Abstract
The gene for a 30-kDa immunodominant antigen, P30, of Mycoplasma agalactiae was cloned from type strain PG2 and expressed in Escherichia coli. P30 is encoded on a monocistronic operon determined by two −10 boxes and a possible −35 region constituting the potential promoter, and a transcription termination site. The gene for the 266-amino-acid protein is preceded by a polypurine-rich region designed as the consensus sequence for a ribosome-binding site. Analysis of the amino acid sequence of P30 revealed the presence of a recognition site for a prokaryotic signal peptidase II at amino acid (aa) 24, indicating that P30 is a transmembrane protein. Moreover, Triton X-114 phase partitioning of M. agalactiae PG2 total antigen revealed that P30 is strongly hydrophobic and hence a possible membrane component. Immunoblot analysis using the monospecific polyclonal anti-P30-His serum indicated that P30 is specific to M. agalactiae. Furthermore, PCR amplification with specific primers for p30 and Southern blot analysis revealed the presence of the gene in all M. agalactiae strains tested and its absence in the other mycoplasma species. Among 27 strains of M. agalactiae studied, 20 strains belonging to the common serotypes A to D, including PG2, expressed P30 or part of it as detected by the monospecific polyclonal anti-P30 antibodies. The other seven strains belonging to the rarely isolated serotypes E to H were negative for P30. The p30 gene was sequenced in 15 strains of M. agalactiae, 10 of which expressed P30 or at least part of it and 5 of which did not express P30. The negative strains carried mutations in both −10 boxes of the promoters. These mutations seem to be responsible for the lack of P30 expression in these strains. Analysis of sera from sheep that were experimentally infected with M. agalactiae revealed that P30 induced a strong and persistent immune response which was still very high two months after infection. In contrast, currently used enzyme-linked immunosorbent assay serology gave only low titers.
Mycoplasma agalactiae is the main causative agent of the contagious agalactia (CA) syndrome of sheep and goats. This syndrome, characterized by agalactia, mastitis, arthritis, and sometimes keratoconjunctivitis, is present all over the world with a particular importance around the Mediterranean basin (6, 11). M. agalactiae causes extensive economical losses particularly in dairy flocks and herds, due to persistent and highly contagious agalactia which makes regular cheese production impossible. From an epidemiological point of view, CA is characterized by an important infection chronicity at the flock level, and long-lasting situations of endemicity at the regional level. A better understanding of mechanisms implicated in the pathogenesis as well as knowledge about the nature of the major antigenic substances of M. agalactiae will be necessary for controlling CA. A few mycoplasmas use a specialized attachment organelle (10) where adhesins are concentrated, and this permits adhesion to the host cells and, then, dissemination into the host (24). In some mycoplasmas, a capsule-like structure has been shown to be located on the external surface of the cell. This structure may be associated with virulence, providing mycoplasmas with the ability to evade the host immune system (20). M. agalactiae possesses a particular ability to modify the surface coat of the membrane, allowing the bacterium to escape the host's immune defense. This is presently the only known mechanism involved in pathogenicity. At present, a great deal is known about the structure and molecular mechanisms of variable surface antigens of some mycoplasmas, including M. agalactiae (28, 33, 35). However, for diagnostic purposes, variable antigens do not permit easy serological identification of mycoplasma isolates and significantly hinder serological subtyping (29). The finding of stable, specific, and strongly immunogenic antigens for M. agalactiae could be a good alternative for detection, identification, and subtyping of this pathogen. So far, one stable and serologically recognized major surface protein of M. agalactiae (P48) has been described as a potential tool for diagnostic studies (26, 27). Recently, a serological grouping of M. agalactiae has been proposed (5). It is based on a dot immunobinding assay using three monoclonal antibodies which show uniform colony immunostainings and no antigenic variability. Analysis of a large number of field isolates from various geographical regions where CA is diagnosed revealed the existence of four main serotypes and a few occasionally occurring serotypes of M. agalactiae.
The present work describes the isolation and analysis of a new immunogenic antigen, tentatively designated P30, from an expression library of the type strain PG2, and its correlation with the four major serotypes A, B, C and D of M. agalactiae.
MATERIALS AND METHODS
Strains and growth conditions.
Mycoplasma strains used in this study are described in Table 1. They were cultured in standard mycoplasma medium at 37°C until stationary growth phase (23). Cells were harvested by centrifugation at 8,000 × g for 15 min, washed with phosphate-buffered saline (PBS) buffer (140 mM NaCl, 2.7 mM KCl, 15 mM KH2PO4, 8 mM Na2HPO4 [pH 7.4]) and resuspended in PBS to a concentration of approximately 109 cells ml−1. All M. agalactiae strains used in this study were confirmed to belong to the species M. agalactiae using the specific uvrC-based PCR as previously described (31).
TABLE 1.
Mycoplasma strains used in this study
| Mycoplasma species | Straina | Origin | Year isolated | Host (comment) | Serotype(s)b | p30c | P30 expression (kDa)d |
|---|---|---|---|---|---|---|---|
| M. agalactiae | PG2 | Spain | 1973 | Goat (type strain) | A | + | 32 |
| 9 | Italy | Sheep | ABCDe | + | 35 | ||
| 190 (Ag1) | Rumania | 1951 | Sheep (vaccine strain) | A | + | 35 | |
| 209 | France | 1983 | Goat | F | + | —g | |
| 3990 | France | H | + | — | |||
| 4021 | France | 1988 | Sheep | A | + | 35 | |
| 4054-1 | France | 1986 | Goat | EFGHe | + | — | |
| 4055 | France | 1987 | Goat | F | + | — | |
| 4210 | France | 1982 | Goat | D | + | 35 | |
| 4212 | France | 1984 | G | + | — | ||
| 4258 | France | Goat | D | + | 35 | ||
| 5225 | Spain | 1991 | Goat | C | + | +f | |
| 5632 | Spain | 1991 | Goat | F | + | − | |
| 5670 | Spain | 1991 | Sheep | ABCDe | + | 35 | |
| 5725 | France | 1990 | Sheep | A | + | 35 | |
| 5826 | Spain | 1992 | Sheep | A | + | 35 | |
| 6833 | Italy | Goat | A | + | 32 | ||
| 6968 | Spain | 1993 | Sheep | D | + | 35 | |
| 7169 | Switzerland | Goat | C | + | 35 | ||
| 7314 | Greece | 1986 | Sheep | A | + | 30+32 | |
| 7327 | Greece | 1987 | Goat | A | + | +f | |
| 7375 | Switzerland | Goat | C | + | 35 | ||
| 7784 | France | 1994 | Sheep | A | + | 35 | |
| 8064 | Ivory Coast | 1989 | Sheep | E | + | — | |
| 8750 | France | 1994 | Sheep | A | + | 35 | |
| 9385 | Portugal | Goat | ABCDe | + | 35 | ||
| 9600 | Portugal | Goat | A | + | 35 | ||
| M. bovis | PG45 | Cattle (type strain) | — | — | |||
| M. capricolum subsp. capricolum | California kid | California | Goat (type strain) | — | — | ||
| M. mycoides subsp. mycoides LC | Y-goat | Australia | Goat (type strain) | — | — | ||
| Mycoplasma sp. bovine group 7 | PG50 | Australia | Cattle (reference strain) | — | — | ||
| M. putrefaciens | KS1 | Goat (type strain) | — | — | |||
| M. ovipneumoniae | Y98 | — | — | ||||
| M. mycoides subsp. capri | PG3 | Goat (type strain) | — | — | |||
| Mycoplasma sp. serogroup 11 | 2D | — | — | ||||
| M. bovigenitalium | PG11 | — | — | ||||
| M. conjunctivae | HCR/581 | — | — | ||||
| M. arginini | G230 | — | — |
Collections: strains were obtained from F. Poumarat, AFSSA, Lyon, France; F. Thiaucourt, CIRAD, Montpellier, France; M. Lambert, AFSSA, Sophia Antipolis, France; and S. Tola, Istituto Zooprofilattico Sperimentale, Sassari, Italy.
According to PCR with primers MagaORF1-L and MagaORF1-R, and by Southern blotting with the p30 DIG-labeled probe.
As obtained by using monospecific polyclonal rabbit serum against recombinant P30.
Serotyped only with monoclonal antibodies 1D4 (5), thus uniquely allowing the differentiation of serotypes A to D from serotypes E to H.
Strains reacted with only anti-P30 serum on colony blots, presumably due to a rather short, truncated form of P30.
—, no amplification or detection.
For genetic manipulation and subcloning, the two Escherichia coli strains XL1-Blue MRF′ and XLOLR (Stratagene, La Jolla, Calif.) were used. E. coli strains were grown in Luria-Bertani (LB) broth at 37°C. The phagemid expression vector pBK-CMV (Stratagene) was propagated in E. coli strain XLOLR. For subcloning, the vector pBluescriptII SK(−) (Stratagene) was propagated in E. coli strain XL1-Blue. The pETHIS-1 expression vector (30) was used for polyhistidine fusion at both N-terminal and C-terminal ends of cloned proteins. The E. coli strain BL21(DE3)pLysE (Novagen, Madison, Wis.) was used for expression of recombinant proteins.
DNA manipulation, construction, and screening of an expression library.
Mycoplasma genomic DNA from strains listed in Table 1 was extracted by the phenol-chloroform technique or by the guanidinium thiocyanate technique (9). DNA of the type strain PG2 was then partially digested by Sau3AI to obtain fragments ranging from 2 to 10 kb. DNA fragments were ligated with BamHI-digested λ Zap Express cloning vector (Stratagene) and packaged with Gigapack II packaging extract (Stratagene). The library was plated according to the manufacturer's protocol using the E. coli host strain XL1-Blue MRF′. Screening of the library was performed following the standard protocol with serum from a naturally infected convalescent sheep (serum PAL 97) used at a dilution of 1:600. The selected positive clones were amplified and excised in vivo into phagemids pBK-CMV using the filamentous helper phage ExAssist (Stratagene) as indicated by the manufacturer.
Nucleotide sequencing and sequence analysis.
DNA sequencing was performed with a DNA Sequenator AB 310 and the Taq Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Norwalk, Conn.). In the first step, oligonucleotide primers containing the T3 and T7 promoter sequences (Table 2) flanking the cloning site of the pBK-CMV vector were used. The sequence was completed by primer walking using synthesized oligonucleotides. The DNA sequence was assembled using the program Sequencher 3.0 (GeneCodes, Ann Arbor, Mich.). The DNA and deduced amino acid sequences were analyzed with the PC/Gene program PROSITE (3). Sequence comparisons with sequences in the GenBank and EMBL databases were made using the BLAST programs BLASTN, BLASTX, and BLASTP (1). Alignments were done with the Wisconsin package (Genetics Computer Group, Inc., Madison, Wis.). The phylogenetic relationship was established with PILEUP from the Genetics Computer Group program package and by further analysis with the Mega 1.02 program (15): corrections were calculated with the Jukes-Cantor algorithm, and a tree was derived by the neighbor-joining method.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequencea | Nucleotide positionb | Annealing tempc (° C) |
|---|---|---|---|
| T3 (universal) | 5′-GCGCGCAATTAACCCTCACTAAAG-3′ | 53.0 | |
| T7 (universal) | 5′-GTAATACGACTCACTATAGGGC-3′ | 48.5 | |
| BKE3P0 | 5′-GATCCTAGTTTAAAAAGGGAAGC-3′ | 50.0 | |
| BKE3P1 | 5′-TTTCAGTTGTAGCTTCACCA-3′ | 2255–2236 | 48.6 |
| BKE3P2 | 5′-TTAAGCAGTTTTAAATAACA-3′ | 110–129 | 46.5 |
| BKE3P3 | 5′-AGGTTCTTCGTCCTCTTCAG-3′ | 1807–1788 | 49.0 |
| BKE3P4 | 5′-TCGCAGGAAAGGAAGCTATC-3′ | 520–539 | 50.4 |
| BKE3P5 | 5′-AAGCGAGCTATCTTCGTGTG-3′ | 1354–1335 | 49.9 |
| BKE3P6 | 5′-TAAGGGCGGCAATTCTACAG-3′ | 893–912 | 50.7 |
| MagaORF1-L | 5′-CAGGGGGATGAACATTTATG-3′ | 130–149 | 49.9 |
| MagaORF1-L2 | 5′-GATCAAATATACAAAATTACAA-3′ | 1–22 | 46.8 |
| MagaORF1-R | 5′-TTACCTCCATCTTTTTCAAC-3′ | 859–840 | 48.0 |
| MagaORF1-R2 | 5′-TAGCTGTTTGCTATTTTTCCAC-3′ | 1155–1134 | 49.7 |
| MagaORF1-R3 | 5′-CACACCTTACTACCAACAAC-3′ | 1065–1046 | 47.6 |
| MagaORF1EcoRIN2 | 5′-GTgaattcAAAATGCAGTGAAGATGACAAA-3′ | 210–239 | 53.4 |
| MagaORF1NotIC | 5′-TTgcggccgcAACAAATCCCATTAGGTTTAG-3′ | 846–817 | 56.4 |
Lowercase letters indicate nucleotides added to create restriction enzyme recognition sites for cloning.
Based on nucleotide sequence AF327858 (2.4-kb contiguous fragment from PG2).
Obtained with the PCR primer annealing temperature calculator developed by J. Boxall (http://www.iacr.bbsrc.ac.uk/res/depts/biochem/old-or-to-move/tcalculator.html) by using the parameters 30% as target GC content and 1,000 bp as target size.
PCR amplification.
PCRs were performed in a DNA thermal cycler Gene Amp 9600 (Applied Biosystems) in a 50-μl reaction mixture [50 mM Tris-HCl, pH 9.2; 1.75 mM MgCl2; 16 mM (NH4)2SO4; a 350 μM concentration of each deoxynucleoside triphosphate] containing a 300 nM concentration of each primer (Table 2), 1.75 U of Taq DNA polymerase (Roche Diagnostics, Rotkreuz, Switzerland), and 100 ng of genomic DNA as the template. The DNAs were amplified for 35 cycles (30 s of denaturation at 94°C, 30 s at the optimal annealing temperature of the primers [Table 2], and 2 min of extension at 68°C) after one step of 2 min at 94°C to ensure denaturation. A mixture of Taq and Pwo DNA polymerase (Expand Long Template PCR System kit; Roche Diagnostics) was used when PCR products were to be cloned. Digoxigenin-11-dUTP (DIG)-labeled probes were produced by PCR as described above in the presence of 50 μM DIG.
Southern blot analysis.
Genomic mycoplasmal DNA was digested with a mix of HindIII and Sau3AI, and fragments were separated by electrophoresis on a 1% agarose gel and then transferred onto a positively charged nylon membrane (Roche Diagnostics). The membrane was briefly rinsed with 1× SSC (150 mM NaCl, 15 mM sodium citrate [pH 7.7]), and the DNA was denatured at 80°C under vacuum for 30 min (2). The membrane was incubated in 10 ml of hybridization buffer—consisting of 5× SSC, 0.02% sodium dodecyl sulfate (SDS), 0.1% N-lauroyl-sarcosine, and 1% blocking reagent (Roche Diagnostics)—at 68°C for 2 h and then incubated in 5 ml of hybridization buffer containing 5 μl of DIG-labeled probe for 15 h at 68°C. It was washed twice for 5 min at room temperature with 2× SSC containing 0.1% SDS and twice for 15 min at room temperature with 0.2× SSC containing 0.1% SDS. The hybridized DIG-labeled probe was detected using phosphatase-labeled anti-DIG antibodies (Roche Diagnostics), as described in the manufacturer's protocol.
Production of polyhistidine-tailed fusion protein P30.
With the aim of constructing a plasmid encoding a polyhistidine-tailed P30 fusion protein, we amplified the full-length p30 gene from genomic DNA of strain PG2 using the primers MagaORF1EcoRIN2 and MagaORF1NotIC (Table 2). The PCR product was digested by EcoRI and NotI for cloning into pETHIS-1. The construct was analyzed by restriction enzyme digestion and partial DNA sequencing and then introduced by transformation into E. coli BL21(DE3)pLysE (Novagen) for expression of the fusion protein.
The production of this fusion protein was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at mid-exponential phase, and incubation continued at 37°C for 3 h. The cells were sedimented by centrifugation at 3,000 × g for 10 min, resuspended in 5 ml of PN buffer (50 mM NaH2PO4, pH 8; 300 mM NaCl), sonicated with a microtip for 4 min with the power output control at 7 and a duty cycle of 50% (1-s pulses) in a Branson Sonifier 250 (Branson Ultrasonics, Danbury, Conn.), and then centrifuged at 15,000 × g for 20 min. The supernatant containing the cytosolic fraction was kept, and the pelleted cell debris was resuspended in 5 ml of PN buffer (insoluble fraction). Analysis of the sonicated fraction on SDS–10% acrylamide gels (16) showed that the induced protein was contained in the pellet. Guanidine hydrochloride was added to a final concentration of 6 M to the insoluble fraction, and the mixture was loaded onto a prewashed volume Ni-nitrilotriacetic acid-agarose column (bed volume, 2.5 ml; Qiagen AG, Basel, Switzerland) and washed once more with 30 ml of PNG buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 6 M guanidine hydrochloride). Step elution of the protein was performed with 10 ml of PNG buffer at pH 7.0, 6.0, 5.5, 5.0, and 4.5, and fractions of 1 ml were collected. The fractions were dialyzed against PN buffer and analyzed on SDS–10% acrylamide gels. The purified fusion protein eluted at pH 4.5 and was dialyzed overnight against PN buffer.
Experimental infections.
Experimental infection with M. agalactiae strain 5725 was done with three clinically, bacteriologically, and serologically negative lactating ewes. Briefly, the ewes were inoculated by subcutaneous route with a culture (titer, 2 × 109 CFU/ml) of the field strain 5725, isolated from a typical outbreak (mastitis, agalactia, some keratitis, and arthritis) in the Atlantic Pyrenees. Every ewe developed mastitis. Hypogalactia began at day 6, and agalactia was observed at day 10 (average data).
Experimental infection with type strain PG2 was performed with four lactating ewes (same selection criteria as above). They were infected by subcutaneous route with a culture whose titer was 1.75 × 1010 CFU/ml. All inoculated ewes developed mastitis. Hypogalactia and agalactia occurred roughly at the same time. In the two models, M. agalactiae was reisolated from affected half-udders.
Sera.
Serum PAL 97 from a naturally infected ewe developing a chronic infection was used for screening the expression library and as a positive control for immunoblots. Serial serum samples were obtained from two ewes inoculated with strain 5725 and two ewes inoculated with type strain PG2, collected prior to inoculation, every two days for 14 days after inoculation, and then weekly for 4 weeks. The last sera tested were collected 2 months after inoculation. All the sera were used to study the immune response of these animals to P30.
Polyclonal monospecific serum directed against polyhistidine-tailed protein P30 was obtained by subcutaneous immunization of rabbits with 50 μg of purified recombinant polyhistidine-tailed P30 protein in 100 μl of PN buffer mixed with 100 μl of Freund's adjuvant (Difco Laboratories, Detroit, Mich.), followed by a booster immunization with the same amount of protein in Freund's incomplete adjuvant (Difco Laboratories) 3 weeks later. The animals were bled 10 days after the booster immunization. Antisera were prepared from the blood samples and stored at −20°C.
Immunoblot analysis and enzyme-linked immunosorbent assay (ELISA).
Total antigen from mycoplasmas or recombinant E. coli was mixed with equal volumes of sample buffer, boiled for 5 min, separated on 12% polyacrylamide gels, and blotted onto nitrocellulose membranes (0.2-μm pore size; Bio-Rad S.A., Hercules, Calif.). The membranes were blocked with 1% milk buffer (100 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% Tween 20; 1% [wt/vol] skim milk powder) for 1 h at room temperature. The membranes were then incubated with either sheep serum (PAL 97) at a dilution of 1:600, sheep experimental serial sera at a dilution of 1:200, or rabbit serum at a dilution of 1:1,000 in milk buffer for 90 min at room temperature. The membranes were then washed and incubated with either monoclonal anti-goat-sheep phosphatase-labeled antibody (Sigma-Aldrich, Saint Quentin Fallavier, France) at a dilution of 1:2,000 or with goat anti-rabbit phosphatase-labeled antibody (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) at a dilution of 1:2,000, respectively, for 90 min at room temperature with shaking. Color reaction was initiated by the addition of nitroblue tetrazolium and bromochloroindolyl phosphate in alkaline substrate buffer (7 mM Na2CO3; 3 mM NaHCO3, pH 9.6; 1 mM MgCl2).
ELISA analysis of sera from the experimentally infected sheep was performed using a commercially available kit, based on total antigen of M. agalactiae (17).
Triton X-114 phase partitioning.
M. agalactiae components were separated into hydrophobic and hydrophilic fractions by a modification of the previously described Triton X-114 partitioning method (7). A 10-ml culture of PG2 was grown until the stationary phase and harvested by centrifugation. The cells were washed three times with ice-cold PBS and resuspended in 1 ml of 0.5% Triton X-114 in PBS. They were left on ice for 1 h, with inverting of the tubes several times every 15 min. After centrifugation at maximum speed at 4°C, the supernatant was gently transferred onto 0.5 ml of sucrose cushion (20 ml of PBS, 1.2 g of sucrose, 100 μl of 11.4% Triton X-114) chilled on ice. The tube was incubated at 37°C for 4 min and then centrifuged at room temperature for 3 min. The Triton X-114 pellet was resuspended in 1 ml of ice-cold PBS, incubated at 37°C for 4 min and then centrifuged at room temperature for 3 min another two times. The final volume of the Triton X-114 pellet was estimated, and 3 volumes of PBS was added. Supernatant hydrophilic phase was transferred into a new tube, and 150 μl of 11.4% Triton X-114 was added. After mixing, the tube was incubated at 37°C for 4 min and then centrifuged at room temperature for 3 min. These steps were repeated twice. Samples from the detergent phase, the aqueous phase, and whole mycoplasma cells were mixed with protein sample buffer, run on SDS–12% acrylamide gels, and blotted onto nitrocellulose. The filter was subsequently used for immunoblotting with the monospecific, polyclonal antibodies directed against P30.
Nucleotide sequence accession number.
The EMBL/GenBank accession number for the nucleotide sequence of the 2,434-bp fragment in plasmid pJFFBF1+E3 is AF327858. The sequences of the p30 genes from 15 M. agalactiae field strains are deposited in EMBL/GenBank under AF327859-73.
RESULTS
Cloning the gene for a 30-kDa antigen of M. agalactiae
The expression library of M. agalactiae type strain PG2 containing approximately 5 × 107 recombinant phage clones was screened with serum PAL 97 from a naturally infected ewe. One predominant reactive phage plaque was selected. The DNA from this plaque was converted into phagemid by in vivo excision. The positive phagemid containing a plasmid with a 5.7-kb insert was named pJFFBF1+E. Using total antigen of the E. coli clone containing pJFFBF1+E, a protein doublet band with apparent molecular masses of 32.5 and 30 kDa, which were jointly designated P30, reacted with serum PAL 97 on immunoblots (data not shown).
In order to localize the gene expressing this protein, EcoRI fragments of pJFFBF1+E were subcloned into the EcoRI site of vector pBluescriptII SK(−), and transformants were analyzed by immunoblotting. The protein doublet band of 32.5 and 30 kDa reacting with the serum PAL 97 was found within the subclone containing plasmid pJFFBF1+E3. Analysis of this plasmid revealed that the gene encoding P30 is localized on an insert of 2.6 kb (Fig. 1).
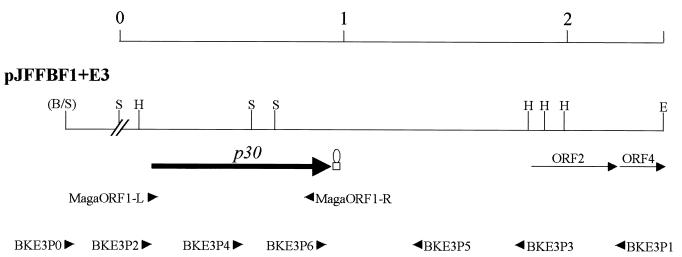
FIG. 1.
Physical map of the p30 locus. The figure shows the physical and genetic map of plasmid pJFFBF1+E3; the p30 gene is represented by a black arrow; the different ORFs are indicated with arrowheads indicating the direction of translation. The putative transcriptional termination site for p30 is indicated by a hairpin. The restriction sites are indicated by the following abbreviations: B, BamHI; S, Sau3AI; P, PstI; H, HindIII; and E, EcoRI. The interrupted line (//) indicates the junction of noncontiguous segments. The EMBL/GenBank accession no. of the DNA sequence is AF327858.
Analysis of the 2.6-kb insert in plasmid pJFFBF1+E3.
The cloned 2.6-kb insert was sequenced using the primer walking method. In order to verify the integrity of the insert in plasmid pJFFBF1+E3, we performed PCR amplifications using genomic DNA of PG2 as a control, with the oligonucleotide primers BKE3P0, BKE3P1, BKE3P2, BKE3P3, BKE3P4, and BKE3P5 (Table 1 and Fig. 1). No amplicon was found after amplification of genomic DNA with primer pairs BKE3P0-BKE3P1, BKE3P0-BKE3P3, or BKE3P0-BKE3P5. However, PCRs with primer pairs BKE3P2-BKE3P1 and BKE3P4-BKE3P1 amplified fragments of 2,145 and 1,735 bp, respectively. Due to the inability of primer BKE3P0 to produce amplicons from genomic DNA (but only from plasmids pJFFBF1+E and pJFFBF1+E3) we assumed that the insert of 2.6 kb was composed of two noncontiguous fragments (Fig. 1). We therefore consider only the 2,434 bp as a contiguous fragment in this study. This fragment presented a complete open reading frame (ORF), p30, and two incomplete ORFs, ORF 2 and ORF 4.
The ORF p30, at nucleotide (nt) 147 to 947, encodes a 266-amino-acid (aa) polypeptide, the protein P30, with a predicted molecular mass of 30 kDa. It is preceded by a potential −35 region at nt 54 to 59 and two possible −10 signal boxes of prokaryotic transcription promoter (nt 75 to 80 and nt 105 to 110) and is followed by a sequence which can form a hairpin with a ΔG of −14.8 kcal/mol, representing a potential rho-independent transcription termination signal. The ORF of P30 is preceded by a consensus sequence for a ribosome binding site (RBS) at nt 131 to 137. It does not contain any TGATrp codons. The predicted amino acid sequence of P30 contains a consensus sequence for a potential recognition site for signal peptidase II. This sequence is localized to aa 1 to 24. The amino acid sequence of the mature protein P30 shows low homology (about 25% identical and 40% similar amino acids) with some variable surface lipoproteins from Mycoplasma bovis (18).
Presence of the p30 gene in different mycoplasma species.
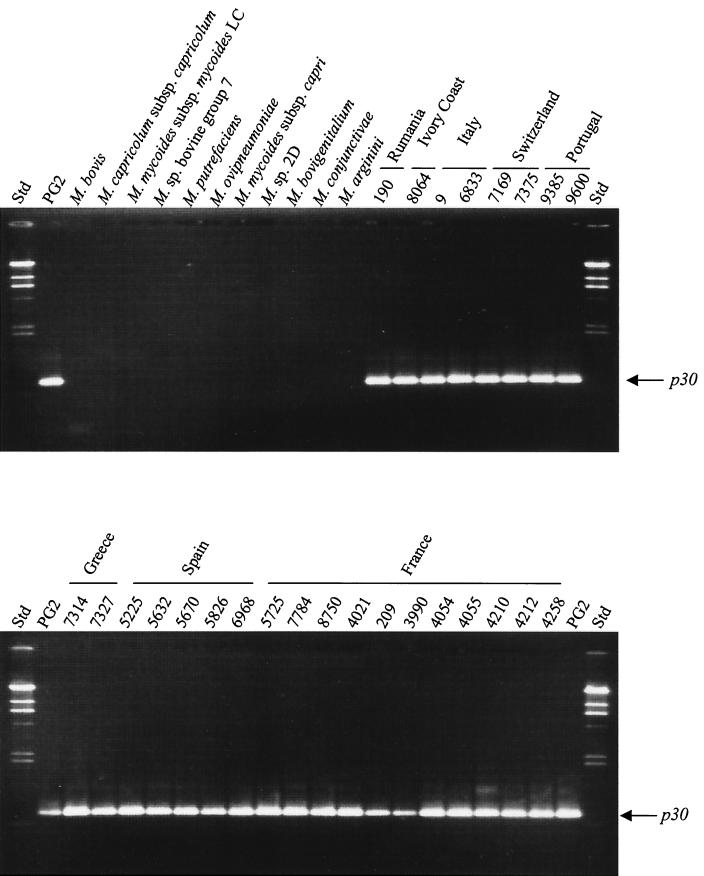
In order to analyze the specificity of the p30 gene for M. agalactiae, we designed a primer pair, MagaORF1-L and MagaORF1-R (Table 2), complementary to the extremities of p30 (Fig. 1). The expected 730-bp fragment was amplified from chromosomal DNA of all M. agalactiae strains used in this study but not from other mycoplasma species tested (Fig. 2 and Table 1). A DIG-labeled probe p30 was used in Southern blot analysis with the genomic DNA from different mycoplasma species cut with HindIII and Sau3AI. M. agalactiae type strain PG2 and all field strains tested showed the two expected bands of 1.1 and 0.5 kb (data not shown). All other mycoplasma species tested showed no hybridization signal (Table 1). These results indicated that the p30 gene is present as a single copy in all strains tested and that it is specific for the species M. agalactiae.
FIG. 2.
Presence of p30 in M. agalactiae strains and other mycoplasma species. Detection of p30 in strains of M. agalactiae and other mycoplasma species by amplification of genomic DNA was performed by PCR with primers MagaORF1-L and MagaORF1-R (Table2). The strains are listed in Table 1. The arrows indicate the p30 amplicons. Std, molecular mass standards (23.1, 9.4, 6.6, 4.4, 2.3, 2.0, and 0.6 kb).
Expression of P30 in different mycoplasma species.
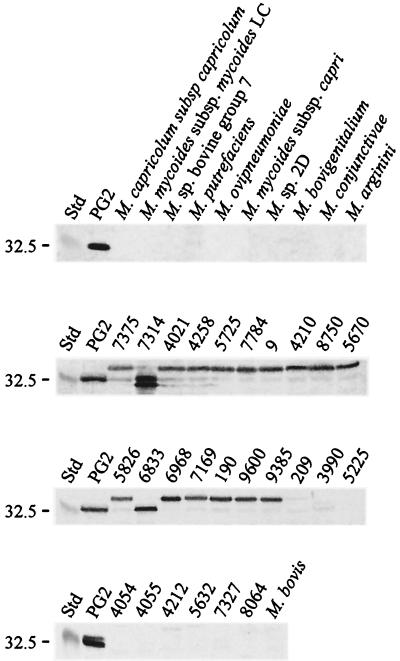
Immunoblots with total antigen of strains of M. agalactiae and other species of mycoplasmas listed in Table 1 were probed with the monospecific rabbit anti-P30 serum directed against purified recombinant polyhistidine-tailed P30. Among the M. agalactiae strains analyzed, different profiles could be observed on immunoblots. The monospecific serum detected the expected 32-kDa protein in type strain PG2 of M. agalactiae and in strain 6833. In most M. agalactiae strains tested (n = 15), a slightly larger protein of 35 kDa reacted with anti-P30. In a single strain, a doublet at 30 to 32 kDa was observed. In nine strains of M. agalactiae the serum did not detect any protein bands on immunoblots (Fig. 3).
FIG. 3.
Distribution of P30 expression in strains of M. agalactiae and of other mycoplasmas. Immunoblot analysis was carried with 10 μg of total antigen per lane. Total antigen of a selection of strains from M. agalactiae and of type and reference strains of other mycoplasmas was separated by SDS–12% PAGE, transferred onto a nitrocellulose membrane, and probed with the antiserum against P30. Std, molecular mass standards (broad-range marker 7708S; New England Biolabs, Inc., Beverly, Mass.).
Colony immunoblotting performed on M. agalactiae strains on solid PPLO medium with anti-P30 serum was positive for all strains that exhibited an immunoreactive protein on immunoblots, as well as with strains 5225 and 7327, which were apparently negative for P30 on immunoblots. These last two strains seem to express only a truncated peptide of P30 (see below) which is too small to be detected on immunoblots. No sectors were observed on colonies of any M. agalactiae strains tested with rabbit anti-P30 antibodies. M. bovis and other closely related mycoplasmas did not reveal a P30 or similar protein, as analyzed with immunoblots using the anti-P30 serum.
Genetic variability of the p30 genes in M. agalactiae
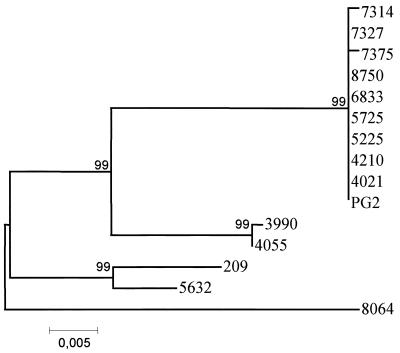
The p30 genes from 15 strains of M. agalactiae (8 strains expressing full-length P30, 2 strains [5225 and 7327] expressing only part of P30, and 5 strains negative for P30 expression) were sequenced with primers MagaORF1-L, MagaORF1-L2, MagaORF1-R, and MagaORF1-R3 (Table 2). Based on the DNA sequences, two distinct groups could be identified. One group (n = 10) comprised of all strains that express full-length P30 (as shown on immunoblots) presented very homogeneous p30 genes. Strains 5225 and 7327, both of which express part of P30, and hence react only on colony immunoblot, also belong to this group. The second group (n = 5) contained the strains that did not express P30 in any form (i.e., negative in both Western and colony immunoblots) and showed a significant polymorphism among the p30 genes (Fig. 4). Major differences which distinguished these two groups were found in the promoter region. Most importantly, these differences included a common transition, T→C, at the end of the first putative −10 box and an A→T transition at the end of the second putative −10 box in all five strains that are fully devoid of P30 expression. These mutations in the putative promoter of P30 seem to be responsible for the lack of expression of P30 in the strains of the second group. Among the strains in the first group expressing all or part of P30, minor differences in the DNA sequences of p30 could explain the phenotypic differences as revealed on immunoblots. The two strains 5225 and 7327 revealed an additional nucleotide in a poly-G tract of the coding sequence, which creates a frameshift leading to a stop codon immediately thereafter, hence resulting in a truncated P30 with only 62 aa. This short peptide could not be detected on our immunoblots but must have reacted on the colony blot system with anti-P30 serum. Strains PG2, 7314, and 6833 showed a slightly lower apparent mass of P30 on immunoblots compared to the majority of field strains (Fig. 3). As deduced from the DNA sequence, these strains revealed a particular difference in the primary structure of P30 approximately 100 aa from the start. In fact, in this region, the residue motif SN was found in the major group of field strains expressing the apparently larger variant of P30 (35 kDa). In contrast, the three strains PG2, 7314, and 6833, which express the smaller P30 variants, possess the residue motif FKY in this region. Furthermore, strain 7314, which showed a 30- to 32-kDa doublet band on immunoblots with anti-P30 serum (Fig. 3), differed from all other strains which expressed P30, showing the residue motif GGNS at aa 60 instead of GET as found in all other P30-producing strains.
FIG. 4.
Phylogenetic grouping of 15 strains of M. agalactiae. Sequence similarity (neighbor-joining) tree based on the comparison of the p30 nucleotide sequences from 15 strains of M. agalactiae. The scale represents the genetic distance in percent.
Interestingly, the p30 genes of the strains that did not express the P30 protein and which have mutations in the putative promoter sequences showed several differences in the silent ORF of P30. These differences are distributed more or less randomly and hence gave a very heterogeneous picture in the phylogenetic grouping seen in Fig. 4.
Immune response of ewes infected with M. agalactiae against recombinant P30.
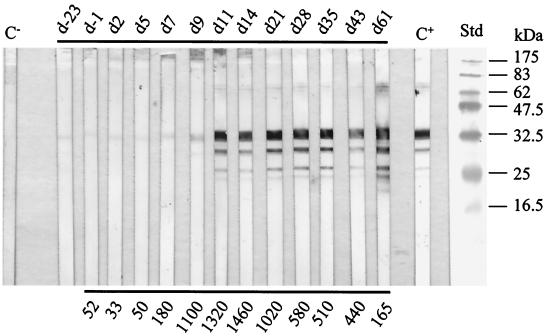
In order to examine the immune response to the recombinant P30 of sera from experimentally infected ewes, immunoblotting was performed with serial serum samples from two ewes infected with M. agalactiae PG2 (ewes 1937 and 1689) and two ewes infected with M. agalactiae strain 5725 (ewes 5B and 10B). The results generated were compared to those of an ELISA which is routinely used for the control of sheep and goats for potential infection by M. agalactiae. The four series of sera all reacted with recombinant P30 at day 11 after inoculation. The reaction with P30 was highest at day 28 after inoculation and persistent until the end of the experiment at day 61 after infection, whereas the titers determined by ELISA had dropped significantly. Figure 5 depicts the immune response against P30 of ewe 1937 infected with strain PG2. The same results were also observed with the sera of the other three infected ewes (not shown).
FIG. 5.
Immune response to P30 of ewe 1937 artificially infected with type strain PG2. SDS–15% PAGE was performed with approximately 1 μg of P30-His per strip. After blotting onto nitrocellulose, serial serum samples were used at a dilution of 1:200. Abbreviations: C−, lamb precolostral serum (negative control); d-23 and d-1, sera collected 23 days and 1 day, respectively, prior to inoculation; dn, serum collected n days after inoculation; C+, serum PAL 97 (anti-M. agalactiae) from a naturally infected ewe, diluted 1:600 (positive control); Std, molecular mass standards. Numbers below the panel indicate the titers in international units as determined by ELISA.
Identification of P30 as membrane protein.
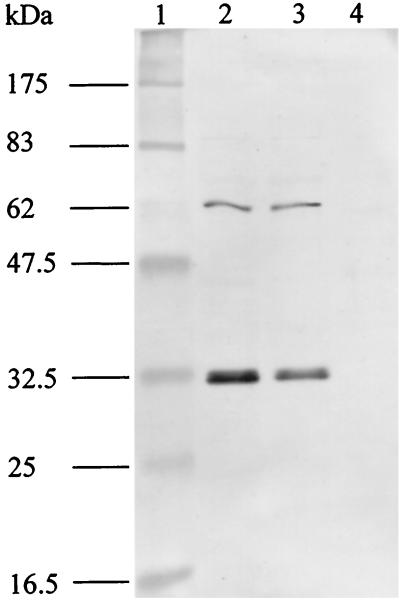
To study the location of P30, total antigen of M. agalactiae type strain PG2 was subjected to a Triton X-114 phase partitioning. This method results in separation of integral hydrophobic membrane proteins (which are incorporated in Triton X-114 micelles) from hydrophilic proteins (which are sequestered in the aqueous phase) (7). Polyclonal monospecific anti-recombinant P30-His serum strongly reacted with a 32-kDa protein and poorly reacted with a 60-kDa protein in both the Triton X-114 phase and total cell proteins, but not with proteins found in the aqueous phase, showing that P30 is an integral membrane protein (Fig. 6).
FIG. 6.
Triton X-114 phase partitioning of M. agalactiae type strain PG2. Mycoplasmas at the end of exponential growth were subjected to Triton X-114 partitioning; 10 μl of fractions and total antigen were separated by SDS–12% PAGE, transferred onto nitrocellulose filter, and probed with monospecific P30 antiserum. We attribute the larger protein band of approximately 60 kDa that is found at low intensity upon reaction with anti-P30 antibodies to formation of a dimeric aggregate of P30, since this band occasionally occurs only with strains that express the protein. Lane1, molecular mass standard (broad-range marker 7708S; 175, 83, 62, 47.5, 32.5, 25, and 16.5 kDa; New England Biolabs); lane 2, total antigen; lane 3, detergent phase; lane 4, aqueous phase.
DISCUSSION
The gene for an immunodominant antigen, P30, was detected from an expression library of genomic DNA of the M. agalactiae PG2 type strain. The gene was subcloned and expressed in E. coli.
Analysis of the DNA sequence of the p30 gene showed that (i) it contains no internal in-frame TGATrp codon and therefore could be entirely expressed in E. coli (32), (ii) it possesses a typical promoter region as reported for most mycoplasmas (8, 25), (iii) it ends with a potential rho-independent termination site of transcription, and (iv) it contains a polypurine-rich nucleotide sequence assigned as a putative RBS. The protein P30 has weak similarities with membrane proteins of M. bovis as found by similarity analysis with the EMBL/GenBank databases. Experiments involving the humoral response to P30 from different ewes artificially infected with PG2 or field strain 5725 indicated that P30 is highly immunogenic and induces an early antibody response which persists for at least 2 months postinfection.
We showed that the p30 gene is specific for the M. agalactiae species by both PCR and Southern blotting. However, immunoblotting performed with 27 strains of M. agalactiae revealed that the protein P30 was not expressed in some of them. In addition, the molecular mass of the protein varied slightly from one strain to another. These minor differences could be correlated to minor variations found in the gene sequences. Such variations, which translate into minor changes in the amino acid sequences of the protein, presumably affect the apparent molecular mass on SDS-polyacrylamide gel electrophoresis (PAGE). The majority of the strains revealed a P30 of approximately 35 kDa on immunoblots reacted with anti-P30. Three strains showed a P30 with an apparently lower molecular mass (32 kDa) and were found to possess a particular motif substitution in the aa sequence. Using the program NNPREDICT (14, 19) to analyze the P30 amino acid sequence of these three strains, we found that the substituted motif is within an α-helix structure, while in all other strains it is found in a region with no predictable structure. This conformational change may affect to some extent the posttranslational processing of the protein, hence leading to a shortened form of P30. Moreover, one of these three strains showed an additional band at approximately 30 kDa on the immunoblot reacted with anti-P30, thus forming a doublet at 30 to 32 kDa. This strain also presented a particular variation of the amino acid sequence in the N-terminal part of P30, which might further affect the conformation of the protein, thus leading to additional processing of the molecule. The appearance of the protein double band could be explained by partial processing of the 32-kDa form. In two other strains, the p30 gene had undergone a mutation leading to a truncated P30 protein of only 62 aa that was, however, still expressed and that reacted with anti-P30 antibodies on colony immunoblotting.
Protein P30 was shown to be membrane located by Triton X-114 partitioning experiments. This location had already been predicted by its amino acid sequence. P30 is not subjected to high-frequency phase variation, as we did not observe sectors when colonies were immunostained with monospecific anti-P30 antiserum. The minor variations in size of P30 detected for certain strains of M. agalactiae are thus not the result of specific mechanisms known for variable antigens (4) but are rather due to punctual mutation events in the coding region of the gene.
Interestingly, some M. agalactiae strains were found which were totally devoid of P30 production but which had retained the entire p30 gene as determined by PCR. Five of these strains were analyzed in detail and found to possess nucleotide substitutions in both possible −10 boxes of the potential transcription promoter of p30. Although very little is known about transcriptional promoters of mycoplasmal genes or operons, DNA sequence analyses very often reveal repeats of −10 boxes as potential promoters in these organisms (13). We therefore interpret the substitutions in both possible −10 boxes of the p30 gene in these strains as the cause of lack of P30 expression, implying that the repeated −10 boxes found upstream of the RBS of p30 play a major role in transcription of this gene.
Comparing the coding sequences of the p30 genes of the M. agalactiae strains analyzed, we detected a significantly higher number of differences among the strains that do not express P30 than in the other strains that express it and which represent a tight cluster (Fig. 4). The high number of mutations in the five strains analyzed which are devoid of P30 expression suggests that this gene has probably been silent for a long time. In fact, silent genes can undergo mutations more frequently than expressed genes since there is no phenotypic selective pressure on silent genes (21, 22). It has to be noted that these five strains were isolated in geographically different regions and in different years.
Using a set of monoclonal antibodies (5), M. agalactiae was subdivided into eight different serotypes, A to H. Among them, serotypes A, B, C, and D, which all have a common epitope reacting with monoclonal antibody 1D4, belonged to the most frequently isolated serotypes of M. agalactiae. The types E, F, G, and H, which showed no reaction with monoclonal antibody 1D4, represented rarely isolated serotypes (5). Our study has shown that all M. agalactiae strains tested which expressed the P30 surface protein belong to the frequently isolated and hence widely distributed serotypes A to D, while the strains which did not express P30 belong to the rarely observed serotypes E to H. However, our experiments indicated that the epitope recognized by the monoclonal antibody 1D4 does not react with the P30 antigen (results not shown). We assume that the rarely observed serotypes E to H might have undergone several mutations which inhibit their ability to express P30 and probably other antigens, such as the yet-unidentified antigen recognized by monoclonal antibody 1D4. These strains appear to be isolated very rarely, representing 6% of a sampling of 246 field strains collected worldwide (5), in contrast to the strains which express P30 and belong to serotypes A to D. Hence, it can be speculated that the loss of P30 expression might have led to a partial loss of the fitness of the strains to cause outbreaks. In the view of the significant antigenic variations observed in M. agalactiae (10, 12, 34), P30 can be considered to be an interesting, stable, and highly specific antigen expressed by the major serotypes of this organism. P30 allows analysis of the serological status of sheep and goats infected by the frequently observed serotypes A to D of M. agalactiae.
In summary, our data indicate that the membrane protein P30 is a stable, immunodominant antigen of M. agalactiae expressed by the main serotypes A to D and that it induces a strong early and persistent immune response in infected animals. P30 does not belong to the cluster of Vpma variable lipoproteins of M. agalactiae, and no particularly similar analogues to P30 have yet been found in related species.
ACKNOWLEDGMENTS
We thank F. Poumarat (AFSSA, Lyon, France), F. Thiaucourt (CIRAD-EMVT, Montpellier, France), and S. Tola (Istituto Zooprofilattico Sperimentale, Sassari, Italy) for the gift of strains. We are also grateful to S. Burr for her editorial help.
We are grateful to Bommeli AG (Liebefeld, Switzerland) for financial support on this project. This study is part of the European COST Action 826 on “ruminant's mycoplasmoses.” It was supported by a Ph.D. fellowship from AFSSA and the Ministry of Agriculture and Fishery, France, by grant C96.0073 of the Swiss Ministry of Education and Science, and by the Swiss Federal Veterinary Office.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. [Google Scholar]
- 3.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1995;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun. 1994;62:5075–5084. doi: 10.1128/iai.62.11.5075-5084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergonier D, DeSimone F, Russo P, Solsona M, Lambert M, Poumarat F. Variable expression and geographic distribution of Mycoplasma agalactiae surface epitopes demonstrated with monoclonal antibodies. FEMS Microbiol Lett. 1996;143:159–165. doi: 10.1111/j.1574-6968.1996.tb08475.x. [DOI] [PubMed] [Google Scholar]
- 6.Bergonier D, Poumarat F. Contagious agalactia of small ruminants: epidemiology, diagnosis and control. Rev Sci Technol. 1996;15:1431–1475. [PubMed] [Google Scholar]
- 7.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 8.Bové J M. Molecular features of mollicutes. Clin Infect Dis. 1993;17(Suppl. 1):S10–S31. doi: 10.1093/clinids/17.supplement_1.s10. [DOI] [PubMed] [Google Scholar]
- 9.Cheng X, Nicolet J, Poumarat F, Regalla J, Thiaucourt F, Frey J. Insertion element IS1296 in Mycoplasma mycoides subsp. mycoides small colony identifies a European clonal line distinct from African and Australian strains. Microbiology. 1995;141:3221–3228. doi: 10.1099/13500872-141-12-3221. [DOI] [PubMed] [Google Scholar]
- 10.Citti C, Rosengarten R. Mycoplasma genetic variation and its implication for pathogenesis. Wien Klin Wochenschr. 1997;109:562–568. [PubMed] [Google Scholar]
- 11.DaMassa A J, Wakenell P S, Brooks D L. Mycoplasmas of goats and sheep. J Vet Diagn Investig. 1992;4:101–113. doi: 10.1177/104063879200400126. [DOI] [PubMed] [Google Scholar]
- 12.Glew M D, Papazisi L, Poumarat F, Bergonier D, Rosengarten R, Citti C. Characterization of a multigene family undergoing high-frequency DNA rearrangements and coding for abundant variable surface proteins in Mycoplasma agalactiae. Infect Immun. 2000;68:4539–4548. doi: 10.1128/iai.68.8.4539-4548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haldimann A, Nicolet J, Frey J. DNA sequence determination and biochemical analysis of the immunogenic protein P36, the lactate dehydrogenase (LDH) of Mycoplasma hyopneumoniae. J Gen Microbiol. 1993;139:317–323. doi: 10.1099/00221287-139-2-317. [DOI] [PubMed] [Google Scholar]
- 14.Kneller D G, Cohen F E, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Tamura K, Nei M. MEGA: Molecular evolutionary genetics analysis, version 1.0. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lambert M, Calamel M, Dufour P, Cabasse E, Vitu C, Pepin V. Detection of false-positive sera in contagious agalactia with a multiantigen ELISA and their elimination with a protein G. conjugate. J Vet Diagn Investig. 1998;10:326–330. doi: 10.1177/104063879801000403. [DOI] [PubMed] [Google Scholar]
- 18.Lysnyansky I, Sachse K, Rosenbusch R, Levisohn S, Yogev D. The vsp locus of Mycoplasma bovis: gene organization and structural features. J Bacteriol. 1999;181:5734–5741. doi: 10.1128/jb.181.18.5734-5741.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClelland J L, Rumelhart D E. Exploration in parallel distributed processing. Cambridge, Mass: MIT Press; 1988. pp. 318–362. [Google Scholar]
- 20.Neyrolles O, Brenner C, Prevost M C, Fontaine T, Montagnier L, Blanchard A. Identification of two glycosylated components of Mycoplasma penetrans: a surface-exposed capsular polysaccharide and a glycolipid fraction. Microbiology. 1998;144:1247–1255. doi: 10.1099/00221287-144-5-1247. [DOI] [PubMed] [Google Scholar]
- 21.Ophir R, Graur D. Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene. 1997;205:191–202. doi: 10.1016/s0378-1119(97)00398-3. [DOI] [PubMed] [Google Scholar]
- 22.Petrov D A, Hartl D L. Pseudogene evolution and natural selection for a compact genome. J Hered. 2000;91:221–227. doi: 10.1093/jhered/91.3.221. [DOI] [PubMed] [Google Scholar]
- 23.Poumarat F, Perrin B, Longchambon D. Identification of ruminant mycoplasmas by dot immunobinding on membrane filtration (MF dot) Vet Microbiol. 1991;29:329–338. doi: 10.1016/0378-1135(91)90140-b. [DOI] [PubMed] [Google Scholar]
- 24.Razin S, Jacobs E. Mycoplasma adhesion. J Gen Microbiol. 1992;138:407–422. doi: 10.1099/00221287-138-3-407. [DOI] [PubMed] [Google Scholar]
- 25.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosati S, Pozzi S, Robino P, Montinaro B, Conti A, Fadda M, Pittau M. P48 major surface antigen of Mycoplasma agalactiae is homologous to a malp product of Mycoplasma fermentans and belongs to a selected family of bacterial lipoproteins. Infect Immun. 1999;67:6213–6216. doi: 10.1128/iai.67.11.6213-6216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosati S, Robino P, Fadda M, Pozzi S, Mannelli A, Pittau M. Expression and antigenic characterization of recombinant Mycoplasma agalactiae P48 major surface protein. Vet Microbiol. 2000;71:201–210. doi: 10.1016/s0378-1135(99)00164-9. [DOI] [PubMed] [Google Scholar]
- 28.Rosengarten R, Wise K S. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science. 1990;247:315–318. doi: 10.1126/science.1688663. [DOI] [PubMed] [Google Scholar]
- 29.Rosengarten R, Yogev D. Variant colony surface antigenic phenotypes within mycoplasma strain populations: implications for species identification and strain standardization. J Clin Microbiol. 1996;34:149–158. doi: 10.1128/jcm.34.1.149-158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaller A, Kuhn R, Kuhnert P, Nicolet J, Anderson T J, MacInnes J I, Segers R P A M, Frey J. Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology. 1999;145:2105–2116. doi: 10.1099/13500872-145-8-2105. [DOI] [PubMed] [Google Scholar]
- 31.Subramaniam S, Bergonier D, Poumarat F, Capaul S, Schlatter Y, Nicolet J, Frey J. Species identification of Mycoplasma bovis and Mycoplasma agalactiae based on the uvrC genes by PCR. Mol Cell Probes. 1998;12:161–169. doi: 10.1006/mcpr.1998.0160. [DOI] [PubMed] [Google Scholar]
- 32.Yamao F, Muto A, Kawauchi Y, Iwami M, Iwagami S, Azumi Y, Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci USA. 1985;82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yogev D, Menaker D, Strutzberg K, Levisohn S, Kirchhoff H, Hinz K H, Rosengarten R. A surface epitope undergoing high-frequency phase variation is shared by Mycoplasma gallisepticum and Mycoplasma bovis. Infect Immun. 1994;62:4962–4968. doi: 10.1128/iai.62.11.4962-4968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yogev D, Rosengarten R, Watson-McKown R, Wise K S. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 1991;10:4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yogev D, Rosengarten R, Wise K S. Variation and genetic control of surface antigen expression in Mycoplasmas—the Vlp system of Mycoplasma hyorhinis. Zentbl Bakteriol-Int J Med Microbiol. 1993;278:275–286. doi: 10.1016/s0934-8840(11)80844-3. [DOI] [PubMed] [Google Scholar]