Abstract
Infections caused by SARS-CoV-2 have brought great harm to human health. After transmission for over two years, SARS-CoV-2 has diverged greatly and formed dozens of different lineages. Understanding the trend of its genome evolution could help foresee difficulties in controlling transmission of the virus. In this study, we conducted an extensive monthly survey and in-depth analysis on variations of nucleotide, amino acid and codon numbers in 311,260 virus samples collected till January 2022. The results demonstrate that the evolution of SARS-CoV-2 is toward increasing U-content and reducing genome-size. C, G and A to U mutations have all contributed to this U-content increase. Mutations of C, G and A at codon position 1, 2 or 3 have no significant difference in most SARS-CoV-2 lineages. Current viruses are more cryptic and more efficient in replication, and are thus less virulent yet more infectious. Delta and Omicron variants have high mutability over other lineages, bringing new threat to human health. This trend of genome evolution may provide a clue for tracing the origin of SARS-CoV-2, because ancestral viruses should have lower U-content and probably bigger genome-size.
Keywords: Coronavirus, Transmission, Mutation, Evolution, Variant, Nucleotide
1. Introduction
The pandemic of COVID-19 (coronavirus disease 2019) has brought over 370 million infection cases and over 5.6 million deaths worldwide by 30 January 2022 [1]. Its causative virus, SARS-CoV-2, has a single-strand genomic RNA of approximately 30,000 nucleotides [2]. Since outbreak of this disease, great efforts have been made to establish fast diagnostic methods [3], [4], to develop effective therapeutic drugs and vaccines [5], [6], [7], [8], to implement strict inspections on transportations of goods, and to enforce certain restrictions on activities of people [9], [10]. All these efforts have helped reduce virus transmission effectively and treat infected people properly [11], [12]. However, with fast emergence of new variants [13], [14], the world is still facing big challenges in controlling this pandemic.
Fast emergence of new variants is largely due to high mutability of SARS-CoV-2 genome, which is prone to mutation though it encodes a proofreading enzyme to prevent replication error [15], [16]. A great number of mutations have been recorded in SARS-CoV-2 genome [17], [18], [19], some of which affect structure and function of viral proteins [20], [21], [22]. Rapid and extensive transmission worldwide has provided more chances for SARS-CoV-2 genome to accumulate mutations. After frequent mutations for around two years, SARS-CoV-2 has diverged greatly and formed many different lineages [23], [24]. While monitoring mutations in specific viral proteins facilitates the development of effective vaccines and antiviral drugs [25], [26], understanding the evolution trend of SARS-CoV-2 genome could help foresee difficulties in controlling spread of the virus [27], [28].
In GISAID (global initiative on sharing all influenza data) database (www.gisaid.gov), SARS-CoV-2 viruses are currently classified into nine clades and five variants of concern (VOCs). The nine clades are named based on presence of specific amino acid at particular site. For examples, clades S and L are named because amino acid 84 of their NSP8 (non-structural protein 8) is serine (S) and leucine (L), respectively. Clades V and G are derived from clade L, in which amino acid 251 of NSP3 protein is V (valine) and amino acid 614 of S (spike) protein is G (glycine), respectively. Clades GH, GK, GR and GV are all derived from clade G. Their names are based on presence of histidine (H), lysine (K), arginine (R) and valine (V) at specific sites, respectively. Clade GRY is derived from clade GR, because tyrosine (Y) is at amino acid position 501 in its S protein. The five VOCs are all derived from G-series clades due to additional nucleotide mutations and deletions/insertions. They are named Alpha, Beta, Gamma, Delta, and Omicron, respectively [29].
The GISAID database has collected over 4.8 million full-genome sequences of SARS-CoV-2 by 31 January 2022. With these sequence resources, tracing mutations in SARS-CoV-2 can be conducted separately for different lineages along certain timelines. Therefore, in this study, we conducted an extensive monthly survey on variations of nucleotide, amino acid and codon numbers in twelve SARS-CoV-2 lineages. Further in-depth analyses reveal that, in all surveyed SARS-CoV-2 lineages, U content has been steadily increased, and genome stability has been slightly reduced. These two changes are considered to make the virus less virulent yet more infectious [30], [31].
2. Materials and methods
2.1. Sampling and processing of genome sequences
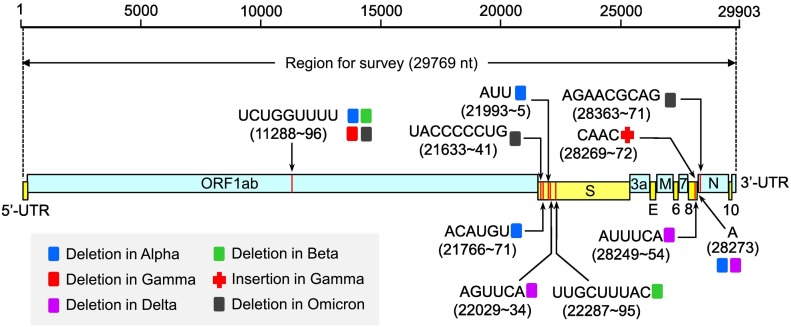
Genome sequences were downloaded from GISAID (www.epicov.org) database for twelve SARS-CoV-2 lineages including seven clades (S, L, V, G, GH, GR and GV) and five variants (Alpha, Beta, Gamma, Delta and Omicron). Filters were set to retrieve sequences with human as host, and having high coverage and complete collection date. Sequences were downloaded separately for different lineages and different months. The downloaded sequences (in Fasta format) were analysed using Alignment Explorer of MEGA X [32] to exclude those having any ambiguous base (e.g. N, R and Y for any base, purine and pyrimidine, respectively). Then, each of them was trimmed to retain 29,769 nucleotides for the seven clades. For Alpha, Beta, Gamma, Delta and Omicron variants, 29,750, 29,751, 29,764, 29,756 and 29,742 nucleotides were retained respectively, because they have various deletions/insertions in the region for survey (Fig. 1 ).
Fig. 1.
Genomic region of SARS-CoV-2 for survey. SARS-CoV-2 genomic sequence of 29,903 nucleotides was used as reference [2]. Viral genomic region corresponding to No. 66 to No. 29834 (totalling 29,769 nucleotides) of the reference sequence was taken for survey. Capital letters pointed to the thin red line indicate deleted nucleotides with deletion site below them. Insertion of “CAAC” and deletion of “A” occur in the intergenic region following ORF8. Abbreviations: UTR (untranslated region), ORF (open reading frame), S (spike), E (envelope), M (matrix) and N (nucelocapsid). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Counting of nucleotide, amino acid and codon numbers
The trimmed sequences were loaded into a computer program to count the numbers of nucleotides in whole survey region and the numbers of amino acids and codons in ORFs (open reading frames). C++ scripts of computer programs are available upon request.
2.3. Measurement of free energy
Free energy of viral genomic sequence was measured using RNAstructure 5.7, which uses a dynamic programming algorithm to predict RNA secondary structures based on the principle of minimizing free energy [33]. The minimum free energy of an analysed viral sequence was used to estimate genome stability of viruses. For each viral sequence, the first 200 nucleotides (i.e. 5′-untranslated region) were loaded into the program directly. The rest of the nucleotides were segmented into 29 pieces of 1000 nucleotides plus the last one of 542–569 nucleotides, and then loaded into the program in order.
2.4. Statistical analysis
SPSS software (version 17.0) was used to conduct independent-sample t-test for comparing the overall variation in nucleotide, amino acid and codon numbers, and for comparing variation of a specific nucleotide at different codon positions. The variation is considered significant when p < 0.05.
3. Results
3.1. Variations in nucleotide numbers
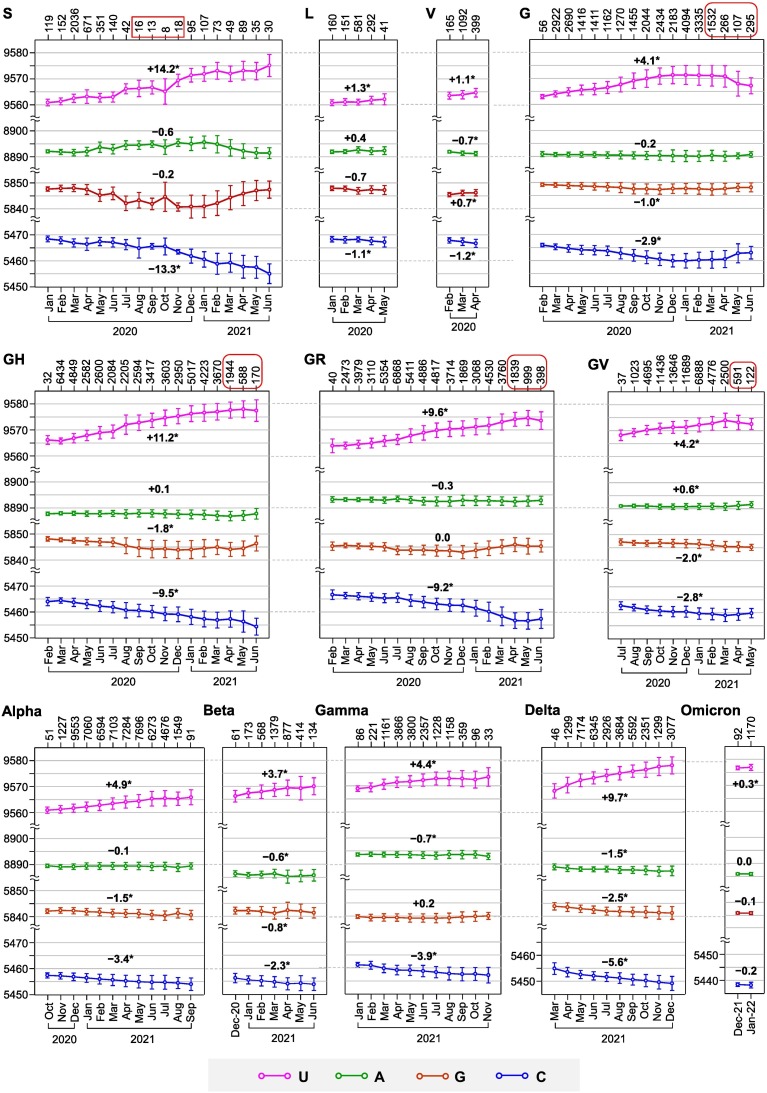
Based on the survey of all 311,260 virus samples, nucleotide numbers of the twelve SARS-CoV-2 lineages were obtained (Fig. 2 ). In all lineages, C numbers are significantly reduced (except Omicron variant), while U numbers are significantly increased. Among them, S clade has the highest variation in both C and U numbers (−13.3 and +14.2), followed by GH clade (−9.5 and +11.2). The C-down and U-up mutations are observed in all lineages, even though the viruses transmitted for only a few months (e.g. L, V and Omicron). G-down and G-up mutations are observed in nine and two lineages respectively, whereas A-down and A-up mutations are observed in eight and three lineages respectively. These data show that mutations of C, G and A into U have occurred in most SARS-CoV-2 lineages.
Fig. 2.
Variations of nucleotide numbers in SARS-CoV-2 lineages. Nucleotide numbers of each month are presented as mean ± standard deviation with sample numbers on top of the graph. Data from sample number below 30 are excluded for analysis, except those for S clade (in red square frame). Values in red round frame indicate obvious decline in sample numbers compared to previous months. Value above or below each line indicates overall variation between the last and the first month. * indicates significant difference (p < 0.05). GK and GRY clades are not surveyed separately, because sequences of these two clades overlap greatly with those of Delta and Alpha variants. Please refer to Table S1 for original data (values in blue colour). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Variations of nucleotide numbers in SARS-CoV-2 lineages. Nucleotide numbers of each month are presented as mean ± standard deviation with sample numbers on top of the graph. Data from sample number below 30 are excluded for analysis, except those for S clade (in red square frame). Values in red round frame indicate obvious decline in sample numbers compared to previous months. Value above or below each line indicates overall variation between the last and the first month. * indicates significant difference (p < 0.05). GK and GRY clades are not surveyed separately, because sequences of these two clades overlap greatly with those of Delta and Alpha variants. Please refer to Table S1 for original data (values in blue colour). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It is to be noted that, reductions of U number from April to June 2021 in G-series clades are due to exclusion of virus samples with genomic deletions. All current SARS-CoV-2 variants are derived from G-series clades [29], most of which involve nucleotide deletions (Fig. 1). From April to June 2021, more and more G-series viruses contained shortened genomes. Correspondently, less and less G-series viral genomes are eligible for survey (Fig. 2, values in red round frame), because our survey compares difference of nucleotide numbers within specific lengths (Fig. 1). Therefore, G-series viruses have evolved in two directions. One direction is followed by the majority of viruses which underwent fast evolution in elevating U-content and reducing genome-size, thus becoming ancestors of the new variants. Viruses under this evolutional direction are not considered as of G-series, because they do not have the required genome length (i.e. 29,769 nucleotides). The other direction is followed by the minority of viruses, which evolved relatively slowly. These viruses were slower in elevating U-content and reducing genome-size. Viruses under this evolutional direction are considered as of G-series. Therefore, their U-content in May/June 2021 is generally lower than that in April 2021. Correspondently, their C-content shows a general trend of increase after April 2021, while G- and A-contents are not changed dramatically (Fig. 2).
3.2. Variations in amino acid and codon numbers
Significant variations in nucleotide numbers have resulted in changes of amino acid numbers at various degrees. Numbers of proline, threonine and alanine are significantly reduced in 7–9 lineages, while those of isoleucine, leucine, phenylalanine, valine and tyrosine are significantly increased in 6–9 lineages (Fig. S1). Correspondently, codon numbers for proline (CCA and CCU), threonine (ACA) and alanine (GCU) are significantly reduced in 6–8 lineages, while those for isoleucine (AUU), phenylalanine (UUU), valine (GUU), tyrosine (UAU) and cysteine (UGU) are significantly increased in 6–8 lineages (Fig. S2). In general, codons with a reduced number are C-rich, and those with an increased number are U-rich, being consistent with C-down and U-up mutational trend shown in Fig. 2.
Among the 20 amino acids, threonine was reduced to the greatest extent in GR clade (−3.4), followed by proline in S clade (−2.9). Isoleucine was increased to the greatest extent in S clade (+2.5) followed by tyrosine (+2.4) in S clade (Fig. S1). Furthermore, 16 amino acids have been significantly changed in S clade. It is understandable because it has undergone transmission for over eighteen months. In lineages that have undergone transmission for over twelve months (i.e. G, GH and GR clades), 10–14 amino acids are changed significantly. Compared to these clades, Alpha, Beta, Gamma and Delta variants have relatively higher mutation rate. After transmission for seven to twelve months, 13–15 amino acids are significantly changed already (Fig. S1).
3.3. Mutation patterns of nucleotides
In order to analyse the mutation patterns of nucleotides, we made a statistic on the number of variations among C, G, A and U at different codon positions (Table S3, data within green frame). Based on these data, overall variations of nucleotides at various codon positions were obtained (Table 1). They can be used to infer mutation patterns involved in genome evolution. For example, in S clade, at codon position 1, only U number is increased by 4.7. It is thus considered that 3.5, 0.3 and 0.9 nucleotides of C, G and A have been mutated into U, respectively. Then, at codon position 2, only C number is reduced by 5.0. It is thus considered that 1.3, 0.1 and 3.6 nucleotides of C have been mutated into G, A and U, respectively. As for codon position 3, 4.7 and 1.2 nucleotides of C and G are reduced, and 1.1 and 4.8 nucleotides of A and U are increased. Thus, it is considered that 5.9 (4.7 + 1.2) nucleotides of C/G have been mutated into U/A (4.8 + 1.1).
Table 1.
Overall variations of nucleotide numbers at different codon positions.
| Lineage | Codon position | C | G | A | U |
|---|---|---|---|---|---|
| S | 1 | −3.5 | −0.3 | −0.9 | +4.7 |
| 2 | −5.0 | +1.3 | +0.1 | +3.6 | |
| 3 | −4.7 | −1.2 | +1.1 | +4.8 | |
| L | 1 | 0.0 | −0.4 | +0.4 | 0.0 |
| 2 | −0.6 | −0.4 | +0.3 | +0.7 | |
| 3 | −0.4 | +0.1 | −0.3 | +0.6 | |
| V | 1 | −0.6 | +0.2 | −0.4 | +0.7 |
| 2 | −0.3 | 0.0 | −0.3 | +0.5 | |
| 3 | −0.3 | +0.5 | −0.1 | −0.1 | |
| G | 1 | −1.2 | −0.6 | −0.1 | +1.9 |
| 2 | −1.3 | +0.4 | −0.1 | +1.0 | |
| 3 | −0.1 | −0.3 | −0.1 | +0.5 | |
| GH | 1 | −1.7 | −0.9 | −0.2 | +2.8 |
| 2 | −4.6 | +0.4 | +0.1 | +4.1 | |
| 3 | −2.2 | −1.0 | +0.4 | +2.8 | |
| GR | 1 | −1.1 | +0.5 | −1.3 | +1.9 |
| 2 | −4.3 | +0.1 | +1.3 | +3.0 | |
| 3 | −2.2 | −0.5 | −0.3 | +2.9 | |
| GV | 1 | −0.8 | −1.0 | −0.1 | +1.8 |
| 2 | −1.1 | 0.0 | 0.0 | +1.2 | |
| 3 | −1.1 | −0.9 | 0.0 | +2.0 | |
| Alpha | 1 | −0.5 | −0.7 | 0.0 | +1.2 |
| 2 | −1.0 | 0.0 | −0.1 | +1.2 | |
| 3 | −1.8 | −0.7 | −0.1 | +2.5 | |
| Beta | 1 | −0.8 | −0.3 | −0.2 | +1.3 |
| 2 | −0.7 | +0.1 | 0.0 | +0.6 | |
| 3 | −1.0 | −0.5 | −0.4 | +1.9 | |
| Gamma | 1 | −0.3 | +0.2 | −0.5 | +0.6 |
| 2 | −2.3 | 0.0 | +0.3 | +2.0 | |
| 3 | −1.3 | 0.0 | −0.2 | +1.5 | |
| Delta | 1 | −0.2 | −2.2 | −0.7 | +3.2 |
| 2 | −2.4 | +0.4 | −0.6 | +2.6 | |
| 3 | −2.0 | +0.5 | −0.2 | +2.7 | |
| Omicron | 1 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 0.0 | 0.0 | 0.0 | 0.0 | |
| 3 | −0.2 | −0.1 | 0.0 | +0.3 |
This table lists increased (+) or reduced (−) number of nucleotides at different codon positions over the survey period. Please refer to Table S3 (values within green frame) for detailed data.
This table lists increased (+) or reduced (−) number of nucleotides at different codon positions over the survey period. Please refer to Table S3 (values within green frame) for detailed data.
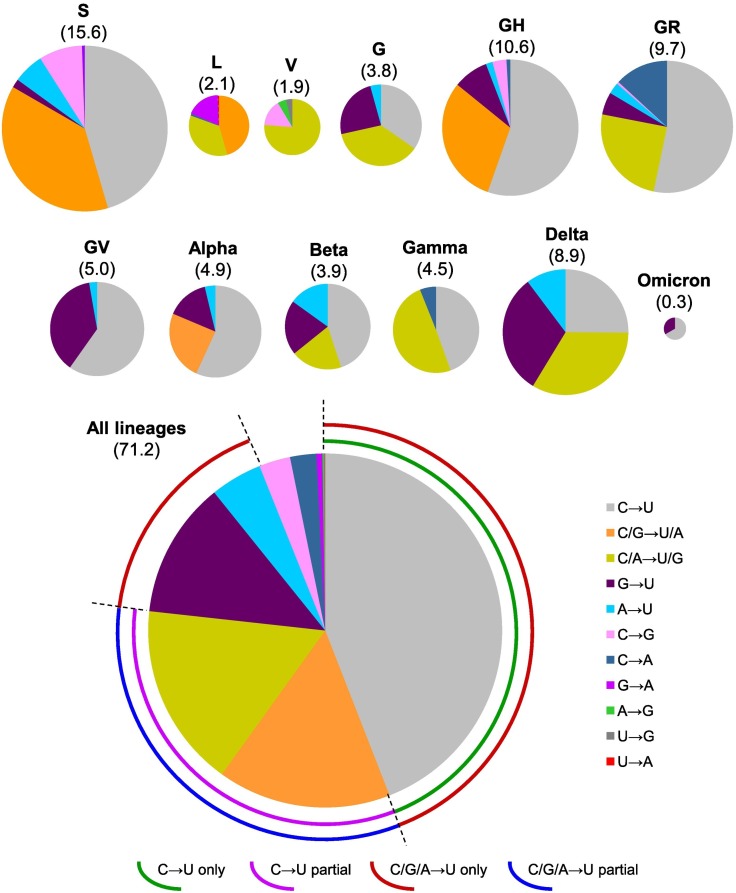
Based on calculations using data listed in Table 1, mutation patterns of nucleotides in all SARS-CoV-2 lineages were obtained (Fig. 3). It is found that, (i) C to U mutation occurs frequently in all lineages except L and V clades. (ii) C/G to U/A mutation mainly occurs in S, L, GH and Alpha lineages. (iii) C/A to U/G mutation occurs predominantly in V clade and Gamma variant. It also occurs frequently in L, G, GR, Beta and Delta lineages. (iv) G to U mutation occurs frequently in G, GV and Delta lineages. (v) G to A and C to A mutations occur specifically in L and GR lineages, respectively. Mutation patterns of all linages (merged data of twelve lineages) show that C is the major target for mutation, and U is the major product of mutation (Fig. 3).
Fig. 3.
Mutation patterns involved in genome evolution of SARS-CoV-2. Size of each pie chart is in scale with total number of nucleotide mutations, which is indicated with number below lineage name. Detailed data for each lineage are listed in Table S3 (values in blue colour, within green frame). Pie chart of all lineages is based on merged data of twelve lineages.
Mutation patterns involved in genome evolution of SARS-CoV-2. Size of each pie chart is in scale with total number of nucleotide mutations, which is indicated with number below lineage name. Detailed data for each lineage are listed in Table S3 (values in blue colour, within green frame). Pie chart of all lineages is based on merged data of twelve lineages.
3.4. Mutations at different codon positions
In order to understand whether nucleotide mutations occur more frequently at codon position 1, 2 or 3, we made a statistics on monthly variations of C, G, A and U at different codon positions (Table S3, data within purple frame). Average monthly variation rate of nucleotides at different codon positions (Fig. S3) shows that only GR, Gamma and Delta lineages have significant difference in C and G mutations between different codon positions. Mutations of all other nucleotides have no significant difference between codon positions. This means that most mutations of C, G and A take place indiscriminately regardless of which positions they are at. This is true even when the mutation leads to formation of a premature stop codon. As we have identified, codon 254 of ORF3a has been mutated from GGA to UGA in Beta variant. Codons 27 and 68 of ORF8 have been mutated from CAA and AAA to UAA in Alpha variant.
3.5. Genome stability of mutated viruses
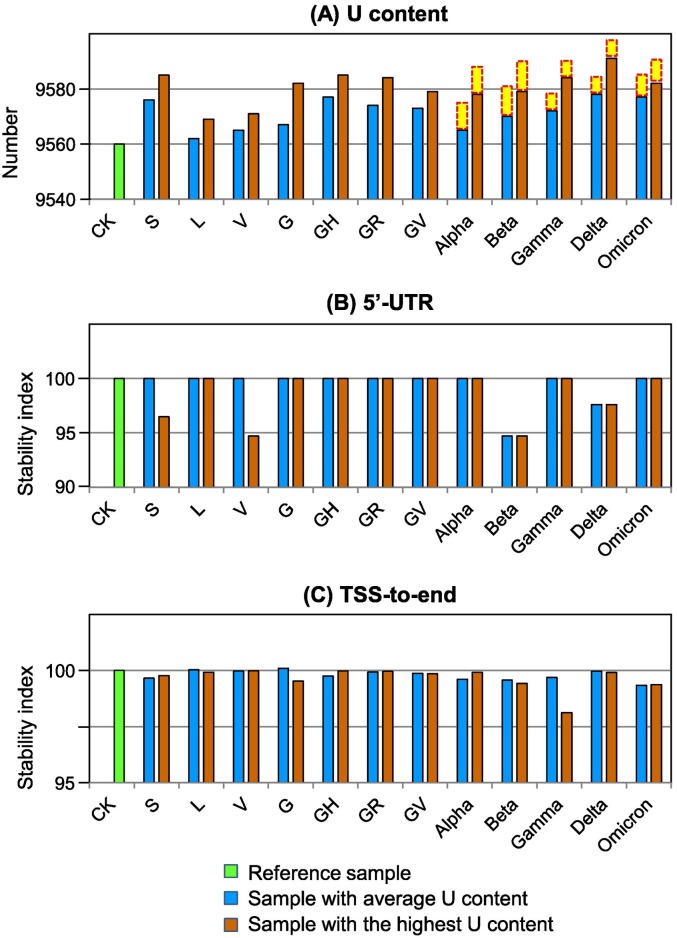
Nucleotide composition in an RNA strand has a close relationship with stability of its secondary structure. An RNA strand with high U + A content will form a secondary structure less stable than that with high C + G content, because C-G and U-A base pairs have three and two hydrogen bonds, respectively. Thus, a viral genome with increased U-content will have lower stability. Such genome can be unfolded more easily for replication. In fact, within the region for survey (totalling 29,769 nucleotides), the highest U number has reached 9591 in individual viruses, being 31 higher over the reference sample. As for the five variants, if the deleted U numbers (Fig. 1) are compensated, their U numbers could be 29–37 higher than the reference sample (Fig. 4A).
Fig. 4.
Genome stability of SARS-CoV-2. For genome stability comparison, two virus samples were taken for calculating free energy of their genomic segments. One of the samples has average U content, while the other has the highest U content. (A) U content in various samples. Yellow block on top of data bar indicates U number lost from deletions (as shown in Fig. 1) in the five variants. (B) and (C) Stability of 5′-UTR (untranslated region) and TSS (translation start site)-to-end region in various samples. Please refer to Table S4 for detailed sample information and free energy values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Genome stability of SARS-CoV-2. For genome stability comparison, two virus samples were taken for calculating free energy of their genomic segments. One of the samples has average U content, while the other has the highest U content. (A) U content in various samples. Yellow block on top of data bar indicates U number lost from deletions (as shown in Fig. 1) in the five variants. (B) and (C) Stability of 5′-UTR (untranslated region) and TSS (translation start site)-to-end region in various samples. Please refer to Table S4 for detailed sample information and free energy values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Stability of 5′-UTR (1–200 nt) is considerably reduced in Beta and Delta variants, and in individual samples with the highest U level in S and V clades (Fig. 4B). It is found that, a C to U mutation occurs at site 142 in S clade, a G-to-U mutation occurs at site 109 in V and Beta lineages, and a G to U mutation occurs at site 145 in Delta variant. This single nucleotide mutation has led to a 3–5% reduction of 5′-UTR stability. Stability of TSS-to-end region is reduced slightly in all samples (Fig. 4C). This slight reduction is understandable, because increased U number only accounts for a very low percentage in the whole genome. Yet, the trends of U-content increase and genome-stability reduction are obvious in all lineages.
4. Discussion
4.1. Diversity of nucleotide mutation
Mutation patterns described in Fig. 2 do not include U, G and A to C. This does not mean their absence in genome evolution of SARS-CoV-2. In fact, monthly variations of codon numbers show that any nucleotide may be mutated to any other nucleotide during transmission of the virus (Table S3, data within purple frame). These observations are consistent with presence of all twelve possible mutation patterns in evolution of SARS-CoV-2 genome [27], [28] and formation of premature stop codons in coding regions of ORF6 and ORF8 [34], [35]. Deletion and insertion are other patterns of nucleotide mutations occurring in SARS-CoV-2 genome [19], [36]. A dozen of deletions and an insertion have contributed to the emergence of Alpha, Beta, Gamma, Delta and Omicron variants, which are 19, 18, 5, 13 and 27 nucleotides shorter than the seven surveyed clades, respectively (Fig. 1). Meanwhile, insertion of 1–8 nucleotides occurred in virus samples of GH, GR and GV clades (data not shown). All these data demonstrate that nucleotide mutations occurring in SARS-CoV-2 genome are greatly diversified.
4.2. Trend of virus evolution
If nucleotide mutations are greatly diversified in SARS-CoV-2, why do current viruses have higher U-content and smaller genome-size? This could be the consequence of natural selection, because a virus with increased U-content and smaller genome-size can be more successful in replication. On one hand, increased U-content in 5′-UTR reduces stability (Fig. 4B) of its IRES (internal ribosome entry site) structure [31], [37]. This makes the virus more cryptic in replication, because less host machinery is recruited to translate viral RNA. On the other hand, higher U-content and smaller genome-size reduce stability of viral RNA (Fig. 4C). This makes the virus more efficient in replication, because less host energy is consumed to disrupt secondary structures of viral RNA. Thus, the virus could become less virulent but more infectious, because more viruses can be replicated from using unit resources.
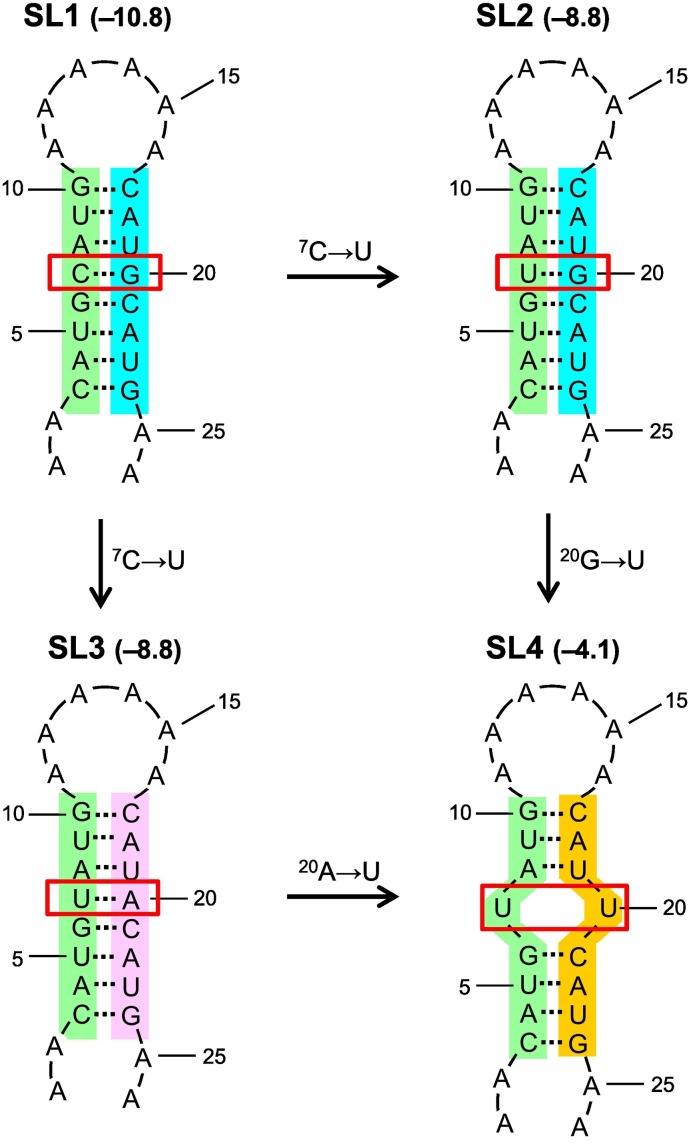
Mutating C, G and A into U is probably a new trend of SARS-CoV-2 genome evolution. Previously, we reported that C to U and G to A mutations allow SARS-CoV-2 to possess a genome with low C + G content. Thus, a potential C-G base-pair formed in genomic RNA could be replaced by a potential A-U base pair [31]. Our current survey reveals that C, G and A to U mutations all occurred in most SARS-CoV-2 lineages (Fig. 2). These data suggest that SARS-CoV-2 has attempted not only to replace a potential C-G base pair with A-U base pair but also to avoid formation of A-U and/or G-U base pairs, because such mutations could reduce viral genome stability to a greater extent (Fig. 5 ).
Fig. 5.
Stability reduction of a stem-loop structure. Shown here is an example of stability reduction of stem-loop structures formed by hypothetical nucleotide sequences. Firstly, stem-loop 1 (SL1) is formed between the green- and cyan-shaded segments. This structure has a free energy of −10.8 kcal/mol. Then, after C at position 7 (7C) is mutated into U, SL2 is formed because U is able to form a canonical base pair with G [33]. Alternatively, SL3 can be formed because the green-shaded segment may pair with another segment (mauve-shaded). Both SL2 and SL3 are less stable than SL1, because they have a free energy of −8.8 kcal/mol. Finally, after 20G in SL2 and 20A in SL3 are mutated into U, stability of the formed stem-loop (SL4) is further reduced, only having a free energy of −4.1 kcal/mol. Red frame highlights the formed or disrupted base pair. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.3. Mutability of Delta and Omicron variants
Delta variant has a high and unique mutability over other lineages, which has probably enabled it to cause many vaccine-breakthrough infections [38], [39]. Its high mutability is reflected in elevating its U number by 9.7 and changing numbers of 14 amino acids significantly within ten months (Figs. 2 and S1). Its unique mutability is reflected in having high number of G and C mutated at codon position 1 and 2 respectively (Fig. S3). Such high and unique mutability could provide a large number of mutated viruses for natural selection against vaccines. This could probably explain why Delta variant caused more and more vaccine-breakthrough infections in many COVID-19 devastated countries [40], [41].
Omicron variant emerged in mid-November 2021 by having high number of amino acid substitutions relative to the earliest SARS-CoV-2 virus [42], [43]. This high mutability has resulted in rapid transmission of the virus and occurrence of a few vaccine breakthroughs [44], [45]. Our survey indicates that Omicron has the smallest genome size among all SARS-CoV-2 lineages. Its genome is 27 nucleotides shorter than the reference one (Fig. 1) and has a considerably lower stability than other SARS-CoV-2 lineages (Fig. 4C). However, because of recent emergence, no sufficient sequence data are available for characterising its nucleotide and amino acid mutations. Thus, whether it has similar mutability like Delta variant awaits future investigation.
In conclusion, human SARS-CoV-2 has evolved to increase U content and reduce genome size. C, G and A to U mutations have all contributed to this U-content increase. Mutations of C, G and A at codon position 1, 2 or 3 have no significant difference in most lineages. Both deletion and insertion are involved in formation of SARS-CoV-2 variants. These results may provide a clue for tracing the origin of SARS-CoV-2, because ancestral viruses should have lower U-content and probably bigger genome-size.
The following are the supplementary data related to this article.
Fig. S1. Overall variations of amino acid numbers in SARS-CoV-2 lineages.
Fig. S2. Overall variations of codon numbers in SARS-CoV-2 lineages.
Fig. S3. Monthly mutation rates of nucleotides at different codon positions.
Monthly data of nucleotide variations in SARS-CoV-2 lineages.
Monthly data of amino acid variations in SARS-CoV-2 lineages.
Monthly data of codon variations in SARS-CoV-2 lineages.
Genome stability data of SARS-CoV-2 lineages.
CRediT authorship contribution statement
Y.W., Q.Y. and K.P.C. conceived the study. Y.W. and X.Y.C. wrote the manuscript. Y.W. compiled the computer program. Y.W., X.Y.C. and L.Y. performed surveys and analyses. All authors reviewed the manuscript.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
The authors are greatly thankful to all contributors of SARS-CoV-2 genome sequences and to GISAID Initiative team for classifying all the sequences into various lineages. This study was supported by National Key Research and Development Program of China (No. 2018YFE0196600) and National Natural Science Foundation of China (No. 31861143051).
Data availability
Data will be made available on request.
References
- 1.WHO . 2022. Weekly Epidemiological Update on COVID-19.www.who.int/publications/m/ [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P., Fang C., Huang D., Huang L.Q., Huang Q., Han Y., Hu B., Hu F., Li B.H., Li Y.R., Liang K., Lin L.K., Luo L.S., Ma J., Ma L.L., Peng Z.Y., Pan Y.B., Pan Z.Y., Ren X.Q., Sun H.M., Wang Y., Wang Y.Y., Weng H., Wei C.J., Wu D.F., Xia J., Xiong Y., Xu H.B., Yao X.M., Yuan Y.F., Ye T.S., Zhang X.C., Zhang Y.W., Zhang Y.G., Zhang H.M., Zhao Y., Zhao M.J., Zi H., Zeng X.T., Wang Y.Y., Wang X.H. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020;7(1):4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai C.K.C., Lam W. Laboratory testing for the diagnosis of COVID-19. Biochem. Biophys. Res. Commun. 2021;538:226–230. doi: 10.1016/j.bbrc.2020.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 6.Gavriatopoulou M., Ntanasis-Stathopoulos I., Korompoki E., Fotiou D., Migkou M., Tzanninis I.G., Psaltopoulou T., Kastritis E., Terpos E., Dimopoulos M.A. Emerging treatment strategies for COVID-19 infection. Clin. Exp. Med. 2021;21(2):167–169. doi: 10.1007/s10238-020-00671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y.C., Dema B., Reyes-Sandoval A. COVID-19 vaccines: breaking record times to first-in-human trials. NPJ Vaccines. 2020;5(1):34. doi: 10.1038/s41541-020-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awadasseid A., Wu Y., Tanaka Y., Zhang W. Current advances in the development of SARS-CoV-2 vaccines. Int. J. Biol. Sci. 2021;17(1):8–19. doi: 10.7150/ijbs.52569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Güner R., Hasanoğlu I., Aktaş F. COVID-19: prevention and control measures in community. Turk. J. Med. Sci. 2020;50(SI-1):571–577. doi: 10.3906/sag-2004-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng F.J., Tu L., Yang Y.S., Hu P., Wang R.S., Hu Q.Y., Cao F., Jiang T.J., Sun J., Xu G.G., Chang C. Management and treatment of COVID-19: the Chinese experience. Can. J. Cardiol. 2020;36(6):915–930. doi: 10.1016/j.cjca.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valle C., Martin B., Touret F., Shannon A., Canard B., Guillemot J.C., Coutard B., Decroly E. Drugs against SARS-CoV-2: what do we know about their mode of action? Rev. Med. Virol. 2020;30(6):1–10. doi: 10.1002/rmv.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asselah T., Durantel D., Pasmant E., Lau G., Schinazi R.F. COVID-19: discovery, diagnostics and drug development. J. Hepatol. 2021;74(1):168–184. doi: 10.1016/j.jhep.2020.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González-Candelas F., Shaw M.A., Phan T., Kulkarni-Kale U., Paraskevis D., Luciani F., Kimura H., Sironi M. One year into the pandemic: short-term evolution of SARS-CoV-2 and emergence of new lineages. Infect. Genet. Evol. 2021;92 doi: 10.1016/j.meegid.2021.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilmjärv S., Abdul F., Acosta-Gutiérrez S., Estarellas C., Galdadas I., Casimir M., Alessandrini M., Gervasio F.L., Krause K.H. Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant. Sci. Rep. 2021;11(1):13705. doi: 10.1038/s41598-021-91662-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79(5):710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moeller N.H., Shi K., Demir Ö., Banerjee S., Yin L., Belica C., Durfee C., Amaro R.E., Aihara H. bioRxiv. 2021. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson B.S., Kolte V., Khan A., Sharma G. Dynamic tracking of variant frequencies depicts the evolution of mutation sites amongst SARS-CoV-2 genomes from India. J. Med. Virol. 2021;93(4):2534–2537. doi: 10.1002/jmv.26756. [DOI] [PubMed] [Google Scholar]
- 18.Giovanetti M., Benedetti F., Campisi G., Ciccozzi A., Fabris S., Ceccarelli G., Tambone V., Caruso A., Angeletti S., Zella D., Ciccozzi M. Evolution patterns of SARS-CoV-2: snapshot on its genome variants. Biochem. Biophys. Res. Commun. 2021;538:88–91. doi: 10.1016/j.bbrc.2020.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bianchi M., Borsetti A., Ciccozzi M., Pascarella S. SARS-Cov-2 ORF3a: mutability and function. Int. J. Biol. Macromol. 2021;170:820–826. doi: 10.1016/j.ijbiomac.2020.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S.Q., Tian C., Liu P.P., Guo D.J., Zheng W., Huang X.Q., Zhang Y., Liu L.J. Effects of SARS-CoV-2 mutations on protein structures and intraviral protein-protein interactions. J. Med. Virol. 2021;93(4):2132–2140. doi: 10.1002/jmv.26597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield COVID-19 Genomics Group. McDanal C., Perez L.G., Tang H.L., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Oosterhout C., Stephenson J.F., Weimer B., Ly H., Hall N., Tyler K.M. COVID-19 adaptive evolution during the pandemic - implications of new SARS-CoV-2 variants on public health policies. Virulence. 2021;12(1):2013–2016. doi: 10.1080/21505594.2021.1960109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amato L., Jurisic L., Puglia I., Lollo V.D., Curini V., Torzi G., Girolamo A.D., Mangone I., Mancinelli A., Decaro N., Calistri P., Giallonardo F.D., Lorusso A., D’Alterio N. Multiple detection and spread of novel strains of the SARS-CoV-2 B.1.177 (B.1.177.75) lineage that test negative by a commercially available nucleocapsid gene real-time RT-PCR. Emerg. Microbes Infect. 2021;10(1):1148–1155. doi: 10.1080/22221751.2021.1933609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice A.M., Morales A.C., Ho A.T., Mordstein C., Mühlhausen S., Watson S., Cano L., Young B., Kudla G., Hurst L.D. Evidence for strong mutation bias toward, and selection against, U content in SARS-CoV-2: implications for vaccine design. Mol. Biol. Evol. 2021;38(1):67–83. doi: 10.1093/molbev/msaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., Meyer B., Li C.L., Marton I., Dym O., Elad N., Lewis M.G., Andersen H., Gagne M., Seder R.A., Douek D.C., Schreiber G., Rudich Y.N. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6(9):1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 27.Roy C., Mandal S.M., Mondal S.K., Mukherjee S., Mapder T., Ghosh W., Chakraborty R. Trends of mutation accumulation across global SARS-CoV-2 genomes: implications for the evolution of the novel coronavirus. Genomics. 2020;112(6):5331–5342. doi: 10.1016/j.ygeno.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto S.P., Day T., Arino J., Colijn C., Dushoff J., Li M., Mechai S., Domselaar G.V., Wu J.H., Earn D.J.D., Ogden N.H. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021;31(14):R918–R929. doi: 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GISAID . 2021. Clade and Lineage Nomenclature Aids in Genomic Epidemiology Studies of Active hCoV-19 Viruses.www.gisaid.org/references/statements-clarifications/ [Google Scholar]
- 30.Chen J.H., Wang R., Wang M.L., Wei G.W. Mutations strengthened SARSCoV-2 infectivity. J. Mol. Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Mao J.M., Wang G.D., Luo Z.P., Yang L., Yao Q., Chen K.P. Human SARS-CoV-2 has evolved to reduce CG dinucleotide in its open reading frames. Sci. Rep. 2020;10(1):12331. doi: 10.1038/s41598-020-69342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delbue S., Alessandro S.D., Signorini L., Dolci M., Pariani E., Bianchi M., Fattori S., Modenese A., Galli C., Eberini I., Ferrante P. Isolation of SARS-CoV-2 strains carrying a nucleotide mutation, leading to a stop codon in the ORF6 protein. Emerg. Microbes Infect. 2021;10(1):252–255. doi: 10.1080/22221751.2021.1884003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeRonde S., Deuling H., Parker J., Chen J. Identification of a novel SARS-CoV-2 strain with truncated protein in ORF8 gene by next generation sequencing. Res. Sq. 2021 doi: 10.21203/rs.3.rs-413141/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patiño-Galindo J.Á., Filip I., Chowdhury R., Maranas C.D., Sorger P.K., AlQuraishi M., Rabadan R. Recombination and lineage-specific mutations linked to the emergence of SARS-CoV-2. Genome Med. 2021;13(1):124. doi: 10.1186/s13073-021-00943-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonenberg N., Pelletier J. Poliovirus translation: a paradigm for a novel initiation mechanism. Bioessays. 1989;11(5):128–132. doi: 10.1002/bies.950110504. [DOI] [PubMed] [Google Scholar]
- 38.McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., Marco A.D., Franko N., Tilles S.W., Logue J., Miranda M.C., Ahlrichs M., Carter L., Snell G., Pizzuto M.S., Chu H.Y., Voorhis W.C.Van, Corti D., Veesler D. bioRxiv. 2021. Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants. [DOI] [PubMed] [Google Scholar]
- 39.Farinholt T., Doddapaneni H., Qin X., Menon V., Meng Q.C., Metcalf G., Chao H., Gingras M.C., Avadhanula V., Farinholt P., Agrawal C., Muzny D.M., Piedra P.A., Gibbs R.A., Petrosino J. Transmission event of SARS-CoV-2 Delta variant reveals multiple vaccine breakthrough infections. BMC Med. 2021;19(1):255. doi: 10.1186/s12916-021-02103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang R., Chen J.H., Hozumi Y., Yin C.C., Wei G.W. ArXiv. 2021. Emerging vaccine-breakthrough SARS-CoV-2 variants. doi: arXiv:2109.04509v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R., Chen J.H., Wei G.W. Mechanisms of SARS-CoV-2 evolution revealing vaccine-resistant mutations in Europe and America. J. Phys. Chem. Lett. 2021;12(49):11850–11857. doi: 10.1021/acs.jpclett.1c03380. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxena S.K., Kumar S., Ansari S., Paweska J.T., Maurya V.K., Tripathi A.K., Abdel-Moneim A.S. Characterization of the novel SARS-CoV-2 omicron (B.1.1.529) variant of concern and its global perspective. J. Med. Virol. 2021 doi: 10.1002/jmv.27524. [DOI] [PubMed] [Google Scholar]
- 43.He X.M., Hong W.Q., Pan X.Y., Lu G.W., Wei X.W. SARS-CoV-2 omicron variant: characteristics and prevention. Med. Comm. 2021;2(4):838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y.C., Zhang L., Li Q.Q., Liang Z.T., Li T., Liu S., Cui Q.Q., Nie J.H., Wu Q., Qu X.W., Huang W.J. The significant immune escape of pseudotyped SARS-CoV-2 variant omicron. Emerg. Microbes Infect. 2022;11(1):1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kannan S.R., Spratt A.N., Sharma K., Chand H.S., Byrareddy S.N., Singha K. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022;126 doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Overall variations of amino acid numbers in SARS-CoV-2 lineages.
Fig. S2. Overall variations of codon numbers in SARS-CoV-2 lineages.
Fig. S3. Monthly mutation rates of nucleotides at different codon positions.
Monthly data of nucleotide variations in SARS-CoV-2 lineages.
Monthly data of amino acid variations in SARS-CoV-2 lineages.
Monthly data of codon variations in SARS-CoV-2 lineages.
Genome stability data of SARS-CoV-2 lineages.
Data Availability Statement
Data will be made available on request.