Abstract
Metabolic adaptations occur with aging but the significance and causal roles of such changes are only partially known. In Drosophila, we find that skeletal muscle aging is paradoxically characterized by increased readouts of glycolysis (lactate, NADH/NAD+) but reduced expression of most glycolytic enzymes. This conundrum is explained by lactate dehydrogenase (LDH), an enzyme necessary for anaerobic glycolysis and whose expression increases with aging. Experimental Ldh overexpression in skeletal muscle of young flies increases glycolysis and shortens life span, suggesting that age-related increases in muscle LDH contribute to mortality. Similar results are also found with overexpression of other glycolytic enzymes (Pfrx/PFKFB, Pgi/GPI). Conversely, hypomorphic mutations in Ldh extend life span, whereas reduction in PFK, Pglym78/PGAM, Pgi/GPI, and Ald/ALDO levels shorten life span to various degrees, indicating that glycolysis needs to be tightly controlled for optimal aging. Altogether, these findings indicate a role for muscle LDH and glycolysis in aging.
Keywords: Aging, Drosophila, Glycolysis, LDH, Life span, Skeletal muscle
Aging is characterized by a number of changes that occur at the molecular, cellular, and organismal levels (1). Whereas some of these age-related changes do not affect the aging process itself, others are causal or conversely are adaptive stress responses that combat aging.
In addition to nutrient-sensing pathways, modulation of energy metabolism by environmental interventions and genetic mutations was found to regulate aging across species (2). For example, mutation of Enigma, an enzyme responsible for fatty acid beta oxidation, extends life span in Drosophila (3), and a plethora of studies has demonstrated that various types of dietary restriction can prevent age-related diseases across species (4,5).
Metabolic profiling of aging has highlighted several metabolites that are biomarkers of aging and that in some cases also modulate life span (6–8), as exemplified by the finding that suppressing the age-dependent accumulation of S-adenosyl-homocysteine promotes longevity in Drosophila (9). However, the significance and causal roles of age-related metabolic changes have been explored only in part.
Glucose and its metabolic utilization via glycolysis are among the most prominent metabolic pathways in animals. However, the impact of glycolysis on aging is incompletely understood. Some studies have shown that reducing glycolysis is beneficial for aging: Glucose restriction extends life span in Caenorhabditis elegans by inducing mitochondrial respiration and by increasing oxidative stress signaling (10,11). Moreover, supplementation of d-glucosamine, a glycolysis inhibitor, extends life span of mice and C. elegans (12). However, other studies suggest that glycolysis is needed for optimal aging. In Drosophila, it was found that aging leads to loss of epigenetic fidelity and a drift in H3K27me3 marks, which impairs the expression of glycolytic enzymes. Glycolysis appears necessary for optimal energy production and redox homeostasis and, consequently, it was found that stimulation of glycolysis promotes longevity (13). Moreover, glycolysis is upregulated and is neuroprotective in amyotrophic lateral sclerosis (14), and depletion of glycolytic enzymes induces senescence in cell culture (15). Altogether, these studies indicate that glycolysis has a complex impact on aging and age-related conditions and that this varies depending on the context and mode of intervention.
Aerobic glycolysis leads to the production of pyruvate in normoxia, whereas anaerobic glycolysis (ie, the transformation of glucose to lactate) is the primary means and glucose/pyruvate utilization in hypoxia. Although anaerobic glycolysis occurs pervasively during normal aging in the human brain and blood cells (16,17) and in C. elegans (18), there is limited understanding of the relative impact of aerobic versus anaerobic glycolysis on aging, especially with regard to skeletal muscle which is a major metabolic tissue in animals.
Skeletal muscle has high metabolic demand and key roles in life-span determination (18–21). Several metabolic pathways have been found to affect aging and life span when modulated specifically in skeletal muscle (22–25). For example, transgenic AMPK overexpression in skeletal muscle of adult flies extends life span while AMPK RNAi reduces it (26). Moreover, life-span extension due to dietary restriction depends, in Drosophila, on a shift toward increased fatty acid synthesis and breakdown in skeletal muscle (27). Although glucose sensing has been found to play key roles in muscle homeostasis (28), relatively little is known on the impact of muscle glycolysis on organismal aging.
Here, we have examined whether aging modulates muscle glycolysis, and whether experimental modulation of glycolysis from young age affects organismal aging in Drosophila. We find that skeletal muscle aging is characterized by increased activity of lactate dehydrogenase (LDH), and that muscle-specific Ldh overexpression from young age shortens life span. Moreover, we find that hypomorphic Ldh mutations extend life span. Altogether, these findings indicate that an age-related increase in muscle LDH decreases survival during aging.
Method
Fly Stocks
Fly stocks used in this study are reported in Table 1 and were obtained from the Transgenic Resource of the DPIM (Boston, MA, USA; https://interfly.med.harvard.edu/transgenic_info.php) and the Bloomington stock center (Bloomington, IN, USA; https://bdsc.indiana.edu/).
Table 1.
Fly Stocks Used
| Name | ID | Use |
|---|---|---|
| w 1118 | Lab collection | Reference strain |
| B3/ O1/ O3 | From Dr. Trudy Mackay | Comparison of Ldh expression |
| MhcF3-Gal4 (F3.580) | BL #38464 (RFP removed) | Adult skeletal muscle expression |
| Mhc-Gal4 | Lab collection | Adult skeletal muscle expression |
| Act5c-GS-Gal4 | From Dr. John Tower | Drug-induced ubiquitous expression |
| Mhc-GS-Gal4 | BL #43641 | Drug-induced skeletal muscle expression |
| UAS-Ldh = P(EP)Ldh | BL #16829 | Ldh overexpression |
| UAS-Pgi | DPIM #2979 | Pgi overexpression |
| UAS-Pfrx | DPIM #1678 | Pfrx overexpression |
| P(EP)Pfk | BL #22605 | Pfk mutant |
| P(EP)Ald | BL #20862 | Ald mutant |
| P(EP)Pglym78 | BL #30063 | Pglym78 mutant |
| P(EP)Pgi | BL #17595 | Pgi mutant |
Note: Bloomington (BL) stock center: https://bdsc.indiana.edu/; Transgenic Resource of the DPIM (Harvard): https://interfly.med.harvard.edu/transgenic_info.php
Survival Analysis
Flies were maintained at 25°C with 60% humidity on 12-hour light/dark cycles with standard cornmeal/soy flour/yeast fly food. The fly food was changed every 2–3 days at which time dead flies found on top of the food were counted. All experiments were done with male flies.
To avoid any contribution of genetic background mutations to the observed phenotypes, UAS- transgenes were backcrossed through 10 generations against w1118 to obtain isogenic male siblings carrying either a UAS- or no transgene (distinguished by eye color: white+ and white−, respectively). Male siblings carrying either a UAS- or no transgene and having the same genetic background were then crossed to homozygous w1118;Mhc-Gal4 (rosy+ white−) females, and the resulting male progenies were sorted (based on eye color) into isogenic transgene-expressing and transgene-nonexpressing cohorts. UAS- transgene expression was confirmed by quantitative real-time polymerase chain reaction (qRT-PCR).
For adult muscle-specific manipulation, heterozygous Mhc-Gal4.F3-580 (BDSC#38464 without Mhc-RFP; here indicated as MhcF3-Gal4) female virgins were crossed to UAS lines to produce progenies that contain Gal4 and UAS- and are isogenic to littermate controls devoid of Gal4. MhcF3-Gal4 is a stronger driver than Mhc-Gal4 due to differences in the Mhc promoter sequences used and because of integration at different sites in the genome.
The GeneSwitch system and the Mhc-GeneSwitch-Gal4 and Act5c-GeneSwitch-Gal4 drivers (29) were also used for the drug-inducible expression of transgenes. In these experiments, transgene expression is induced by adding RU486 (dissolved in ethanol) to a final concentration of 50 µM to the fly food; ethanol alone was used for the mock treatment of uninduced controls.
For experiments with EP lines, such lines (marked by white+) were backcrossed through 10 generations against w1118 to obtain isogenic homozygous (white++) and heterozygous (white+) mutants and wild-type controls (white−). Such lines reduce expression of the respective glycolytic enzymes (ie act as hypomorphic mutants) because the EP transposon is inserted in the 5′UTR and intron of transcripts (Pfk), promoter region and 5′UTR (Pglym78, Pgi, Ald), and promoter region (Ldh) of the target gene. Reduction in the expression of the respective glycolytic enzymes was confirmed by qRT-PCR. In addition, because the EP element carries UAS sequences, the EP line for Ldh was also used for achieving Ldh overexpression in experiments with Gal4 drivers.
qRT-PCR and Oligonucleotide Sequences
qRT-PCR was done as previously (30). Specifically, total RNA was extracted from Drosophila thoraces using Trizol (Life Technologies, Carlsbad, CA, USA). For qRT-PCR, cDNAs were reverse-transcribed from 500 ng total RNA with the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). qRT-PCR was performed with SYBR Green and a CFX96 apparatus (Bio-Rad), and α-Tubulin84B was used as a reference gene. The qRT-PCR oligonucleotide sequences here used are reported in Table 2.
Table 2.
qRT-PCR Oligo Sequences
| Gene Name | NCBI Accession # | Forward Sequence | Reverse Sequence |
|---|---|---|---|
| Tub84B | 40848 | GTTTGTCAAGCCTCATAGCCG | GGAAGTGTTTCACACGCGAC |
| Hex-A | 45875 | GTGCTGGTGAAGATGACCCA | GCACTTCGGAATCCTGTCCA |
| Hex-C | 44008 | TCTCGCTGAAAGGTCACCAC | ATGCGGTCTTCATGTCGTGT |
| Pgi | 35886 | CTTCCTCCGCTCAACCAGG | GCAGGCGCAGGCTGTATTTT |
| Pfk | 36060 | CGAGGGTCATCCCATCACAG | CCATGCGGCAAGCCAAAATA |
| Ald | 43183 | AAGAACACCCCCGAGGAGAT | ACCTCCAGACAGGAAGGTCA |
| Tpi | 43582 | GTGCATTGCATCAGTAACGCT | GGTGGTTCCGATGATACGGG |
| Gapdh1 | 35728 | ACTCGACTCACGGTCGTTTC | CTCCACCACATACTCGGCTC |
| Gapdh 2 | 32545 | GATTTCCTCAGCGACACCCA | CCATTCTACCGCGCCCTAAT |
| Pgk | 33461 | CGTGTGGAACGGACCCCC | AGTGCCTCCGTGTTCCACTT |
| Pglym78 | 43447 | GAGTTCCCCCAGTTCGAGTC | GCGTCCTCAGAAAGGTTGTCTA |
| Pglym87 | 46246 | CTGCCCGTTCTCGAAGTCAT | CGGACTCTCCATGACGAACC |
| Eno | 33351 | CAAATCAGCAGTCAGCCAGC | CATCTCGTTGCTGTCACAAGT |
| Pyk | 42620 | AAGGTTCTTGGCGAGAAGGG | TGATCTCGTCCAGGTTGTGC |
| Ldh | 45880 | GCGAGGATCCAGAGAAGTGG | CAATGCCATGTTCGCCCAAA |
Lactate Assays
Lactate assays were performed using the Cayman L-Lactate Assay kit (Ann Arbor, MI, USA; #700510). In brief, Drosophila thoraces were homogenized with a bullet blender in phosphate buffer saline (31), and proteins were precipitated with 1 volume of 0. 5 M metaphosphoric acid, followed by centrifugation at 10 000 g for 5 minutes at 4°C. The supernatant was collected and assayed following the recommended procedures for the Cayman L-Lactate Assay kit. Lactate concentration was estimated based on sample interpolation into the standard curves.
LDH Activity
LDH activity was measured with the Sigma Lactate Dehydrogenase Activity Assay Kit (St. Louis, MO, USA; #MAK066). For this assay, Drosophila thoraces were homogenized with a bullet blender in the provided assay buffer, and tissue homogenates were briefly centrifuged to remove cuticle remnants. Subsequently, the supernatant was assayed following the assay kit protocol.
COX Activity
Cytochrome C oxidase (COX) activity was measured using the Biovision Cytochrome Oxidase Activity Colorimetric Assay (Milpitas, CA, USA; #K287-100). For this assay, Drosophila thoraces were homogenized in the Biovision Cell Lysis Buffer (#1067) using a Dounce homogenizer, and tissue homogenates were subsequently processed following recommended procedures.
NAD+/NADH Levels
Nicotinamide adenine dinucleotide (NAD+)/NADH levels were estimated with the Biovision NAD/NADH Quantitation colorimetric kit (#K337). Drosophila thoraces were homogenized with a bullet blender in the provided extraction buffer, and proteins were removed by precipitation with 1 volume of 0.5 M metaphosphoric acid, followed by centrifugation at 10 000 g for 5 minutes at 4°C. The cleared supernatants were collected and assayed by following the recommended protocol to estimate total NADt and NAD+ levels. The NAD quantity was determined by subtracting the amount of NADH (supernatant sample after decomposition of NAD at 60°C for 30 minutes) from total NADt (sample without decomposition of NAD).
Statistical Analysis
All experiments were performed with biological replicates unless otherwise indicated. The unpaired two-tailed Student’s t-test was used to compare the means of 2 independent groups to each other. One-way analysis of variance with Tukey’s post hoc test was used for multiple comparisons of more than 2 groups of normally distributed data. The “n” for each experiment can be found indicated in the figure legends and represents individual flies for survival analyses and batches of 10 thoraces each for biochemical assays and qRT-PCR. Bar graphs present the mean ± SEM or ± SD, as indicated in the figure legend. Throughout the figures, asterisks indicate the significance of the p value: *p < .05, **p < .01, ***p < .001. A significant result was defined as p < .05. Statistical analyses were done with Excel and GraphPad Prism (San Diego, CA, USA). Statistical analysis of survival data was done using OASIS (32).
Results
Glycolysis Increases in Drosophila Skeletal Muscle With Aging
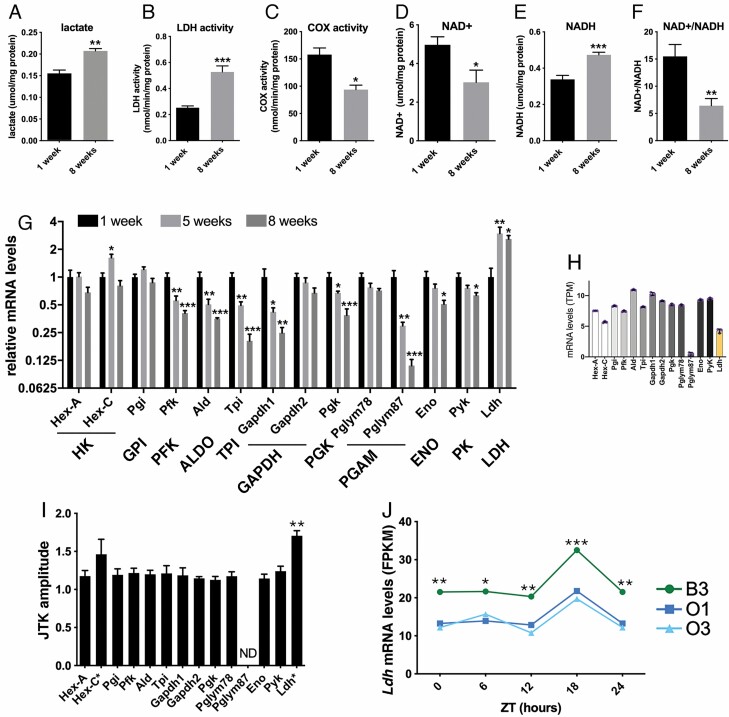
To examine whether aging affects glycolysis in Drosophila skeletal muscle, we have examined the levels of lactate, the end-product of anaerobic glycolysis, in skeletal muscle (thoraces) of old (8 weeks) versus young (1 week) flies. Consistent with an age-related increase in anaerobic glycolysis, lactate levels were significantly higher in the skeletal muscle of old flies (Figure 1A), in parallel with an increase in LDH activity (Figure 1B). Conversely, COX activity, which is indicative of mitochondrial oxidative phosphorylation, decreased with aging (Figure 1C), consistent with previous findings that mitochondrial function declines with aging (8).
Figure 1.
Glycolysis increases in Drosophila skeletal muscle with aging. (A) The levels of lactate, an end-product of glycolysis, increase in skeletal muscle of old (8 weeks) versus young (1 week) flies (n = 3). (B) The activity of lactate dehydrogenase (LDH) increases with aging (n = 4–10). (C) Cytochrome C oxidase (COX) activity decreases with aging, indicative of decreased mitochondrial function (n = 3). (D) NAD+, which is utilized in its oxidized state as a cofactor in glycolysis, declines with aging (n = 8). (E) NADH, which is a product of glycolysis, increases with aging (n = 8). (F) The NAD+/NADH ratio declines with aging (n = 8). (G) Expression of 8 out of 14 glycolytic enzymes declines with aging in skeletal muscle with the exception of Ldh, whose expression increases with aging (n = 4). (H) RNA-seq reads (TPM) of glycolytic enzymes in Drosophila skeletal muscles (n = 3). Apart Pglym87, Ldh is the glycolytic enzyme with the lowest expression. Note that Pglym78 and Pglym87 are paralogs, as for Hex-A and Hex-C. (I and J) Ldh is the glycolytic enzyme that has the highest amplitude of daily cyclic expression (n = 3). Ldh mRNA levels are lower in long-lived O1 and O3 fly strains compared to the parental B3 Drosophila strain (n = 3) at 1 week of age.
NAD+ is utilized in its oxidized state as a cofactor and is converted to NADH as a byproduct of glycolysis. Consistent with an increase in glycolysis with aging, NAD+ and the NAD+/NADH ratio declines (Figure 1D and F) whereas NADH increases with aging (Figure 1E).
To examine the mechanistic basis for glycolytic changes with aging, we profiled the mRNA levels of glycolytic enzymes and found that 8 out of 14 have decreased expression in the skeletal muscle of old flies (Figure 1G). Specifically, the mRNA levels of Pfk/PFK, Ald/ALDO, Tpi/TPI, Gapdh1/GAPDH, Pgk/PGK, Pglym87/PGAM, Eno/ENO, and Pyk/PK decline progressively in skeletal muscle during aging. Apart from Hex-C/HK expression, which transiently increases at 5 weeks, Ldh is the only glycolytic enzyme that is transcriptionally upregulated with aging (Figure 1G).
Analysis of RNA-seq reads (TPM) revealed that Ldh is the glycolytic enzyme with the lowest expression, apart from glycolytic enzymes (Hex-C and Pglym87) related to more abundantly expressed paralogs, that is, Hex-A and Pglym78 (Figure 1H). Altogether, these analyses indicate that Ldh mRNA levels are relatively low compared to other glycolytic enzymes and that therefore age-related changes are likely to affect lactate production, consistent with our finding that lactate levels and LDH activity increase in aging muscle (Figure 1A and B).
Another key feature that differentiates LDH from other glycolytic enzymes is that LDH is the most cyclic of them. Specifically, analysis of the Drosophila muscle circadian transcriptome (33) indicates that Ldh is the glycolytic enzyme that has the highest amplitude of daily cyclic expression (Figure 1I and J). This suggests that age-related changes in LDH activity may affect the circadian regulation of glycolysis (34).
To start to assess whether age-related increases in LDH activity affect survival during aging, we compared Ldh mRNA levels in long-lived Drosophila strains (O1 and O3) that were experimentally selected to have a long life span compared to a control B3 strain (33,35). Consistent with an association between muscle Ldh expression and life span, Ldh mRNA levels are lower in long-lived O1 and O3 fly strains compared to the B3 strain (Figure 1J).
Muscle-Restricted Ldh Overexpression Reduces Life Span
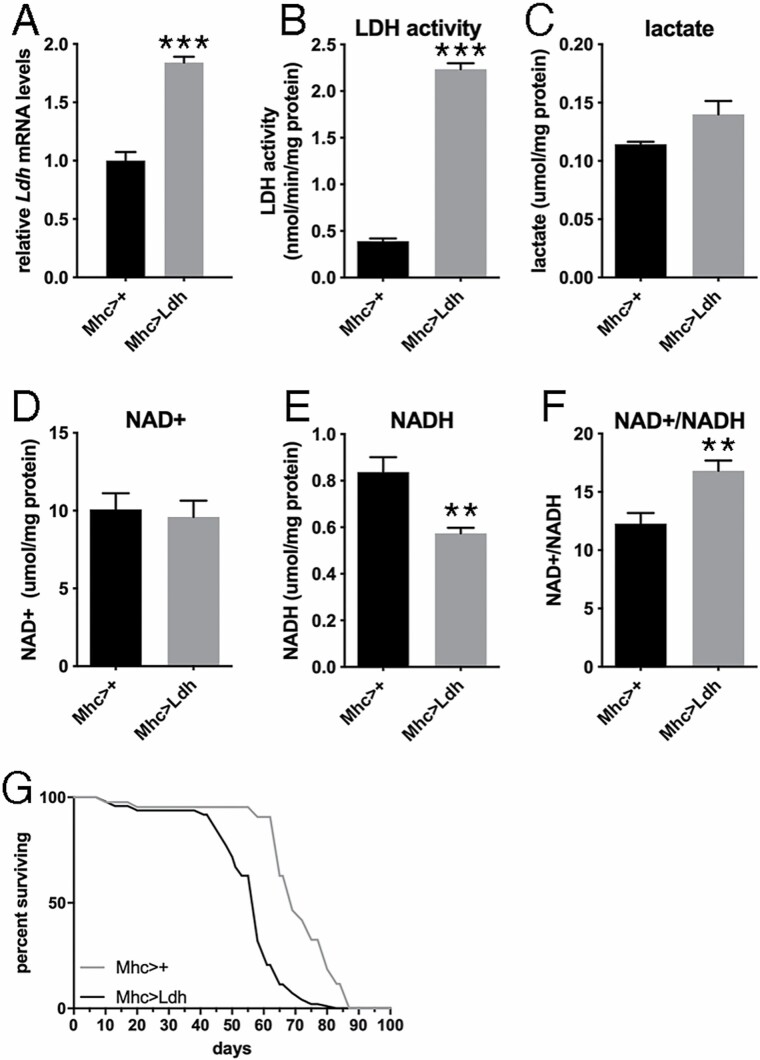
To determine whether an age-related increase in muscle Ldh expression affects survival during aging, we experimentally increased Ldh expression from young age specifically in skeletal muscle by using the UAS/Gal4 system and the muscle-specific Mhc-Gal4 driver. Importantly, we utilized an EP line that allows a Gal4-dependent ~2-fold increase in Ldh mRNA levels (Figure 2A), which is similar to the ~2.5-fold age-induced upregulation in Ldh mRNA levels that is found with aging (Figure 1G). In parallel with increased Ldh expression, young flies with Ldh overexpression displayed increased Ldh activity (Figure 2B) although this led to a marginal (nonsignificant) increase in lactate levels (Figure 2C). Whereas NAD+ levels do not change in response to Ldh overexpression (Figure 2D), NADH levels decline (Figure 2E) and the NAD+/NADH ratio increases (Figure 2F), which are likely explained by the utilization of NADH by LDH to convert pyruvate into lactate.
Figure 2.
Muscle-restricted overexpression of lactate dehydrogenase (Ldh) increases glycolysis and shortens life span in Drosophila. (A–C) Ldh overexpression in skeletal muscle significantly increases Ldh mRNA levels (A; n = 3) and LDH activity (B; n = 4) and marginally affects lactate levels (C; n = 4). (D–F) Ldh overexpression does not affect NAD+ (D; n = 4), reduces NADH levels (E; n = 4), and increases the NAD+/NADH ratio (F; n = 6). In A–F, the mean + SEM is indicated. (G) Muscle-restricted Ldh overexpression, driven by Mhc-Gal4, shortens life span, compared to isogenic controls; Mhc>+ (n = 43) and Mhc>Ldh (n = 97), p < .0001.
Survival analysis revealed that muscle-restricted Ldh overexpression with Mhc-Gal4 (Mhc>Ldh) significantly shortens life span (Figure 2G), compared to isogenic controls (Mhc>+). Altogether, these findings indicate that mimicking age-induced upregulation of Ldh in skeletal muscle reduces life span.
Induction of Glycolysis in Skeletal Muscle and Ubiquitously Shortens Life Span
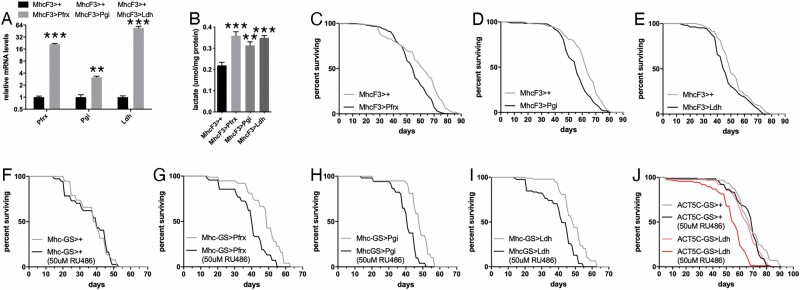
In addition to Ldh, we next tested the impact of overexpression of 2 other glycolytic enzymes: Pfrx/PFK (phosphofructokinase, also known as Pfk) and Pgi/GPI (glucose-6-phosphate isomerase, also known as phosphoglucose isomerase). For these studies, we utilized the MhcF3-Gal4, which is specific for skeletal muscle but drives stronger expression than Mhc-Gal4. As expected, transgene overexpression led to higher Pfrx, Pgi, and Ldh mRNA levels, compared to isogenic controls (Figure 3A) and to significantly higher lactate levels (Figure 3B).
Figure 3.
Muscle-restricted overexpression of glycolytic enzymes increases glycolysis and shortens life span in Drosophila. (A) Overexpression of Pfrx, Pgi, and Ldh in skeletal muscle with MhcF3-Gal4 correspondingly increases Pfrx, Pgi, and Ldh mRNA levels (n = 3). (B) Muscle-restricted overexpression of Pfrx, Pgi, and Ldh increases lactate levels in muscle (n = 3). In A and B, the mean + SEM is indicated. (C–E) Overexpression of Pfrx (C), Pgi (D), and Ldh (E) in skeletal muscle with MhcF3-Gal4 shortens life span. In C, MhcF3>+ (n = 239) and MhcF3>Pfrx (n = 319), p = .0035. In D, MhcF3>+ (n = 185) and MhcF3>Pgi (n = 180), p < .0001. In E, MhcF3>+ (n = 103) and MhcF3>Ldh (n = 105), p = .0002. (F–I) Compared to uninduced controls, drug-induced, muscle-restricted overexpression of Pfrx (G), Pgi (H), and Ldh (I) with Mhc-GS-Gal4 shortens life span compared to mock-induction of no transgene (F). In F, Mhc-GS>+ is n = 38 (−RU486) and n = 37 (+RU486), p = .9104. In G, Mhc-GS>Pfrx is n = 75 (−RU486) and n = 69 (+RU486), p < .0001. In H, Mhc-GS>Pgi is n = 58 (−RU486) and n = 51 (+RU486), p < .0001. In I, Mhc-GS>Ldh is n = 93 (−RU486) and n = 85 (+RU486), p < .0001. (J) Drug-induced, general tissue overexpression of Ldh with Act5c-GS-Gal4 shortens life span, compared to uninduced controls and mock no-transgene induction. In J, Act5c-GS>Ldh is n = 73 (−RU486) and n = 135 (+RU486), p < .0001; Act5c-GS>+ is n = 156 (-RU486) and n = 178 (+RU486).
Survival analyses revealed that Pfrx, Pgi, and Ldh overexpression reduces life span compared to no-transgene expression (Figure 3C–E). To further corroborate these findings, we next tested the impact of drug-induced Pfrx, Pgi, and Ldh transgenic overexpression with the muscle GeneSwitch Mhc-GS-Gal4 driver (29). Administration of 50 μM RU486 (dissolved in ethanol and added to the fly food) did not significantly affect life span compared to mock-treated control flies (Mhc-GS>+) that were fed food with ethanol but without RU486 (Figure 3F). However, RU486-induced expression of Pfrx, Pgi, and Ldh in skeletal muscle significantly reduced life span (Figure 3G–I). We next examined whether ubiquitous Ldh overexpression with the drug-inducible Act5c-GS-Gal4 regulates life span and found that Ldh overexpression shortens life span compared to uninduced controls and to mock RU486 treatment (Figure 3J).
Altogether, these survival analyses demonstrate that muscle-restricted and ubiquitous induction of glycolysis reduces life span.
Hypomorphic Mutations in Glycolytic Enzymes Have Distinct Outcomes on Life Span
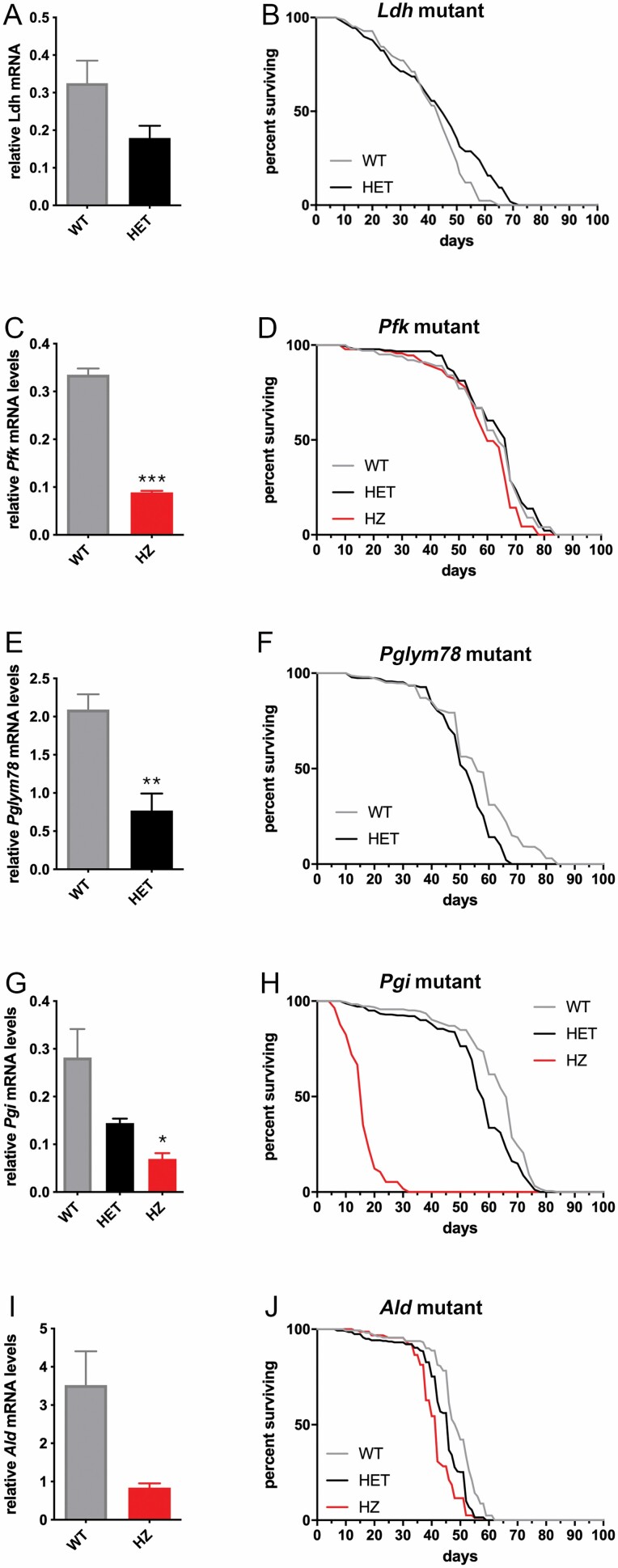
To further test the impact of glycolysis on aging, we next examined the outcome on life span of mutations in glycolytic enzymes. Because complete loss of glycolysis is likely lethal, we identified hypomorphic mutations that only partially reduce the levels of Ldh, Pfk, Pglym78, Pgi, and Ald glycolytic enzymes due to insertion of P{EP} artificial transposons into the promoter and/or 5′-UTR regions of glycolytic enzymes (see Method).
Hypomorphic Ldh mutants reduce Ldh mRNA levels to ~55% of normal levels (Figure 4A) and extend survival late in life, that is, in middle and old-aged flies (Figure 4B). These findings indicate that, converse to muscle Ldh overexpression, partial Ldh loss extends life span.
Figure 4.
Hypomorphic mutations in glycolytic enzymes affect life span to different extents. (A and B) Hypomorphic Ldh mutants reduce Ldh mRNA levels (A) and extend survival late in life, that is, in middle-aged and old flies (B); +/+ (n = 83) and +/Ldh (n = 108), p = .006. (C and D) Hypomorphic Pfk mutants reduce Pfk mRNA levels (C) but have minimal or no effect on life span (D). In C, n (WT) = 4 and n (HZ) = 2. In D, n (WT) = 100, n (HET) = 181, and n (HZ) = 91, p = .386 in WT vs HZ. (E and F) Heterozygous Pglym78 mutants reduce Pglym78 mRNA levels (E; n = 4) but have minimal effect on life span (F); n (WT) = 300 and n (HET) = 233, p < .0001. (G and H) Homozygous Pgi mutants strongly reduce Pgi mRNA levels (G; n = 2) and life span (H); n (WT) = 185 and n (HET) = 241, n (HZ) = 57, p < .0001 in WT vs HZ. (I and J) Decline in Ald expression decreases Ald mRNA levels (I; n = 2) and life span (J); n (WT) = 161, n (HET) = 278, and n (HZ) = 156, p < .0001 in WT vs HZ. In A, C, E, G, and I, the mean + SEM is indicated. WT = wild-type (isogenic control); HET = heterozygous; HZ = homozygous.
Homozygous hypomorphic Pfk mutants reduce Pfk mRNA levels to ~25% of normal levels (Figure 4C) but do not affect life span (Figure 4D), suggesting that Pfk levels are normally not rate-limiting for glycolysis and/or life-span determination. Similarly, heterozygous Pglym78 mutants reduce Pglym78 mRNA levels to ~40% of normal levels (Figure 4E) but reduce life span only marginally (Figure 4F).
Despite a ~50% reduction in Pgi mRNA levels (Figure 4G), heterozygous Pgi mutations have a limited impact on life span (Figure 4H). However, a stronger reduction in Pgi mRNA levels due to homozygous mutations leads to life-span shortening (Figure 4G and H). Lastly, mutations in Ald decrease Ald mRNA levels (Figure 4I) and reduce life span (Figure 4J), although to a lower extent compared to Pgi (Figure 4G and H). Altogether, these findings indicate that hypomorphic, whole-body mutations in glycolytic enzymes have distinct outcomes on life span.
Discussion
Glycolysis is a key metabolic pathway that sustains physiologic homeostasis but that also drives pathological metabolic adaptations (36,37). In aging, glycolysis has been reported to counteract or to contribute to age-related degeneration, depending on the tissue and context analyzed (10–15). Therefore, much remains to be learned about the complex interaction between glycolysis, metabolism, and aging in different organ systems.
In this study, we have found that skeletal muscle aging is characterized in Drosophila by an increase in lactate levels and that this is due to age-related increases in LDH activity and expression. Conversely, the expression of other glycolytic enzymes declines with aging. Altogether, these findings suggest that whereas anaerobic glycolysis is limited to hypoxic states in skeletal muscles of young animals, it may more pervasively occur in the skeletal muscle of old animals. Interestingly, it was previously reported that declining mitochondrial function with aging leads to a pseudohypoxic state of skeletal muscle characterized by nuclear accumulation of HIF-1α under normoxic conditions in mice (38), suggesting that several age-related changes may promote anaerobic glycolysis even in the presence of oxygen. In agreement with these findings, lactate levels have been reported to increase with aging in mouse skeletal muscle, suggesting that there is an increase in anaerobic glycolysis during muscle aging (39). However, LDH activity has been reported to decline during aging in other studies that have analyzed older models (40,41): this may reflect the preferential age-related atrophy and loss of glycolytic myofibers compared to other myofiber types that rely primarily on oxidative metabolism (22,42).
Based on the results of our study in Drosophila, it is possible that LDH expression in glycolytic myofibers (such as mouse type 2b and human type 2x myofibers) indeed contributes to their preferential atrophy and degeneration with aging in mice and humans (22,42). Because there are no myofiber types in Drosophila as defined in higher organisms (43), we propose that Drosophila may represent a useful model organism to define the impact of muscle glycolysis and oxidative phosphorylation in aging irrespective of the role of these metabolic pathways in defining myofiber types.
Although our study is centered on skeletal muscle, it is possible that increased activity of LDH is detrimental across tissues during aging. In agreement with this scenario, it has been recently reported that Ldh overexpression in the brain fuels neurodegeneration and shortens life span in Drosophila (44).
By analyzing hypomorphic mutants of several glycolytic enzymes, we have found that they shorten life span to different degrees (Pglym78, Pgi, and Ald), have no meaningful effect (Pfk), or increase late survival (Ldh), compared to their isogenic controls (Figure 4). These findings suggest that certain glycolytic enzymes are more likely to affect glycolysis and life span in response to perturbation, perhaps because of their putative lower levels or enzymatic activities which make them rate-limiting or dispensable. For example, the production of ATP and of many glycolytic metabolites can still be accomplished without Ldh via oxidative metabolism of pyruvate. Therefore, the finding that partial Ldh loss extends life span may be explained by the fact that this intervention reduces anaerobic glycolysis but does not affect aerobic glycolysis. However, Pgi is essential for the formation of pyruvate and of many glycolytic metabolites and this might explain why Pgi loss had the greatest negative effect on life span. Thus, the specific roles of the glycolytic enzymes that are impacted may determine the overall outcome on aging and life span.
Although glycolytic enzymes have prominent metabolic roles, there is extensive evidence that they can also play signaling functions that are unrelated to their enzymatic activities (45–47). On this basis, the different outcomes on life span of hypomorphic mutants of distinct glycolytic enzymes (Figure 4) may also be explained by the differential impact of their respective signaling functions on life span.
Because we have found that muscle-specific and ubiquitous overexpression of Ldh and other glycolytic enzymes shortens life span (Figures 2 and 3), a decline in glycolysis was expected to lead to converse changes, that is, to extend life span, which was not the case apart for Ldh loss (Figure 4). This suggests that moderate repression of anaerobic glycolysis might be beneficial but that tight control of aerobic glycolysis is also needed for ensuring optimal survival. However, a recent study on LDH in Alzheimer’s disease has found that both strong increases and decreases in LDH are detrimental in Drosophila (48), again suggesting that the level of modulation and tissue where glycolysis is perturbed might result in different outcomes on life span. Altogether, detrimental effects of decreases and increases in glycolytic activity may arise from multiple impacts of such interventions on signaling and metabolic homeostasis.
Another important factor to consider when examining the outcome of glycolysis-based interventions on life span is the time at which glycolysis is perturbed. Specifically, whereas overexpression of Ldh and other glycolytic enzymes was induced in adulthood in our experiments (Figures 2 and 3), hypomorphic mutants (Figure 4) reduce the activity of glycolytic enzymes from development and presumably across all tissues, which may have a different impact on life span compared to adult-onset and tissue-restricted modulation of glycolysis. Future studies shall address whether the timepoint of repression or induction of glycolysis is a determinant of life span.
We have found that Ldh overexpression in skeletal muscle and in the whole organism reduces life span (Figures 2 and 3), suggesting that age-related increases in muscle Ldh expression and activity are detrimental and contribute to mortality during aging. These findings reinforce the notion that modulation of metabolic pathways in skeletal muscle affects organismal survival (23). The mechanisms connecting muscle LDH activity with life span are likely multifaceted and may include regulation of systemic metabolism as well as initiation of muscle systemic signaling via myokines (24,49,50). For example, because Ldh is the glycolytic gene with the highest cyclic amplitude of expression in skeletal muscle (Figure 1), age-related increases in LDH activity may also contribute to derangement of circadian oscillations in muscle metabolism (33,51,52), which have been found to have profound systemic consequences (51,52). Interestingly, lactate can also be released by skeletal muscle, in particular following exercise, and it has been proposed that circulating lactate may coordinate the metabolic and redox statuses of skeletal muscle with those of other tissues. On this basis, an age-induced increase in LDH activity and lactate levels may affect organism survival via systemic lactate signaling.
Conclusions
In summary, we have defined a causal role for increases in muscle LDH activity and lactate in life-span determination. Moreover, by modulating the levels of several glycolytic enzymes, we have found that glycolysis needs to be tightly controlled to sustain optimal survival during aging. Altogether, this study highlights a key role for skeletal muscle glycolysis in aging.
Supplementary Material
Acknowledgments
We thank the Bloomington stock center, the Transgenic Resource of the DPIM (Harvard), and Dr. John Tower for fly stocks.
Funding
This work was supported by research grants to F.D. from the Ellison Medical Foundation, Glenn Foundation for Medical Research, American Federation for Aging Research, Hartwell Foundation, American Parkinson Disease Association, and the National Institute on Aging of the National Institutes of Health (NIH; R01AG055532 and R56AG063806). L.C.H. was supported by a Glenn/AFAR Postdoctoral Fellowship for Translational Research on Aging. Research at St. Jude Children’s Research Hospital is supported by the ALSAC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict of Interest
None declared.
Author Contributions
Conceptualization: L.C.H. and F.D.; investigation: L.C.H.; writing—original draft: F.D.; writing—review and editing: L.C.H. and F.D.; data curation: L.C.H.; formal analysis: L.C.H.; visualization: L.C.H.; funding acquisition: F.D.; resources: L.C.H. and F.D.; and supervision: F.D.
Data Availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplementary tables. Primary data are provided in Supplementary Table 1, and the statistical analysis of life-span data with OASIS is reported in Supplementary Table 2.
References
- 1. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. López-Otín C, Galluzzi L, Freije JMP, Madeo F, Kroemer G. Metabolic control of longevity. Cell. 2016;166(4):802–821. doi: 10.1016/j.cell.2016.07.031 [DOI] [PubMed] [Google Scholar]
- 3. Mourikis P, Hurlbut GD, Artavanis-Tsakonas S. Enigma, a mitochondrial protein affecting lifespan and oxidative stress response in Drosophila. Proc Natl Acad Sci U S A. 2006;103(5):1307–1312. doi: 10.1073/pnas.0510564103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kapahi P, Kaeberlein M, Hansen M. Dietary restriction and lifespan: lessons from invertebrate models. Ageing Res Rev. 2017;39:3–14. doi: 10.1016/j.arr.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 2008;7(4):478–490. doi: 10.1111/j.1474-9726.2008.00400.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Avanesov AS, Ma S, Pierce KA, et al. Age- and diet-associated metabolome remodeling characterizes the aging process driven by damage accumulation. Elife. 2014;3:e02077. doi: 10.7554/eLife.02077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hunsberger HC, Greenwood BP, Tolstikov V, Narain NR, Kiebish MA, Denny CA. Divergence in the metabolome between natural aging and Alzheimer’s disease. Sci Rep. 2020;10:12171. doi: 10.1038/s41598-020-68739-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rallis C, Mülleder M, Smith G, Au YZ, Ralser M, Bähler J. Amino acids whose intracellular levels change most during aging alter chronological life span of fission yeast. J Gerontol A Biol Sci Med Sci. 2021;76(2):205–210. doi: 10.1093/gerona/glaa246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parkhitko AA, Binari R, Zhang N, Asara JM, Demontis F, Perrimon N. Tissue-specific down-regulation of S-adenosyl-homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev. 2016;30(12):1409–1422. doi: 10.1101/gad.282277.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6(4):280–293. doi: 10.1016/j.cmet.2007.08.011 [DOI] [PubMed] [Google Scholar]
- 11. Onken B, Kalinava N, Driscoll M. Gluconeogenesis and PEPCK are critical components of healthy aging and dietary restriction life extension. PLoS Genet. 2020;16(8):e1008982. doi: 10.1371/journal.pgen.1008982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weimer S, Priebs J, Kuhlow D, et al. D-Glucosamine supplementation extends life span of nematodes and of ageing mice. Nat Commun. 2014;5:3563. doi: 10.1038/ncomms4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma Z, Wang H, Cai Y, et al. Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. Elife. 2018;7:1–33. doi: 10.7554/eLife.35368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manzo E, Lorenzini I, Barrameda D, et al. Glycolysis upregulation is neuroprotective as a compensatory mechanism in ALS. Elife. 2019;8:1–20. doi: 10.7554/eLife.45114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kondoh H, Lleonart ME, Gil J, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65(1):177–185. [PubMed] [Google Scholar]
- 16. Goyal MS, Vlassenko AG, Blazey TM, et al. Loss of brain aerobic glycolysis in normal human aging. Cell Metab. 2017;26(2):353–360.e3. doi: 10.1016/j.cmet.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ravera S, Podestà M, Sabatini F, et al. Discrete changes in glucose metabolism define aging. Sci Rep. 2019;9(1):10347. doi: 10.1038/s41598-019-46749-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Demontis F, Patel VK, Swindell WR, Perrimon N. Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Rep. 2014;7(5):1481–1494. doi: 10.1016/j.celrep.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143(5):813–825. doi: 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunt LC, Schadeberg B, Stover J, et al. Antagonistic control of myofiber size and muscle protein quality control by the ubiquitin ligase UBR4 during aging. Nat Commun. 2021;12(1):1418. doi: 10.1038/s41467-021-21738-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rai M, Coleman Z, Curley M, et al. Proteasome stress in skeletal muscle mounts a long-range protective response that delays retinal and brain aging. Cell Metab. 2021;33(6):1137–1154.e9. doi: 10.1016/j.cmet.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demontis F, Piccirillo R, Goldberg AL, Perrimon N. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis Model Mech. 2013;6(6):1339–1352. doi: 10.1242/dmm.012559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Demontis F, Piccirillo R, Goldberg AL, Perrimon N. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell. 2013;12(6):943–949. doi: 10.1111/acel.12126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rai M, Demontis F. Systemic nutrient and stress signaling via myokines and myometabolites. Annu Rev Physiol. 2016;78:85–107. doi: 10.1146/annurev-physiol-021115-105305 [DOI] [PubMed] [Google Scholar]
- 25. Hakimi P, Yang J, Casadesus G, et al. Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J Biol Chem. 2007;282(45):32844–32855. doi: 10.1074/jbc.M706127200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stenesen D, Suh JM, Seo J, et al. Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell Metab. 2013;17(1):101–112. doi: 10.1016/j.cmet.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katewa SD, Demontis F, Kolipinski M, et al. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab. 2012;16(1):97–103. doi: 10.1016/j.cmet.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunt LC, Xu B, Finkelstein D, et al. The glucose-sensing transcription factor MLX promotes myogenesis via myokine signaling. Genes Dev. 2015;29(23):2475–2489. doi: 10.1101/gad.267419.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98(22):12596–12601. doi: 10.1073/pnas.221303298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiao J, Kavdia K, Pagala V, et al. An age-downregulated ribosomal RpS28 protein variant regulates the muscle proteome. G3 (Bethesda). 2021;11(7):jkab165. doi: 10.1093/g3journal/jkab165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rai M, Curley M, Coleman Z, et al. Analysis of proteostasis during aging with western blot of detergent-soluble and insoluble protein fractions. STAR Protoc. 2021;2(3):100628. doi: 10.1016/j.xpro.2021.100628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han SK, Lee D, Lee H, et al. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget. 2016;7(35):56147–56152. doi: 10.18632/oncotarget.11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hunt LC, Jiao J, Wang YD, et al. Circadian gene variants and the skeletal muscle circadian clock contribute to the evolutionary divergence in longevity across Drosophila populations. Genome Res. 2019;29(8):1262–1276. doi: 10.1101/gr.246884.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thurley K, Herbst C, Wesener F, et al. Principles for circadian orchestration of metabolic pathways. Proc Natl Acad Sci U S A. 2017;114(7):1572–1577. doi: 10.1073/pnas.1613103114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rose MR. Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution. 1984;38(5):1004–1010. doi: 10.1111/j.1558-5646.1984.tb00370.x [DOI] [PubMed] [Google Scholar]
- 36. Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26(9):877–890. doi: 10.1101/gad.189365.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang BL. Glucose, glycolysis, and neurodegenerative diseases. J Cell Physiol. 2020;235(11):7653–7662. doi: 10.1002/jcp.29682 [DOI] [PubMed] [Google Scholar]
- 38. Gomes AP, Price NL, Ling AJ, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155(7):1624–1638. doi: 10.1016/j.cell.2013.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Houtkooper RH, Argmann C, Houten SM, et al. The metabolic footprint of aging in mice. Sci Rep. 2011;1:134. doi: 10.1038/srep00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaczor JJ, Ziolkowski W, Antosiewicz J, Hac S, Tarnopolsky MA, Popinigis J. The effect of aging on anaerobic and aerobic enzyme activities in human skeletal muscle. J Gerontol A Biol Sci Med Sci. 2006;61(4):339–344. doi: 10.1093/gerona/61.4.339 [DOI] [PubMed] [Google Scholar]
- 41. Washington TA, Healey JM, Thompson RW, Lowe LL, Carson JA. Lactate dehydrogenase regulation in aged skeletal muscle: regulation by anabolic steroids and functional overload. Exp Gerontol. 2014;57:66–74. doi: 10.1016/j.exger.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 42. Talbot J, Maves L. Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip Rev Dev Biol. 2016;5(4):518–534. doi: 10.1002/wdev.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Piccirillo R, Demontis F, Perrimon N, Goldberg AL. Mechanisms of muscle growth and atrophy in mammals and Drosophila. Dev Dyn. 2014; 243( 2): 201–215. doi: 10.1002/dvdy.24036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Long DM, Frame AK, Reardon PN, et al. Lactate dehydrogenase expression modulates longevity and neurodegeneration in Drosophila melanogaster. Aging (Albany NY). 2020;12(11):10041–10058. doi: 10.18632/aging.103373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Snaebjornsson MT, Schulze A. Non-canonical functions of enzymes facilitate cross-talk between cell metabolic and regulatory pathways. Exp Mol Med. 2018;50(4):1–16. doi: 10.1038/s12276-018-0065-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30(3):142–150. doi: 10.1016/j.tibs.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 47. Huangyang P, Simon MC. Hidden features: exploring the non-canonical functions of metabolic enzymes. Dis Model Mech. 2018;11:1–15. doi: 10.1242/dmm.033365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Niccoli T, Kerr F, Snoeren I, et al. Activating transcription factor 4-dependent lactate dehydrogenase activation as a protective response to amyloid beta toxicity. Brain Commun. 2021;3(2):fcab053. doi: 10.1093/braincomms/fcab053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pedersen BK, Akerström TC, Nielsen AR, Fischer CP. Role of myokines in exercise and metabolism. J Appl Physiol (1985). 2007;103(3):1093–1098. doi: 10.1152/japplphysiol.00080.2007 [DOI] [PubMed] [Google Scholar]
- 50. Robles-Murguia M, Rao D, Finkelstein D, Xu B, Fan Y, Demontis F. Muscle-derived Dpp regulates feeding initiation via endocrine modulation of brain dopamine biosynthesis. Genes Dev. 2020;34(1–2):37–52. doi: 10.1101/gad.329110.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ehlen JC, Brager AJ, Baggs J, et al. Bmal1 function in skeletal muscle regulates sleep. Elife. 2017;6:1–14. doi: 10.7554/eLife.26557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harfmann BD, Schroder EA, Kachman MT, Hodge BA, Zhang X, Esser KA. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle. 2016;6:12. doi: 10.1186/s13395-016-0082-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplementary tables. Primary data are provided in Supplementary Table 1, and the statistical analysis of life-span data with OASIS is reported in Supplementary Table 2.