Abstract
We introduce a label-free surface-enhanced Raman scattering (SERS) biosensing platform equipped with metallic nanostructures that can identify the efficacy of Oxford-AstraZeneca (AZD1222) vaccine in vaccinated individuals using non-invasive tear samples. We confirmed the hypothesis that the tears of people who receive the AZD1222 vaccine may be similar to those of adenovirus epidemic keratoconjunctivitis patients since the Oxford-AstraZeneca vaccine is derived from a replication-deficient ChAdOx1 vector of chimpanzee adenovirus. Additionally, we confirmed the potential of the three markers for estimating the vaccination status via analyzing the signals emanating from antibodies or immunoglobulin G by-product using our label-free, SERS biosensing technique with a high reproducibility (<3% relative standard deviation), femtomole-scale limit of detection (1 × 10−14 M), and high SERS response of >108. Therefore, our label-free SERS biosensing nanoplatforms with long-term storage and robust stability will enable rapid and robust monitoring of the vaccine presence in vaccinated individuals.
Keywords: Label-free SERS biosensing platform, Near- and far-field properties, Tear fluids, Stability and robustness, Oxford-AstraZeneca vaccine
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in increased mortality in elderly person and patients with underlying diseases. Enforced physical distancing and large-scale shutdowns due to high infectivity of SARS-CoV2 have caused significant social and economic challenges worldwide (Ewer et al., 2021). As the number of infected individuals continues to increase despite preventive measures, the importance of population immunity is being emphasized to overcome the pandemic, with COVID-19 vaccination being of crucial importance (Voysey et al., 2021). The ChAdOx1 nCoV-19 vaccine (Oxford-AstraZeneca COVID-19, AZD1222) consists of a replication-deficient chimpanzee adenoviral vector ChAdOx1, carrying the full-length SARS-CoV-2 spike protein, nCoV-19 (Ewer et al., 2021; Voysey et al., 2021). When the vaccine enters the body, it induces the production of antibodies targeting the spike protein, triggering an immune response. In humans, antibody testing can be performed using the Emergency Use Authorization (EUA) serology test (The Centers for Disease Control and Prevention, 2021) on blood samples, but it is a cumbersome and time-consuming procedure. Moreover, the U.S. Food and Drug Administration currently does not recommend antibody testing to confirm immunity status against COVID-19 in fully vaccinated people (U.S. Food and Drug Administration, 2021). Therefore, developing a non-invasive, fast, and convenient monitoring method to determine the vaccination status is imperative. Recent studies (de Souza et al., 2006; You et al., 2013) have reported the presence of SARS-CoV-2 in tear fluids (Muyldermans et al., 2021) that also contain various biomarker proteins. The SARS-CoV-2 spike protein specific antibodies appear in tear fluids following immune response to AZD1222 vaccination (Folegatti et al., 2020; Hu et al., 2014). This leads to variations in the biochemical composition of tears among individuals of different vaccination status, reflected by changes in their spectroscopic waveforms (Choi et al., 2014; von Thun und Hohenstein-Blaul et al., 2013).
There are various methods for SARS-CoV-2 antibody testing, including the EUA serology test, but most are based on detecting the presence of immunoglobulin G (IgG) antibodies (Carter et al., 2020). Enzyme-linked immunosorbent assays, lateral flow immunoassays, and chemiluminescent immunoassays have excellent specificity but their results are sensitive to reaction time, surrounding environment, and enzymatic components and detection antibodies. Furthermore, since tears contain a lower concentration of biomarkers compared to blood (Anderson and Anderson, 2002), alternative biosensing techniques with greater sensitivity and robust stability are required. Surface-enhanced Raman scattering (SERS) is a promising sensing technology that can detect target molecules present in extremely low amounts using a bottom-up nanofabrication technique (Laing et al., 2017; Langer et al., 2020; Zong et al., 2018). Raman spectroscopy uses light to generate a structural fingerprint of molecules in biological samples via identifying their vibrational modes, without the need for complex pretreatment steps. However, Raman scattering signals originating from feeble molecular interaction vibrations should be amplified in an enhanced resonance of metal surface plasmons by SERS effect. The degree of SERS enhancement depends on the size, shape, and spacing of the plasmonic nanomaterials and the nanostructure-absorbed excitation wavelengths (Álvarez-Puebla, 2012; Lee et al., 2019). A highly sensitive SERS platform has a lower the limit of detection, while a label-free biosensing approach based on metallic nanostructures increases the stability of immunoassays (Kim et al., 2018, 2021; Lee et al., 2021).
Here, we propose a label-free optical SERS biosensing platform to evaluate the efficacy status of a vaccine in the body using the tear fluids of individuals fully vaccinated with AZD1222. Three different gold nanostructures compatible with three representative excitation wavelengths used in Raman spectroscopy were synthesized, and the optical properties of these SERS biosensing platforms, in which gold nanostructures were uniformly distributed over a large area without the coffee-ring effect on the cellulose chips, were compared. The SERS performance of the optical biosensing platform was maximized by selecting the optimal Raman excitation wavelength and measurement condition set-up based on the optical properties of the nanoplatforms. The tears were collected according to the established clinical protocol in the hospital, and those of people receiving the AZD1222 vaccine were collected two weeks after vaccination. All tears were analyzed based on label-free SERS spectra using an optical biosensing nanoplatforms without any pretreatment step. Hence, a 785 nm excitation wavelength-optimized, robust gold nanostructure-based SERS biosensing nanoplatform was applied to identify the presence of AZD1222 after vaccination.
2. Results and discussion
2.1. Near- and far-field properties
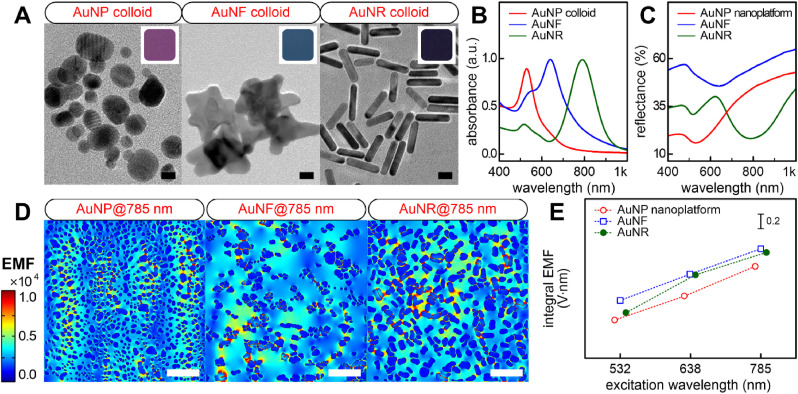
To ensure compatibility with the Raman laser excitation source and localized surface plasmon resonance (LSPR) peak wavelength, three gold nanostructures exhibiting LSPR bands with maximum absorption peaks at 532, 638, and 785 nm, respectively, were selected. Fig. 1 A shows the nanoscale morphologies and colors of the three colloidal nanoparticles. First, although spherical gold nanoparticles (AuNPs) with good optical properties at 532-nm laser excitation were well deposited on the substrate via deep coating, a uniform SERS effect could not be obtained owing to the low surface coverage (Fig. S1). Therefore, we developed an independent fabrication method in which the AuNPs were directly synthesized on the cellulose material, with abundant nanoparticles and adjacent nanogap (Kim et al., 2015, 2016). The synthesized AuNPs (12 ± 4 nm) were spherical and exhibited the maximum absorption peak at approximately 529 nm (Fig. 1B). Second, the anisotropic gold nanoflower (AuNF) was selected for 638-nm laser excitation. This nanostructure was synthesized using a facile one-step seedless method with only gold ions and a Good's buffer reducing agent (Chandra et al., 2016; Xie et al., 2008). The optical properties of the AuNF colloids can be controlled using varying concentrations of the precursor and the pH of the reactants at room temperature (Fig. S2, Table S1). The AuNFs (44 ± 11 nm) were synthesized by the optimized synthesis conditions (Fig. S3) that exhibited a maximum LSPR peak at approximately 640 nm. Lastly, for 785-nm laser excitation, gold nanorod (AuNR) colloids were prepared based on a previous study wherein the LSPR peak was optimized by tuning the aspect ratio of the rod shape and controlling the amount of AgNO3 molecules (Kim et al., 2021). AuNRs with an aspect ratio of 3.6 (18 ± 5 nm length and 5 ± 2 nm width) and exhibiting a maximum LSPR peak at approximately 789 nm, were synthesized. These results demonstrate that the LSPR bands of the three preferred nanoparticles were in good agreement with the three representative Raman laser sources. A dense and uniform distribution of nanoparticles on the substrate can be achieved by controlling the surface charge of the substrate (Fig. S4). The minimum reflectance wavelengths of each nanoplatform were measured at 530 nm for AuNPs, 637 nm for AuNFs, and 788 nm for AuNRs (Fig. 1C), and were similar to the maximum LSPR peaks of each colloidal nanoparticle. This result indicates that each nanoparticle was well-distributed over a large area of the two-dimensional cellulose matrix without being aggregated. The far-field properties of the three nanoplatforms were tuned to match three laser excitation sources widely used in SERS applications, and each laser source-optimized nanoplatform was expected to show high performance.
Fig. 1.
(A) Transmission electron microscopy images and (B) UV–Vis absorbance spectra of three nanoparticles. Inset indicates planar color images. Scale bar = 10 nm. (C) UV–Vis reflectance spectra of three nanoplatforms. (D) The 785 nm-excited EMF enhancement map within the corresponding ROIs. Scale bar = 200 nm. (E) Comparative analysis of integral near-field enhancements calculated from the three nanoplatforms depending on three Raman laser excitation sources.
It is important to ensure that the near-field responses of nanoplatforms with far-field optical properties are well matched to the excitation source. We confirmed that the near-field properties of three isolated nanoparticles were closely related to their far-field properties (Fig. S6). Electromagnetic field (EMF) enhancement of anisotropic nanoparticles was localized near the tip and rod ends by finite element analysis (FEA) models (D. S. Indrasekara et al., 2015; Harder et al., 2021). Plasmon coupling of FEA models for three nanoplatforms was investigated based on three laser sources (Fig. S7). This realistic simulation was performed within regions of interest (ROIs) spanning an area of 1 μm × 1 μm in accordance with the focal spot size of our Raman measurement conditions. Despite the low surface coverage of the AuNF nanoplatform (Table S4), it showed greater near-field plasmon coupling properties than the others. This indicates that the geometry as well as density of the particles have a direct influence on the EMF enhancement. In addition, near-field coupling between particles occurs when the interparticle distance is smaller than the particle diameter; thus, the plasmon resonance is red-shifted to a higher wavelength by reducing the particle separation of the light (Pinchuk and Schatz, 2008). However, the maximum EMF enhancement was observed at the 785 nm incident wavelength regardless of the nanoplatform (Fig. 1D). The near-field coupling from nanoparticle clusters with hot spots significantly affects the SERS response and LSPR property rather than the far-field properties originating from the particle monomer. Although the maximum enhanced EMF directly correlated with the SERS intensity, the EMF within the focal spot of the incident laser was dominant. The 785 nm-excited Raman laser source also led to the highest integral EMF enhancement regardless of the nanoplatform (Figs. 1E and S9). Overall, the near-field plasmon coupling of the AuNFs was superior to that of the others (Table S5). Although the hotspots can increase the maximum EMF due to the geometry of bumpy AuNFs, the wide adjacent interface due to the longitudinal AuNR configuration may contribute to the overall near-field coupling in the focal area of interest. The well-distributed AuNR nanoplatform also produced strong EMF enhancement at 785 nm, similar to the AuNFs. Collectively, these computational results recommend the use of anisotropic nanoplatforms (AuNRs and AuNFs) for further applications. Additionally, the near-field plasmon coupling of the nanoplatform (substrate) significantly affected the EMF exhibiting the SERS response, but not the LSPR optical reflectance signature (far-field properties).
2.2. SERS performance
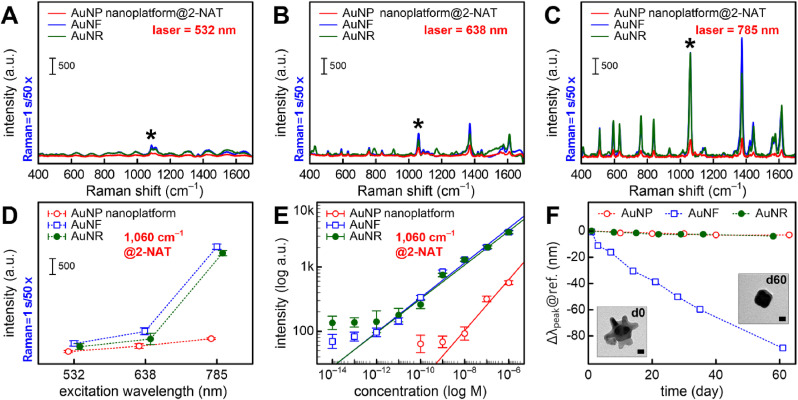
Although the far-field properties of nanoplatforms well matched the three representative excitation wavelengths, there was a significant difference with the calculated near-field properties. To further study the wavelength-dependent behavior of the near-field in nanoplatforms, we investigated the variations in Raman intensity exhibiting a unique signature to evaluate the SERS activity depending on the three excitation sources. A 2-NAT molecule (1 μM) containing a thiol group chemisorbed on the gold surface was used as a Raman probe reporter. Overall, the 532-nm wavelength-excited SERS intensity was weak, but the prominent peaks of the 2-NAT molecule were observed with all excitation sources (Fig. 2 A–C). Quantitative analysis using the 1060 cm−1 characteristic peak (Fig. 2D) showed that all three platforms had higher SERS responses at the 785 nm excitation; the 785 nm-excited anisotropic nanoplatforms showed high reproducibility (Fig. S11) and improved SERS enhancement ability (6-fold increase compared to isotropic nanoplatform). This indicates that the selection of the most suitable Raman laser wavelength is dominated by a hotspot-driven near-field SERS response rather than a far-field optical response (Kurouski et al., 2016; Wustholz et al., 2010). Even a SERS biosensing platform based on a single nanostructure requires the selection of a suitable Raman laser wavelength depending on whether the Raman probe or analyte is in the internal structure or on the external surface (Lim et al., 2011; Lin et al., 2018; Yoon et al., 2012). Furthermore, the results of large-area 2D Raman imaging (10 × 10 mapping) showed that compared to other wavelengths, the 785 nm-excited nanoplatforms had an excellent reproducibility (<5% relative standard deviation (RSD)) and intense SERS enhancement regardless of the nanogeometry (Fig. S12). The sensitivity of each nanoplatform was evaluated based on the relationship between the intensities of the 2-NAT Raman molecule at 1060 cm−1 and at concentrations ranging from 10−14 to 10−6 M (Figs. 2E and S13). The 785 nm-excited anisotropic nanoplatforms achieved a limit of detection of 1 × 10−14 M (Table S6) and a limit of quantification between 10−12 and 10−6 M (R2 = 0.98). Several proteins are expressed in the human body, with as high collective concentration as approximately 80 mg/mL in the serum and 10 mg/mL in tears, while few proteins are present in very low concentrations ranging from just fg/mL to μg/mL (Walker and Hall, 1990; Zhou et al., 2012). Because the proposed AuNF and AuNR nanoplatforms can detect protein in this concentration range (10−14 to 10−4 M), they have potential for label-free biosensing clinical applications. The SERS substrate enhancement factor (SEF) of the anisotropic nanoplatforms was 3.4 × 108 for the AuNFs and 8.4 × 108 for the AuNRs (Fig. S14, Table S7). Owing to their excellent SERS performance, such as high reproducibility (<3% RSD), femtomole-scale limit of detection (1 × 10−14 M), and high SERS response (>108 SEF), the anisotropic nanoplatforms including AuNF and AuNR nanogeometries with a 785 nm laser excitation source have the potential for biosensing applications in clinical settings.
Fig. 2.
(A–C) SERS spectra of the 10−6 M 2-NAT Raman molecule for the three nanoplatforms depending on the three different Raman laser excitation sources. The asterisk indicates the characteristic peak of 2-NAT Raman probe reporter. Plots of Raman intensities for 2-NAT Raman molecule at 1060 cm−1 peak depending on (D) Raman laser excitation sources and (E) concentrations between 10−14 M and 10−6 M with the 785 nm Raman laser source used in the three nanoplatforms. (F) Stability testing of each nanoplatform over two months. Insets indicate the AuNF geometries immediately (d0) and 60 days (d60) after AuNF fabrication. Scale bar = 10 nm.
Fig. 2F shows the variations in reflectance of each nanoplatform exposed to air for more than two months. The AuNF nanoplatform showed a pronounced blue shift in the reflectance peak (Δλpeak@ref.) over time compared with the others that showed little change. We observed that the sharp tips of most AuNF particles disappeared 60 days after exposure to air (Fig. S15). However, the colloidal AuNF particles in the growth solution showed little change in the LSPR absorption peak (Δλpeak@abs. in Fig. S16A) over time (Chandra et al., 2016). It is likely that the anisotropic sharp portions of the AuNFs were blunted by oxidative etching (Qin et al., 2016) caused by the air exposure, leading to a pronounced blue shift of the reflectance peak. Consequently, the low stability of AuNF particles in contact with air led to an extreme decrease in the SERS response of the AuNF nanoplatform over time compared to others (low reproducibility of 23% RSD and low SERS intensity with a 3-fold decrease) (Figs. S16B–D). Therefore, the 785 nm Raman laser-excited AuNR nanoplatform, with its excellent SERS performance and robust stability, may be more suitable for on-site SERS biosensing applications.
2.3. Bioapplications
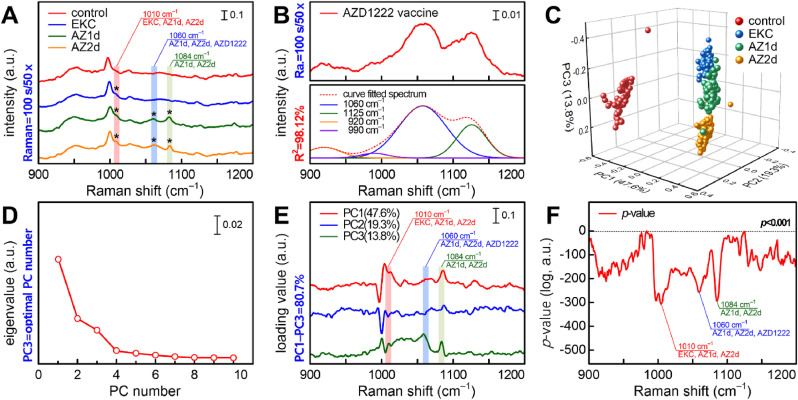
Fig. 3A shows the SERS spectra of human tear fluid samples obtained from four control, epidemic keratoconjunctivitis (EKC), first dose (AZ1d), second dose (AZ2d) groups. We observed three significant differential SERS peaks (1010, 1060, and 1084 cm−1) among the four groups (Table S8). The 1010 cm−1 peak assigned to the β-sheet structure of the IgG molecule with phenylalanine (Hu et al., 2014; Talari et al., 2015) was observed in the EKC, AZ1d, and AZ2d groups (Fig. S17). The tears of COVID-19 convalescent people have reportedly significantly higher anti-SARS-CoV-2 IgG and IgA levels compared to those of healthy individuals (Muyldermans et al., 2021). Therefore, the 1010 cm−1 peak may have been caused by IgG binding to the capsid protein of adenovirus in EKC. The 1060 cm−1 peak was observed only in the AZ1d and AZ2d groups (Fig. 3B). This peak is assigned to the PO2 − stretching of DNA (Talari et al., 2015), and is expected to be derived from the vector DNA encoding the SARS-CoV-2 spike protein in the AZD1222 vaccine. Although why is vector DNA detected in the tears of the AZD1222-vaccinated subjects is not entirely clear, it may be released from the host cells destroyed by cytotoxic T cells that recognize the spike protein. There are two possible explanations for the presence of the 1084 cm−1 peak observed only in the AZ1d and AZ2d groups. First, this peak can be attributed to tryptophan present in the IgG light chain that is responsible for the binding and biological activity of IgG (Iizumi et al., 2018; Talari et al., 2015; Wei et al., 2007). Since this peak was only observed in AZD1222-vaccinated individuals, this may be derived from the immunoglobulins directed against the SARS-CoV-2 spike protein. Second, the 1084 cm−1 peak could also be assigned to the PO2 − stretching of DNA (Talari et al., 2015) which is released from the host cells that are attacked and lysed by the cytotoxic T cells. Therefore, the three significant SERS peaks could be used as spectral markers to identify the effectiveness of the Oxford-AstraZeneca COVID-19 vaccine, as well as its correlation with the adenovirus EKC disease.
Fig. 3.
Clinical application of the 785 nm Raman laser-excited AuNR nanoplatform to evaluate the effectiveness of the Oxford-AstraZeneca vaccine using human tear samples. (A) SERS spectra of tear samples obtained from four different study groups. The asterisk indicates the significant SERS peaks. (B) SERS spectrum of AZD1222 solution and its Gaussian decomposition. (C) Cluster plot of the four groups with a percentage of variances of PC1 = 47.6%, PC2 = 19.3%, and PC3 = 13.8%. (D) Scree plot of the eigenvalues. (E) PC1–PC3 loading profiles calculated from the principal component analysis algorithm. (F) P-value profiles of a one-way analysis of variance test for SERS spectra of the four groups.
Fig. 3C shows the results of four different groups using the principal component analysis algorithm. The selection of the principal component (PC) number is important for classification as it reflects the difference in SERS spectral information among groups. In this study, PC1–PC3 were selected using the scree plot (Fig. 3D), and their cumulative percent variance was 80.7%. As shown in Fig. 3C, the 3D cluster plot using PC1 to PC3 indicated a high classification performance (close to 100%) in all groups. Moreover, a clear difference was observed between the control and the experimental groups, including the EKC group, and various overlaps were noted among the EKC, AZ1d, and AZ2d groups, however they could still be classified. Fig. 3E shows the loading profiles of PC1 to PC3, where the larger absolute values led to the greater classification contribution to the Raman shift. We confirmed that the clinically significant SERS peaks related to the AZD1222 vaccine (1010, 1060, and 1084 cm−1) were also detected in the loading profiles. This provided the evidence that the classification shown in Fig. 3C was based on clinically significant SERS spectral information. The statistical significance of the classification among the four groups was verified using the p value of a one-way analysis of variance. The Raman shifts of the four prominent negative peaks according to the p-value (Fig. 3F) were consistent with those of the three SERS spectral markers for AZD1222 and EKC groups, except for the 1003 cm−1 peak that was assigned to the ring breathing mode of phenylalanine. This clearly indicates that our classification has a confidence of at least 99.9% (p < 0.001) for the three markers, with significant differences among the four groups. Therefore, a 785 nm excitation wavelength-optimized, robust, AuNR-based SERS biosensing nanoplatform is a potential candidate to monitor the immune profile of individuals vaccinated with AZD1222 and other vaccines using tear samples and their distinct SERS spectral markers.
3. Conclusions
We investigated the immune status of individuals vaccinated by the AZD1222 using their tear samples and a label-free SERS biosensing nanoplatform. Three gold nanostructures with spherical (AuNP), flower-shaped (AuNF), and rod-shaped (AuNR) geometries were synthesized to match the three representative Raman lasers, and the near- and far-field optical properties of each nanostructure and nanoplatform were compared. The AuNFs with multiple anisotropic tips had the maximal EMF properties, but because of their oxidation-induced degradation following air exposure, the more stable AuNR nanoplatforms were selected for on-site SERS biosensing applications. In addition, this biosensing platform with metallic nanomaterials is amenable to long-term storage with robust stability. Therefore, we anticipate that the optimized label-free SERS nanoplatform will allow a rapid and in mass monitoring of the vaccine presence in vaccinated individuals. There are some limitations to this study, including limited sample size and single vaccination efficacy, and further studies on different types of COVID-19 vaccine and cross vaccination are necessary to expand the applications of this platform.
CRediT authorship contribution statement
Wansun Kim: Conceptualization, experiments, Writing – original draft. Soogeun Kim: Conceptualization, bioapplications, writing review. Jisang Han: Writing – original draft, sample preparation, writing review. Tae Gi Kim: Sample preparation, evidence-based clinical review and writing review. Ayoung Bang: method Synthesis and methodology. Hyung Woo Choi: method Synthesis and methodology. Gyeong Eun Min: writing Clinical review and writing review. Jae-Ho Shin: Sample preparation, clinical review, and English editing. Sang Woong Moon: Clinical review and writing review. Samjin Choi: Conceptualization, sample preparation, manuscript drafting, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the National Research Foundation of Korea (2018M3A9E8078812, 2019R1A2C2089239, 2019R1C1C1007917, 2021R1A2C4002700, and 2021R1I1A1A01044700) and a grant from Kyung Hee University in 2019 (KHU -20192442).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114079.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Álvarez-Puebla R.A. J. Phys. Chem. Lett. 2012;3:857–866. doi: 10.1021/jz201625j. [DOI] [PubMed] [Google Scholar]
- Anderson N.L., Anderson N.G. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra K., Culver K.S.B., Werner S.E., Lee R.C., Odom T.W. Chem. Mater. 2016;28:6763–6769. [Google Scholar]
- Choi S., Moon S.W., Shin J., Park H., Jin K. Anal. Chem. 2014;86:11093–11099. doi: 10.1021/ac5025478. [DOI] [PubMed] [Google Scholar]
- de Souza G.A., Godoy L.M.F., Mann M. Genome Biol. 2006;7:1–11. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer K.J., et al. Nat. Med. 2021;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- Folegatti P.M., et al. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder R.A., Wijenayaka L.A., Phan H.T., Haes A.J. J. Raman Spectrosc. 2021;52:497–505. doi: 10.1002/jrs.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Brendle S., Balogh K., Bywaters S., Christensen N. J. Gen. Virol. 2014;95:2030–2037. doi: 10.1099/vir.0.064154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizumi K., Kawasaki H., Shigenaga A., Tominaga M., Otsu A., Kamo A., Kamata Y., Takamori K., Yamakura F. J. Clin. Biochem. Nutr. 2018;63:197–204. doi: 10.3164/jcbn.18-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrasekara A.S.D.S., Thomas R., Fabris L. Phys. Chem. Chem. Phys. 2015;17:21133–21142. doi: 10.1039/c4cp04517c. [DOI] [PubMed] [Google Scholar]
- Kim W., Kim Y.-H., Park H.-K., Choi S. ACS Appl. Mater. Interfaces. 2015;7:27910–27917. doi: 10.1021/acsami.5b09982. [DOI] [PubMed] [Google Scholar]
- Kim W., Lee J.-C., Shin J.-H., Jin K.-H., Park H.-K., Choi S. Anal. Chem. 2016;88:5531–5537. doi: 10.1021/acs.analchem.6b01123. [DOI] [PubMed] [Google Scholar]
- Kim W., Lee Sung Ho, Ahn Y.J., Lee Seung Ho, Ryu J., Choi S.K., Choi S. Biosens. Bioelectron. 2018;111:59–65. doi: 10.1016/j.bios.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Kim S., Joo J., Kim W., Bang A., Choi H.W., Moon S.W., Choi S. Sensor. Actuator. B Chem. 2021;343 [Google Scholar]
- Kurouski D., Large N., Chiang N., Greeneltch N., Carron K.T., Seideman T., Schatz G.C., Van Duyne R.P. Analyst. 2016;141:1779–1788. doi: 10.1039/c5an01921d. [DOI] [PubMed] [Google Scholar]
- Laing S., Jamieson L.E., Faulds K., Graham D. Nat. Rev. Chem. 2017;1 [Google Scholar]
- Langer J., et al. ACS Nano. 2020;14:28–117. doi: 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Lee Y.H., Koh C.S.L., Phan-Quang G.C., Han X., Lay C.L., Sim H.Y.F., Kao Y.-C., An Q., Ling X.Y. Chem. Soc. Rev. 2019;48:731–756. doi: 10.1039/c7cs00786h. [DOI] [PubMed] [Google Scholar]
- Lee W., Kang B., Yang H., Park M., Kwak J.H., Chung T., Jeong Y., Kim B.K., Jeong K. Nat. Commun. 2021;12:159. doi: 10.1038/s41467-020-20413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.-K., Jeon K.-S., Hwang J.-H., Kim H., Kwon S., Suh Y.D., Nam J.-M. Nat. Nanotechnol. 2011;6:452–460. doi: 10.1038/nnano.2011.79. [DOI] [PubMed] [Google Scholar]
- Lin L., Zhang Q., Li X., Qiu M., Jiang X., Jin W., Gu H., Lei D.Y., Ye J. ACS Nano. 2018;12:6492–6503. doi: 10.1021/acsnano.7b08224. [DOI] [PubMed] [Google Scholar]
- Muyldermans A., Bjerke M., Demuyser T., De Geyter D., Wybo I., Soetens O., Weets I., Kuijpers R., Allard S.D., Piérard D., Raus P.P.M. BMJ Open Ophthalmol. 2021;6 doi: 10.1136/bmjophth-2021-000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchuk A.O., Schatz G.C. Mater. Sci. Eng. B. 2008;149:251–258. [Google Scholar]
- Qin F., Zhao T., Jiang R., Jiang N., Ruan Q., Wang J., Sun L., Yan C., Lin H. Adv. Opt. Mater. 2016;4:76–85. [Google Scholar]
- Talari A.C.S., Movasaghi Z., Rehman S., Rehman I.U. Appl. Spectrosc. Rev. 2015;50:46–111. [Google Scholar]
- The Centers for Disease Control and Prevention Interim guidelines for COVID-19 antibody testing. 2021. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html URL.
- U.S. Food and Drug Administration . 2021. Antibody (Serology) Testing for COVID-19.https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/antibody-serology-testing-covid-19-information-patients-and-consumer URL. [Google Scholar]
- von Thun und Hohenstein-Blaul N., Funke S., Grus F.H. Exp. Eye Res. 2013;117:126–137. doi: 10.1016/j.exer.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Voysey M., et al. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker H.K., Hall W.D. third ed. Butterworth-Heinemann; Boston: 1990. Clinical Methods: the History, Physical, and Laboratory Examinations. [PubMed] [Google Scholar]
- Wei Z., Feng J., Lin H.-Y., Mullapudi S., Bishop E., Tous G.I., Casas-Finet J., Hakki F., Strouse R., Schenerman M.A. Anal. Chem. 2007;79:2797–2805. doi: 10.1021/ac062311j. [DOI] [PubMed] [Google Scholar]
- Wustholz K.L., Henry A.-I., McMahon J.M., Freeman R.G., Valley N., Piotti M.E., Natan M.J., Schatz G.C., Van Duyne R.P. J. Am. Chem. Soc. 2010;132:10903–10910. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- Xie J., Zhang Q., Lee J.Y., Wang D.I.C. ACS Nano. 2008;2:2473–2480. doi: 10.1021/nn800442q. [DOI] [PubMed] [Google Scholar]
- Yoon J.H., Lim J., Yoon S. ACS Nano. 2012;6:7199–7208. doi: 10.1021/nn302264f. [DOI] [PubMed] [Google Scholar]
- You J., Willcox M.D., Madigan M.C., Wasinger V., Schiller B., Walsh B.J., Graham P.H., Kearsley J.H., Li Y. Tear fluid protein biomarkers, 1st ed. Adv. Clin. Chem. 2013 doi: 10.1016/B978-0-12-800096-0.00004-4. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Zhou L., Zhao S.Z., Koh S.K., Chen L., Vaz C., Tanavde V., Li X.R., Beuerman R.W. J. Proteonomics. 2012;75:3877–3885. doi: 10.1016/j.jprot.2012.04.053. [DOI] [PubMed] [Google Scholar]
- Zong C., Xu M., Xu L.-J., Wei T., Ma X., Zheng X.-S., Hu R., Ren B. Chem. Rev. 2018;118:4946–4980. doi: 10.1021/acs.chemrev.7b00668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.