Abstract
Background
We assessed the safety and immunogenicity of two recombinant DNA vaccines for COVID-19: GX-19 containing plasmid DNA encoding the SARS-CoV-2 spike protein, and GX-19N containing plasmid DNA encoding the SARS-CoV-2 receptor-binding domain (RBD) foldon, nucleocapsid protein, and plasmid DNA encoding the spike protein.
Methods
Two open-label non-randomised phase 1 trials, one of GX-19 and the other of GX-19N were done at two hospitals in South Korea. We enrolled healthy adults aged 19–49 years for the GX-19 trial and healthy adults aged 19–54 years for the GX-19N trial. Participants who tested positive by serological testing for SARS-CoV-2 were excluded. At 4-week intervals, the GX-19 trial participants received two vaccine doses (either 1·5 mg or 3·0 mg), and the GX-19N trial participants received two 3·0 mg doses. The vaccines were delivered intramuscularly using an electroporator. The participants were followed up for 52 weeks after first vaccination. Data collected up to day 57 after first vaccination were analysed in this interim analysis. The primary outcome was safety within 28 days after each vaccination measured in the intention-to-treat population. The secondary outcome was vaccine immunogenicity using blood samples collected on day 43 or 57 after first vaccination measured in the intention-to-treat population. The GX-19 (NCT044445389) and GX-19N (NCT04715997) trials are registered with ClinicalTrials.gov.
Findings
Between June 17 and July 30, 2020, we screened 97 individuals, of whom 40 (41%) participants were enrolled in the GX-19 trial (20 [50%] in the 1·5 mg group and 20 [50%] in the 3·0 mg group). Between Dec 28 and 31, 2020, we screened 23 participants, of whom 21 (91%) participants were enrolled on the GX-19N trial. 32 (52%) of 61 participants reported 80 treatment-emergent adverse events after vaccination. All solicited adverse events were mild except one (2%) case of moderate fatigue in the 1·5 mg GX-19 group; no serious vaccine-related adverse events were detected. Binding antibody responses increased after second dose of vaccination in all groups (p=0·0002 in the 1·5 mg GX-19 group; p<0·0001 in the 3·0 mg GX-19; and p=0·0004 for the spike protein and p=0·0001 for the RBD in the 3·0 mg GX-19N group).
Interpretation
GX-19 and GX-19N are safe and well tolerated. GX-19N induces humoral and broad SARS-CoV-2-specific T-cell responses. GX-19N shows lower neutralising antibody responses and needs improvement to enhance immunogenicity.
Funding
The Korea Drug Development Fund, funded by the Ministry of Science and ICT, Ministry of Trade, Industry, and Energy, and Ministry of Health and Welfare.
Introduction
To overcome the COVID-19 pandemic, the development of safe and effective vaccines is crucial. Several vaccines using different platforms have been developed.1 Most currently authorised vaccines for emergency use or in development target the SARS-CoV-2 spike protein, which binds to the angiotensin-converting enzyme 2 receptor to enter the host cell.
Despite the successful clinical efficacy of the vaccines currently approved or authorised for emergency use,2, 3, 4, 5 the emergence of new variants possessing mutations in the receptor-binding domain (RBD) of the spike protein has resulted in reduced neutralisation by therapeutic antibodies and vaccine-induced immune serum samples;6 this is in line with the reduced clinical efficacies of the currently authorised vaccines.7 Although new vaccines have been developed against these variants (eg, mRNA-1273.351 produced by Moderna, Cambridge, MA, USA; NCT04785144), SARS-CoV-2 variants might rapidly evolve into escape variants through mutations in the RBD.
Research in context.
Evidence before this study
We searched PubMed for research articles published from the inception of the database to Nov 9, 2021, using the terms “COVID-19” or “SARS-CoV-2”, “vaccine”, and “clinical trial”. No language or data restrictions were applied. We also searched the ClinicalTrials.gov registry and WHO draft landscape of COVID-19 candidate vaccines for ongoing trials of COVID-19 vaccines from the inception of both databases to Nov 9, 2021. 15 DNA-based vaccines, including the vaccines reported here, are in ongoing clinical trials. Four of which are in phase 3 clinical trials. Of these, safety and immunogenicity results were reported from only two phase 1 trials of DNA vaccines against SARS-CoV-2 (INO-4800 and ZyCoV-D). INO-4800 and ZyCoV-D targeted the spike protein of SARS-CoV-2. Both had favourable safety and tolerability and were immunogenic, eliciting humoral or cellular immune responses.
Added value of this study
These are the first in human phase 1 trials of recombinant DNA vaccines for COVID-19 containing the coding regions of the spike protein or the spike and nucleocapsid proteins in healthy adults. The trials showed that GX-19 and GX-19N are safe and well tolerated, and GX-19N can induce both humoral and cellular responses. A two-dose vaccination of 3·0 mg GX-19N (on days 1 and 29) induced significant humoral and cellular responses. The neutralising geometric mean titres in individuals vaccinated with GX-19N were lower than those of human convalescent serum samples. However, the GX-19N group showed increased T-cell responses, similar to those analysed using convalescent peripheral blood mononuclear cells that were drawn from the convalescent patients but with older age distribution. Furthermore, GX-19N induced both SARS-CoV-2 spike protein-specific T-cell responses and broad nucleocapsid protein-specific T-cell responses, which were also specific to other SARS-CoV-2 variants.
Implications of all the available evidence
GX-19N contains a plasmid encoding both the spike and nucleocapsid proteins. It showed broad SARS-CoV-2-specific T-cell responses, which might allow cross-reactivity with emerging SARS-CoV-2 variants. Because the antibody responses induced by GX-19N were shown to be probably weaker than those induced by commercial vaccines, a strategy to strengthen its immunogenicity is required if vaccine-induced T-cell response is not enough for protection against SARS-CoV-2. On the basis of these safety and immunogenicity findings, GX-19N was selected for phase 2 immunogenicity trials.
GX-19 is a candidate recombinant DNA vaccine that contains a plasmid encoding SARS-CoV-2 spike proteins S1 and S2.8 The plasmid vector (pGX27) used for the human papillomavirus DNA vaccine undergoing clinical trials was used to develop GX-19.9 In our preclinical trials, we developed five prototype DNA vaccines expressing the SARS-CoV-2 spike protein: 1) S1S2full (GX-19F), 2) S1S2ΔTM/IC (GX-19A), 3) S1 (GX-19B), 4) S1S2ΔTM/IC with the CD40 ligand (GX-19C), and 5) S1 with the CD40 ligand (GX-19D; appendix p 2). On the basis of vaccine-induced immune responses observed for some of these constructs,8 binding and neutralisation antibody responses, and interferon-γ (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assay responses evaluated in mice and monkeys, we selected GX-19A (henceforth referred to as GX-19) as the prototype vaccine for the clinical trial (appendix pp 3–4).
During the phase 1 clinical trial with GX-19, T-cell immunity was identified as an important factor for COVID-19 disease protection.10, 11, 12, 13, 14 Because the nucleocapsid protein is more conserved and stable than the spike protein,15 and strong nucleocapsid protein-specific T-cell responses were observed in patients with COVID-19,16 the nucleocapsid protein gene was chosen to be added to the vaccine to induce T-cell response with wider coverage against emerging variants.17 We generated GX-19N, a next generation DNA vaccine that contained a 1:2 mix ratio of GX-19 (1 mg) and GX-21 (2 mg; a construct expressing the RBD protein fused with the T4 fibritin C-terminal foldon and nucleocapsid protein; appendix pp 2, 5–6). These vaccines have been evaluated in mouse and monkey models in preclinical studies that showed higher neutralising antibody responses, and cellular immune responses were expected following vaccination with GX-19N (appendix p 7). We aimed to assess the safety and immunogenicity of GX-19 and GX-19N in humans up to 57 days after the first vaccine dose.
Methods
Study design and participants
We did two open-label, non-randomised phase 1 trials to evaluate the safety, tolerability, and immunogenicity of GX-19 and GX-19N after intramuscular vaccination in healthy adults at two hospitals in Seoul, South Korea: Severance Hospital and Gangnam Severance Hospital. The GX-19 trial was a dose-escalation study done before the GX-19N trial.
Healthy adult participants aged 19–49 years were included in the GX-19 trial and healthy adults aged 19–54 years were included in the GX-19N trial; participants were recruited through local advertisements. Full details of the eligibility criteria are described in the GX-19 and GX-19N trial protocols (appendix pp 8–12), in brief patients with immune dysfunction and those who tested positive by serological testing for SARS-CoV-2 were excluded. We report protocol permitted interim analyses with the results until 57 days after the first vaccination, based on which the decision on whether to proceed to phase 2 trials will be made. Written informed consent was obtained from all participants. The trials were done according to the principles of the Declaration of Helsinki and Good Clinical Practice. This study was approved in South Korea by the Korea Ministry of Food and Drug Safety (reference 20200261849) and the Institutional Review Board of Severance Hospital (4-2020-1220).
Procedures
The vaccines were produced in accordance with the current good manufacturing practices and supplied by BINEX, Incheon, South Korea, a contract manufacturing organisation. The vaccines were delivered using an electroporator (Elimtek, Seongnam-si, Korea) to maximise cellular DNA uptake by generating a high-voltage pulse after drug injection.
Participants in the GX-19 trial were assigned (1:1:1) to receive a 1·5 mg, 3·0 mg, or 4·0 mg doses; they received two injections of the trial vaccine 4 weeks apart. Participants in the 4·0 mg group received the vaccines using a needle-free injector. Due to the difference in vaccine delivery device, only the interim results in the 1·5 mg and 3·0 mg dose groups are reported here. The GX-19N trial was subsequently done on the basis of results obtained in the GX-19 trial. Briefly, the 3·0 mg dosing regimen exhibited excellent efficacy in terms of safety and immunogenicity compared with the 1·5 mg regimen in the GX-19 phase 1 trial, as observed in the preclinical results of GX-19N (appendix p 13). Therefore, we selected a dose of 3·0 mg to expedite the clinical development of GX-19N.
In the GX-19 trial, participants were first enrolled in the 1·5 mg (0·38 mL) group. After safety and tolerability data up to 7 days after vaccination from the first three participants in this group were reviewed by the independent data safety monitoring committee, participants were enrolled in the 3·0 mg (0·75 ml) group. In the GX-19N trial, all participants were enrolled in a single dose group to receive 3·0 mg (0·75 mL) GX-19N. In both trials, the vaccines were administered intramuscularly into the deltoid muscle on days 1 and 29, and the participants were followed up for 52 weeks following their first vaccination. To assess the incidence of adverse events, daily follow-up by telephone was done for up to 7 days after the first vaccination, and on the eighth day safety was assessed by site visits.
Follow-up visits were scheduled on days 8, 29, 43, and 57, and weeks 24 and 52 after first vaccination during which safety data were collected. The participants were instructed to list any adverse events over approximately 4 weeks in a diary distributed at the time of the first and second vaccination. To assess immunogenicity for this interim analysis report, blood was sampled at baseline, and at 29, 43, and 57 days after first vaccination (appendix pp 14–17).
Local and systemic reactions after each vaccination were monitored for 28 days. Serious adverse events were recorded throughout the follow-up period. The solicited adverse events reported were graded according to guidelines issues by the South Korean Ministry of Food and Drug Safety. The severity of unsolicited adverse events was assessed according to Common Terminology Criteria for Adverse Events (version 5.0) and Medical Dictionary for Regulatory Activities (version 24.0) classification and relatedness to the vaccine. Adverse events of special interest—including acute respiratory distress syndrome, pneumonitis, enhanced disease following immunisation, acute cardiac injury, arrhythmia, septic shock-like syndrome, and acute kidney injury—were also assessed. Laboratory adverse events were graded using site-specific toxicity tables, adapted from the South Korean Ministry of Food and Drug Safety guidelines for assessing the severity of adverse events in vaccine clinical trials.
Binding antibody responses against the SARS-CoV-2 spike and RBD proteins before the vaccination and on days 43 and 57 after first vaccination were assessed by ELISA. The ELISA kits were developed in-house for the spike protein and purchased from Bionote (Hwaseong-si, South Korea) for the RBD protein (appendix p 18). Neutralising antibody responses were assessed only in the 3·0 mg GX-19N group based on the results obtained in the binding antibody responses. Vaccine-induced neutralising activity was assessed using live wild-type SARS-CoV-2 (Wuhan-hu-1 [SARS-CoV-2/human/KOR/KCDC03-NCCP43326/2020] from the National Culture Collection for Pathogen, Cheongju-si, South Korea); a focus reduction neutralisation testing (FRNT) assay; and a SARS-CoV-2 S-pseudotyped murine leukaemia virus retrovirus-based single round of infection neutralisation assay (PsVNA; appendix pp 18–19). The 50% neutralisation titres were reported as the interpolated reciprocal of the dilutions yielding 50% reductions in viral foci. The FRNT assay was done on serum samples collected before and on days 43 and 57 after first vaccination, whereas PsVNA was done before and on day 43 after first vaccination. T-cell responses were evaluated before and on day 43 after first vaccination with a direct ex-vivo IFN-γ ELISPOT assay with peripheral blood mononuclear cells. In these assays, peripheral blood mononuclear cells were stimulated overnight with overlapping peptides encoding the full length sequences of the spike protein (for GX-19) and the spike and nucleocapsid proteins (for GX-19N; appendix pp 19–20). To selectively evaluate vaccine-induced SARS-CoV-2-specific T-cell responses in participants who did not show enhanced T-cell responses based on the results of the IFN-γ ELISPOT assay, the T-SPOT assay (Oxford Immunotec, Abingdon, UK), which can measure the SARS-CoV-2-specific T-cell response (not specific to other betacoronaviruses), was used. For this assay, the peripheral blood mononuclear cells were stimulated with overlapping peptides spanning the spike and nucleocapsid proteins from which specific sequences with high homology to endemic (non-SARS-CoV-2) coronaviruses had been removed. We also examined whether the alpha (B.1.1.7), beta (B.1.351), gamma (p.1), delta (B.1.617.2), and lambda (C·37) SARS-CoV-2 variants displayed any mutations in the T-cell epitope sequences recognised by GX-19N-induced nucleocapsid protein-specific T cells. To identify 15-mer peptides containing nucleocapsid protein-specific minimal epitopes in participants who received GX-19N-vaccination, we used the IFN-γ ELISPOT assay with a matrix of overlapping peptides spanning the entire nucleocapsid protein or 15-mer individual peptides with peripheral blood mononuclear cells from participants who exhibited strong nucleocapsid protein-specific T-cell responses following GX-19N immunisation. Subsequently, the amino acids sequences of GX-19N and those of SARS-CoV-2 variants were compared.
To compare vaccine-induced neutralising antibody and cellular responses with those induced in response to natural SARS-CoV-2 infection, we analysed blood specimens collected from a separate group of convalescent patients within 3 months of PCR-based diagnosis of COVID-19; these patients had consented to use of these samples for comprehensive research purposes. FRNT or PsVNA and ELISPOT assays were used to evaluate the neutralising antibody and T-cell responses.
Outcomes
The primary outcome was the safety and tolerability of the vaccines based on the frequency, characteristics, and severity of adverse events within 28 days of each vaccination. The secondary outcome was the immunogenicity of the vaccines. Immunogenicity endpoints in this interim analysis included changes in SARS-CoV-2 spike protein and RBD specific binding antibody concentrations compared with baseline concentrations within 57 days in all participants in both the GX-19 and GX-19N trials (on days 1, 43, 57 days after first vaccination) and changes in SARS-CoV-2 neutralising antibody concentrations within 57 days in participants in GX-19N trial (on days 1, 43, and 57 for the FRNT assay and days 1 and 43 after first vaccination for PsVNA). Immunogenicity endpoints also included determining the SARS-CoV-2-specific T-cell responses of GX-19 and GX-19N measured on days 1 and 43 after first vaccination. Some of the outcomes of the full trial were not examined in this interim analysis, including immune cell proportion, immunophenotype, and functional analysis of antigen-specific CD8 T cells after GX-19N vaccination.
Statistical analyses
Continuous variables were assessed using descriptive statistics, and categorical variables are presented as the frequency and percentage, with a 95% two-sided CI when appropriate. When theoretical hypothesis testing was required, parametric methods with normal distribution and non-parametric methods with non-normal distribution were used for continuous values. χ2 and Fisher's exact tests were used for categorical values. Student's t-test, Wilcoxon matched-pairs signed-rank test, and Mann–Whitney's U test were used to compare the continuous values. If missing values occurred at a certain point or the participant discontinued before completing the study the available raw data were used in the analysis. We did not determine the sample size based on a statistical power calculation because the study was exploratory and descriptive in nature and did not involve the testing of statistical hypotheses.
For the safety assessment, adverse events are presented as the number and percentage of participants who had adverse events and the number of actual cases with a 95% CI. Adverse events were classified by type, severity, and causality for each group, and the frequency and percentage of each were calculated.
For immunological analysis, the geometric mean titres (GMTs) of binding and neutralising antibodies before and after vaccination were assessed and a 95% CI was presented. The proportion of participants with an increase in their binding and neutralising antibodies titres of four-times or more compared with the baseline concentrations were also presented. The protocol immunological outcomes were meant to be measured as seroconversion rate and geometric mean fold rise. However, the results of other SARS-CoV-2 vaccine trials published during the study period in which the GX-19 and GX-19N trials were done showed that the definition of seroconversion or antibody responder varied from study to study. Referring to previous vaccine studies,18, 19, 20 we decided not to use seroconversion rate or geometric mean fold rise as defined in the protocol in this interim analysis. T-cell responses were expressed as the number of spot-forming units per 106 cells measured using IFN-γ ELISPOT or T-SPOT assays before and after vaccination. The participants with at least a two-times increase in the number of spots or more than 100 spots per 106 peripheral blood mononuclear cells increase in IFN-γ ELISPOT assay on day 43 compared with day 1 were regarded as responders. Post-hoc analyses were done to compare neutralising antibody responses and T-cell responses with those of patients with COVID-19 who were convalescent. Analyses were done using IBM SPSS Statistics (version 22.0) and GraphPad Prism (version 8.4.1), and 95% CIs were calculated using the Clopper-Pearson method; p values less than 0·05 were considered significant. The GX-19 (NCT044445389) and GX-19N (NCT04715997) trials are registered with ClinicalTrials.gov.
Role of the funding source
The funder of the study had no role in data collection and analysis, the interpretation of the findings, writing of the report, or study design.
Results
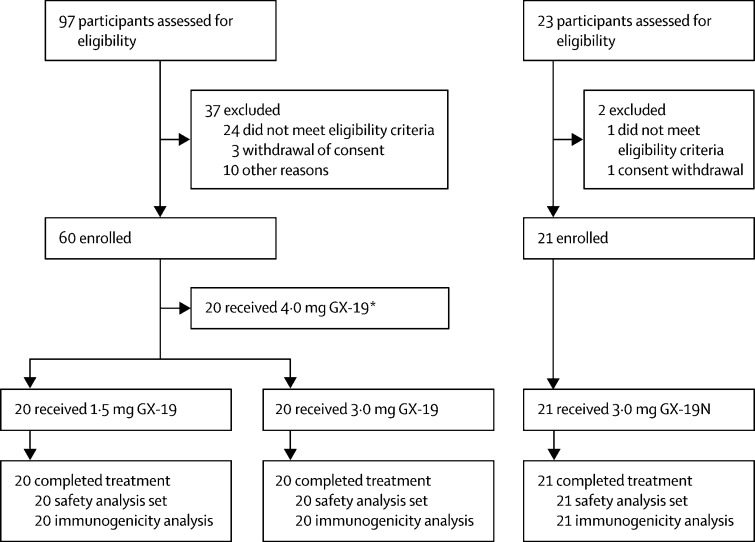
Between June 17, and July 30, 2020, 97 individuals were screened for inclusion in the GX-19 trial, of whom 40 (41%) were enrolled on the trial. 20 (50%) individuals received two doses of 1·5 mg GX-19 and 20 received two doses of 3·0 mg GX-19 (50%). Between Dec 28 and 31, 2020, 23 individuals were screened for inclusion in the GX-19N trial, of whom 21 (91%) were enrolled and received two doses of 3·0 mg GX-19N (figure 1 ).
Figure 1.
Trial flow diagram
*Data not reported in this study.
All 61 (100%) participants across the two trials received their second vaccine dose, continued to attend the scheduled trial visits until day 57, and were included in both the safety and immunogenicity analyses. Similar demographic characteristics were observed in all three groups (table 1 ). Across both trials mean age was 37·4 years (SD 8·3); 29 (48%) participants were female and 32 (52%) were male. All participants were Korean. The demographic characteristics of 54 patients with COVID-19 who provided convalescent blood specimens are reported in table 1.
Table 1.
Descriptive statistics of demographic characteristics
| GX-19 1·5 mg group (n=20) | GX-19 3·0 mg group (n=20) | GX-19N 3·0 mg group (n=21) | Patients with COVID-19*(n=28) | Patients with COVID-19†(n=20) | Patients with COVID-19‡(n=6) | ||
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Male | 10 (50%) | 13 (65%) | 9 (43%) | 26 (93%) | 13 (65%) | 1 (17%) | |
| Female | 10 (50%) | 7 (35%) | 12 (57%) | 2 (7%) | 7 (35%) | 5 (83%) | |
| Ethnicity | |||||||
| Asian | 20 (100%) | 20 (100%) | 21 (100%) | 28 (100%) | 20 (100%) | 6 (100%) | |
| Age, years | 35·5 (7·7) | 37·3 (8·8) | 39·3 (8·3) | 42·6 (15·6) | 66·7 (10·7) | 65·2 (8·5) | |
| Baseline height, cm | 165·5 (7·0) | 169·3 (8·2) | 169·6 (7·0) | .. | .. | .. | |
| Baseline weight, kg | 66·9 (7·8) | 70·6 (11·6) | 67·8 (10·7) | .. | .. | .. | |
| Baseline BMI, kg/m2 | 24·4 (2·0) | 24·5 (2·3) | 23·6 (2·5) | 24·6 (1·8)§ | 24·1 (4·6)¶ | 27·1 (4·0)‖ | |
| Childbearing potential | 10 (50%) | 6 (30%) | 12 (57%) | 0 | 0 | 0 | |
| Smoking history | |||||||
| Non-smoker | 14 (70%) | 15 (75%) | 11 (52%) | .. | .. | .. | |
| Former smoker | 6 (30%) | 5 (25%) | 10 (48%) | .. | .. | .. | |
| Alcohol history | |||||||
| No | 11 (55%) | 11 (55%) | 3 (14%) | .. | .. | .. | |
| Ex-drinker** | 9 (45%) | 9 (45%) | 18 (86%) | .. | .. | .. | |
| No history of drug use | 20 (100%) | 20 (100%) | 21 (100%) | .. | .. | .. | |
| From COVID-19 diagnosis to sampling, days | .. | .. | .. | 59 (48–77) | 42 (39–50) | 32 (27–38) | |
| Score on ordinal scale†† | |||||||
| 4–5 | .. | .. | .. | 25 (89%) | 8 (40%) | 4 (67%) | |
| 6 | .. | .. | .. | 1 (4%) | 8 (40%) | 0 | |
| 7 | .. | .. | .. | 2 (7%) | 4 (20%) | 2 (33%) | |
Data are n (%), Mean (SD), or Median (IQR). BMI=body-mass index.
Convalescent patients with COVID-19 provided plasma for the comparison of neutralising response in the focus reduction neutralisation test.
Convalescent patients with COVID-19 provided plasma for the comparison of neutralising response in the pseudovirus neutralisation assay.
Convalescent patients with COVID-19 provided peripheral blood mononuclear cells for evaluating the T cell response in IFN-γ enzyme-linked immunosorbent spot assay.
n=four (14%) of 28 patients.
n=20 (100%) of 20 patients.
n=six (100%) of six patients.
Participants who had ever drank alcohol, but had stopped alcohol consumption at least 3 months before the study.
National Institute of Allergy and Infectious Diseases ordinal scale was used; the ordinal score is the patient's worst score during COVID-19; scores on the ordinal scale are as follows: 1=not hospitalised, no limitations of activities; 2=not hospitalised, limitation of activities, home oxygen requirement, or both; 3=hospitalised, not requiring supplemental oxygen and no longer requiring ongoing medical care (used if hospitalisation was extended for infection-control reasons); 4=hospitalised, not requiring supplemental oxygen but requiring ongoing medical care (COVID-19–related or other medical conditions); 5=hospitalised, requiring any supplemental oxygen; 6=hospitalised, requiring non-invasive ventilation or use of high-flow oxygen devices; and 7=hospitalised, receiving invasive mechanical ventilation or extracorporeal membrane oxygenation.
32 (52%) participants reported 80 treatment-emergent adverse events after vaccination—12 (60%) of 20 participants from the 1·5 mg GX-19 group, eight (40%) of 20 participants from the 3·0 mg GX-19 group, and 12 (57%) of 21 participants from the 3·0 mg GX-19N group (table 2 ). 72 (90%) of the 80 adverse events were mild; six (8%) events were moderate, and two (2%) were severe. None of the participants had serious drug-related adverse events or adverse events of special interest (appendix p 21). None of the participants discontinued the trial due to adverse events.
Table 2.
Incidence of treatment-emergent adverse events by system organ class and preferred term
| GX-19 1·5 mg group (n=20) | GX-19 3·0 mg group (n=20) | GX-19N 3·0 mg group (n=21) | |
|---|---|---|---|
| Participants with treatment-emergent adverse events | 12 (60%) | 8 (40%) | 12 (57%) |
| General disorders and administration site conditions | 11 (55%) | 4 (20%) | 6 (29%) |
| Chills | 0 | 0 | 1 (5%) |
| Fatigue | 3 (15%) | 0 | 0 |
| Injection site erythema | 2 (10%) | 0 | 1 (5%) |
| Injection site oedema | 1 (5%) | 0 | 0 |
| Injection site pain | 6 (30%) | 3 (15%) | 6 (29%) |
| Injection site pruritus | 5 (25%) | 2 (10%) | 0 |
| Nervous system disorders | 4 (20%) | 0 | 2 (10%) |
| Cubital tunnel syndrome | 0 | 0 | 1 (5%) |
| Headache | 3 (15%) | 0 | 1 (5%) |
| Paresthesia | 1 (5%) | 0 | 0 |
| Musculoskeletal and connective tissue disorders | 1 (5%) | 1 (5%) | 2 (10%) |
| Back pain | 0 | 0 | 1 (5%) |
| Myalgia | 1 (5%) | 0 | 1 (5%) |
| Rotator cuff syndrome | 0 | 1 (5%) | 0 |
| Somatic dysfunction | 0 | 1 (5%) | 0 |
| Gastrointestinal disorders | 2 (10%) | 1 (5%) | 0 |
| Dyspepsia | 1 (5%) | 0 | 0 |
| Haematochezia | 1 (5%) | 0 | 0 |
| Stomatitis | 0 | 1 (5%) | 0 |
| Skin and subcutaneous tissue disorders | 0 | 1 (5%) | 3 (14%) |
| Dermatitis | 0 | 1 (5%) | 0 |
| Pruritus | 0 | 0 | 1 (5%) |
| Rash | 0 | 0 | 2 (10%) |
| Urticaria | 0 | 0 | 1 (5%) |
| Infections and infestations | 0 | 2 (10%) | 1 (5%) |
| Appendicitis | 0 | 1 (5%) | 0 |
| Folliculitis | 0 | 0 | 1 (5%) |
| Vaginal infection | 0 | 1 (5%) | 0 |
| Investigations | 0 | 0 | 1 (5%) |
| Alanine aminotransferase increased | 0 | 0 | 1 (5%) |
| Aspartate aminotransferase increased | 0 | 0 | 1 (5%) |
| Reproductive system and breast disorders | 0 | 2 (10%) | 1 (5%) |
| Dysmenorrhoea | 0 | 1 (5%) | 1 (5%) |
| Premenstrual syndrome | 0 | 1 (5%) | 0 |
| Hepatobiliary disorders | 0 | 0 | 1 (5%) |
| Cholecystitis acute | 0 | 0 | 1 (5%) |
| Psychiatric disorders | 1 (5%) | 0 | 0 |
| Depression | 1 (5%) | 0 | 0 |
| Eye disorders | 1 (5%) | 1 (5%) | 0 |
| Blepharospasm | 0 | 1 (5%) | 0 |
| Visual impairment | 1 (5%) | 0 | 0 |
Adverse events defined according to Medical Dictionary for Regulatory Activities (version 24.0).
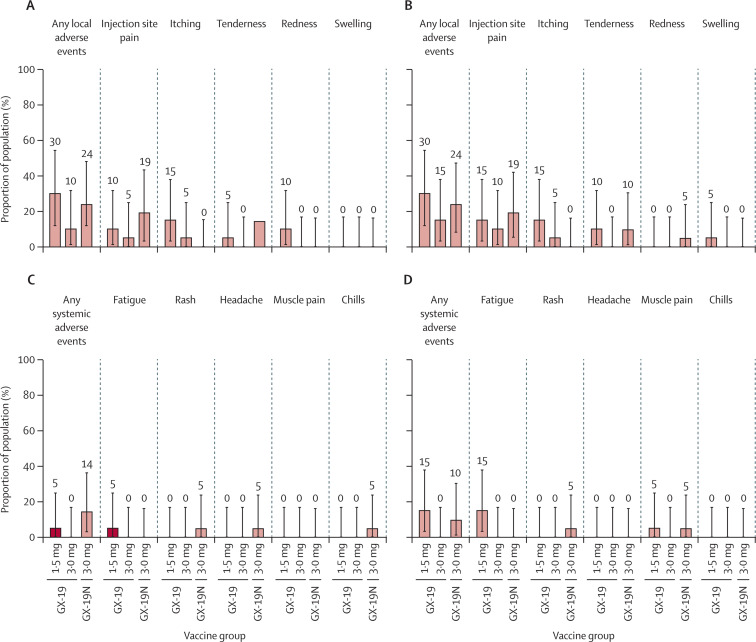
The first vaccination caused local adverse events in six (30%) of 20 participants in the 1·5 mg GX-19 group, two (10%) of 20 participants in the 3·0 mg GX-19 group, and five (24%) of 21 participants in the 3·0 mg GX-19N group. After both doses, solicited local adverse events were reported by six (30%) participants in the 1·5 mg GX-19 group, three (15%) in the 3·0 mg GX-19 group, and five (24%) in the 3·0 mg GX-19N group. Solicited systemic adverse events were reported by one (5%) participant in the 1·5 mg GX-19 group and two (10%) in the 3·0 mg GX-19N group after first vaccination; and by three (15%) participants in the 1·5 mg GX-19 group and two (10%) in the 3·0 mg GX-19N group after both doses. There were no reported solicited systemic adverse events in the 3·0 mg GX-19 group. The most common solicited adverse events were injection site pain, itching, tenderness, and fatigue (figure 2 ). All solicited adverse events developed within 2 days of vaccination and most events resolved within 3 days of onset. All solicited adverse events were mild except for one (2%) case of moderate fatigue reported in the 1·5 mg GX-19 group. There was no increase in the number of solicited adverse events related to the vaccine dose nor was there an increase in the frequency of adverse events with the second vaccination compared with those observed after the first vaccination in all groups (figure 2).
Figure 2.
Solicited adverse events reported after vaccination
The percentage of participants with adverse events during the 28-day post-vaccination period in each vaccine group is plotted for solicited local adverse events after dose one (A) and dose two (B) and systemic adverse events after dose one (C) and dose two (D). There were no severe events. Error bars are 95% CIs.
28 unsolicited adverse events were reported by 16 (26%) participants. 22 (79%) events were mild and they distributed similarly across the groups (four [25%] participants in the 1·5 mg GX-19 group, six [38%] in the 3·0 mg GX-19 group, and six [38%] in the 3·0 mg GX-19N group). However, two severe unsolicited adverse events were reported: one case of acute appendicitis in the 3·0 mg GX-19 group and one case of acute cholecystitis in the 3·0 mg GX-19N group, but these events were unlikely to be related to the vaccine. A vaccine-related adverse event (transient mild paresthesia) was reported by one (1%) participant in the 1·5 mg GX-19 group. Changes in laboratory variables (mild and transient elevation in aspartate aminotransferase and alanine aminotransferase concentrations) were observed in one (1%) participant in the 3·0 mg GX-19N group.
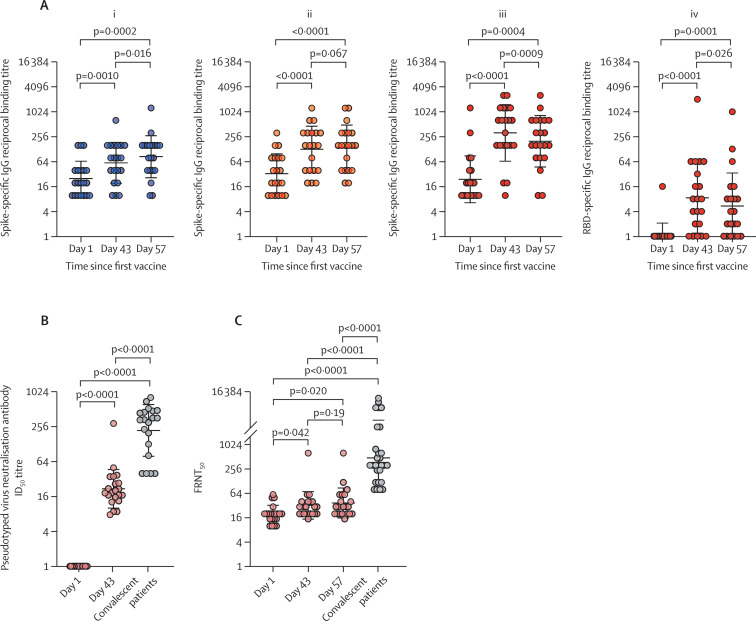
The GMTs of binding antibodies to the spike protein increased after first vaccination by day 57 in both the 1·5 mg GX-19 and 3·0 mg GX-19 groups (figure 3 ). In the GX-19N group, the GMT of antibody responses to the spike protein increased from 24·38 (95% CI 13·49–44·05) at baseline to 201·59 (15·32–383·83) at day 57 (p=0·0004). The GMT of antibodies to RBD proteins also increased from 1·14 (0·87–1·50) at baseline to 5·38 (2·33–12·45) at day 57 (p=0·0001). Of note, the GMTs for the spike protein decreased from 320·00 (95% CI 156·93–652·53) at day 43 to 201·59 (105·32–385·83) at day 57 (p=0·0009). The GMT of antibodies for the RBD protein also decreased from 8·55 (3·48–10·97) at day 43 to 5·38 (2·33–12·45) at day 57 (p=0·026; figure 3A; appendix p 22). The GMTs of spike protein binding antibodies on day 57 were 85·74 (95% CI 49·67–148·00) in the 1·5 mg GX-19 group, 144·20 (80·81–257·32) in the 3·0 mg GX-19 group, and 201·59 (105·32–385·83) in the 3·0 mg GX-19N group. The proportions of participants with an increase in spike protein or RBD protein antibody concentrations after vaccination by four times or more were 11 (55%) of 20 participants in the 1·5 mg GX-19 group, 13 (65%) of 20 in the 3·0 mg GX-19 group, and 17 (81%) of 21 in the 3·0 mg GX-19N group.
Figure 3.
Vaccine induced binding and neutralisation antibody responses
(A) SARS-CoV-2 spike-specific and RBD-specific IgG antibodies measured by ELISA for the 1·5 mg (i) and 3·0 mg (ii) groups of the GX-19 trial and SARS-CoV-2 spike-specific (iii) and RBD-specific (iv) IgG antibodies measured by ELISA of the GX19N trial. GX-19N-induced neutralisation antibody responses were determined by pseudotyped virus neutralization assay (B) and FRNT (C). Each circle indicates the reciprocal binding antibody titre, ID50 or FRNT50 titre of each serum sample. The p-values were calculated with the Wilcoxon matched-pairs signed-rank test. FRNT=focus reduction neutralisation. test ID50=50% inhibitory dilution.
On the basis of binding antibody responses, neutralisation responses were only analysed in the 3·0 mg GX-19N group. No participant had detectable PsVNA responses before the vaccination; however, a significant increase in PsVNA responses was observed after vaccination. The GMTs of the 50% inhibitory dilution (ID50) values in PsVNA were 1·00 (95% CI 1·00–1·00) before the first vaccination and 21·87 (15·30–31·07) on day 43. An increase in the 50% neutralisation titre by four-times or more was observed in all participants with PsVNA. However, the GMT of the ID50 values on day 43 were lower (p<0·0001) than that of the serum samples from 20 convalescent patients (GMT of 222·20 [95% CI 137·40–359·26]; figure 3B). Using FRNT, a significant increase in the neutralising antibody GMT post-vaccination was observed in the 3·0 mg GX-19N group by days 43 and 57 compared with day 1: neutralising antibody GMTs were 19·45 (95% CI 15·39–24·58) on day 1, 32·47 (22·71–46·41) on day 43, and 37·26 (25·29–54·90) on day 57. These corresponded to 16·91 IU/mL on day 43 and 19·41 IU/mL on day 57 (appendix p 23) using the WHO international standard serum samples (National Institute for Biological Standards and Control 20–136 [neutralising antibody titre 1000 IU/mL]) as a reference. Five (24%) of 21 participants had an increase in antibody concentrations by four times or more by day 57. The neutralising antibody GMTs by day 57 were significantly lower (p<0·0001) than those of the serum samples from 28 participants who were convalescent (GMT 487·45; figure 3C).
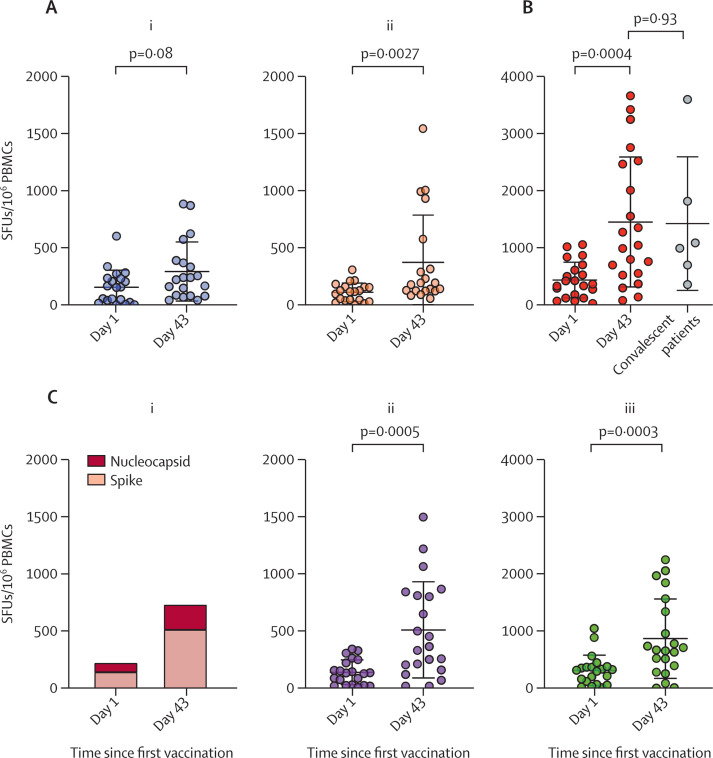
In the GX-19 trial, the 3·0 mg GX-19 group showed significantly enhanced spike protein-specific T-cell responses following vaccination (p=0·0027). Specifically, 11 (55%) of 20 participants in the 1·5 mg GX-19 group and 11 (55%) of 20 in the 3·0 mg GX-19 group had augmented spike protein-specific T-cell responses on day 43 (figure 4A ; appendix p 24). In the GX-19N trial, participants had significantly enhanced T-cell responses to spike protein, increasing from 274·90 SFU per 106 cells (95% CI 176·36–373·44) at baseline to 1016·53 (633·27–1399·78) at day 43 (p=0·0005). These participants also had an increased response to nucleocapsid protein: increasing from 156·07 (95·37–216·76) at baseline to 433·53 (95% CI 275·26–591·80) at day 43 (p=0·0003; (figures 4B and 4C; appendix p 24). 15 (71%) of 21 participants had vaccine-induced T-cell responses after vaccination (appendix p 25), and the magnitude of GX-19N-induced T-cell responses was similar to those observed in the convalescent peripheral blood mononuclear cells from six convalescent patients with COVID-19 (p=0·93; figure 4B). Furthermore, of the five (24%) participants (01SN001, 01SN004, 01SN006, 02SN008, and 02SN011) who did not show enhanced T-cell responses based on the results of the IFN-γ ELISPOT assay, the T-SPOT assay using peripheral blood mononuclear cells from four participants (01SN001, 01SN004, 01SN006, and 02SN008) showed that three participants had increased GX-19N-induced SARS-CoV-2 spike protein or nucleocapsid protein specific T-cell responses (appendix pp 26–27).
Figure 4.
Vaccine elicited T-cell responses to SARS-CoV-2 antigens
Antigen-specific T-cell responses on days 1 and 43 were measured by IFN-γ ELISPOT assay using PBMCs stimulated with overlapping peptides spanning the full length of the spike protein for GX-19 and the spike protein and nucleocapsid protein for GX-19N. Individual data points are shown as a dot plot with lines showing the mean with standard deviation. (A) Spike protein specific T-cell responses on days 1 and 43 in participants who received 1·5 mg (i) and 3·0 mg (ii) GX-19. (B) Sum of spike protein and nucleocapsid protein specific T-cell response in participants who received GX-19N. (C) Sum of spike protein and nucleocapsid protein specific (i), spike protein specific (ii), and nucleocapsid protein specific (iii) T-cell responses on days 1 and 43 in participants who received GX-19N. PBMCs=peripheral blood mononuclear cells. SFU=spot-forming-unit.
In four (19%) participants who had strong nucleocapsid protein-specific T-cell responses following GX-19N vaccination, the T-cell responses to multiple 15-mer peptides were induced, suggesting the induction of broad nucleocapsid protein-specific T-cell responses (appendix pp 26–27). We found that the amino acid sequences of 15-mer peptides containing T-cell epitopes identified in participants who received the GX-19N vaccine were identical (except for one 15-mer peptide) to those of SARS-CoV-2 variants (ie, alpha, beta, gamma, delta, and lambda; appendix pp 26–27).
Discussion
We report the findings from two phase 1 clinical trials of recombinant DNA vaccines—GX-19 and GX-19N—administered to healthy adults. Both were safe, and GX-19N induced significant humoral and cellular responses. The neutralising antibody GMTs in individuals vaccinated with GX-19N were lower than those who provided convalescent blood specimens. The GX-19N group showed increased T-cell responses and induced both SARS-CoV-2 spike protein-specific T-cell responses and broad nucleocapsid protein-specific T-cell responses, which were also specific to SARS-CoV-2 variants.
GX-19 and GX-19N were well tolerated and had an acceptable safety profile without serious vaccine-related adverse events. The overall incidence of solicited adverse events in this study was lower than that observed for the currently approved vaccines or vaccines authorised for emergency use against SARS-CoV-2.2, 3, 4 The safety profile in this study was similar to trials of another DNA vaccine candidate for COVID-19: INO-4800.21
The GX-19N group had stronger cellular responses than the GX-19 groups. Studies have suggested the role of SARS-CoV-2-specific T cells in reducing COVID-19 severity in patients with asymptomatic or mild COVID-19 in the absence of antibodies.10, 11, 12, 22 A non-human primate study suggests the contribution of CD8+ T cells on sterilising immunity.23 T-cell epitope vaccination in the absence of neutralising antibodies has been shown to protect mice from SARS-CoV-2 infection.24
Furthermore, GX-19N not only induced spike protein-specific T-cell responses, but it also induced broad nucleocapsid protein-specific T-cell responses, which were also specific to SARS-CoV-2 variants. A key feature of GX-19N is that it contains a plasmid encoding nucleocapsid protein, which is highly conserved (with >90% amino acid homology and fewer mutations over time) in diverse SARS-CoV-2 variants. To our knowledge, this is the first study reporting a phase 1 clinical trial of a SARS-CoV-2 DNA vaccine candidate that includes a non-spike protein-encoding DNA sequence. Although escaping neutralising antibody-based immunity is easily achieved via a few RBD mutations, escaping T cell-based immunity is far more complex because T-cell epitopes are more abundant throughout the whole antigen.14 Therefore, considering the growing evidence regarding the protective role of T cells against severe disease, GX-19N—which induces broad T-cell immunity—might be advantageous for long-term protection against current and future SARS-CoV-2 variants.
On the basis of the results of the binding and neutralising antibody response assay, GX-19N-induced antibody responses to SARS-CoV-2 antigens are probably weaker than those induced in response to natural COVID-19 infection or commercialised mRNA-based or adenoviral vector-based vaccines,18, 19, 20 although the immune responses are not directly compared in the same setting. Previous studies on DNA vaccines have determined that the number of doses required for effective vaccination is dependent on the antigen and virus types and how they interact with the immune system.25 One to two doses of the DNA vaccine are enough to elicit sufficient neutralising antibody responses to the influenza virus, whereas three to four doses are needed to produce protective humoral responses against HIV.26 In a preclinical study on a plasmid DNA vaccine for SARS-CoV-2, the magnitude of binding and neutralising antibody responses was the highest after three doses.25 Future studies on humoral immunogenicity after another booster dose of GX-19N are required.
GX-19N is required in larger amounts (3·0 mg per dose) than mRNA vaccines (up to 100 μg per dose), in addition to disposables and power supply for the electroporation-mediated delivery; these costs might influence the mass distribution of the vaccine. However, plasmid DNA is produced from Escherichia coli host cells, whereas mRNA is produced by in-vitro transcription and capping using enzymes before the purification.27 Thus, a high cultivation yield of transformed E coli and a simplified plasmid purification would enable the mass production of DNA vaccines. Because DNA vaccines are stable for several years under conditions of refrigeration (2–8°C), and for several months at room temperature, the mass transport and storage of DNA vaccines would not generate high cost and quality problems. In addition, portable electroporators are in development for the large-scale distribution of electroporation-adjuvanted DNA vaccine.
This study is a phase 1 interim analysis. We cannot report persistent safety outcomes or vaccine-induced immune responses, and the results do not permit a vaccine efficacy assessment. The interpretation of the results of this trial is also restricted by the open-label, non-randomised design and small population size. The participants in this trial were not ethnically diverse, and participants more susceptible to SARS-CoV-2, such as older individuals and those with comorbidities, were not included. Patients who donated the convalescent blood samples were not enrolled in this vaccine study, and variables that could affect the immune responses—including age distribution—were not controlled. However, this study has several strengths. To our knowledge, this is the first trial to assess the safety and immunogenicity of a DNA vaccine that encodes both the spike protein and the highly conserved nucleocapsid protein. Given the concerns regarding the decreased efficacy of the current COVID-19 vaccines against SARS-CoV-2 variants, we also did an additional detailed assessment of T-cell responses and analysed the reactivity of vaccine-induced T-cell responses to SARS-CoV-2 variants (appendix p 15).
Considering the reduction of neutralising antibody titres and the emergence of SARS-CoV-2 escape variants, the protection strategy of the vaccine involves relieving symptoms and preventing the occurrence of severe disease rather than preventing infection. Inequitable access to the vaccines currently approved or authorised for emergency use owing to the restricted supply for global demand is a crucial hurdle to establishing herd immunity, which can be resolved by long-lasting and broadly protective vaccines. The antibody responses induced by GX-19N were shown to be probably weaker than those induced by commercialised vaccines. Because antibody responses play a role in blocking viral entry, strategies for improving GX-19N-induced antibody responses are required for enhanced protection against COVID-19. This study shows that GX-19N induced broad and robust T-cell responses to spike and nucleocapsid proteins, which need to be evaluated for their symptom-preventing efficacy in countries where neutralising antibody-escape variants are widespread. On the basis of these safety and immunogenicity findings, the GX-19 trial was discontinued and GX-19N was selected as a vaccine candidate for phase 2 immunogenicity trials (NCT04715997).
This online publication has been corrected. The corrected version first appeared at thelancet.com/microbe on April 12, 2022
Data sharing
Deidentified participant data will be made available when the trial is complete in June, 2022, upon request directed to the corresponding authors. Study protocols and raw immunoassay data can be shared via proposal sent to the corresponding authors.
Declaration of interests
YSS, Y-JC, JWY, JWW, and YCS are employees and stakeholders of Genexine. All other authors declare no competing interests. YSS, Y-JC, JWY, JWW, and YCS report grants from Korea Drug Development Fund during the conduct of the study. YCS has a patent (10-2020-0176874) issued for a novel vaccine composition for preventing and treating coronavirus.
Acknowledgments
Acknowledgments
This research was supported by the Korea Drug Development Fund funded by the Ministry of Science and ICT, Ministry of Trade, Industry, and Energy, and Ministry of Health and Welfare (HQ20C0016, South Korea). The pathogen resources (NCCP43326) for this study were provided by the National Culture Collection for Pathogens. We thank the study participants, members of the trial management groups, site research staff, and trial steering committee.
Contributors
JYC, S-HP, and YCS were the coprincipal investigators of this trial. JYC, S-HP, and YCS conceptualised the trial and led the research. JYA, JL, and YSS were the cofirst authors of this manuscript and wrote the original draft. JWW conceptualised the trial and study protocol. YGS, KHL, and JYC recruited participants, collected data, and did follow-up assessments. MS, E-CS, and JWW verified the data. JYA, Y-JC, and JYC did the safety analysis and interpreted the data. JL, SHS, J-WO, H-YS, MK, J-EK, JWY, and E-CS did the immunogenicity tests. JYA, JL, YSS, J-WO, JWW, E-CS, S-HP, and JYC analysed the immunogenicity data. All authors contributed to manuscript reviewing and editing and approved the final version. All authors contributed to article preparation and the decision to submit for publication. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Material
References
- 1.WHO COVID-19 vaccine tracker and landscape. 2021. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- 2.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 7.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N Engl J Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo YB, Suh YS, Ryu JI, et al. Soluble spike DNA vaccine provides long-term protective immunity against SARS-CoV-2 in mice and nonhuman primates. Vaccines (Basel) 2021;9:307. doi: 10.3390/vaccines9040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi YJ, Hur SY, Kim TJ, et al. A phase II, prospective, randomized, multicenter, open-label study of GX-188E, an HPV DNA vaccine, in patients with cervical intraepithelial neoplasia 3. Clin Cancer Res. 2020;26:1616–1623. doi: 10.1158/1078-0432.CCR-19-1513. [DOI] [PubMed] [Google Scholar]
- 10.Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Candia P, Prattichizzo F, Garavelli S, Matarese G. T cells: warriors of SARS-CoV-2 infection. Trends Immunol. 2021;42:18–30. doi: 10.1016/j.it.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarke A, Sidney J, Kidd CK, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J Virol. 2020;94:e00647–e00720. doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y, Mentzer AJ, Liu G, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbott TR, Dhamdhere G, Liu Y, et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrade VM, Christensen-Quick A, Agnes J, et al. INO-4800 DNA vaccine induces neutralizing antibodies and T cell activity against global SARS-CoV-2 variants. NPJ Vaccines. 2021;6:121. doi: 10.1038/s41541-021-00384-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang A, Bange E, Han N, et al. CD8 T cells compensate for impaired humoral immunity in COVID-19 patients with hematologic cancer. Res Sq. 2021 doi: 10.21203/rs.3.rs-162289/v1. published online Feb 2. (preprint). [DOI] [Google Scholar]
- 20.McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang Z, Lai X, Sun J, et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J Exp Med. 2021;218 doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulligan MJ, Lyke KE, Kitchin N, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 24.Folegatti PM, Ewer KJ, Aley PK, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almansour I, Macadato NC, Alshammari T. Immunogenicity of multiple doses of pDNA vaccines against SARS-CoV-2. Pharmaceuticals. 2021;14:39. doi: 10.3390/ph14010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Lu S. DNA immunization. Curr Protoc Microbiol. 2013;31:18.3.1–18.3.24. doi: 10.1002/9780471729259.mc1803s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: challenges and bottlenecks. Vaccine. 2021;39:2190–2200. doi: 10.1016/j.vaccine.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified participant data will be made available when the trial is complete in June, 2022, upon request directed to the corresponding authors. Study protocols and raw immunoassay data can be shared via proposal sent to the corresponding authors.