruses and SARS-CoV-2 in Community and Health Care Workers. J. Infect. Dis. 2021;224:70–80. doi: 10.1093/infdis/jiab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Oxford COVID Vaccine Trial Group Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe. 2021;29:1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Klompas M. Understanding Breakthrough Infections Following mRNA SARS-CoV-2 Vaccination. JAMA. 2021;326:2018–2020. doi: 10.1001/jama.2021.19063. [DOI] [PubMed] [Google Scholar]
- Knierman M.D., Lannan M.B., Spindler L.J., McMillian C.L., Konrad R.J., Siegel R.W. The Human Leukocyte Antigen Class II Immunopeptidome of the SARS-CoV-2 Spike Glycoprotein. Cell Rep. 2020;33:108454. doi: 10.1016/j.celrep.2020.108454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koup R.A., Donis R.O., Gilbert P.B., Li A.W., Shah N.A., Houchens C.R. A government-led effort to identify correlates of protection for COVID-19 vaccines. Nat. Med. 2021;27:1493–1494. doi: 10.1038/s41591-021-01484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. A correlate of protection for SARS-CoV-2 vaccines is urgently needed. Nat. Med. 2021;27:1147–1148. doi: 10.1038/s41591-021-01432-4. [DOI] [PubMed] [Google Scholar]
- Krammer F. Correlates of protection from SARS-CoV-2 infection. Lancet. 2021;397:1421–1423. doi: 10.1016/S0140-6736(21)00782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna B.A., Lim E.Y., Mactavous L., NIHR Bioresource Team. Lyons P.A., Doffinger R., Bradley J., Smith K.G.C., Sinclair J., Matheson N.J., et al. Retrospective diagnosis of SARS-CoV-2 infection in patients with Long COVID by measuring specific T cell mediated IL-2 release. SSRN. 2021 doi: 10.2139/ssrn.3947817. Preprint at. [DOI] [Google Scholar]
- Kruse M., Dark C., Aspden M., Cochrane D., Competiello R., Peltz M., Torres L., Wrighton-Smith P., Dudek M. Performance of the T-SPOT(R).COVID test for detecting SARS-CoV-2-responsive T cells. Int. J. Infect. Dis. 2021;113:155–161. doi: 10.1016/j.ijid.2021.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leshchinsky M., Ken Dror S., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat. Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lind Enoksson S., Bergman P., Klingström J., Boström F., Da Silva Rodrigues R., Winerdal M.E., Marits P. A flow cytometry-based proliferation assay for clinical evaluation of T-cell memory against SARS-CoV-2. J. Immunol. Methods. 2021;499:113159. doi: 10.1016/j.jim.2021.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Ojalvo D., Camara C., Lopez-Granados E., Nozal P., Del Pino-Molina L., Bravo-Gallego L.Y., Paz-Artal E., Pion M., Correa-Rocha R., Ortiz A., et al. Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in naive and COVID-19 recovered individuals. Cell Rep. 2021;36:109570. doi: 10.1016/j.celrep.2021.109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Vogels C.B.F., Yildirim I., Rothman J.E., Lu P., Monteiro V., Gehlhausen J.R., Campbell M., Silva J., Tabachnikova A., et al. Yale SARS-CoV-2 Genomic Surveillance Initiative Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature. 2021;600:523–529. doi: 10.1038/s41586-021-04085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Gallo M., Esperalba J., Pujol-Borrell R., Sandá V., Arrese-Muñoz I., Fernández-Naval C., Antón A., Cardona V., Labrador-Horrillo M., Pumarola T., Hernandéz-González M. Commercialized kits to assess T-cell responses against SARS-CoV-2 S peptides. A pilot study in health care workers. Med. Clin. 2021 doi: 10.1016/j.medcli.2021.09.013. Published online September 25, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey D., Berrent D., Akrami A., Assaf G., Davis H., Harris K., McCorkell L., Ring A.M., Schulz W.L., Wei H., et al. Change in Symptoms and Immune Response in People with Post-Acute Sequelae of SARS-Cov-2 Infection (PASC) After SARS-Cov-2 Vaccination. medRxiv. 2021 doi: 10.1101/2021.07.21.21260391. Preprint at. [DOI] [Google Scholar]
- Massey D., Berrent D., Krumholz H. Breakthrough Symptomatic COVID-19 Infections Leading to Long Covid: Report from Long Covid Facebook Group Poll. medRxiv. 2021 doi: 10.1101/2021.07.23.21261030. Preprint at. [DOI] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi B., Lotan R., Kalkstein N., Peretz A., Perez G., Ben-Tov A., Chodick G., Gazit S., Patalon T. Correlation of SARS-CoV-2-breakthrough infections to time-from-vaccine. Nat. Commun. 2021;12:6379. doi: 10.1038/s41467-021-26672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan K., Jagannathan P., Altamirano J., Maldonado Y.A., Bonilla H.F., Jacobson K.B., Parsonnet J., Andrews J.R., Shi R.-Z., Boyd S., et al. Long term accuracy of SARS-CoV-2 Interferon-gamma release assay and its application in household investigation. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac045. Published January 25, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan K., Jagannathan P., Pham T.D., Pandey S., Bonilla H.F., Jacobson K., Parsonnet J., Andrews J.R., Weiskopf D., Sette A., et al. Interferon-gamma release assay for accurate detection of SARS-CoV-2 T cell response. Clin. Infect. Dis. 2020;73:e3130–e3132. doi: 10.1093/cid/ciaa1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessl J., Sekine T., Buggert M. T cell immunity to SARS-CoV-2. Semin. Immunol. 2021;55:101505. doi: 10.1016/j.smim.2021.101505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbe A., Kronsteiner B., Skelly D.T., Pace M., Brown A., Adland E., Adair K., Akhter H.D., Ali M., Ali S.E., et al. Oxford Immunology Network Covid-19 Response T Cell Consortium. Oxford Protective T Cell Immunology for COVID-19 (OPTIC) Clinical Team T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat. Commun. 2021;12:2055. doi: 10.1038/s41467-021-21856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega N., Ribes M., Vidal M., Rubio R., Aguilar R., Williams S., Barrios D., Alonso S., Hernández-Luis P., Mitchell R.A., et al. Seven-month kinetics of SARS-CoV-2 antibodies and role of pre-existing antibodies to human coronaviruses. Nat. Commun. 2021;12:4740. doi: 10.1038/s41467-021-24979-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C., Kos F., Bui H.H., Peters B., Pasquetto V., Glenn J., Palmore T., Sidney J., Tscharke D.C., Bennink J.R., et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc. Natl. Acad. Sci. USA. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K., Chiu Y., Huang E., Chen M., Wang J., Lai I., Singh S., Shaw R.M., MacCoss M.J., Yee C. Mass spectrometric identification of immunogenic SARS-CoV-2 epitopes and cognate TCRs. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2111815118. e2111815118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Karosiene E., Dhanda S.K., Jurtz V., Edwards L., Nielsen M., Sette A., Peters B. Determination of a Predictive Cleavage Motif for Eluted Major Histocompatibility Complex Class II Ligands. Front. Immunol. 2018;9:1795. doi: 10.3389/fimmu.2018.01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Olliaro P. Rolling out COVID-19 antigen rapid diagnostic tests: the time is now. Lancet Infect. Dis. 2021;21:1052–1053. doi: 10.1016/S1473-3099(21)00152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone L., Petruccioli E., Vanini V., Cuzzi G., Najafi Fard S., Alonzi T., Castilletti C., Palmieri F., Gualano G., Vittozzi P., et al. A whole blood test to measure SARS-CoV-2-specific response in COVID-19 patients. Clin. Microbiol. Infect. 2021;27:286.e7–286.e13. doi: 10.1016/j.cmi.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels K.B., Pritchard E., Matthews P.C., Stoesser N., Eyre D.W., Vihta K.D., House T., Hay J., Bell J.I., Newton J.N., et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat. Med. 2021;27:2127–2135. doi: 10.1038/s41591-021-01548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida F., Cassaniti I., Paolucci S., Percivalle E., Sarasini A., Piralla A., Giardina F., Sammartino J.C., Ferrari A., Bergami F., et al. SARS-CoV-2 vaccine breakthrough infections with the alpha variant are asymptomatic or mildly symptomatic among health care workers. Nat. Commun. 2021;12:6032. doi: 10.1038/s41467-021-26154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e114. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2021;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.T., Lim J.M., Le Bert N., Kunasegaran K., Chia A., Qui M.D., Tan N., Chia W.N., de Alwis R., Ying D., et al. Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. J. Clin. Invest. 2021;131:e152379. doi: 10.1172/JCI152379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Dercon Q., Harrison P.J. Six-month sequelae of post-vaccination SARS-CoV-2 infection: a retrospective cohort study of 10,024 breakthrough infections. medRxiv. 2021 doi: 10.1101/2021.10.26.21265508. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo N., Navalpotro D., Martínez-Serrano M., Moreno M., Grosson F., Tur I., Remedios Guna M., Soriano P., Tornero A., Gimeno C. Commercial interferpon-gamma release assay to assess the immune response to first and second doses of mRNA vaccine in previously COVID-19 infected versus uninfected individuals. Diagn. Microbiol. Infect. Dis. 2022;102:115573. doi: 10.1016/j.diagmicrobio.2021.115573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevethan R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front. Public Health. 2017;5:307. doi: 10.3389/fpubh.2017.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S., Klaeger S., Sarkizova S., Pearlman L.R., Chen D.Y., Gallagher K.M.E., Bauer M.R., Taylor H.B., Dunn W.A., Tarr C., et al. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell. 2021;184:3962–3980.e17. doi: 10.1016/j.cell.2021.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelba H., Worbs D., Harter J., Pieper N., Kyzirakos-Feger C., Kayser S., Seibold M., Bartsch O., Ködding J., Biskup S. A Highly Specific Assay for the Detection of SARS-CoV-2-Reactive CD4+ and CD8+ T Cells in COVID-19 Patients. J. Immunol. 2021;206:580–587. doi: 10.4049/jimmunol.2000811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets generated and analyzed in this study will be shared by the lead contact upon reasonable request. Additional supplemental items are available from Mendeley Data at https://doi.org/10.17632/s6gpnrmxg2.1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.