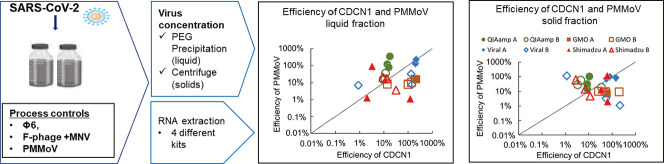
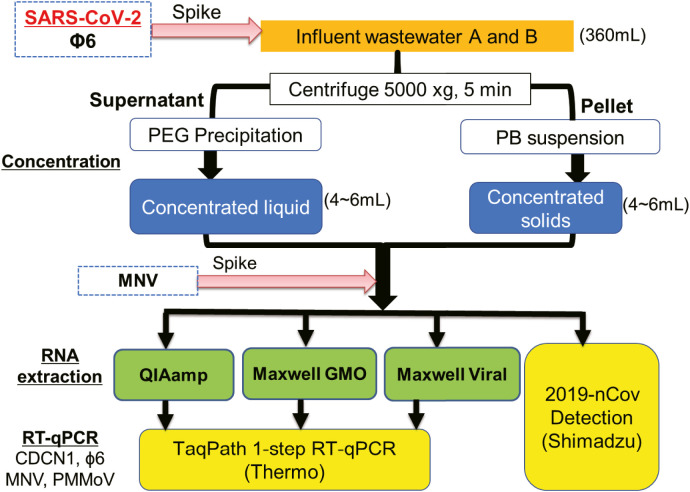
Abstract
Since SARS-CoV-2 RNA in wastewater is often present at low concentration or under detection limit, ensuring the reliability of detection processes using appropriate process controls is essential. The objective of this study was to evaluate applicability and limitations of candidate surrogate viruses as process controls under combinations of different virus concentration and RNA extraction methods. Detection efficiency of SARS-CoV-2 spiked in wastewater was compared with those of candidate surrogate viruses of bacteriophage ϕ6, pepper mild mottle virus (PMMoV), F-specific coliphage (F-phage), and murine norovirus (MNV). After inactivated SARS-CoV-2 and ϕ6 were spiked in two different wastewaters, the viruses in solid and liquid fractions of wastewater were concentrated by centrifuge and polyethylene glycol (PEG) precipitation, respectively. Viral RNA was extracted by using QIAamp Viral RNA Mini Kit and 3 other commercially available extraction kits, then quantified by reverse transcription-quantitative PCR using CDCN1 assay. Regardless of extraction kits, SARS-CoV-2 was consistently detected with good efficiency from both liquid (11–200%) and solid fractions (7.1–93%). Among the candidate process controls, PMMoV was widely detected at good efficiencies from both liquid and solid fractions regardless of selection of RNA extraction kits. F-phage and MNV also showed good detection efficiencies in most combinations of wastewater fractions and RNA extraction kits. An enveloped virus ɸ6 was found often undetected or to have very low detection efficiency (0.1–4.2%) even when SARS-CoV-2 spiked in wastewater was detected with good efficiency. Consequently, PMMoV is widely applicable as process control for detection of SARS-CoV-2 either in liquid fractions concentrated by PEG precipitation, or in solid fractions concentrated by centrifuge.
Keywords: Wastewater-based epidemiology (WBE), Pepper mild mottle virus (PMMoV), Solid fraction, Polyethylene glycol (PEG) precipitation, Pseudomonas phage ϕ6, Murine norovirus (MNV)
Graphical abstract
1. Introduction
The coronavirus infectious disease 2019 (COVID-19) pandemic since 2020 caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has enormous damage to global health and world economics. Wastewater-based epidemiology (WBE) is considered as an efficient approach for early warning of COVID-19 outbreak in the sewershed (Medema et al., 2020; Peccia et al., 2020). Recently, detection of SARS-CoV-2 RNA in wastewater has been reported in many countries (Kumar et al., 2020; Ahmed et al., 2020a; Medema et al., 2020; Peccia et al., 2020; Randazzo et al., 2020; Tomasino et al., 2021; Haramoto et al., 2020; Hata et al., 2021; Wannigama et al., 2021; F. Ahmed et al., 2021). For detection of SARS-CoV-2 RNA in wastewater, concentration of the virus is applied before viral RNA extraction and detection by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay. The typical detection limit for SARS-CoV-2 RNA in wastewater by the currently available methodology is 103 to 104 copies/L. However, concentrations of SARS-CoV-2 RNA in wastewater are reportedly often under the detection limit or only at 104 to 106 copies/L even at the maximum (Kumar et al., 2021, Wurtzer et al., 2020; F. Wu et al., 2020). Therefore, introduction of process control is also important for viruses present in a high concentration, especially when its quantification is aimed. The purpose of the process control is to identify potential underestimation and false negative due to loss of virion and its RNA, and/or inefficient RT-qPCR. For process control, efficiency of concentration, nucleotide extraction and detection by RT-qPCR are checked by using surrogate viruses and nucleic acid (Haramoto et al., 2018). Murine norovirus (MNV), poliovirus (PV) and MS2 coliphage are often used as process controls in detection of enteric viruses in wastewater, e.g. norovirus, human adenovirus, enteroviruses including PV, etc. (Kazama et al., 2017; Hata et al., 2014; Mäde et al., 2013).
SARS-CoV-2 is an enveloped virus wrapped with the phospholipid layer, while most of enteric viruses with the outer layer of protein capsid. Hence, SARS-CoV-2 is considered to have different efficiency in concentration and RNA extraction when compared with those of non-enveloped enteric viruses. Therefore, process controls which are popularly used for enteric virus detection are possibly inappropriate for SARS-CoV-2 detection. According to the recent studies, SARS-CoV-2 and murine hepatitis virus (MHV), a non-human coronavirus, are abundant also in the solid fraction of wastewater unlike non-enveloped enteric viruses (Kitamura et al., 2021; Ahmed et al., 2020b). Thus, concentration methods including the solid phase possibly have better efficiency for enveloped viruses. Moreover, efficiency in RNA extraction was reportedly different between an enveloped virus of Pseudomonas phage ϕ6 and a non-enveloped virus of MS2 (Torii et al., 2021). Therefore, appropriate selection of process control is the key issue to ensure reliability of SARS-CoV-2 detection from wastewater. So far, various viruses have been applied for process control in detection of SARS-CoV-2; e.g. MHV, Pseudomonas phage ϕ6, MS2 coliphage and PMMoV (Ahmed et al., 2020b; Torii et al., 2021). However, no systematic comparison of candidate process control in detection of SARS-CoV-2 in wastewater has been conducted yet.
The objective of this study was to clarify applicability and limitations of candidate process controls in detection of SARS-CoV-2 in wastewater. First, detection efficiency of SARS-CoV-2 and its variance was clarified under different conditions of the concentrated fractions of wastewater (i.e. liquid or solid fraction) and selection of RNA extraction kits. Then, applicability of process controls of ϕ6, PMMoV, F-phage and MNV was evaluated by comparing their detection efficiencies with SARS-CoV-2. Appropriate process control in SARS-CoV-2 was suggested regarding detection efficiency in the applied virus concentration and RNA extraction methods.
2. Materials and methods
Detection efficiencies of SARS-CoV-2 and candidate process controls were compared under different conditions of virus concentration, RNA extraction and RT-qPCR (Fig. 1 ). An enveloped virus ϕ6 and popularly used surrogate viruses of PMMoV, F-phage and MNV were chosen as candidate process controls. After the solid fraction of wastewater was collected and concentrated by centrifugation, the liquid fraction was subjected to concentration by polyethylene glycol (PEG) precipitation. For each of the concentrated fractions, three commercially available RNA extraction kits and one direct RT-PCR kit with different purification procedures were tested to compare efficiencies of RNA extraction and following RT-qPCR detection of SARS-CoV-2 and candidate process controls.
Fig. 1.
Flow diagram of concentration, RNA extraction and Detection for the sample processing. QIAamp = QIAamp Viral RNA Mini Kit, Maxwell GMO = Maxwell RSC PureFood GMO and Authentication Kit, Maxwell viral TNA = Maxwell RSC Viral Total Nucleic Acid (TNA) Purification Kit.
2.1. Wastewater
Wastewater samples with different quality were collected from two wastewater treatment plants (WWTP) located in Japan. Wastewater A had 11.9 mg/L of total organic carbon (TOC), 17.9 mg/L of total nitrogen (TN) and 1.6 g/L of suspended solids (SS); wastewater B had 7.4 mg/L of TOC, 10.3 mg/L of TN and 2.2 g/L of SS. Excitation-emission matrix (EEM) was analyzed by fluorescence spectrophotometer (Spectrofluorometer, JASCO, Japan) (Fig. S1). The wastewater samples were transported on ice and stored at 4 °C until analyses. All samples were processed within six hours after the sampling. The wastewater samples were confirmed to be negative for SARS-CoV-2 by RNA extraction with QIAamp Viral Mini Kit and RT-qPCR quantification using CDCN1 assay with TaqPath 1-Step RT-qPCR Kit as procedures described below.
2.2. SARS-CoV-2 and process controls
Inactivated SARS-CoV-2 (NATrol SARS-CoV-2 Stock, ZeptoMetrix, USA) was used as the seed strain of SARS-CoV-2 in wastewater. 360 mL of wastewater was divided into 40 mL in 9 conical tubes. 40 μL of inactivated SARS-CoV-2 stock solution (1.0 × 106 copies/mL) was added into each of 40-mL wastewater to have the final concentration of 1.0 × 106 copies/L. Pseudomonas phage ϕ6 and PMMoV were used as whole process controls (WPC); F-phage was used as a process control of virus concentration; murine norovirus (MNV) was used as a molecular process control (MPC). PMMoV and F-phage were not spiked into the wastewater samples because they are usually present at a sufficiently high concentration in municipal wastewater. Pseudomonas phage ϕ6 stock solution, prepared in advance as described below, and was spiked 40 μL into each of 40-mL wastewater to have the final concentration of 5 × 107 PFU/L. After spiking SARS-CoV-2 and ϕ6, wastewaters were incubated at 4 °C for 1.5–2 h until the liquid-solid partitioning of spiked viruses reached equilibrium (Ye et al., 2016). A reference virus solution was prepared by spiking 40 μL of inactivated SARS-CoV-2 and 40 μL of Pseudomonas phage ϕ6 into 40 mL of MilliQ water to have the equal virus concentration as the wastewater samples. The original concentrations of F-phage and PMMoV in each wastewater were analyzed as described below.
2.3. Pseudomonas phage Φ6 culture stock
Pseudomonas phage ϕ6 (NBRC 105899, NITE) was propagated using Pseudomonas syringae (NBRC14084, NITE) as the host bacteria strain by the soft agar propagation method (Sinclair et al., 1976). The host culture, Pseudomonas syringae, was prepared by adding 0.5 mL of the seed culture into 30 mL of 2% Luria-Bertani (LB) broth, then incubated at 25–30 °C for 24 h with agitation at 150 rpm. After incubation, 2 mL of the host culture was gently mixed in 30 mL soft agar medium containing 0.6% agar in 2% LB broth. On the petri dish prepared with the bottom agar containing 1.5% agar in 2% LB broth, 5 mL of the soft agar containing the host bacteria was poured and solidified. Then, 100 μL of the ϕ6 strain was spread on the top soft agar, and incubated at 25 °C for 24 h with agitation at 150 rpm. After incubation, the top soft agar was collected and purified by centrifugation at 10,000 ×g, 4 °C for 15 min. The supernatant was filtrated with 0.22-μm sterilized filter (SLGPR33RS, Merck, Japan) to remove the host cells. The concentration of propagated ϕ6 stocks was approximately 5 × 109 PFU/mL according to plaque assay.
2.4. Concentration of viruses
Viruses in the liquid and solid fraction of each wastewater were concentrated by PEG precipitation and centrifuge, respectively. First, 40 mL of the wastewater sample was subjected to centrifugation at 5000 ×g for 5 min at 4 °C to separate into liquid and solid fraction (Fig. 1). The supernatant (liquid fraction) was transferred to another conical tube for concentration process by PEG precipitation (Hata et al., 2021). The remaining pellet (solid fraction) was resuspended with 500 μL of phosphate buffer (PB) solution (concentrated solid fraction). The supernatant was added with 4 g of PEG-8000 (Promega, USA) and 2.3 g of NaCl to the final concentrations of 10% PEG and 1 M NaCl, respectively. After incubation at 4 °C overnight with gentle agitation, the mixture was centrifuged at 10,000 ×g for 30 min. After discarding supernatant, the PEG pellet was resuspended in 500 μL of 10 mM PB solution (concentrated liquid fraction). The concentrates by each method were collected into one tube to make one uniform concentrate sample. The final concentration factor was 44- to 76-fold in the solid fraction and 57- to 62-fold in the liquid fraction. Immediately after the concentration processing, F-phage in each of concentrated liquid and solid fractions was quantified by the conventional plaque assay as described below. The virus concentrate was stored at −80 °C until RNA extraction.
2.5. RNA extraction
Three commercially available RNA extraction kits and one direct RT-PCR kit were used in this study: QIAamp Viral RNA Mini Kit (QIAGEN, USA), Maxwell RSC Viral Total Nucleic Acid Purification Kit (Promega, USA), Maxwell RSC PureFood GMO and Authentication Kit (Promega, USA), and nCoV-2019 Novel Coronavirus Detection Kit (Shimadzu Corporation, Japan). The Shimadzu kit is the direct RT-PCR kit designed for SARS-CoV-2 detection in water and environment samples. RNA extraction was triplicated for each of virus concentrates (N = 3). As molecular process control (MPC), 2 μL of MNV stock (105 TCID50/mL) was spiked into the concentrate sample before extraction. With QIAamp kit, 140 μL of a concentrated sample seeded with 2 μL of MNV was extracted and purified according to manufacturer instruction to obtain 60 μL of total RNA extract. With Maxwell Viral TNA kit, 200 μL of a concentrated sample seeded with 2 μL of MNV was extracted and purified to obtain 50 μL of total RNA extract. With Maxwell GMO kit, 200 μL of concentrated sample with 2 μL of MNV was extracted and purified to obtain 100 μL of total RNA extract. The MNV was not spiked in extraction by Shimadzu kit, which uses only 5 μL of a sample for direct processing until RT-qPCR. The concentration factor by the three RNA extraction kit were 2.3-fold with QIAamp, 4.0-fold with Maxwell Viral TNA and 2.0-fold with Maxwell GMO. The quantity and purity of total RNA extract were assessed by measuring the optical density (OD) at wavelengths of 230 nm, 260 nm and 280 nm using the Eppendorf μCuvette Biophotometer (Eppendorf, Hamburg, Germany). Absorbance ratio at 260/280 nm and 260/230 nm was used to assess the purity of the RNA extract. The RNA extracts were stored at −80 °C until RT-qPCR assay. With Shimadzu kit, 5 μL of a concentrated sample was vortexed with 5 μL of Sample Treatment Reagent according to the instruction manual. The mixture was then incubated at 90 °C for 5 min for the purpose of RNA extraction. The RNA extract by Shimadzu kit was directly processed without purification step for RT-qPCR assay supplied in the same kit.
2.6. RT-qPCR
SARS-CoV-2 and candidate process controls of ɸ6, PMMoV and MNV in the RNA extracts were quantified by one-step RT-qPCR assays. For quantification of SARS-CoV-2, CDCN1 premix primer and probe (IDT, USA) were used. Primer and probe information is provided in Supplementary Material (Table S1). TaqPath 1-Step RT-qPCR Master Mix, CG (Thermo Fisher Scientific) was used for quantification of the target viruses in RNA extracts by QIAamp, Maxwell Viral and Maxwell GMO kits. Five microliter of RNA extract was used as a template for each RT-qPCR assay. RNA extract by Shimadzu kit was processed for RT-qPCR assay supplied in the nCoV-2019 Novel Coronavirus Detection Kit (Shimadzu Corporation, Japan). For quantification of SARS-CoV-2, the multiplex primer-probe set including CDCN1 and CDCN2 which were supplied in the kit was used according to the instruction manual. Quantification of PMMoV and ɸ6 were performed by using the same Shimadzu kit but the primer-probe sets were replaced to ones for the target virus. Thermal cycling condition for the TaqPath kit was 2 min at 25 °C for uracil-DNA glycosylase incubation, 15 min at 50 °C for reverse transcription, 2 min at 95 °C for activation of the Taq DNA polymerase, and 45 cycles of 3 s at 95 °C and 30 s at 60 °C for CDCN1; 40 cycles of 15 s at 95 °C and 60 s at 60 °C for ɸ6, PMMoV and MNV. For Shimadzu kit, thermal cycling condition was 10 min at 42 °C for reverse transcription, 1 min at 95 °C for initial PCR activation and 45 cycles of 5 s at 95 °C and 30 s at 60 °C for CDCN1; 40 cycles of 15 s at 95 °C and 60 s at 60 °C for ɸ6 and PMMoV. All the RT-qPCR assays for the SARS-CoV-2, ɸ6, PMMoV and MNV were performed in triplicate including no template control (NTC) and positive standards. A standard curve was generated from 10-fold serial dilution of synthetic standard ssDNA (Eurofin Genomics, Japan) (Table S2). In the CDCN1 standard, 20 bp of dummy sequence was inserted to enable identification of contamination with standards in PCR assay. To obtain the standard curves for all assays, a 10-fold dilution series of standard was prepared; 1.0 × 101 to 1.0 × 103 copies/reaction for CDCN1 and ɸ6; 1.0 × 102 to 1.0 × 106 copies/reaction for MNV and 1.0 × 103 to 1.0 × 106 copies/reaction for PMMoV. The coefficient of determination (R2) for CDCN1, ɸ6, PMMoV and MNV was over 0.995 with TaqPath kit assay and over 0.985 with Shimadzu kit assay. The limit of detection (LOD) was assumed as 1 copy/reaction. The limit of quantification (LOQ) was determined as copy number in the lowest standard, i.e. 1.0 × 101 copies/reaction for CDCN1 and ɸ6, 1.0 × 102 copies/reaction for MNV, and 1.0 × 103 copies/reaction for PMMoV.
2.7. Concentration efficiency of F-phage
F-phage in a wastewater sample was quantified by plaque assay before and after virus concentration. Salmonella enterica serovar Typhimurium WG49 was used as the host strain, in accordance with the ISO standard 10705-1 (Anon, 1995) as described previously (Mooijman et al., 2002). Concentration efficiency of F-phage was determined from the concentrations before and after the concentration process by the following Eq. (1).
| (1) |
where, R F-phage: concentration efficiency of F-phage, Cc (PFU/mL): the F-phage concentration in the virus concentrate, V c (μL): total volume of the concentrate, V 0 (mL): total volume of the raw wastewater sample (before the virus concentration), C 0 (PFU/μL): the F-phage concentration in the raw wastewater sample.
2.8. Detection efficiencies of SARS-CoV-2, ɸ6, PMMoV, and MNV
Detection efficiencies of SARS-CoV-2, ɸ6, and PMMoV, were calculated from the proportion of the observed concentration detected by RT-qPCR to the reference concentration in the raw sample. The observed concentration in the wastewater sample was determined by the following Eq. (2):
| (2) |
where, C (copies/L): the observed concentration of the target virus in the sample, X t (copies): copy number of the target virus in a template detected by RT-qPCR, V t (μL): template volume, V e (μL): total volume of RNA extract, V c ′ (μL): volume of the concentrate taken for RNA extraction, V c (μL): total volume of the concentrate, V 0 (L): total volume of the original wastewater sample before concentration.
The observed concentration of MNV in the extracted sample was determined by the following Eq. (3)
| (3) |
Detection efficiencies of SARS-CoV-2, ɸ6, PMMoV and MNV were calculated by the following Eq. (4).
| (4) |
where, R: detection efficiency of the target virus, C 0: the reference concentration of the target virus in the original sample (copies/L).
The reference concentrations of SARS-CoV-2 and ɸ6 were determined from the reference virus solution, where the equal amount of the virus was spiked in MilliQ water (as described in the Section 2.2). The reference virus solution in water was directly extracted with QIAamp (without concentration step), then quantified by one-step RT-qPCR with TaqPath 1-Step RT-qPCR kit, as described above (in the Section 2.6). The reference concentration of MNV was determined by spiking 2 μL of the MNV stock in 140 μL of nucleotide-free water, then extracted with QIAamp and quantified by one-step RT-qPCR with TaqPath 1-Step RT-qPCR kit, as described above (in the Section 2.6). The reference concentration of PMMoV was determined by RT-qPCR assay of the raw wastewater sample without the concentration process. The raw wastewater sample was directly subjected to RNA extraction with QIAamp kit, then quantified by one-step RT-qPCR with TaqPath 1-Step RT-qPCR kit, as described above (in the Section 2.6).
3. Results and discussion
3.1. Concentration and quality of RNA extract
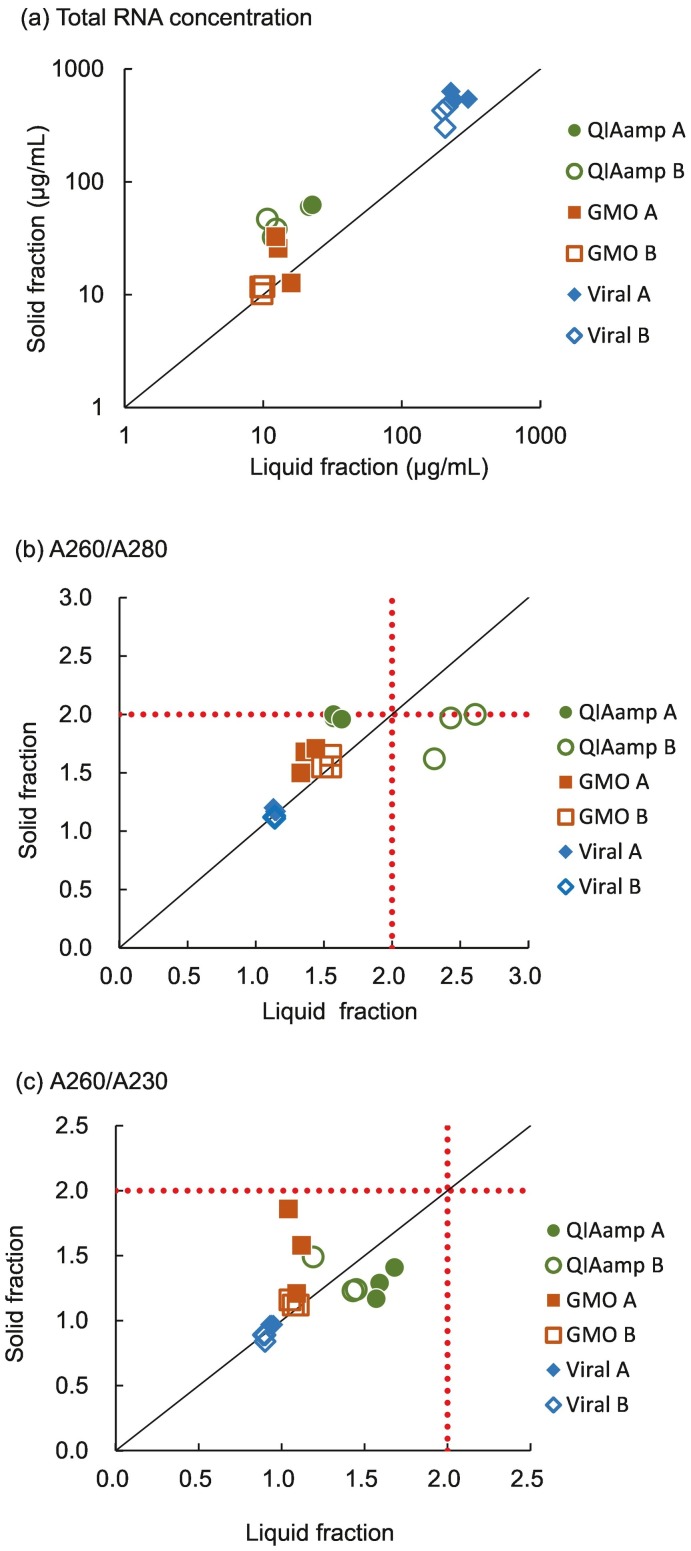
The observed total RNA yield was higher with solid fractions (11.2–62.3 μg/mL) than liquid fractions (9.8–15.5 μg/mL) in most (17 of 18) of the combinations of wastewater sample and extraction kit (Fig. 2a). This is consistent with the previous study reporting that RNA yield was higher in solid fractions than liquid phase (Tomasino et al., 2021). Higher RNA yields in solid fraction were probably because of increased amount of biomass (Lever et al., 2015). Among the three extraction kits, Maxwell Viral kit showed higher observed yield than Maxwell GMO and QIAamp kit, regardless of the sample fractions. However, RNA concentration with Maxwell Viral kit was possibly overestimated because RNA purity was lower than the other two kits, i.e. A260/A280 and A260/230 were remarkably lower with Maxwell Viral kit (1.14–1.18 and 0.87–0.97, respectively) than those with other kits (1.38–2.45 and 1.07–1.61) (Fig. 2b and c). RNA yield and purity was not significantly different between solid and liquid fractions but rather dependent on the extraction kits. RNA purity in terms of A260/A280 was high in the order of QIAamp > Maxwell GMO > Maxwell Viral, although the RNA purity did not affect detection efficiency of SARS-CoV-2 in wastewater (as described in the next section). These differences of the RNA purity among the extraction kits are probably caused by difference of purification mechanisms. QIAamp purifies RNA by using a spin column equipped with RNA-binding silica membrane, while Maxwell GMO and Viral kits purifies by using RNA-binding magnetic beads. Maxwell GMO kit uses “cellulose-coated” magnetic beads, while Maxwell Viral TNA kit uses “silica-coated” magnetic beads to bind nucleic acids. Since cellulose-based beads have more binding capacity of nucleic acids per surface area than silica-based beads, cellulose-based beads have less unspecific binding of organic contaminants. Moreover, cetyltrimethylammonium bromide (CTAB) is added in extraction step of Maxwell GMO kit. Since CTAB enhances removal of proteins by forming complexes with proteins (Tan and Yiap, 2009). Hence, extracts by Maxwell GMO kit had better RNA quality than Maxwell Viral kit. All the extracts were subjected to RT-qPCR quantification of SARS-CoV-2 and the candidate process controls, since the sufficient RNA was extracted.
Fig. 2.
(a) Total RNA concentration (μg/mL), (b) RNA quality by A260/A280, and (c) A260/A230 by concentrated liquid and solids. The concentrates from wastewaters A and B were extracted by QIAamp Viral RNA Mini Kit (QIAamp), Maxwell RSC PureFood GMO and Authentication Kit (GMO), Maxwell RSC Viral Total Nucleic Acid Purification Kit (Viral). Dotted lines are the quality values of the pure RNA.
3.2. Detection efficiency of SARS-CoV-2
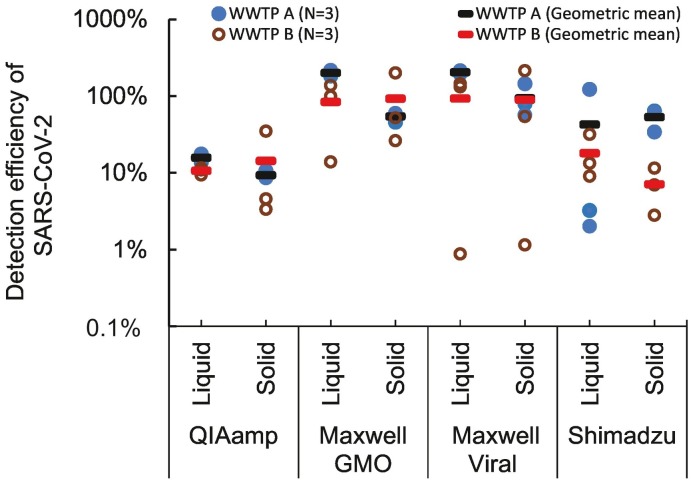
SARS-CoV-2 spiked in wastewater was detected as positive with all fractions and extraction kits. With both liquid and solid fractions, average detection efficiency was higher than 10% except when solid fraction of wastewater A and B was extracted with QIAamp kit (average efficiency =9.3%) and Shimadzu kit (average efficiency = 7.1%), respectively (Fig. 3 ). Detection efficiencies of SARS-CoV-2 ranged 11–204% from the liquid fraction, and 7.1–94% from the solid fraction. The detection efficiencies were not significantly different between the solid and liquid fractions when extracted with the same extraction kit. These results suggest that SARS-CoV-2 are present both in liquid and solid fraction of wastewater, and that the partitioned proportion to liquid and solid phase is not clearly different. This is consistent with the past studies reporting that SARS-CoV-2 was detected in both liquid and solid fractions (Kitamura et al., 2021; Tomasino et al., 2021, Michael-Kordatou et al., 2020, M. Juel et al., 2021, Forés et al., 2021). Meanwhile, Kitamura et al. (2021) reported that SARS-CoV-2 was consistently detected at higher concentration from the solid fraction than from liquid fraction. In Kitamura et al. (2021), the centrifuge condition was 1840 ×g for 30 min, while it was 5000 ×g for 5 min in this study. Therefore, longer time duration of centrifuge condition possibly enhances efficiency in solid fraction.
Fig. 3.
Detection efficiency of SARS-CoV-2 spiked in wastewater. Liquid fractions of wastewaters were concentrated by PEG precipitation; solid fractions by centrifuge. Extraction from each concentrate was triplicated. QIAamp = QIAamp Viral RNA Mini Kit, Maxwell GMO = Maxwell RSC PureFood GMO and Authentication Kit, Maxwell viral TNA = Maxwell RSC Viral Total Nucleic Acid (TNA) Purification Kit.
Detection efficiency of SARS-CoV-2 in wastewater and its stability was more dependent on selection of extraction kits than concentrated fractions of wastewater. Relatively higher detection efficiency was observed with Maxwell GMO and Viral kits than QIAamp and Shimadzu kit. However, the efficiency with Maxwell Viral was sometimes as low as 1%. The efficiencies with Shimadzu kit were also fluctuated. Virus detection efficiency of wastewater is affected by (i) loss in virus concentration (Ye et al., 2016), (ii) loss in RNA extraction (Read, 2001) and (ii) inhibition of PCR (Ahmed et al., 2022). From our results, loss in virus concentration was not critical since the high efficiency was observed both from the liquid and solid fractions. Besides, loss in extraction and/or PCR inhibition was probably more important because the detection efficiency depended on extraction kits. Difference in purification mechanisms of the extraction kits probably affected recovery efficiency of RNA and the residue inhibitory substances. Maxwell GMO kit had more stable detection efficiency than Maxwell Viral kit because of improvement in purification, such as CTAB dose and nucleotide-binding beds. Shimadzu kit could be more sensitive to PCR inhibition, because it has no purification step but designed to directly carry over RNA extract to PCR detection. Dilution of the template may be effective to eliminate PCR inhibition (Hata et al., 2021, Hata et al., 2017).
Wastewater quality was another factor affecting detection efficiency of SARS-CoV-2. Fluctuations of efficiency among replicates were larger in wastewater B than those from wastewater A. Standard deviations of efficiency were larger with wastewater B than wastewater A, especially when extracted with Maxwell Viral kits. From liquid fractions of wastewater A and B, the standard deviations of detection efficiencies were ±7.9% and ± 65% with Maxwell Viral, ±14% and ± 52% with Maxwell GMO, respectively. Similarly, from solid fractions of wastewater A and B, the standard deviations of detection efficiencies were ±0.93% and ± 15% with QIAamp, ±37% and ± 90% with Maxwell Viral, and ± 6.1% and ± 76% with Maxwell GMO, respectively. Larger standard deviations in solid than liquid fractions and with wastewater B than A are possibly originated from heterogeneity of the concentrate. Since the purification procedures with Maxwell Viral and GMO were machine-automated, their fluctuation was not due to handling errors by the operator.
3.3. Applicability of candidate process controls
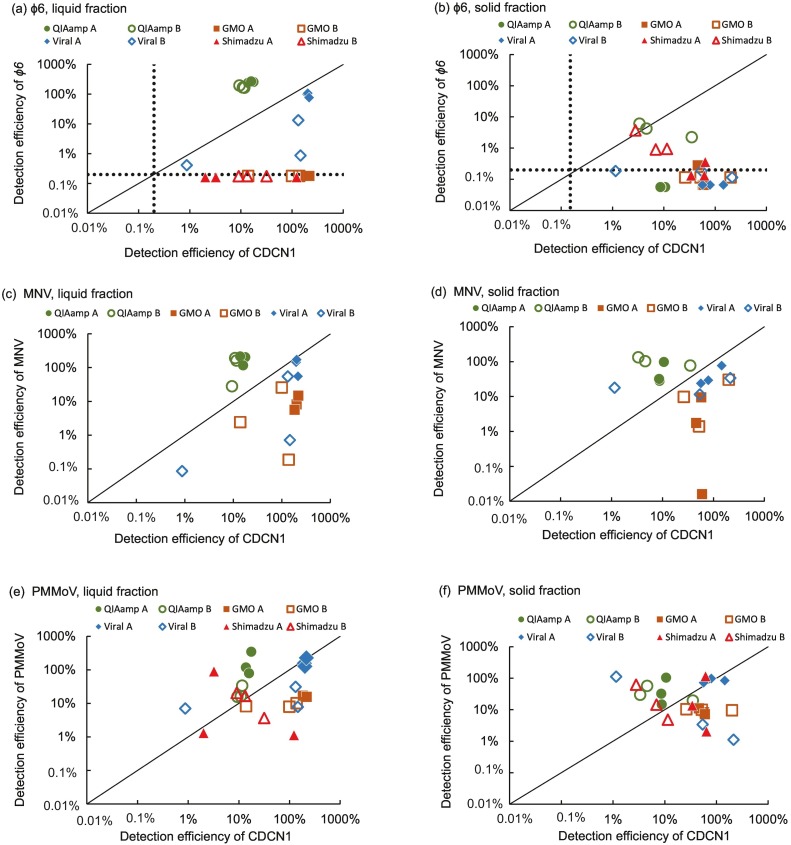
ɸ6 is an enveloped virus expected as an appropriate process control in detection of SARS-CoV-2. However, ɸ6 spiked in wastewater was not detected (<0.20%) in some cases even when detection efficiency of SARS-CoV-2 was higher than 10%. Detection of ɸ6 was quite unstable depending on combinations of sample fraction, RNA extraction kit and wastewater sample. Therefore, application of ɸ6 as surrogate virus needs prior confirmation of detection efficiency under the applied conditions of concentration, RNA extraction and RT-qPCR. In liquid fractions, ɸ6 were detected only with QIAamp and Maxwell Viral kits (Fig. 4a and b). Among the two extraction kits, QIAamp showed good efficiency (216.7 ± 39.8%) for both wastewater samples while Maxwell Viral had lower and fluctuated efficiency (4.9 ± 6.0%) for wastewater B. In solid fractions, ɸ6 were detected in more cases than liquid fraction but mostly with very low detection efficiency (<1%). With QIAamp and Maxwell Viral kits, detection efficiency of liquid fraction was significantly higher form the liquid than the solid fraction of wastewater (p = 0.003). These results suggest that ɸ6 tends to be partitioned in liquid fraction rather than solid fraction of wastewater. Ye et al. (2016) reported virus detection efficiency with liquid fraction was higher than solid fraction. ɸ6 was presented at least 94% of the virions partitioned into the liquid fraction, no more than 0.8% of virions partitioned to the solids (Lee et al., 2016). Meanwhile, ɸ6 were mostly under detection limit or had very low efficiency with Maxwell GMO and Shimadzu kits. Low detection efficiency of ɸ6 in wastewater was also reported by Torii et al. (2022) and 1.4–3.0% in Torii et al. (2021) (0.071–0.51% and 1.4–3.0% respectively), in which the liquid fraction of wastewater were processed by PEG-precipitation and QIAamp. In this study, ɸ6 was not detected or showed low detection efficiency in many cases where SARS-CoV-2 was detected. In such cases, true positives of SARS-CoV-2 could be misinterpreted as false positives; or true negative may be misinterpreted as false negative if only ɸ6 was used for process control. In this sense, ɸ6 is suggested to be applied together with another surrogate virus or only in the case where it is confirmed to be reliably detected.
Fig. 4.
Detection efficiency of CDCN1 and the surrogate viruses of ɸ6, MNV and PMMoV by different combination of concentrated fraction and extraction kits. Dotted line shows limit of detection (LOD).
F-phage and MNV is generally applied in combination for process control. F-phage is used as a control for the concentration process; MNV is used as a control for molecular processes of RNA extraction and RT-qPCR (Haramoto et al., 2018). Advantages of F-phage as the concentration process control are no need of spiking in wastewater and possible evaluation of the concentration efficiency solely. In this study, F-phage concentrations before virus concentration steps were 2.3 and 2.6 log10 PFU/mL in wastewater A and B, respectively. These concentrations were close to the typical range which was reported in previous studies as 2.8–4.5 log10 PFU/mL (Hata et al., 2013, Zhang and Farahbakhsh, 2007, Zanetti et al., 2010). Recovery efficiency of F-phage was higher in concentrate of liquid (13%) than in concentrate of solids (1.1–1.7%) (Table 1 ). These results suggests that F-phage were present both in liquid and solid fraction of wastewater, but with higher proportion in the liquid fraction. Good recovery efficiency of F-phage was also reported in Hata et al. (2021), where liquid fraction was concentrated by PEG precipitation (10–260%). Consequently, F-phage is applicable as a process control of virus concentration efficiency for both solid and liquid fractions, although efficiency in solid fraction can be lower than 10%. It should be also noted that F-phage is not applicable when a wastewater sample has been disinfected or inactivated before concentration. Heat inactivation of a wastewater sample is sometimes suggested to secure safety in sample transportation and storage (Bivins et al., 2020). However, F-phage cannot be quantified by plaque assay if wastewater is inactivated.
Table 1.
Recovery of F-phage.
| Wastewater | F- phage |
Volume (mL) | Fractions | Virus concentration | F-phage in concentrate |
Volume of concentrate |
Concentration factor | Recovery |
|---|---|---|---|---|---|---|---|---|
| (PFU/mL) | (PFU/mL) | (mL) | ||||||
| A | 199 | 360 | Liquid | PEG ppt.a | 1650 | 5.8 | 62 | 13% |
| Solid | Centrifuge | 170 | 4.8 | 76 | 1.1% | |||
| B | 388 | 360 | Liquid | PEG ppt.a | 2900 | 6.3 | 57 | 13% |
| Solid | Centrifuge | 290 | 8.3 | 44 | 1.7% |
PEG ppt. = PEG precipitation.
MNV were successfully detected with all extraction kits. Average detection efficiency of MNV was more than 10% in most of the cases. Average detection efficiencies of MNV spiked in concentrated liquids were 10–179%, except when the liquid fraction of wastewater B was extracted with Maxwell GMO (9.0% efficiency on average) (Fig. 4c and d). Average detection efficiencies of MNV spiked in concentrated solid fractions were 14–104%, except when the solid fraction of wastewater A was extracted with Maxwell GMO (3.8% efficiency on average). Moreover, very low efficiency (0.02%) was found when the solid fraction of wastewater A was extracted with Maxwell GMO. Low detection efficiency of MNV was sometimes observed in the past studies (Hata et al., 2021, Hata et al., 2011, Hata et al., 2017; Torii et al., 2021). The possible reason for such low efficiency of the spiked MNV is presence of RT-PCR inhibitory substances and loss in RNA extraction. In this study, detection efficiency of MNV was always higher than that of CDCN1 with QIAamp kit, although it was significantly lower than CDCN1 when extracted with Maxwell GMO or Viral kits (p < 0.05). Therefore, MNV is applicable as molecular process control in most cases to ensure efficiency of SARS-CoV-2 in RNA extraction step. However, it should be also noted that low detection efficiency is sometimes observed. It is recommended to check the detection efficiency with the selected extraction kits and wastewater samples. If the low efficiency is caused by presence of PCR inhibitory substances, dilution of RNA extract is sometimes effective to improve the efficiency (Hata et al., 2021, Hata et al., 2017).
PMMoV is as a whole process control in detection of enteric viruses in wastewater (Shirasaki et al., 2017, Shirasaki et al., 2018, Shirasaki et al., 2020; Papp et al., 2020). Advantage of PMMoV as the whole process control is its high abundance in wastewater, which allows its quantification without spiking in a wastewater sample. In this study, PMMoV was detected in all cases where SARS-CoV-2 was detected. The observed PMMoV concentrations in wastewater A and B were 8.8 and 8.9 log10 copies/L, respectively. The PMMoV concentrations in the wastewater were in the typical range which was reported in previous studies, i.e. 5.6–10.0 log10 copies/L (Ahmed et al., 2020c). Average detection efficiency of PMMoV was 12–102% from liquid fractions, and 9.4–62% from solid fractions (Fig. 4e and f). The detection efficiency of PMMoV was consistent with the past studies. Torii et al. (2022) reported detection efficiency ranged 7.8–89% in liquid fraction concentrated by PEG precipitation. Graham et al. (2021) reported that detection efficiency of PMMoV was 8.0–30% in liquid fraction processed by PEG precipitation, and 6.0–17% in the solid fraction. In this study no significant difference in detection efficiency were observed between liquid and solid fractions, independent of the applied extraction kits. Hence, the PMMoV was partitioned and detectable in both liquid and solid fractions in wastewater. The lowest detection efficiency was 1.1%, which was observed in two cases, the liquid fraction of wastewater A with Shimadzu kit, and solid fraction of wastewater B with Maxwell GMO kit. However, there were no cases where PMMoV was under detection limit. Consequently, PMMoV was widely applicable as surrogate virus in detection of SARS-CoV-2 in wastewater. PMMoV was always detected among extraction kits and both liquid and solid fractions of wastewater. Since PMMoV were present at detectable level in both liquid and solid fractions in wastewater, it is probably applicable in either case where liquid or solid fraction of wastewater is analyzed. Moreover, PMMoV were detected with at least 1% efficiency regardless of the RNA extraction kits which have different purification mechanisms. Therefore, PMMoV is applicable for a variety of extraction kits by setting 1% efficiency as criteria.
3.4. Appropriate process controls for detection of SARS-CoV-2 in wastewater
The purpose of process control is to identify false negatives and false positives due to process failure as well as to identify inefficient process by checking detection efficiency (Haramoto et al., 2018). Therefore, requirements of a process control are that (i) the surrogate virus is not detected (or have low detection efficiency) when the target virus is not detected by process failure (confirmation of true negative) and that (ii) the surrogate virus is detected with good efficiency when the target virus are successfully detected and quantified. In this study, SARS-CoV-2 spiked in wastewater was detected in both fractions of wastewater and with any combinations of the extraction kits. Therefore, the tested requirement for the surrogate virus is detection with good efficiency in all combinations of wastewater fraction and extraction kits. F-phage had detection efficiency higher than 10% from liquid fraction, while efficiency from solid fraction was approximately 1%. MNV had detection efficiency higher than 10% with most of the cases, although its detection efficiency was even below 1% in some cases. PMMoV as candidate surrogate virus is successfully detected with higher efficiency than 10% in most of the cases, and with higher efficiency than 1% in any combinations of concentrated fractions and extraction kits. In this sense, combination of F-phage and MNV or PMMoV had good applicability as the process control. ɸ6 was not widely applicable because its detection only in the limited cases even when the spiked SARS-CoV-2 was positively detected. In this study, SARS-CoV-2 in liquid fraction of wastewater was concentrated by PEG precipitation and those in solid fraction were concentrated by centrifugation. Some studies reported that solid fraction is an effective source for the detection compared with liquid fraction (Kitamura et al., 2021). However, no significant difference in efficiency was found between the liquid and solid fractions in this study. This is still consistent with Kitamura et al. (2021), in which SARS-CoV-2 were also detected in the liquid fraction. F-phage was used as process control for virus concentration efficiency in the detection of SARS-CoV-2 in wastewater by Hata et al., 2021. In detection of enteric virus in wastewater, only liquid fraction of wastewater is often analyzed. In this study, efficiency of F-phage in solid fraction was also evaluated; however, the efficiency was as low as 1–2%. As a result, F-phage is applicable for process control in concentration of the liquid fraction rather than the solid fraction. However, it is still applicable to solid fraction if 1% of the efficiency is acceptable. MNV was also widely detected in most extraction kits; however, its detection efficiency was sometimes below 10%. RNA extraction kits tested in this study contains protease for extraction of nucleotide from virions. For RNA purification, QIAamp kit basically uses nucleic acid (or RNA) capturing membrane, while Maxwell kits uses magnetic beads to capture nucleotide. (Shimadzu kit has no purification step.) As discussed above, low detection efficiency of MNV was sometimes observed in the past studies (Hata et al., 2021, Hata et al., 2011, Hata et al., 2017; Torii et al., 2021). Therefore, it should be noted that efficiency of MNV might become low affected by wastewater quality and selection of extraction kits. PMMoV is also popularly used as a process control of enteric viruses in wastewater. Liquid fraction of wastewater is often analyzed in detection of enteric viruses. However, PMMoV were detected also in the solid fraction of wastewater (Kitamura et al., 2021; Graham et al., 2021). In this study, detection efficiency of PMMoV in solid fractions was not significantly different from that in liquid fractions. Therefore, PMMoV have good applicability as a process control when solid fraction is used for detection of SARS-CoV-2. Moreover, efficiency of extraction replicates was mostly higher than 10% or at least 1%, independent of extraction kits. Thus, PMMoV is expectedly have stable detection efficiency when detection procedures of SARS-CoV-2 in wastewater are successfully performed. Consequently, PMMoV is an appropriate process control for detection of SARS-CoV-2 in either of liquid or solid fractions of wastewater with a variety of RNA extraction kits using different purification mechanisms.
4. Conclusions
SARS-CoV-2 spiked in wastewater was detected with good efficiency in both liquid (11–200%) and solid (7.1–93%) fractions. Thus, SARS-CoV-2 in wastewater was partitioned and present at similar extent in both of liquid and solid fractions of wastewater. Detection efficiency of SARS-CoV-2 was more dependent on selection of RNA extraction kits. Heterogeneity of the wastewater concentrate also possibly affected fluctuation of efficiency. Among the candidate surrogate viruses for process control, PMMoV was most widely applicable for both of concentration methods and a variety of extraction kits with different purification mechanisms. F-phage and MNV were also applicable in most combinations of concentration methods and RNA extraction kits. An enveloped virus ɸ6 was found often undetected or to have very low efficiency even when SARS-CoV-2 in wastewater was detected. Application of ɸ6 as a surrogate virus is suggested in combination with another surrogate virus, or after their detection is confirmed under the applied concentration and RNA extraction methodology.
CRediT authorship contribution statement
Md. Alamin: Investigation, Formal analysis, Writing – original draft. Shohei Tsuji: Investigation, Formal analysis. Akihiko Hata: Methodology, Writing – review & editing. Hiroe Hara-Yamamura: Supervision, Resources, Formal analysis, Writing – review & editing. Ryo Honda: Supervision, Conceptualization, Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: this study was supported by Japan Science and Technology Agency (JST) MIRAI Program (Grant No. JPMJMI18DC), JST CREST (Grant No. JPMJCR20H1) and partly sponsored by the joint research project with Shimadzu Corporation. Maxwell RNA extraction kits were provided by Promega K.K., Japan.
Acknowledgments
This study was financially supported by JST MIRAI Program (Grant No. JPMJMI18DC), JST CREST (Grant No. JPMJCR20H1), Grants by Hiramoto-Gumi Inc. and I-Tech Muramoto Co. Ltd., Shimadzu Corporation. We acknowledge Technical Service Department of Promega K.K., Japan for their provision and technical support of RNA extraction kits.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.153737.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., Hossen F., Hossain M.S., Islam M.S., Uddin M.M., Islam M.N., Bahadur N.M., Didar-ul-Alam M., Reza H.M., Jakariya M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: variation along the sewer network. Sci. Total Environ. 2021;776 doi: 10.1016/j.scitotenv.2021.145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Blackall L.L., Bofill-Mas S., Bosch A., Brandão J., Choi P.M., Ciesielski M., Donner E., D’Souza N., Farnleitner A.H., Gerrity D., Gonzalez R., Griffith J.F., Gyawali P., Haas C.N., Shanks O.C.… Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739(June) doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Kitajima M., Tandukar S., Haramoto E. Recycled water safety: current status of traditional and emerging viral indicators. Curr. Opin. Environ. Sci. Health. 2020;16:62–72. doi: 10.1016/j.coesh.2020.02.009. [DOI] [Google Scholar]
- Anon . International Organization for Standardization; Geneva, Switzerland: 1995. Water Quality. Detection and Enumeration of Bacteriophages – Part 1. Enumeration of F-Specific RNA Bacteriophages. ISO 10705-1. [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forés E., Bofill-Mas S., Itarte M., Martínez-Puchol S., Hundesa A., Calvo M., Borrego C.M., Corominas L.L., Girones R., Rusiñol M. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total Environ. 2021;768 doi: 10.1016/j.scitotenv.2020.144786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Inaba M., Katayama H., Furumai H. Characterization of natural organic substances potentially hindering RT-PCR-based virus detection in large volumes of environmental water. Environ. Sci. Technol. 2017;51(23):13568–13579. doi: 10.1021/acs.est.7b00306. [DOI] [PubMed] [Google Scholar]
- Hata A., Katayama H., Kojima K., Sano S., Kasuga I., Kitajima M., Furumai H. Effects of rainfall events on the occurrence and detection efficiency of viruses in river water impacted by combined sewer overflows. Sci. Total Environ. 2014;468–469:757–763. doi: 10.1016/j.scitotenv.2013.08.093. [DOI] [PubMed] [Google Scholar]
- Hata A., Kitajima M., Katayama H. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J. Appl. Microbiol. 2013;114(2):545–554. doi: 10.1111/jam.12051. [DOI] [PubMed] [Google Scholar]
- Hata A., Katayama H., Kitajima M., Visvanathan C., Nol C., Furumai H. Validation of internal controls for extraction and amplification of nucleic acids from enteric viruses in water samples. Appl. Environ. Microbiol. 2011;77(13):4336–4343. doi: 10.1128/AEM.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juel M.A.I., Stark N., Nicolosi B., Lontai J., Lambirth K., Schlueter J., Gibas C., Munir M. Performance evaluation of virus concentration methods for implementing SARS-CoV-2 wastewater based epidemiology emphasizing quick data turnaround. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama S., Miura T., Masago Y., Konta Y., Tohma K., Manaka T., Liu X., Nakayama D., Tanno T., Saito M., Oshitani H., Omura T. Environmental surveillance of norovirus genogroups I and II for sensitive detection of epidemic variants. Appl. Environ. Microbiol. 2017;83(9) doi: 10.1128/AEM.03406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021;763 doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Alamin M., Kuroda K., Dhangar K., Hata A., Yamaguchi H., Honda R. Potential discharge, attenuation and exposure risk of SARS-CoV-2 in natural water bodies receiving treated wastewater. Npj Clean Water. 2021;4(1):1–11. doi: 10.1038/s41545-021-00098-2. [DOI] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.T., Pruden A., Marr L.C. Partitioning of viruses in wastewater systems and potential for aerosolization. Environ. Sci. Technol. Lett. 2016;3(5):210–215. doi: 10.1021/acs.estlett.6b00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M.A., Torti A., Eickenbusch P., Michaud A.B., Šantl-Temkiv T., Jørgensen B.B. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front. Microbiol. 2015;6(MAY) doi: 10.3389/fmicb.2015.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäde D., Trübner K., Neubert E., Höhne M., Johne R. Detection and typing of norovirus from frozen strawberries involved in a large-scale gastroenteritis outbreak in Germany. Food Environ. Virol. 2013;5(3):162–168. doi: 10.1007/s12560-013-9118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8(5) doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooijman K.A., Bahar M., Muniesa M., Havelaar A.H. Optimisation of ISO 10705–1 on enumeration of F-specific bacteriophages. J. Virol. Methods. 2002;103(2):129–136. doi: 10.1016/S0166-0934(02)00004-6. [DOI] [PubMed] [Google Scholar]
- Papp K., Moser D., Gerrity D. Viral surrogates in potable reuse applications: evaluation of a membrane bioreactor and full advanced treatment. J. Environ. Eng. 2020;146(2):1–11. doi: 10.1061/(ASCE)EE.1943-7870.0001617. [DOI] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(October) doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S.J. Recovery efficiencies of nucleic acid extraction kits as measured by quantitative LightCyclerTM PCR. J. Clin. Pathol. Mol. Pathol. 2001;54(2):86–90. doi: 10.1136/mp.54.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki N., Matsushita T., Matsui Y., Koriki S. Suitability of pepper mild mottle virus as a human enteric virus surrogate for assessing the efficacy of thermal or free-chlorine disinfection processes by using infectivity assays and enhanced viability PCR. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116409. [DOI] [PubMed] [Google Scholar]
- Shirasaki N., Matsushita T., Matsui Y., Yamashita R. Evaluation of the suitability of a plant virus, pepper mild mottle virus, as a surrogate of human enteric viruses for assessment of the efficacy of coagulation–rapid sand filtration to remove those viruses. Water Res. 2018;129:460–469. doi: 10.1016/j.watres.2017.11.043. [DOI] [PubMed] [Google Scholar]
- Shirasaki N., Matsushita T., Matsui Y., Murai K. Assessment of the efficacy of membrane filtration processes to remove human enteric viruses and the suitability of bacteriophages and a plant virus as surrogates for those viruses. Water Res. 2017;115:29–39. doi: 10.1016/j.watres.2017.02.054. [DOI] [PubMed] [Google Scholar]
- Sinclair J.F., Cohen J., Mindichi L. The isolation of suppressible nonsense mutants of bacteriophage φ6. Virology. 1976;75(1):198–208. doi: 10.1016/0042-6822(76)90018-0. [DOI] [PubMed] [Google Scholar]
- Tan S.C., Yiap B.C. DNA, RNA, and protein extraction: the past and the present. J. Biomed. Biotechnol. 2009;2009 doi: 10.1155/2009/574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasino M.P., Semedo M., Vieira e Moreira P., Ferraz E., Rocha A., Carvalho M.F., Magalhães C., Mucha A.P. SARS-CoV-2 RNA detected in urban wastewater from Porto, Portugal: method optimization and continuous 25-week monitoring. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Oishi W., Zhu Y., Thakali O., Malla B., Yu Z., Zhao B., Arakawa C., Kitajima M., Hata A., Ihara M., Kyuwa S., Sano D., Haramoto E., Katayama H. Comparison of five polyethylene glycol precipitation procedures for the RT-qPCR based recovery of murine hepatitis virus, bacteriophage phi6, and pepper mild mottle virus as a surrogate for SARS-CoV-2 from wastewater. Sci. Total Environ. 2022;807 doi: 10.1016/j.scitotenv.2021.150722. (October 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Furumai H., Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2021;756(xxxx) doi: 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannigama D.L., Amarasiri M., Hurst C., Phattharapornjaroen P., Abe S., Hongsing P., Rad S.M.A.H., Pearson L., Saethang T., Luk-in S., Kueakulpattana N., Storer R.J., Ounjai P., Jacquet A., Leelahavanichkul A., Chatsuwan T. Tracking COVID-19 with wastewater to understand asymptomatic transmission. Int. J. Infect. Dis. 2021;108:296–299. doi: 10.1016/j.ijid.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in wastewater, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee L., Armas F., Kauffman K. SARS-CoV-2 titers in wastewater are higher than expected. MSystems. 2020;5(July):1–9. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zanetti F., De Luca G., Sacchetti R. Performance of a full-scale membrane bioreactor system in treating municipal wastewater for reuse purposes. Bioresour. Technol. 2010;101(10):3768–3771. doi: 10.1016/j.biortech.2009.12.091. [DOI] [PubMed] [Google Scholar]
- Zhang K., Farahbakhsh K. Removal of native coliphages and coliform bacteria from municipal wastewater by various wastewater treatment processes: implications to water reuse. Water Res. 2007;41(12):2816–2824. doi: 10.1016/j.watres.2007.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material