Abstract
In this study we explored the molecular mechanism of RdRp (Non-Structural Protein, NSP12) interaction with its co-factors NSP7 and NSP8 which is the main toolbox for RNA replication and transcription of SARS-CoV-2 and SARS-CoV. The replication complex is a heterotetramer consists of one NSP12, one NSP7 and two NSP8. Extensive molecular dynamics (MD) simulations were applied on both the heterotetramer complexes to generate the conformations and were used to estimate the MMPBSA binding free energy (BFE) and per-residue energy decomposition of NSP12-NSP8 and NSP12-NSP7 and NSP7-NSP8 complexes. The BFE of SARS-CoV-2 heterotetramer complex with its corresponding partner protein was significantly higher as compared to SARS-CoV. Interface hotspot residues were predicted using different methods implemented in KFC (Knowledge-based FADA and Contracts), HotRegion and Robetta web servers. Per-residue energy decomposition analysis showed that the predicted interface hotspot residues contribute more energy towards the formation of complexes and most of the predicted hotspot residues are clustered together. However, there is a slight difference in the residue-wise energy contribution in the interface NSPs on heterotetramer viral replication complex of both coronaviruses. While the overall replication complex of SARS-CoV-2 was found to be slightly flexible as compared to SARS-CoV. This difference in terms of structural flexibility/stability and energetic characteristics of interface residues including hotspots at PPI interface in the viral replication complexes may be the reason of higher rate of RNA replication of SARS-CoV-2 as compared to SARS-CoV. Overall, the interaction profile at PPI interface such as, interface area, hotspot residues, nature of bonds and energies between NSPs, may provide valuable insights in designing of small molecules or peptide/peptidomimetic ligands which can fit into the PPI interface to disrupt the interaction.
Keywords: Protein-protein interaction, Hotspot residues, Interacting interface, Binding energy, Per-residue energy decomposition, MD simulation
1. Introduction
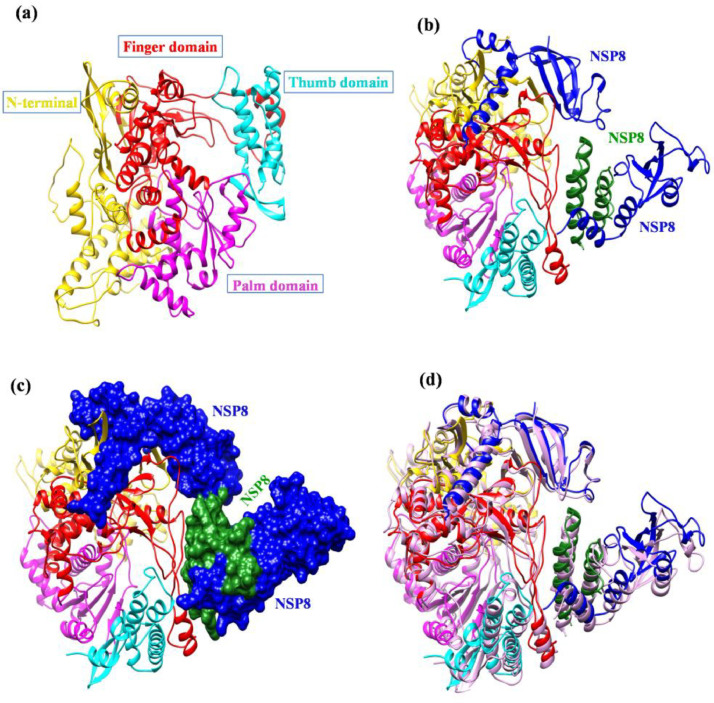
Coronaviruses are positive-strand RNA viruses which belong to the family of Coronaviridae. Beta-subtype coronaviruses have become a big threat to public health, in the past two decades it caused three major outbreaks: the Severe Acute Respiratory Syndrome-associated Coronavirus (SARS-CoV) in 2003, the Middle East Respiratory Syndrome-associated Coronavirus (MERS-CoV) in 2012, and presently, the Severe Acute Respiratory Syndrome associated Coronavirus 2 (SARS-CoV-2) [1,2]. Among these three CoV, SARS-CoV-2 is responsible for current pandemic of coronavirus disease 2019 (COVID-19), with more than 254 million confirmed infected cases and over five million deaths globally, leading to social, societal, and economic. Though there is a progress in vaccine development, there is no highly effective therapeutic agent against SARS-CoV, MERS-CoV or SARS-CoV-2. To combat the current and future coronavirus outbreaks, novel therapeutics are desperately needed. Coronaviruses RNA genome encodes for some specific viral components such as RNA dependent RNA polymerase (RdRp, NSP12), replicase, spike, envelop and nucleocapside proteins among these, NSP12 is the central component of replication and transcription machinery of coronaviruses. The replication complex consists of one NSP12, one NSP7 and two NSP8 (Fig. 1 ). The NSP7 and NSP8 act as cofactors which drive the functional activity of NSP12, by increasing the binding of NSP12 to the template-primer RNA [3]. Thus, there is a need to obstruct this NSPs interaction to inhibit the replication to have control over the SARS-CoV-2 infection.
Fig. 1.
Cartoon representation of SARS-CoV-2NSP12(red)in complex withthecofactorsNSP8 and NSP7(PDB ID: 6M71). (a) Heterotetramer complex, consists of one NSP12 chain interacting with one NSP7 (blue) and there are two NSP8 (green) one is interacting with NSP12 and another is interacting with NSP7 (b) NSP12-NSP7 complex (c) NSP12-NSP8 complex.
The characterization of the protein-protein interaction (PPI) sites is an essential step towards identifying drug targets in order to design potential drugs to obstruct the protein-protein interactions that forms protein complexes [4], [5], [6], [7], [8], [9]. Understanding of PPI is critical to decipher the molecular contacts which are important for recognition and the physical basis of affinity [10]. The PPI interfaces are generally less conserved than active sites, PPI inhibitors are also commonly considered to have a greater opportunity for being selective to obstruct PPI [11,12]. Amino acid residues present at PPI, interact with each other, where some of these residues contribute highly to stabilizing energy of the protein-protein complex, provide specificity at their binding sites [13] and thus these residues are termed as hotspots. Identifying these hotspot residues within the protein-protein interfaces can help us in better understanding of protein-protein interactions and may also help researchers to modulate protein-protein binding [14]. The hotspots are those upon alanine mutation results in significant increase in binding free energy (ΔΔG) of 1.5 kcal/mol, other studies have considered the ΔΔG of at least 2.0 kcal mol−1 [15], [16], [17], [18]. While null-spots exist in the surrounding regions of the hotspots and protect them from solvent exposure [19]. Hotspot residues exist in clusters and are well conserved and more buried in comparison to other interface residues in the protein-protein complex. Amino acids like Tyr, Arg-and Trp-amino acids have a greater tendency in being a hotspot, while Leu, Thr, Ser, and Val-are less likely to act as a hotspot [20], [21], [22]. Identification of hotspots are helpful in studying protein dimer and also aid in the identification of probable binding sites for other binding partners [23]. Therefore, identifying the hotspot residues within the interfaces of NSPs of replication complex of SARS-CoV-2 can be helpful in better understanding the PPI and may be helpful to modulate interacting interface area.
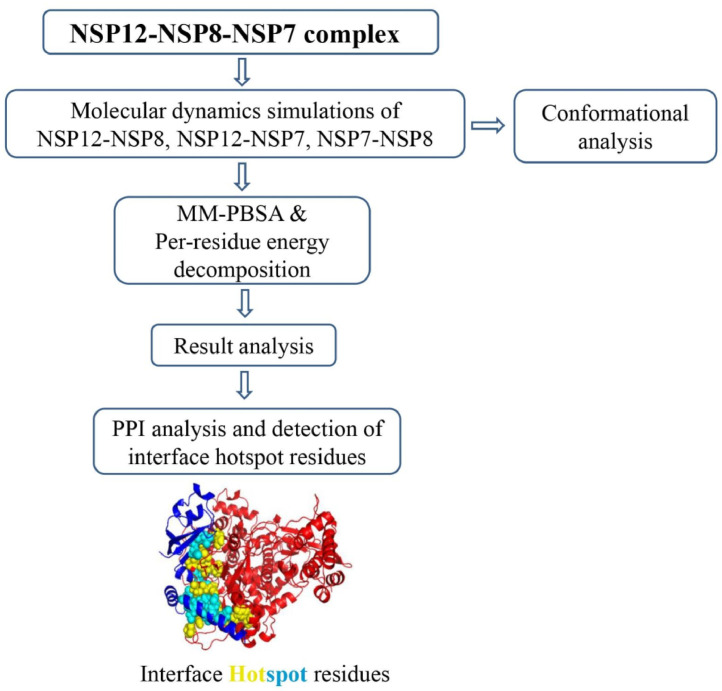
Experimental methods such as Alanine scanning mutagenesis (ASM) have been used extensively to identify hotspot residues at protein-protein interfaces where residues are systematically replaced with alanine, to measure the binding free energy difference [24]. However, this method is time-consuming and expensive. Thereby we have used computational methods which are freely available web-based services including KFC [25], HotRegion [26], and Robetta server [27,28] to determine the probable hotspots at the NSPs interface. Predicting the hotspot residues using a single method might give inaccurate results, so these three servers along with the per-residue energy contribution were used to improve the accuracy of the predicted hotspot result. The sheer amount of work has been done towards the identification of hotspot residues at PPI interface through computational methods [29], [30], [31], [32], [33]. In the current study, the viral replication complexes were subjected to Molecular Dynamics (MD) simulation. MD simulation is the feasible tool to obtain the dynamic in formation in protein-protein interfaces as protein-protein interactions are dynamic in nature and adopt different conformations. MD simulation allows the transient pockets and buried hotspot residues to emerge on the protein surfaces and these transient areas and hotspots could be targeted with small molecules [34], [35], [36], [37], [38]. The average structure extracted from last 10 ns MD trajectory MD were used to calculate the total binding free energy contribution using Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA) method and followed by per-residue energy decomposition [39]. The same conformer was used to study PPI interaction profile using PDBsum server [40].The overall workflow of the study demonstrated in Fig. 2 .
Fig. 2.
Schematic representation of the work flow of the study.
Our findings provide significant insights into the interface area, bonded and non-bonded interactions, interface residues and potential hotspot residues across PPIinvolved in the heterotetramer replication complex of SARS-CoV-2 and SARS-CoV.This information may be helpful in targeting the interacting interface area of SARS-CoV-2 replication complexes in order to control the viral replication and transcription. And also, the current study highlights the potential unknown location of hotspot residues, which could help researchers in performing ASM experimentally. ASM is time-consuming and expensive process; hence, by identifying the possible locations of hot spots with the help of in silico methods, researchers will be able to perform alanine mutations only onthose amino acid locations identified by in silico methods.
2. Materials and methods
2.1. Preparation of complex structures
The three-dimensional (3-D) viral replication complex (NSP12-NSP8-NSP7) of SARS-CoV-2 and SARS-CoV was downloaded from RCSB Protein Data Bank (PDB ID: 6M71 and 6NUR, respectively) [41,42]. The replication complex is a heterotetramer, consists of one NSP12 polypeptide chain (Chain A), one NSP7 (Chain C) and two NSP8 chains (Chain B and D) [41,42]. Each heterotetramer complexes were imported in UCSF Chimera [43] and solvent molecules (crystal water), salt, ions and other heteroatoms were removed. The arrangement of heterotetramer complex is demonstrated in Fig. 1.
2.2. Molecular dynamics simulations and binding free energy
The heterotetramer complex NSP12-NSP7-NSP8(2)of SARS-CoV-2 and SARS-CoVwere subjected to MD simulations withGromacs 5.0.4 package [44] using CHARM force field and SPC water model [45]. MD simulations were performed under periodic boundary conditions (PBCs) with a cubic box maintaining a distance of 2.0 nm between the PPI complex and the boundary edges. The complexes were solvated with SPC water molecules and the system was neutralized by adding the counter ions into the solvated box depending on the charge of the system. Then, the systems were subjected to 1000 steepest descent minimization followed by 2000 conjugate-gradient minimization for initial energy minimization to avoid a further structural clash in the solvated system. Further, the whole system was subjected to 5000 steepest descent minimizations followed by 6000 conjugate-gradient minimizations with a maximum step size of 0.01fs for final energy minimization. The minimized systems were then subjected to position restraint equilibrations. Then, the systems were heated under canonical ensemble from 0 to 303 K for 500 ps using a modified Berendsen thermostat [46,47]. Later, the systems were equilibrated for 1 ns under isothermal-isobaric conditions (with a constant pressure of 1.0 bar). Finally, production run of 100 ns was done with no restraints followed by an integration time step of 0.2 ps. The coordinates were saved every 2 ps under constant conditions of 300 K temperature and 1 atm pressure. The LINCS algorithm [48] was used to restrain the bond lengths and the long-range electrostatics were calculated using the particle mesh Ewald (PME) [49], while the SETTLE algorithm [50] was employed to constrain the geometry of water molecules. Molecular mechanics-Poisson-Boltzmann surface area (MMPBSA) method using g_mmpbsa package, was employed to calculate the binding free energies of protein-drug complexes using the last 10 ns MD trajectory.
The total binding free energy from MM-PBSA calculation incorporates explicit solvation model with the calculations of electrostatic contribution to the solvation and the non-polar contribution to estimate the binding free energy ∆G binding.
The binding free energy of the complex could be calculated using Eq. (1)
| (1) |
The MM-PBSA approach incorporates the following equations:
| (2) |
| (3) |
| (4) |
Where ΔEMM is the molecular mechanics energy of the system in a vacuum, ΔGPBSA is the solvation free energy, TΔS is the entropy. EMM can be split into internal energy (Eint), van der Waals forces (EvdW) and electrostatic energy (ΔEele) and ΔGPBSA is the sum total of polar solvation free energy of generalized born model (GPB), and the non-polar/surface solvation free energy (Gsurf). The entropy calculation was neglected in the above calculation as the study mainly focusedon calculating only relative binding energy contribution of each amino acid to the formation of protein complex. The last 10 ns trajectory of the original 100 ns trajectory (i.e. 90 ns to 100 ns) was used for the MMPBSA calculation. MmPbSaStat.py program was used to calculate the binding energies and MmPbSaDecomp.py was used to extract the residue-specific contributions towards protein-protein binding [51].
2.3. Protein-protein interaction and Hotspot residue identification
The PPI profiles for the heterotetramer complexes were analyzed using PDBsum server. PDBsum is a web server provides a largely pictographic summary of the important information on macromolecular structure. It includes images of the structure, annotated plots of each protein chain's secondary structure, detailed structural analyses, summary PROCHECK results and schematic diagrams of protein-protein, protein-ligand and protein-DNA interaction. The Hotspot residues have been identified using different computational methods which are freely available online services viz., KFC2 [25], Hotregion [26] and Robetta server [27,28]. KFC2 server is a machine learning based tool that utilizes in silico alanine scanning mutagenesis, considering hydrogen bonds, atomic contacts and residue sizes for hotspot identification [25]. Hotregion database is a structure-based hotspot prediction method which predicts hotspot residues using algorithms based on structural neighborhoods (Euclidian and Voronoi), and then selects optimal features using random forest and sequential backward elimination algorithms [26]. To calculate the interaction free energy, Robetta server includes different parameters such as implicit solvation and hydrogen bonding, packing interactions, solvation interactions, and Lennard-Jones interactions. The Robetta server can accurately predict 79% of hotspot residues with a cutoff value of 1.0 kcal/mol [27,28].
3. Results and discussion
3.1. Molecular dynamics simulation and MMPBSA analysis
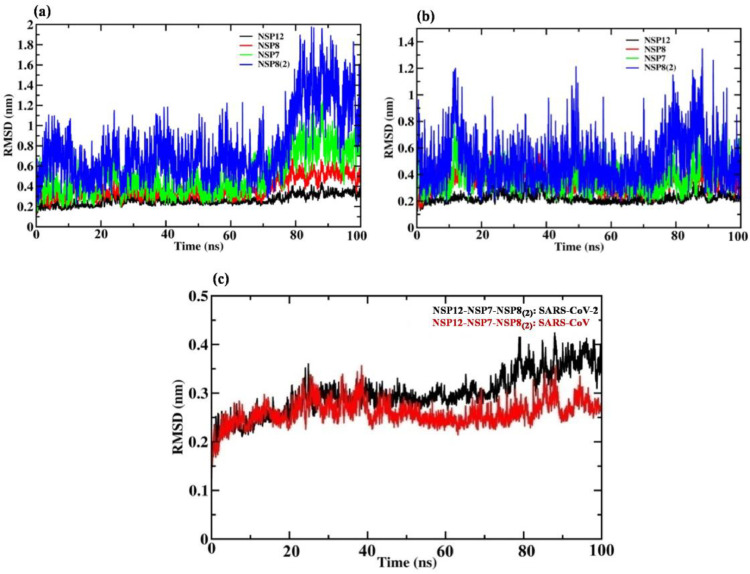
To confirm the structural stability of the selected PPI complexes, MD simulations were carried out for a period of 100 ns. The MD trajectories were used to assess their dynamic behavior including stability, flexibility, and binding affinity by measuring RMSD, RMSF and energy profiles. The SARS-CoV-2, NSP12 attain stability with RMSD value of 0.2 nm till 70 ns, after that from 70 to 100 ns there is slight rise in the RMSD value near 2.5 nm, shown in Fig. 3 (a). While NSP12 of SARS-CoV retains the smoothness in the graph throughout the 100 ns with the RMSD value of 0.2 nm, shown in Fig. 3(b). SARS-CoV-2 NSP7, the RMSD graph observed to be stable till 70 ns then there is a slightly rise in the graph and attain the stability around 0.8 nm RMSD value, shown in Fig. 3(a). Whereas, SARS-CoV NSP7 maintains the smoothness in the graph with the RMSD value of 0.4 nm throughout the 100 ns MD simulation shown in Fig. 3(b). SARS-CoV-2 and SARS-CoV NSP8, the RMSD value fluctuate in between 0.2 to 0.4 nm, attain the stability at the end of the 100 ns MD simulation, shown in Fig. 3(a) and (b). The other NSP8 of SARS-CoV-2 which interacts with NSP7, shows more fluctuation in the RMSD value, ranges between 0.6 to 1.4 nm, while the SARS-CoV NSP8 shows less fluctuation and retain the smoothness in the graph with the RMSD value of in between 0.4 to 0.5 nm, shown in Fig. 3(a) and (b). Overall, the heterotetramer replication complex of SARS-CoV-2 is slightly flexible as compared to SARS-CoV as shown in Fig. 3(c).
Fig. 3.
Protein backbone root mean square deviation (RMSD) graphs of NSPs involved in the formation of heterotetramer replication complex of (a) SARS-CoV-2 and (b) SARS-CoV along the 100 ns MD simulation. (c) RMSD of overall heterotetramer along the 100 ns MD simulation, black and red color represented the SARS-CoV-2 and SARS-CoV respectively.
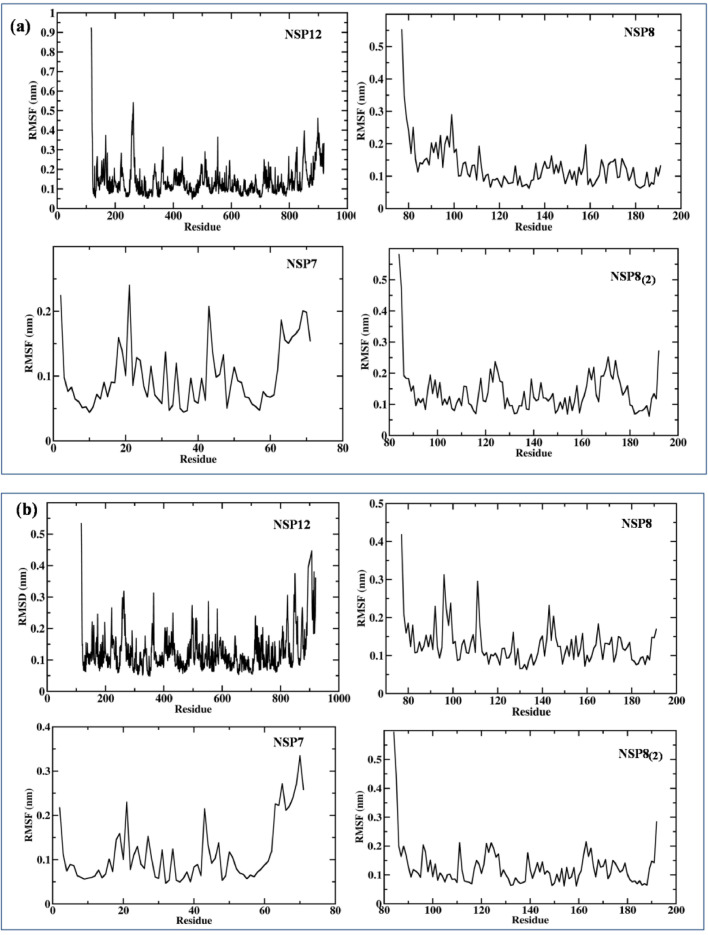
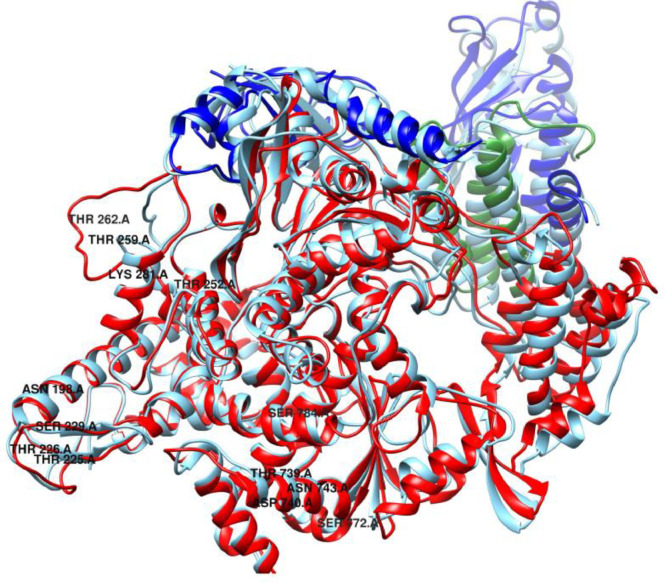
From the RMSF result obtained, it was observed that more fluctuation was present at N-terminal region (117–397) of SARS-CoV-2 NSP12 when it is in complex with its cofactor NSP8 and NSP7 as compared to SARS-CoV, shown in Fig. 4 . The N-terminal region consists of two sub-domains, NiRAN (117–250) and interface domain (251–398). The interface domain act as a protein interaction junction, interacting with NiRAN domain, RdRp domain and the second subunit of NSP8 [42]. The fluctuation may be due to mutations occurred at both the N terminal domain of SARS-CoV-2 viz 198Ala (CoV-Asp), Thr225 (CoV-Val), Thr226 (CoV-Ala), Ser229 (CoV-Cys) and the mutations at interface domain are Thr252 (CoV-Ala), Thr259 (CoV-Ala), Thr262 (CoV-Ala), Lys281(CoV-Cys281). The mutations are depicted in Fig. S1. The fluctuation in the interface domain may enhance the binding of NSP8 which may influence the polymerase activity. We could not see any significant difference between the fluctuations in the cofactors (NSP8 and NSP7) of SARS-CoV-2 and SARS-CoV.
Fig. 4.
Root Mean Square Fluctuation (RMSF) graph of NSPs of heterotetramer viral replication complex of (a) SARS-CoV-2 and (b) SARS-CoV during the 100 ns MD simulation.
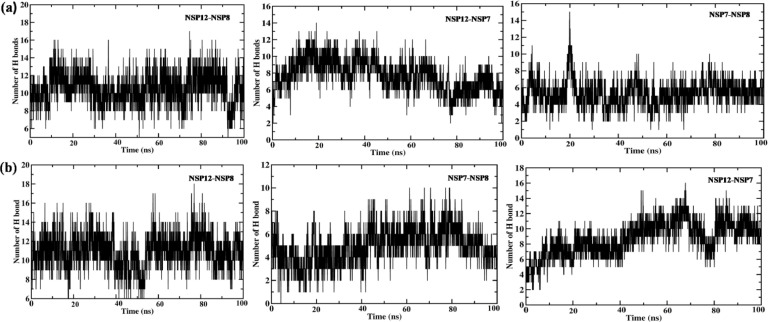
However, we can say that the mutations at N-terminal domain of SARS-CoV-2 NSP12 and binding of cofactors significantly influences the stability and flexibility of the NSP12 of SARS-CoV-2. In addition, the formation of H bond bonds across NSP12-NSP8, NSP12-NSP7 and NSP7-NSP8 throughout the simulation is depicted in Fig. 5 To see the conformational difference in the structure of heterotetramer replication complex of SARS-CoV-2 and SARS-CoV, we superimposed the last 10 ns average structures using UCSF Chimera, we can clearly see the difference in the structure with the RMSD value of 2.5 Ả. The difference in the conformation of heterotetramer along with mutation in SARS-CoV-2 NSP12 depicted in Fig. 6 .
Fig. 5.
Hydrogen bonds observed at PPI interface of NSP12 & NSP8, NSP12 &NSP7 and NSP7-NSP8 of (a) SARS-CoV-2 and (b) SARS-CoV along the 100 ns MD simulation.
Fig. 6.
Superimposed cartoon representation of SARS-CoV-2 (red) NSP12 with SARS-CoV NSP12. The highlighted residues (in black) are the mutated residues of NSP12 of SARS-CoV-2. Both the superimposed structures are the average conformer extracted from last 10 ns MD trajectory.
The BFE calculations of NSP12 with its co-factors (NSP8 and NSP7) and NSP7-NSP8 heterodimer complex were done using MM-PBSA. The MM-PBSA results are summarized in Table 1 . In comparison, the BFE for the SARS-CoV-2, NSP12-NSP8 (−574.82 kcal/mol), NSP12-NSP7 (−205.07 kcal/mol) and NSP7-NSP8 (−294.81 kcal/mol) is much higher than that for SARS-CoV (−433.49, −143.31, −223.95 kcal/mol) respectively. The higher contribution of non-polar interaction energy, i.e. van der Waal's energy (ΔEvdW) + non-polar solvation energy (SASA) into the ΔGbind suggested that hydrophobic interaction plays a crucial role towards the formation of protein-protein complexes. The calculated values of ΔGbind components signify, van der Waal's energy (ΔEvdW) and electrostatic energy (ΔEelec) as driving force of the protein-protein interactions.
Table 1.
MM-PBSA analysis of NSP12-NSP8, NSP12-NSP7 and NSP7-NSP8 complexes. The energy terms (in kcal/mol) were calculated from the data obtained from last 10 ns trajectory.
| Human CoV | Complexes | van der Waal energy | Electrostatic energy | Polar Solvation energy | SAS Aenergy | Binding energy |
|---|---|---|---|---|---|---|
| SARS-CoV-2 | NSP12-NSP8 | −1061.23 | −573.42 | 1179.21 | −119.38 | −574.82 |
| SARS-CoV | −1040.93 | −637.82 | 1360.69 | −115.43 | −433.49 | |
| SARS-CoV-2 | NSP12-NSP7 | −316.53 | −395.20 | 544.54 | −37.87 | −205.07 |
| SARS-CoV | −266.606 | −795.26 | 956.63 | −38.07 | −143.31 | |
| SARS-CoV-2 | NSP7-NSP8 | −514.66 | −247.94 | 526.29 | −58.49 | −294.81 |
| SARS-CoV | −482.52 | −260.75 | 576.68 | −57.35 | −223.95 |
3.2. Protein-protein interaction profile
The interface statistics of the initial structure (before MD) of heterotetrameric NSP12-NSP7-NSP8(2) complex of SARS-CoV-2 and SARS-CoV was analysed using PDBSum and compared with average complex structure extracted from last 10 ns (90–100 ns) trajectory. The interaction profile is summarized in Table 2 . In the initial SARS-CoV-2 NSP12-NSP8 and NSP12-NSP7 complex, the total number of interface residues observe to be 49 (in NSP12) and 46 (in NSP8) and across NSP12-NSP7 interface, 13 (in NSP12) and 15 (NSP7)however at the end of the simulation, the average structure extracted from last 10 ns MD trajectory shows less interacting residue that is 28 for NSP12 and NSP8 and at NSP12-NSP7 interface is 13 (in NSP12) and 15 (in NSP7). The interface statistics contains number of interacting residues, interface area, H-bond, salt bridges, non-bonded contacts are summarized in Table 2. Although the SARS-CoV-2 complexes have shown high binding free energy in all the three complexes, the salt bridges were lost at the end of the simulation in the NSP12-NSP8 and NSP12-NSP7 complex (Table 2), however in case of SARS-CoV salt bridges are retained at NSP12-NSP8 and NSP12-NSP7 interface and there are more no. of H-bonds as compared SARS-CoV-2. But NSP8-NSP7 of SARS-CoV2 has more no. of interactions (H-bonds, salt-bridges, non-bonded contacts)
Table 2.
Comparison of the interface statistics of the initial structure (before MD) of NSP12-NSP8, NSP12-NSP8 and NSP7-NSP8 complexes of SARS-CoV-2 and SARS-CoV with the average complex structure extracted from last 10 ns trajectory. For each complexe, interface statistics obtained from PDBsum server.
| Human CoV | Time | PPI complexes | No. of interface residues | Interface area (Ӑ2) |
No. of salt bridges | No. of H-bonds |
No. of non- bonded contacts |
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | Initial structure | NSP12 (A) | 49 | 2412 | 3 | 13 | 228 |
| NSP8 (B) | 46 | 2524 | |||||
| Average Str. from last 10 ns |
NSP12 (A) | 28 | 2386 | – | 7 | 83 | |
| NSP8 (B) | 28 | 2453 | |||||
| SARS-CoV | Initial structure | NSP12 (A) | 48 | 2388 | 3 | 14 | 210 |
| NSP8 (B) | 44 | 2499 | |||||
| Average Str. from last 10 ns |
NSP12 (A) | 36 | 2294 | 2 | 13 | 113 | |
| NSP8 (B) | 30 | 2397 | |||||
| SARS-CoV-2 | Initial structure | NSP12 (A) | 13 | 696 | – | 4 | 59 |
| NSP7 (C) | 15 | 729 | |||||
| Average Str. from last 10 ns |
NSP12 (A) | 8 | 712 | – | 4 | 30 | |
| NSP7 (C) | 11 | 717 | |||||
| SARS-CoV | Initial structure | NSP12 (A) | 14 | 683 | – | 4 | 57 |
| NSP7 (C) | 14 | 710 | |||||
| Average Str. from last 10 ns |
NSP12 (A) | 13 | 748 | 1 | 9 | 73 | |
| NSP7 (C) | 15 | 773 | |||||
| SARS-CoV-2 | Initial structure | NSP7 (C) | 27 | 1296 | 1 | 7 | 126 |
| NSP8 (D) | 24 | 1314 | |||||
| Average Str. from last 10 ns |
NSP7 (C) | 18 | 1185 | – | 3 | 51 | |
| NSP8 (D) | 17 | 1175 | |||||
| SARS-CoV | Initial structure | NSP7 (C) | 27 | 1288 | 1 | 7 | 117 |
| NSP8 (D) | 23 | 1298 | |||||
| Average Str. from last 10 ns |
NSP7 (C) | 12 | 1151 | – | 2 | 44 | |
| NSP8 (D) | 16 | 1142 |
However, the theory of importance of salt bridges is only partially true at least in the perspective of protein-protein interfaces [52], after the introduction of continuum electrostatic models which numerically solve the Poisson-Boltzmann equation for the system of protein-solvent [53]. Though there are more number interactions in case of SARS-CoV NSP12-NSP8, NSP12-NSP7, the per residue energy contribution is slightly more in many of the interface residues of SARS-CoV-2 NSP12-NSP7 and NSP12-NSP8 as compared to SARS-CoV (Tables 3 , 4 , 6 and 7). According to the experimental study, binding of NSP7-NSP8 heterodimer to the index finger loop of NSP12 is responsible for the stabilization of NSP12 region which is involved in RNA binding and second NSP8 subunit plays a crucial role in polymerase activity. This indicates the importance of NSP7-NSP8 heterodimer interacting with NSP12 through NSP7 interface and this is essential for efficient RdRp activity in replication process [54], [55], [56], [57]. In agreement with the experimental evidence, our MM-PBSA result justify that the strong high binding affinity between all NSPs in NSP12 heterotetramer complex of SARS-CoV-2 may be the reason of high rate of replication in SARS-CoV-2 as compared to SARS-CoV.
Table 3.
List of interacting residues at protein-protein interacting interface of SARS-CoV-2 NSP12 (Chain A) and NSP8 (Chain B), interface hotspot residues are predicted using three computational methods implemented in KFC server, HotRegion database and Robetta web server. The predicted hotspot residues contribute reasonably good binding energy which may help in the complexation process. The per-residue energy decomposition analysis was carried out using the last 10 ns MD trajectory.
| Residues | KFC | HotRegion | Robetta ΔΔG (kcal/mol) | Per-residue energy contribution (kcal/mol) |
|---|---|---|---|---|
| LEU-271A | HS | – | 2.16 | −14.06 |
| LYS-272A | – | – | – | −15.99 |
| TYR-273A | HS | HS | 0.75 | −4.67 |
| PRO-323A | – | – | – | −1.01 |
| THR-324A | – | – | 0.52 | 1.72 |
| SER-325A | – | – | – | −1.56 |
| PHE-326A | – | – | – | 3.48 |
| GLY-327A | – | – | – | 0.71 |
| PRO-328A | HS | – | – | −2.39 |
| LEU-329A | HS | HS | 0.60 | −9.92 |
| VAL-330A | HS | HS | 0.98 | −6.56 |
| ARG-331A | – | – | – | −28.16 |
| LYS-332A | – | – | – | −15.04 |
| VAL-335A | – | HS | – | −1.29 |
| ASP-336A | – | – | – | −15.88 |
| VAL-338A | – | – | – | −6.05 |
| PRO-339A | – | – | – | −1.58 |
| PHE-340A | HS | HS | 1.60 | −8.65 |
| VAL-341A | HS | HS | 0.72 | −7.46 |
| SER-343A | – | – | – | −0.13 |
| THR-344A | – | – | – | 0.63 |
| HIS-355A | – | – | – | −1.29 |
| LEU-366A | – | – | – | −5.89 |
| PHE-368A | – | – | 1.50 | −7.42 |
| LEU-371A | HS | HS | 1.81 | −9.57 |
| LEU-372A | – | – | – | −1.91 |
| TYR-374A | – | – | – | −1.12 |
| ALA-375A | – | – | – | −4.15 |
| PRO-378A | HS | – | – | −2.82 |
| ALA-379A | HS | HS | – | −1.64 |
| MET-380A | HS | HS | 1.33 | −11.68 |
| HIS-381A | – | – | – | 1.2 |
| ALA-382A | HS | HS | – | −0.38 |
| ALA-383A | HS | HS | – | −2.39 |
| SER-384A | – | – | – | 1.13 |
| GLY-385A | – | – | – | 0.69 |
| ASN-386A | – | – | – | −4.11 |
| LEU-387A | HS | HS | 2.81 | −19.69 |
| LEU-388A | HS | HS | 0.93 | −9.96 |
| LEU-389A | HS | HS | 1.59 | −13.88 |
| ASP-390A | – | – | – | 48.78 |
| LYS-391A | – | – | – | −60.14 |
| ARG-392A | – | HS | – | −54.46 |
| PHE-396A | – | – | – | −6.79 |
| SER-397A | – | – | – | 1.31 |
| VAL-398A | HS | HS | 0.84 | −2.42 |
| ALA-399A | HS | – | – | −3.18 |
| ALA-400A | – | – | – | −3.36 |
| LEU-401A | – | – | – | −2.86 |
| ASN-403A | – | – | – | 3.77 |
| ASN-404A | – | – | – | −2.07 |
| VAL-405A | HS | HS | 1.56 | −11.44 |
| PRO-505A | – | – | – | −2.81 |
| PHE-506A | – | – | – | −5.69 |
| TRP-509 | – | – | 2.29 | |
| LEU-514A | – | – | – | −5.26 |
| TYR-515A | HS | HS | – | −4.35 |
| ASP-517A | – | – | – | −39.5 |
| MET-666A | – | HS | – | −3.00 |
| ARG-80B | – | – | −84.85 | |
| VAL-83B | HS | HS | 1.14 | −13.05 |
| THR-84B | – | – | – | −2.6 |
| ALA-86B | – | – | – | −4.49 |
| MET-87B | HS | HS | 1.85 | −23.47 |
| MET-90B | HS | HS | 1.03 | −18.15 |
| LEU-91B | HS | HS | 2.32 | −15.16 |
| PHE-92B | – | HS | 1.09 | −9.56 |
| MET-94B | – | – | – | −10.42 |
| LEU-95B | HS | HS | 1.91 | −13.45 |
| ASN-108B | – | – | – | 1.23 |
| ASP-112B | – | – | – | 62.09 |
| GLY-113B | – | – | – | 0.60 |
| CYS-114B | HS | HS | −0.12 | −11.26 |
| VAL-115B | HS | HS | 0.55 | −6.58 |
| PRO-116B | HS | – | – | −21.44 |
| LEU-117B | HS | HS | 2.77 | −25.50 |
| ASN-118B | HS | – | 0.74 | 4.82 |
| PRO-121B | HS | – | – | −13.70 |
| ALA-125B | – | – | – | −4.57 |
| LYS-127B | – | – | – | −24.51 |
| LEU-128B | HS | HS | 1.40 | −12.3 |
| MET-129 | HS | – | 0.91 | −15.01 |
| VAL-130B | HS | HS | 0.84 | −9.71 |
| VAL-131B | HS | HS | 0.64 | −11.73 |
| PRO-133B | – | – | – | −8.7 |
| PRO-183B | – | – | – | −4.87 |
| ILE-185B | HS | HS | 1.43 | −10.92 |
| ARG-190B | – | – | – | −37.68 |
*HS: Hotspot.
Table 4.
List of interacting residues at protein-protein interacting interface of SARS-CoV-2 NSP12 (chain A) and NSP7 (chain C), interface hotspot residues are predicted using three computational methods implemented in KFC server, HotRegion database and Robetta web server. The predicted hotspot residues contribute reasonably good binding energy which may help in the complexation process. The per-residue energy decomposition analysis was carried out using the last 10 ns MD trajectory.
| Residues | KFC | HotRegion | Robetta ΔΔG (kcal/mol) | Per-residue energy contribution (kcal/mol) |
|---|---|---|---|---|
| PHE-415A | – | – | - | −7.64 |
| TYR-420A | – | HS | 1.14 | −5.11 |
| LEU-437A | – | HS | – | −3.82 |
| PHE-440A | HS | HS | 2.16 | −15.72 |
| PHE-441A | HS | HS | 0.28 | −1.11 |
| PHE-442A | HS | HS | 2.34 | −16.34 |
| ALA-443A | HS | HS | – | −8.69 |
| GLN-444A | – | – | – | −0.52 |
| ASP-5C | – | – | – | 108.29 |
| LYS-7C | – | – | – | −81.99 |
| CYS-8C | HS | HS | −0.03 | −12.40 |
| VAL-11C | HS | HS | 1.56 | −14.08 |
| VAL-12C | – | – | – | −6.82 |
| LEU-14C | HS | HS | 0.83 | −7.35 |
| TRP-29C | – | – | – | −4.24 |
| VAL-33C | – | HS | 1.19 | −7.76 |
| HIS-36C | HS | HS | 2.52 | −3.55 |
| ASN-37C | – | – | 2.52 | 0.004 |
| LEU-40C | HS | HS | 1.83 | −11.06 |
*HS: Hotspot.
Table 6.
List of interacting residues at protein-protein interacting interface of SARS-CoV NSP12 (chain A) and NSP8 (chain B), interface hotspot residues are predicted using three computational methods implemented in KFC server, HotRegion database and Robetta web server. The predicted hotspot residues contribute reasonably good binding energy which may help in the complexation process.The per-residue energy decomposition analysis was carried out using the last 10 ns MD trajectory.
| Residues | KFC server | HotRegion Database | Robetta ΔΔG (kcal/mol) | Per-residue energy contribution (kcal/mol) |
|---|---|---|---|---|
| LEU-271A | HS | HS | 2.47 | −16.33 |
| TYR-273A | HS | HS | 1.03 | −4.23 |
| THR-324A | – | – | – | 1.33 |
| PHE-326A | – | – | – | 2.55 |
| PRO-328A | HS | – | −2.07 | |
| LEU-329A | HS | HS | 0.61 | −9.3 |
| VAL-330A | HS | HS | 0.97 | −7.15 |
| ARG-331A | – | – | 1.14 | −25.63 |
| LYS-A332A | – | – | – | 2.29 |
| PRO-339A | – | – | – | −4.73 |
| PHE-368A | HS | HS | 2.33 | −14.1 |
| LEU-371A | HS | HS | 1.84 | −9.73 |
| PRO-378A | HS | – | −2.86 | |
| ALA-379A | HS | HS | – | −1.93 |
| MET-380A | HS | HS | 0.62 | −10.38 |
| ALA-382A | HS | HS | – | −2.41 |
| ALA-383A | HS | HS | – | −5.97 |
| SER-384A | HS | – | −0.11 | −5.28 |
| ASN-386A | – | – | – | −4.32 |
| LEU-387A | HS | HS | 2.89 | −20.34 |
| LEU-388A | HS | HS | 0.80 | −9.37 |
| LEU-389A | HS | HS | 1.64 | −14.76 |
| ASP-390A | – | – | – | 43.6 |
| LYS-391A | – | – | – | −54.42 |
| ARG-392A | – | – | – | −49.17 |
| VAL-398A | HS | HS | 0.75 | −2.74 |
| ASN-403A | – | – | – | 2.83 |
| PRO-505A | HS | – | – | −4.02 |
| TRP-509A | - | – | 1.74 | −7.92 |
| LEU-514A | - | – | 1.07 | −7.58 |
| TYR-515A | HS | HS | 0.75 | −3.87 |
| ASP-517A | – | – | – | −19.43 |
| SER-518A | – | – | – | 2.61 |
| SER-520A | – | – | – | 0.09 |
| ASP-523A | – | – | – | −11.73 |
| MET-666A | – | HS | 0.26 | −2.09 |
| LYS-79B | – | – | – | −60.76 |
| ARG-80B | HS | – | 2.69 | −76.29 |
| LYS-82B | – | – | −59.94 | |
| VAL-83B | – | HS | 1.54 | −20.54 |
| THR-84B | – | – | – | −1.48 |
| ALA-86B | – | – | – | −7.2 |
| MET-90B | – | – | – | −12.07 |
| LEU-91B | HS | HS | 1.83 | −15.12 |
| MET-94B | HS | HS | 0.83 | −15.12 |
| LYS-97B | – | – | – | −49.7 |
| LEU-98B | HS | HS | 1.96 | −14.25 |
| ASP-99B | – | – | – | 47.86 |
| LEU-103B | HS | HS | 0.85 | −5.72 |
| ASN-104B | – | – | – | 1.45 |
| ILE-106B | – | – | – | −0.8 |
| ALA-110B | – | – | – | −5.3 |
| ASP-112B | – | – | – | 85.61 |
| CYS-114B | HS | HS | −0.19 | −12.31 |
| VAL-115B | HS | HS | 0.46 | −6.33 |
| PRO-116B | HS | – | −23.49 | |
| LEU-117B | HS | HS | 2.50 | −25.74 |
| ASN-118B | HS | 0.59 | 2.88 | |
| ILE-119B | HS | HS | 1.57 | −12.93 |
| ILE-120B | HS | HS | 1.30 | −11.02 |
| PRO-121B | HS | – | – | −13.32 |
| ALA-125B | – | – | – | −5.42 |
| LYS-127B | – | – | – | −39.46 |
| LEU-128B | HS | HS | 1.41 | −12.82 |
| MET-129B | HS | – | 0.96 | −14.6 |
| VAL-130B | HS | HS | 0.81 | −10.02 |
| VAL-131B | HS | HS | 0.77 | −12.36 |
| PRO-133B | – | – | – | −8.27 |
| TRP-154B | – | HS | 0.49 | −2.55 |
| PRO-183B | – | – | – | −7.7 |
| ARG-190B | – | – | – | −54.64 |
*HS: Hotspot.
Table 7.
List of interacting residues at protein-protein interacting interface of SARS-CoV NSP12 (chain A) and NSP7 (chain C), interface hotspot residues are predicted using three computational methods implemented in KFC server, HotRegion database and Robetta web server. The predicted hotspot residues contribute reasonably good binding energy which may help in the complexation process. The per-residue energy decomposition analysis was carried out using the last 10 ns MD trajectory.
| Residues | KFC server | HotRegion Database | Robetta ΔΔG (kcal/mol) | Per-residue energy contribution (kcal/mol) |
|---|---|---|---|---|
| LYS-411A | – | – | – | 16.08 |
| PHE-415A | – | – | 1.10 | −8.13 |
| TYR420A | HS | HS | 1.79 | −3.74 |
| PHE-429A | – | HS | 0.35 | −3.88 |
| GLU431A | – | – | – | −14.44 |
| GLU436A | – | – | 3.02 | 6.5 |
| LEU437A | HS | HS | 0.30 | −6.51 |
| PHE-440A | HS | HS | 2.33 | −13.09 |
| PHE-442A | HS | HS | 2.00 | −15.61 |
| ALA443A | HS | HS | – | −3.17 |
| GLN-444A | – | – | – | −0.24 |
| ALA550A | – | – | – | 1.06 |
| ASN552A | – | – | – | 0.27 |
| LYS-2C | – | – | – | −180.18 |
| MET-3C | – | HS | −8.59 | |
| SER-4C | – | HS | 1.24 | −8.58 |
| ASP-5C | – | – | – | 104.23 |
| LYS-7C | HS | – | – | −78.31 |
| CYS-8C | HS | HS | 0.04 | −11.8 |
| VAL-11C | – | – | – | −10.08 |
| VAL-12C | – | – | – | −5.96 |
| LEU-14C | – | HS | 0.32 | −4.04 |
| GLU-23C | – | – | – | 65.03 |
| TRP-29C | – | HS | 2.01 | −4.03 |
| VAL-33C | – | HS | 1.10 | −7.5 |
| HIS-36C | HS | HS | 1.38 | 9.78 |
| ASN-37C | – | – | 0.45 | 5.13 |
| LEU-40C | HS | HS | 1.50 | −8.86 |
| LEU-41C | – | – | – | −8.69 |
*HS: Hotspot.
3.3. Hotspot residue detection and per-residue energy contribution
To understand the mechanism of molecular interaction in PPI, identification of hotspot residues is useful in obstructing PPI [11], [12]. Here, four different computational methods (KFC server, HotRegion database, Robetta server and per-residue energy decomposition) were used to predict the hotspot residues across PPI interface of viral replication complex (NSP12-NSP8, NSP12-NSP7, NSP8-NSP7). Analysis of results from all the four methods will help to improve the accuracy of the predicted hotspots, and the hotspot information derived can be used further for designing PPI inhibitors. The results obtained from the four methods are summarized in Table 3, Table 4, Table 5, Table 6, Table 7, Table 8 , for SARS-CoV-2 and SARS-CoV heterotetramer replication complex respectively. Comparison of the results from for different methods suggests that most of the predicted hotspot residues contribute high binding energy and when mutated to alanine in Robetta server, ∆∆G values to be > 1 kcal/mol or close to 1 kcal/mol.
Table 5.
List of interacting residues at protein-protein interacting interface of SARS-CoV-2 NSP7 (chain C) and NSP8 (chain D), interface hotspot residues are predicted using three computational methods implemented in KFC server, HotRegion database and Robetta web server. The predicted hotspot residues contribute reasonably good binding energy which may help in the complexation process. The per-residue energy decomposition analysis was carried out using the last 10 ns MD trajectory.
| Residues | KFC | HotRegion | Robetta ΔΔG (kcal/mol) | Per-residue energy contribution (kcal/mol) |
|---|---|---|---|---|
| CYS-8C | – | – | – | −1.89 |
| THR-9C | HS | – | 0.88 | −2.13 |
| VAL-11C | – | – | – | −0.61 |
| VAL-12C | – | HS | 1.14 | −12.48 |
| LEU-13C | HS | HS | – | −6.69 |
| VAL-16C | HS | HS | 1.14 | −9.49 |
| LEU-35C | – | HS | – | −3.06 |
| PHE-49C | – | HS | 1.95 | −12.86 |
| MET-52C | HS | HS | 0.63 | −7.81 |
| VAL-53C | HS | HS | 1.70 | −16.16 |
| SER-54C | – | – | 0.76 | 1.89 |
| LEU-56C | HS | HS | 2.10 | −14.12 |
| SER-57C | HS | – | 0.88 | 1.18 |
| LEU-59C | HS | HS | – | −3.4 |
| LEU-60C | HS | HS | 1.83 | −10.61 |
| SER-61C | – | – | – | −4.86 |
| ALA-65C | – | – | – | −1.09 |
| VAL-66C | – | HS | 0.37 | −4.64 |
| ASP-67C | – | – | – | 8.64 |
| LYS-70C | – | – | – | 8.12 |
| LEU-71C | HS | HS | 1.60 | 39.38 |
| MET-87D | – | – | −9.63 | |
| GLN-88D | – | – | 2.42 | −2.16 |
| THR-89D | – | – | 1.19 | |
| LEU-91D | HS | HS | 2.63 | −18.55 |
| PHE-92D | HS | HS | 2.50 | −16.48 |
| MET-94D | – | – | 0.72 | −11.81 |
| LEU-95D | HS | HS | 1.42 | −12.29 |
| ARG-96D | – | – | – | −13.58 |
| ASN-100D | – | – | – | 0.66 |
| LEU-103D | HS | HS | 1.70 | −12.59 |
| ILE-106D | HS | HS | 1.80 | −10.37 |
| ILE-107D | HS | HS | 0.99 | −7.41 |
| PRO-116D | HS | HS | – | −3.9 |
| LEU-117D | – | – | – | −2.78 |
| ILE-119D | HS | – | 1.70 | −17 |
| ILE-120D | – | – | – | −9.65 |
| LEU-122D | – | – | 1.08 | −9.22 |
*HS: Hotspot.
Table 8.
List of interacting residues at protein-protein interacting interface of SARS-CoV NSP8 (chain A) and NSP7 (chain C), interface hotspot residues are predicted using three computational methods implemented in KFC server, HotRegion database and Robetta web server. The predicted hotspot residues contribute reasonably good binding energy which may help in the complexation process. The per-residue energy decomposition analysis was carried out using the last 10 ns MD trajectory.
| Residues | KFC server | Hotregion database | Robetta | Per-residue energy contribution |
|---|---|---|---|---|
| VAL-6C | – | HS | 0.34 | −5.82 |
| CYS-8C | – | – | – | −1.69 |
| THR-9C | HS | – | 0.96 | −3.02 |
| VAL-11C | – | – | – | −0.45 |
| LEU-13C | HS | HS | 0.55 | −6.27 |
| VAL-16C | – | HS | 1.22 | −9.6 |
| MET-52C | HS | HS | 0.49 | −6.23 |
| PHE-49 | – | HS | 2.01 | −13.20 |
| VAL-53C | HS | – | 1.39 | −13.75 |
| SER-54C | HS | HS | 0.81 | −0.17 |
| LEU-56C | HS | HS | 2.15 | −14.83 |
| SER-57C | HS | – | 0.87 | −0.38 |
| LEU-60C | HS | HS | 1.38 | −8.98 |
| SER-61C | – | – | – | −4.07 |
| LEU-71C | HS | HS | 1.59 | 30.13 |
| MET-87D | – | – | – | −9.09 |
| LEU-91D | HS | HS | 2.56 | −18.59 |
| PHE-92D | HS | HS | 1.96 | −12.27 |
| MET-94D | – | – | 0.86 | −12.85 |
| LEU-98D | HS | HS | 2.01 | −14.52 |
| ASN-100D | – | – | – | −0.08 |
| ALA-102D | – | – | – | −3.25 |
| LEU-103D | HS | HS | 2.14 | −12.94 |
| ILE-106D | HS | HS | 1.26 | −7.7 |
| ALA-110D | HS | HS | – | −0.05 |
| ARG-111D | – | – | – | −8.77 |
| PRO-116D | HS | – | – | −2.89 |
| LEU-117D | – | – | – | −2.42 |
| ASN-118D | – | – | – | 0.48 |
| ILE-119D | HS | – | 1.46 | −16.33 |
| ILE-120D | HS | HS | 0.63 | −7.97 |
*HS: Hotspot.
3.4. Analysis of per-residue energy contribution
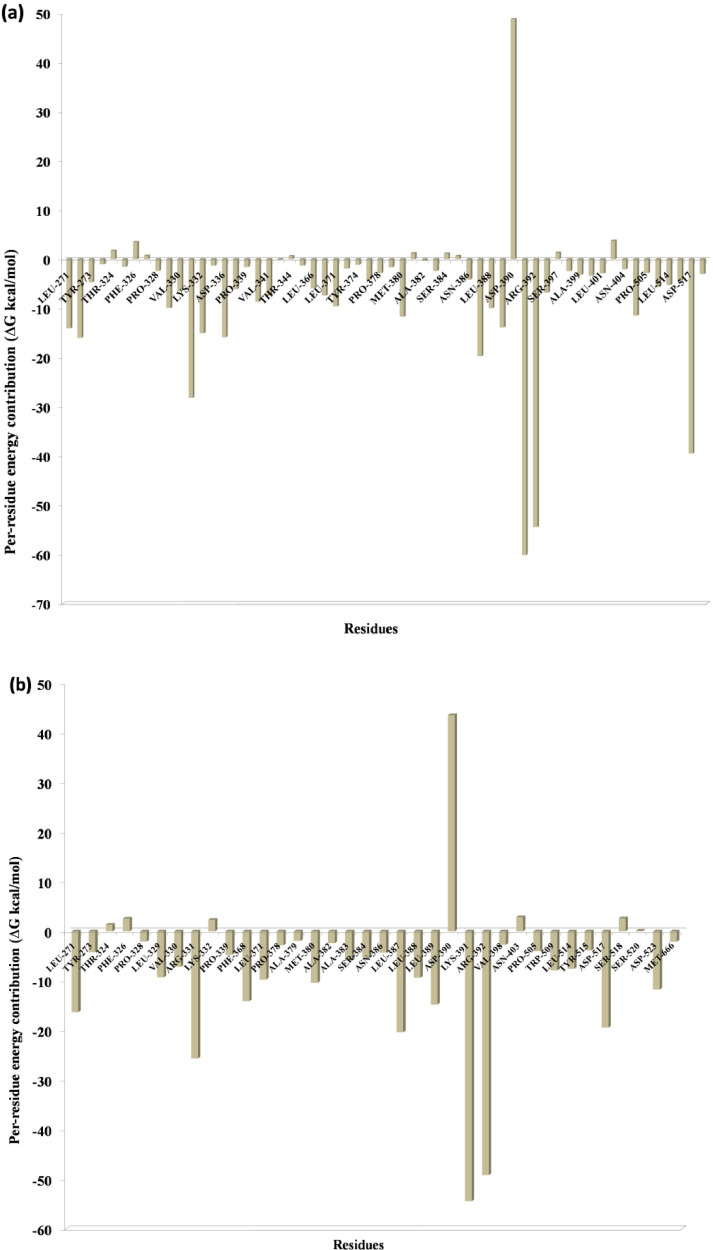
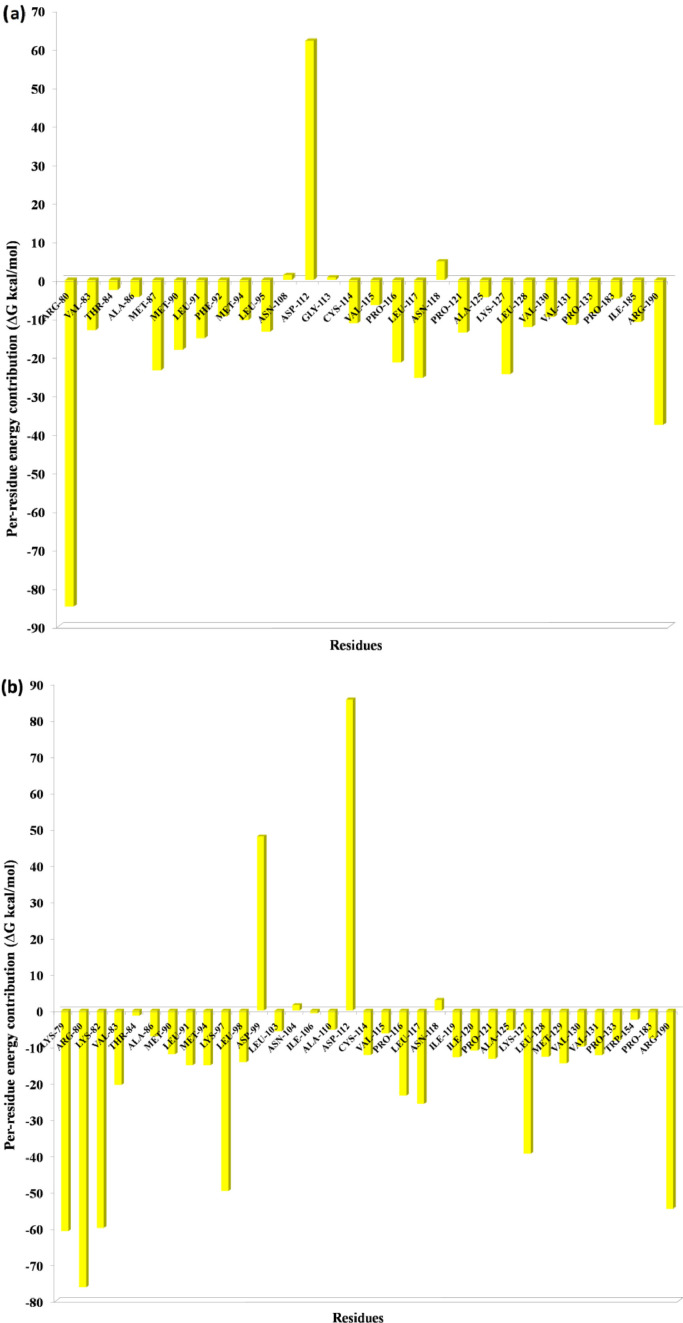
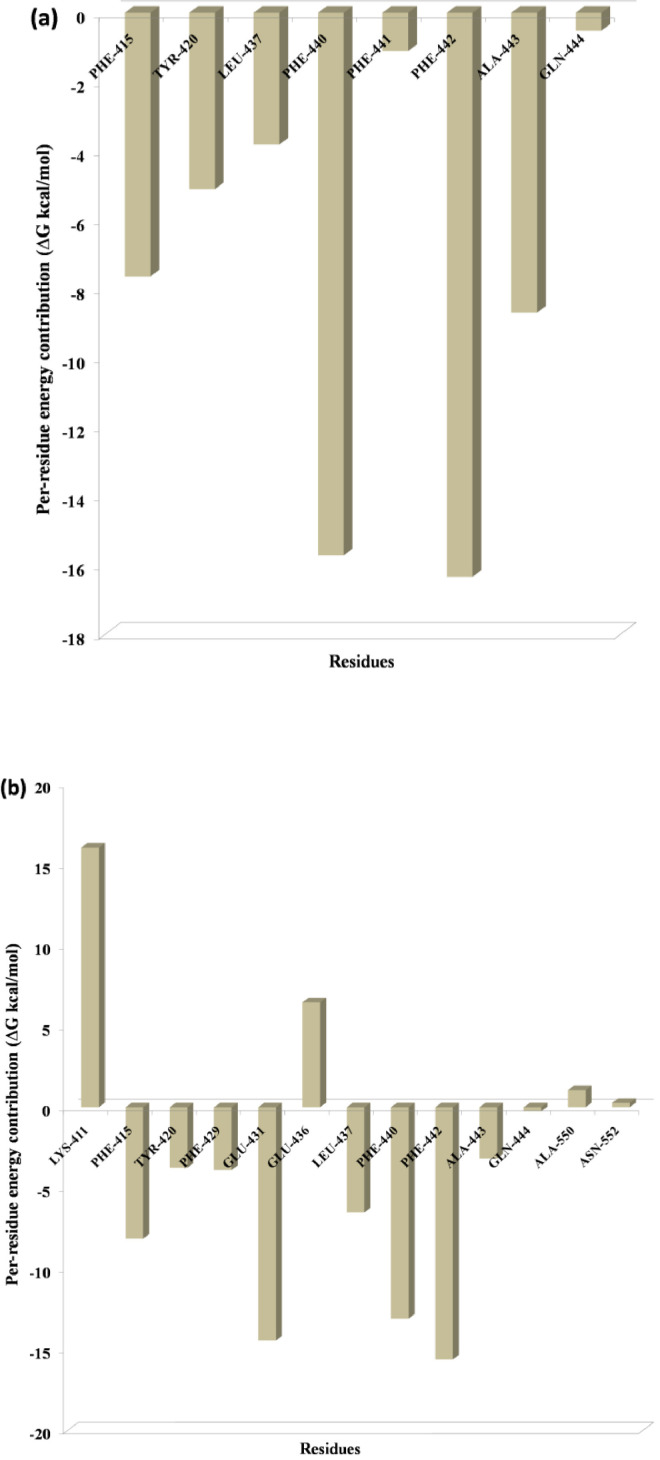
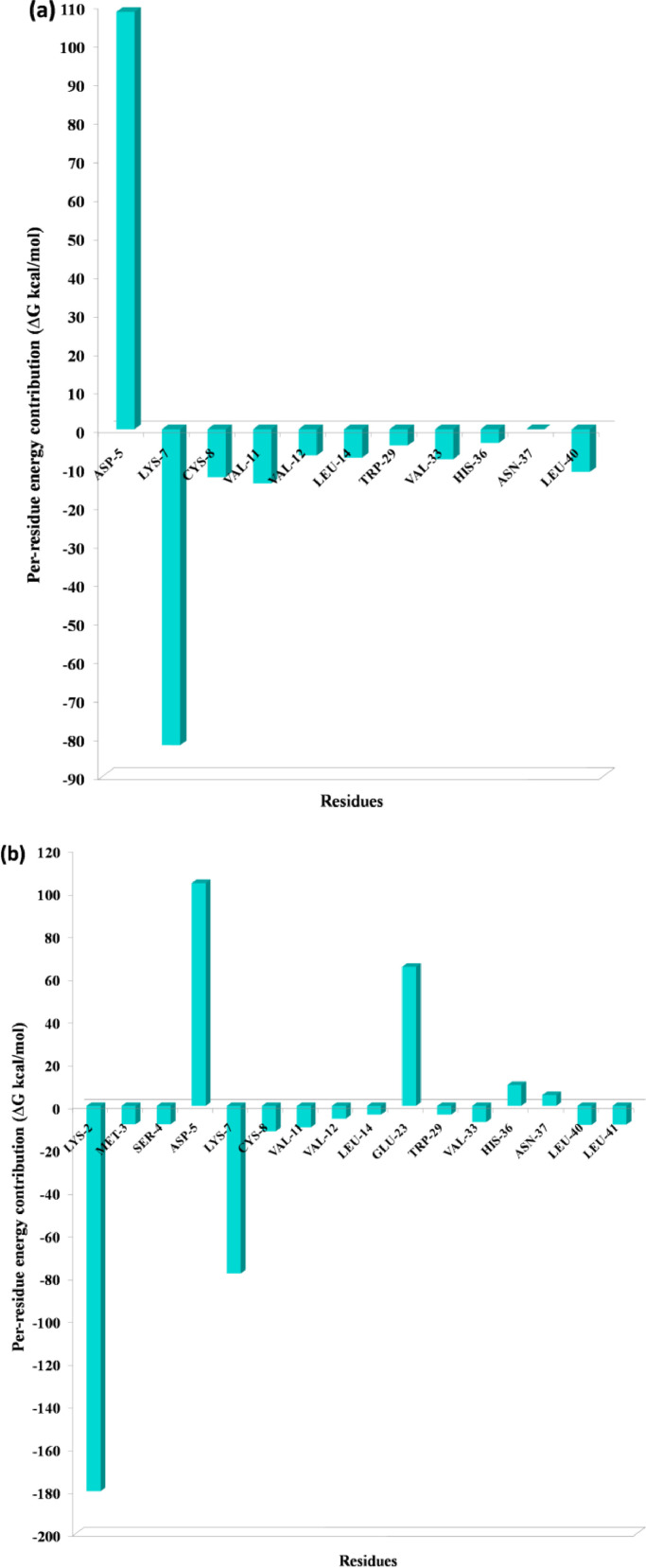
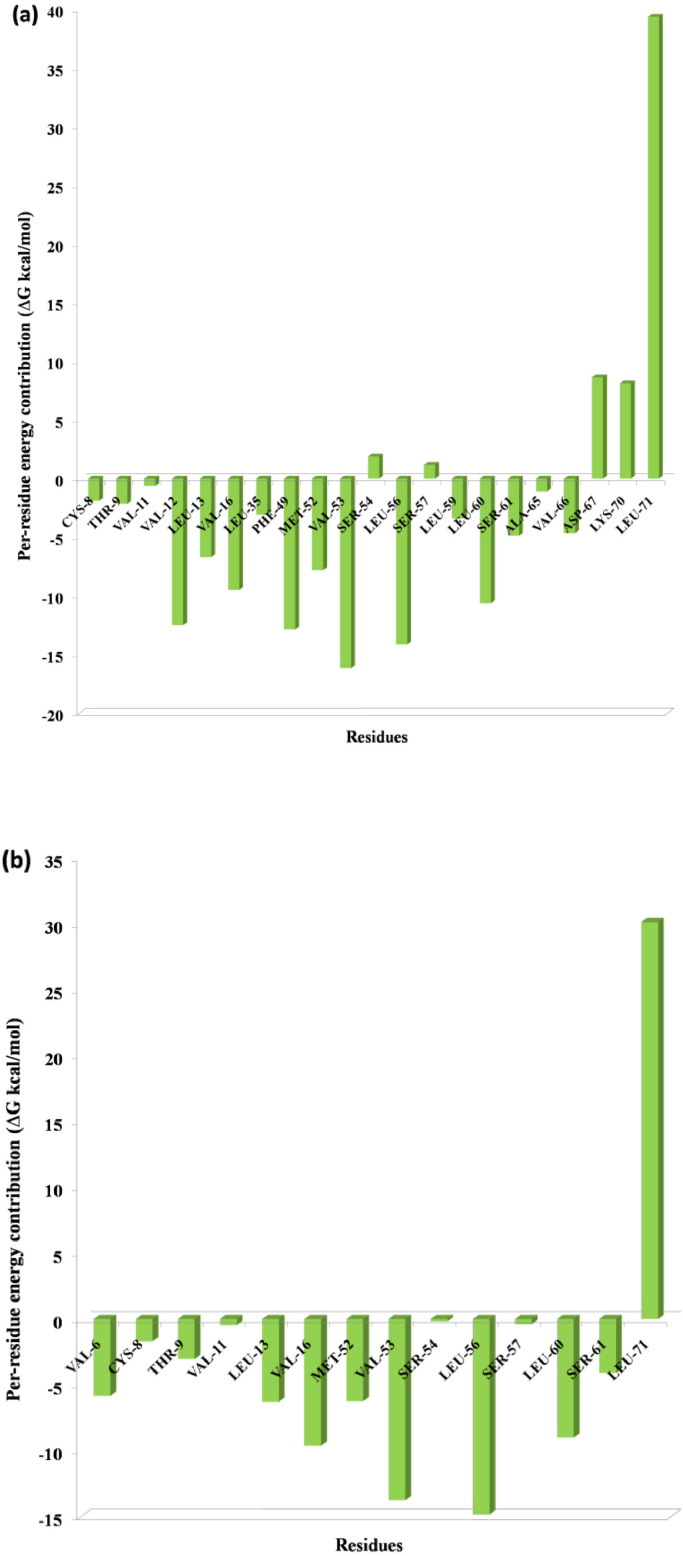
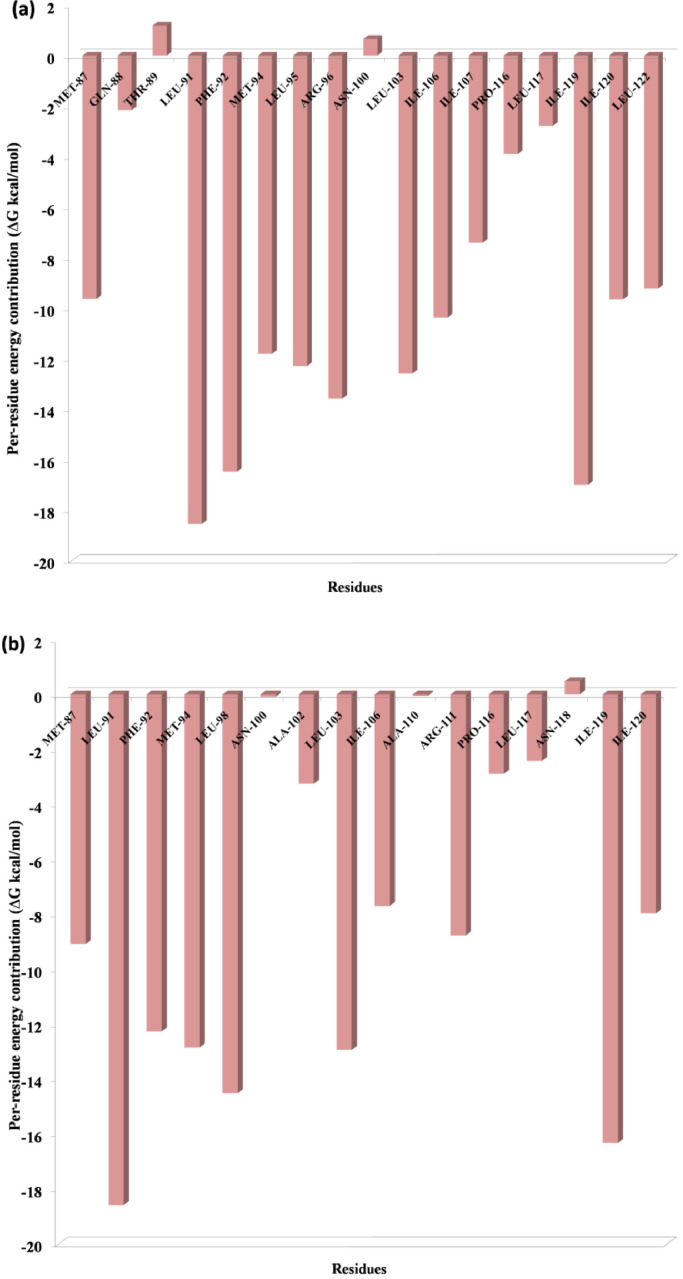
One of the most significant properties of PPI interface is that the energy is not uniformly distributed. Some of the interface residues have the greatest impact on binding energy in the protein complex and those residues are considered to be the hotspot residue [15,17]. To validate our predicted hotspot residues across NSP12 heterotetramer PPI interface we carried out per-residue energy decomposition. The detailed energy contributions of each interface residue across PPI interface are presented in Table 3, Table 4, Table 5, Table 6, Table 7, Table 8 and Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11, Fig. 12 . The hotspots are presented in boldface in Table 3, Table 4, Table 5, Table 6, Table 7, Table 8. Studies have shown that hotspots tend to cluster near the center of the interface [20], [21], [22]. Our predicted hotspot residues are found to be clustered and mostly in the center of the PPI interface. In SARS-CoV-2 and SARS-CoV NSP12-NSP8 interface, the common hotspot residues at NSP12 (Chain A) interface are Leu271, Tyr273, Leu329, Val330, Phe368, Leu371, Met380, Leu387, Leu388, Leu389 and for NSP8 residues Val83, Leu91, Val117, Leu128, Val130, Val131 are designate as common hotspot residue. In SARS-CoV-2 and SARS-CoV NSP12-NSP7 interface, NSP12 (Chain A) residues Tyr420, Phe440, Phe442 and for NSP7 Cys8, Val33, His36, Leu40 are designate as common hotspot residues. In SARS-CoV-2 and SARS-CoV NSP7-NSP8 heterodimer, in NSP8 interface Leu91, Phe92, Leu103, Ile106, Ile119 and in NSP7 interface Val16, Phe49, Leu56, Leu60 and Leu71 are designated as hotspot residues.
Fig. 7.
Residue-wise energy contribution analysis of NSP12 of (a) SARS-CoV2 and (b) SARS-CoV when it in complex with NSP8. The per-residue energy decomposition was calculated from the last 10 ns MD trajectory.
Fig. 8.
Residue-wise energy contribution of the NSP8 (a) SARS-CoV-2 and (b) SARS-CoV when it is in complex with NSP12. The per-residue energy decomposition was calculated from the last 10 ns MD trajectory.
Fig. 9.
Residue-wise energy contribution of the NSP12 (a) SARS-CoV-2 and (b) SARS-CoV when it is in complex with NSP7. The per-residue energy decomposition was calculated from the last 10 ns MD trajectory.
Fig. 10.
Residue-wise energy contribution of NSP7 (a) SARS-CoV-2 and (b) SARS-CoV when it is in complex with NSP12. The per-residue energy decomposition was calculated from the last 10 ns MD trajectory.
Fig. 11.
Residue-wise energy contribution of NSP7 (a) SARS-CoV-2 and (b) SARS-CoV when it is in complex with NSP8. The per-residue energy decomposition was calculated from the last 10 ns MD trajectory.
Fig. 12.
Residue-wise energy contribution of NSP8 (a) SARS-CoV-2 and (b) SARS-CoV when it is in complex with NSP7. The per-residue energy decomposition was calculated from the last 10 ns MD trajectory.
Our predicted hotspots contain mostly Tyr, Pro, Phe, Val, Leu-and Arg-amino acids and in literature these residues found to have a tendency in being a hotspot [20], [21], [22]. The hotspots are presented in boldface in Table 3, Table 4, Table 5, Table 6, Table 7, Table 8 and encircled in Figs. S2–S4 and represented as spheres in 3-D form in Fig. 13 . Hotspot residues are known to be enriched in forming H-bonding and salt bridges [13]. Few of our predicted hotspots are also involved in the formation of H-bonds and salt bridges across NSP12-NSP8 interface of SARS-CoV-2 and SARS-CoV, three predicted hotspot residues (Val330, Leu387, Leu389) at NSP12 interface involved in the formation of H-bond and at NSP8 interface, three residues namely Val117, Val131, Met 129 formed H-bond. In SARS-CoV NSP8 Arg80 formed one salt bridge and one H-bond. Across NSP12-NSP7 interface, three residues (Tyr420, Phe441, Ala443) of SARS-CoV-2 NSP12 involved in the H-bond formation whereas one predicted hotspot residue (Tyr420) of SARS-CoV NSP12 involved in the formation of H-bond. At SARS-CoV-2 NSP7 interface two residues (His36, Asn37) and at SARS-CoV NSP7 interface three residues (Ser4, Trp29, His36) involved in H-bond formation. The H-bonds depicted in blue lines in Figs. S2–S4. The detailed H-bond atomic interactions between the residues are tabulated in Tables S1–S8.
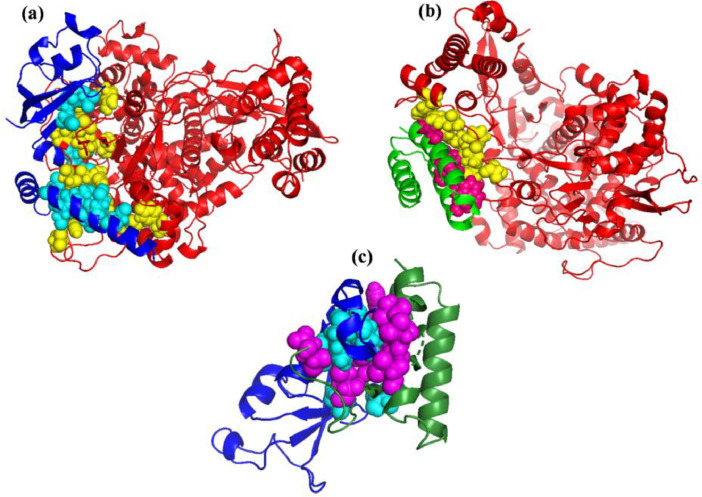
Fig. 13.
Cartoon view of the (a) NSP12-NSP8 (b) NSP12-NSP7 and (c) NSP7-NSP8 complexes of SARS-CoV-2 interacting interface hotspot residues represented in spheres. The NSP12 is represented in red, NSP8 in navy blue and NSP7 in green color. Hotspot residues are represented in spheres, yellow (in NSP12), cyan (in NSP8) and pink (in NSP7). The represented structures are the average conformer extracted from last 10 ns MD trajectory.
A recent study has suggested that in SARS-CoV-2 NSP7-NSP8 heterodimer interface, mutation of NSP7 F49A, M52A and L56A, leads to decrease of RdRp efficiency and in the current study per-residues energy contribution for these three residues found to be −12.86, −7.81, and −14.12, kcal/mol respectively (Table 5), and for SARS-CoV NSP7 residue energy contribution to be −13.20, −6.23, and −14.83 (Table 8) respectively. Mutation of NSP8 F92A leads to a decrease of RdRp efficiency to various extents, along with F49A, M52A, L56A triple mutation at NSP7 leads to stronger effect than individual mutation [54]. The per residue energy contribution of NSP8 F92 to be −16.48 (Table 5) and SARS-CoV is −12.27 (Table 8) at NSP7-NSP8 interface. Among these residues, our computational study has identified NSP7 Phe49, Met52 and NSP8 Phe92 to be as hotspot residues across NSP7-NSP8 complex of SARS-CoV-2.
NSP7 C8G and V11A hamper the association of both NSP7-NSP8 and NSP8-NSP7-NSP12 complex. The same study identified, mutation of NSP8 interface residues viz., NSP8 F92A, M90A and M94A leads to even more severe reduction of RdRp efficiency because these three residues at NSP8 involved in association of both NSP7-NSP8 complex and NSP12-NSP8 complexes [54].
At NSP12-NSP7 interface, the energy contribution of SARS-CoV-2 NSP7 C8 and V11 when it is in complex with NSP12 to be −12.40 and −14.08 (Table 4) respectively and for SARS-CoV to be −11.80 and −10.08 (Table 7) respectively. At NSP7-NSP8 interface, for SARS-CoV-2 NSP7 C8 and V11per-residue energy contribution found to be 1.89 and −0.61 respectively and −1.69 and −0.45 respectively. The C8 and V11, residue contributing more energy across NSP12-NSP7 interface then NSP7-NSP8 interface which is in good agreement with the experimental findings.
In the interface of SARS-CoV-2 NSP12-NSP8, NSP8 F92, M90, and M94, the residue energy contributions are −9.59, −18.15 and −10.42 (Table 3), whereas in SARS-CoV, residue energy contribution for NSP8 M90, and M94 to be −12.07 and −15.12 (Table 6) respectively, however we did not see interaction of NSP8 F92 with NSP12 and all the three servers did not consider F92. These know important residue has shown significant energy contribution at the PPI interface and M90 identified as hotspot in our study. Over all the per-residue energy contribution found to be slightly more in the NSPs interface of SARS-CoV-2 as compared to SARS-CoV.
The same study has suggested that in SARS-CoV-2 NSP7, Asn37 serve as a H-bond donor in NSP12-NSP8-NSP7 complex but not in NSP7-NSP8 heterotetramer. When Asn37 mutated to Val-it does not affect the stability of NSP7-NSP8 heterotetramer, however it leads to the disruption of NSP12-NSP7-NSP8 complex and compromise the replication efficacy of NSP12-NSP7-NSP8 complex [54]. In the initial complex SARS-CoV-2 NSP7 Asn37 formed one H-bond and one non-covalent contact with Ala443, when we analysed the PPI in the average conformer from last 10 ns trajectory of heterotetramer, NSP7 Asn37 formed two H-bond with NSP12 Ala443 and one non-covalent interaction with NSP12 Phe442 and one non-covalent interaction with NSP12 Ala443.
Whereas, initial structure of SARS-CoV NSP7, Asn37 formed one H-bond and one non-bonded contact with Ala443 while at the end of the simulation NSP7 Asn37 does not form any H-bond while it formed non-bonded interaction with NSP12 Asn552 and Phe442. The residue energy contribution of NSP7 Asn37 of SARS-CoV-2 and SARS-CoV are 0.004 and 5.13 kcal/mol respectively. We found a contrasting result, in hotspot residue prediction; KFC and Hotregion database could not predict SARS-CoV-2 NSP7 Asn37 as hotspot residue, however interestingly, in Robetta sever, ASM shows ∆∆G value to be 2.52 kcal/mol and 0.45 kcal/mol for SARS-CoV-2 and SARS-CoV respectively. We can say that NSP7 Asn37 play an important role towards the formation of NSP12-NSP7 complex but may be less important in SARS-CoV.
There are few residues at SARS-CoV-2 NSP12-NSP8 interface, identified as probable hotspots viz., NSP12 (Chain A) interface, Leu271, Tyr273, Pro328, Leu329, Val330, Phe340, Val341, Phe368, Leu371, Met380, Leu387, Leu388, Leu389, Arg392, Val398, Val405 and at NSP8 (Chain B) interface, Val83, Met87, Met90, Leu91, Phe92, Leu95, Cys114, Val115, Leu117, Met129, Val130, Val131, Ile185). Across NSP12-NSP7 interface, in NSP12 (Chain A)residues Tyr420, Phe440, Phe441, Phe442, Ala443 identified as hotspot and at NSP7 (Chain C) interface, Cys8, Val11, Leu14, Val33, His36,Asn37, Leu40). Across NSP7-NSP8 interface, atNSP7(Chain C) interface, Val12, Leu13, Val16, Phe49, Leu56, Leu60, Leu71) and at NSP8(2) (Chain D) interface, Leu91, Phe92, Leu95, Leu103, Ile106, Ile107, Ile119, Leu122) (Table 3, Table 4, Table 5, Table 6, Table 7, Table 8). The residues in boldface are newly predicted hotspot residues through in silico methods in this study. The predicted hotspot residue may play key roles on the stability of the NSPs association in the heterotetramer replication complex.
4. Conclusions
In the present study, we have carried out a comparative computational study on the heterotetramer viral replication complex of SARS-CoV-2 and SARS-CoV. The heterotetramer complex of SARS-CoV-2 and SARS-CoV were subjected to MD simulations followed by MM-PBSA and per-residue decomposition energy calculations were employed to investigate the binding mechanism and to analyze the energetic difference in both the complexes. The overall heterotetramer complex of SARS-CoV-2 was found to be slightly flexible and not rigid as that of SARS-CoV. Though there are a smaller number of molecular interactions in terms of H-bond, salt bridges and non-bonded contact in NSP12-NSP8 and NSP12-NSP7 complex of SARS-CoV-2, the binding free energy between NSP12-NSP8, NSP12-NSP7 and NSP7-NSP8 were found to be high in SARS-CoV-2 as compared to SARS-CoV. The per-residue energy contributions of the interacting interface residues of these complexes were comparatively higher in SARS-CoV-2 as compared to SARS-CoV. Detailed interaction profile of NSP12-NSP8, NSP12-NSP7 and NSP7-NSP8 were analyzed using PDBsum server. Additionally, interface hotspot residues were predicted using different web servers along with per-residue energy decomposition analysis. Most of the predicted hotspot residues at NSPs interface have more energy contributions. This difference in terms of structural flexibility, stability and energetic of the interface residues and hotspots residue of viral replication complex may be the reason of high rate of RNA replication in SARS-CoV-2 as compared to SARS-CoV. We see that, few of the experimentally identified key interface residue identified as hotspots in our study. There are many other residues we identified as hotspots for which there are no experimental ASM done yet. Therefore, our study may pave a direction to predict the potential unknown location of hotspots across PPI interface of SARS-CoV-2 viral replication complex (NSP12-NSP7-NSP8(2)), which could help researchers in performing ASM in the wet lab. As ASM is expensive and time-consuming, therefore, by identifying the possible locations of hotspots with the help of in silico methods, the researchers can perform alanine mutations only in those amino acid locations which are identified as hotspot by in silico methods. Additionally, the predicted hotspot will also help in designing the small molecule or peptide/peptidomimetic to disrupt the PPI.
Credit author statement
GNS has conceived and designed the study, HS has made the plan and execution of the work and involved in the interpretation of the results. EJ has involved in all the calculations and analysis. HS and EJ has prepared the preliminary draft of the manuscript, which was thoroughly checked and modified by GNS.
Declaration of Competing Interest
No conflict of interest.
Acknowledgments
GNS thanks J C Bose fellowship of DST New Delhi. HS thanks for an institutional Post-Doctoral Fellowship of CSIR-NEIST, EJ thanks DST for INSPIRE fellowship.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.132602.
Appendix. Supplementary materials
References
- 1.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell. Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.teVelthuis A., Arnold J., Cameron C., van den Worm S., Snijder E. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38(1):203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blazer L., Neubig R. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsycho. Pharmacol. Rep. 2009;34(1):126–141. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 5.Gurung A., Bhattacharjee A., Ajmal Ali M., Al-Hemaid F., Lee J. Binding of small molecules at interface of protein-protein complex - a newer approach to rational drug design,Saudi. J. Biol. Sci. 2017;24(2):379–388. doi: 10.1016/j.sjbs.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuenemann M., Sperandio O., Labbé C., Lagorce D., Miteva M., Villoutreix B. In silico design of low molecular weight protein-protein interaction inhibitors: overall concept and recent advances. Prog. Biophys. Mol. Biol. 2015;119(1):20–32. doi: 10.1016/j.pbiomolbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Panwar D., Rawal L., Ali S. Molecular docking uncovers TSPY binds more efficiently with eEF1A2 compared to eEF1A1. J. Biomol. Struct. Dyn. 2015;33(7):1412–1423. doi: 10.1080/07391102.2014.952664. [DOI] [PubMed] [Google Scholar]
- 8.Rognan D. Rational design of protein–protein interaction inhibitors. Med. Chem. Commun. 2015;6:51–60. [Google Scholar]
- 9.Xu J., Xu J., Chen H. Interpreting the structural mechanism of action for MT7 and human muscarinic acetylcholine receptor 1 complex by modeling protein-protein interaction. J. Biomol. Struct. Dyn. 2012;30(1):30–44. doi: 10.1080/07391102.2012.674188. [DOI] [PubMed] [Google Scholar]
- 10.Pallara C., Jiménez-García B., Pérez-Cano L., Romero-Durana M., Solernou A., Grosdidier S., Pons C., Moal I., Fernandez-Recio J. Expanding the frontiers of protein-protein modeling: from docking and scoring to binding affinity predictions and other challenges. Proteins. 2013;81(12):2192–2200. doi: 10.1002/prot.24387. [DOI] [PubMed] [Google Scholar]
- 11.Jin L., Wang W., Fang G. Targeting protein-protein interaction by small molecules. Annu. Rev. Pharmacol. Toxicol. 2014;54:435–456. doi: 10.1146/annurev-pharmtox-011613-140028. [DOI] [PubMed] [Google Scholar]
- 12.Cesa L.C., Mapp A.K., Gestwicki J.E. Direct and propagated effects of small molecules on protein-protein interaction networks. Front. Bioeng. Biotechnol. 2015;3:119. doi: 10.3389/fbioe.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keskin O., Ma B., Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345(5):1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 14.González-Ruiz D., Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr. Med. Chem. 2006;13(22):2607–2625. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- 15.Bogan A., Thorn K. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 16.Cheung L., Kanwar M., Ostermeier M., Konstantopoulos K. A hot-spot motif characterizes the interface between a designed ankyrin-repeat protein and its target ligand. Biophys. J. 2012;102(3):407–416. doi: 10.1016/j.bpj.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clackson T., Wells J. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196) doi: 10.1126/science.7529940. 383-383. [DOI] [PubMed] [Google Scholar]
- 18.Thorn K., Bogan A. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. J. Bioinform. 2001;17(3):284–295. doi: 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- 19.I. Moreira, P. Fernandes, M. Ramos. Hot spots-a review of the protein-protein interface determinant amino-acid residues, Proteins 68(4) (2007) 803–812. [DOI] [PubMed]
- 20.Caffrey D., Somaroo S., Hughes J., Mintseris J., Huang E. Are protein-protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci. 2004;13(1):190–202. doi: 10.1110/ps.03323604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.S. Lockless, R. Ranganathan. Evolutionarily conserved pathways of energetic connectivity in protein families, Science 286(5438) (1999) 295–299. [DOI] [PubMed]
- 22.Schreiber G., Fersht A.R. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 1995;248(2):478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 23.Thornton J. The Hans neurath award lecture of the protein society: proteins- a testament to physics, chemistry, and evolution. Protein Sci. 2001;10(1):3–11. doi: 10.1110/ps.90001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunningham B., Wells J. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244(4908):1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 25.Darnell S., LeGault L., Mitchell J. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36(Web Server issue):265–269. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cukuroglu E., Gursoy A., Keskin O. HotRegion: a database of predicted hot spot clusters. Nucleic Acids Res. 2012;40(D1):829–833. doi: 10.1093/nar/gkr929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D. Kim, D. Chivian, D. Baker. Protein structure prediction and analysis using the Robetta server, Nucleic Acids Res. 32(Web Server issue) (2004) 526–531. [DOI] [PMC free article] [PubMed]
- 28.Kortemme T., Kim D., Baker D. Computational alanine scanning of protein-protein interfaces. Sci. STKE. 2004;2004(219) doi: 10.1126/stke.2192004pl2. pl2-pl2. [DOI] [PubMed] [Google Scholar]
- 29.Sarvagalla S., Cheung C., Tsai J., Hsieh H., Coumar M. Disruption of protein–protein interactions: hot spot detection, structure-based virtual screening and in vitro testing for the anti-cancer drug target–surviving. RSC Adv. 2016;6(38):31947–31959. [Google Scholar]
- 30.S. Sarvagalla, T. Lin, S. Kondapuram, C. Cheung, M. Coumar. Survivin-caspase protein-protein interaction: experimental evidence and computational investigations to decipher the hotspot residues for drug targeting, J. Mol. Struct. 1229, (2021) 129619.
- 31.Jha V., Rameshwaram R N., Janardhan S., Raman R., Sastry G.N., Sharma V., Subba Rao J., Kumar D., Mukhopadhyay S. Uncovering structural and molecular dynamics of ESAT-6: β2M interaction: asp53 of human β2-microglobulin is critical for the ESAT-6: β2M complexation. J. Immunol. 2019;203(7):1918–1929. doi: 10.4049/jimmunol.1700525. [DOI] [PubMed] [Google Scholar]
- 32.Badrinarayan P., Sastry G.N. Specificity rendering 'hot-spots' for aurora kinase inhibitor design: the role of non-covalent interactions and conformational transitions. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0113773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faisal H.N., Katti K.S., Katti D.R. Differences in interactions within viral replication complexes of SARS-CoV-2 (COVID-19) and SARS-CoV coronaviruses control RNA replication ability. JOM. 2021:1684–1695. doi: 10.1007/s11837-021-04662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arkin M., Randal M., DeLano W., Hyde J., Luong T., Oslob D J., Raphael D., Taylor L., Wang J., McDowell R., Wells J., Braisted A. Binding of small molecules to an adaptive protein-protein interface. Proc. Natl. Acad. Sci. U. S. A. 2003;100(4):1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma R., Sagurthi S.R., Sastry G.N. Elucidating the preference of dimeric over monomeric form for thermal stability of Thermus thermophilus isopropylmalate dehydrogenase: a molecular dynamics perspective. J. Mol. Gr. Modell. 2020;96 doi: 10.1016/j.jmgm.2020.107530. [DOI] [PubMed] [Google Scholar]
- 36.Sharma R., Sastry G.N. Deciphering the dynamics of non-covalent interactions affecting thermal stability of a protein: molecular dynamics study on point mutant of Thermus thermophilus isopropylmalate dehydrogenase. PloS one. 2015;10(12) doi: 10.1371/journal.pone.0144294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyrisch S., Helms V. What induces pocket openings on protein surface patches involved in protein-protein interactions? J. Comput. Aided Mol. Des. 2009;23:73–86. doi: 10.1007/s10822-008-9239-y. [DOI] [PubMed] [Google Scholar]
- 38.Eyrisch S., Medina-Franco J., Helms V. Transient pockets on XIAP-BIR2: toward the characterization of putative binding sites of small-molecule XIAP inhibitors. J. Mol. Model. 2012;18(5):2031–2042. doi: 10.1007/s00894-011-1217-y. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava H.K., Sastry G.N. Efficient estimation of MMGBSA-based BEs for DNA and aromatic furan amidino derivatives. J. Biomol. Struct. Dyn. 2013;31(5):522–537. doi: 10.1080/07391102.2012.703071. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. PDBsum new things. Nucleic Acids Res. 2009;37(Database issue):355–359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirchdoerfer R., Ward A. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen E., Goddard T., Huang C., Couch G., Greenblatt D., Meng E., Ferrin T. E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Abraham M., Murtola T., R.Schulz S.Páll, Smith J., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 45.Fuhrmans M., Sanders B., Marrink S., Alex H. Effects of bundling on the properties of the SPC water model. Theor. Chem. Acc. 2010;125:335–344. [Google Scholar]
- 46.Berendsen H., Postma J., van Gunsteren W., DiNola A., Haak J. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 47.Berendsen H., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;95:43–56. [Google Scholar]
- 48.Hess B., Bekker H., Berendsen H., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463. [Google Scholar]
- 49.T. Darden, D. York, L. Pedersen. Particle mesh Ewald: an N, log (N) method for Ewald sums in large systems, J. Chem. Phys. 98(1993)10089e10092.
- 50.Miyamoto S., Kollman P. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 51.Kumari R., Kumar R. Open-source drug discovery consortium, Lynn A. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 52.McCoy A., Epa V., Colman P. Electrostatic complementarity at protein/protein interfaces. J. Mol. Biol. 1997;268(2):570–584. doi: 10.1006/jmbi.1997.0987. [DOI] [PubMed] [Google Scholar]
- 53.Li G., Zhang X., Cui Q. Free Energy Perturbation Calculations with Combined QM/MM Potentials Complications, Simplifications, and Applications to Redox Potential Calculations. J. Phys. Chem. B. 2003;107:8643–8653. [Google Scholar]
- 54.Biswal M., Diggs S., Xu D., Khudaverdyan N., Lu J., Fang J., Song J. Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP. Nucleic Acids Res. 2021;(10):5956–5966. doi: 10.1093/nar/gkab370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeLano W. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 56.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., …, Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18(1):1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hillen H., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.