Abstract
Transcription-mediated amplification (TMA) is an isothermal, autocatalytic target amplification method which has the potential to detect less than 50 hepatitis C virus (HCV) RNA copies/ml (10 IU/ml). The TMA assay was used to assess the presence of residual HCV RNA in plasma from patients treated with polyethylene glycol-modified interferon α-2a (peginterferon α-2a) who showed a virologic relapse after the end of therapy. Stored end-of-treatment and end-of-follow-up plasma samples from 177 of 267 patients treated with peginterferon α-2a (S. Zeuzem et al., N. Engl. J. Med. 343:1666–1672, 2000) were available for retesting by TMA. Plasma samples from patients in the same study who exhibited virologic relapse after treatment with standard interferon α-2a served as controls. Virologic response during the trial was defined as HCV RNA that was undetectable using a PCR-based test system with a sensitivity of 50 IU/mL (Cobas Amplicor HCV version 2.0) and was compared with TMA-based retesting results (VERSANT HCV RNA Qualitative Assay). Residual HCV RNA was detected in 4 of 60 cases (7%) by the TMA technology in end-of-treatment plasma samples from patients who relapsed after receiving peginterferon α-2a and in 6 of 18 patients (33%) following therapy with standard interferon α-2a. For peginterferon α-2a-treated patients with sustained virologic response, HCV RNA was detectable by TMA in end-of-treatment samples in 3 of 78 cases but in none of the end-of-follow-up samples. For all end-of-treatment and end-of-follow-up plasma samples of virologic nonresponders, a complete concordance between the PCR-based assay and TMA was observed. In conclusion, in patients with virologic relapse after the end of therapy, according to PCR, who were treated with peginterferon α-2a or standard interferon α-2a, residual HCV RNA was detectable in end-of-treatment samples by the TMA-based assay in 7 or 33% of cases, respectively. The lower rate of residual HCV RNA detection by TMA for patients treated with peginterferon α-2a than that for patients treated with standard interferon α-2a may be due to the maintained antiviral pressure of the long-acting peginterferon α-2a at the end-of-treatment visit.
Chronic hepatitis C virus (HCV) infection is one of the major causes of the development of cirrhosis and hepatocellular carcinoma (3, 18). The current standard treatment of HCV infection with alpha interferon (IFN-α) in combination with ribavirin leads to virologic end-of-treatment response in approximately 50% of cases. At the end of a 24-week follow-up period after completion of therapy, approximately 40% of patients achieve a sustained virologic response (13, 17). Thus, in about one of five patients who achieve an end-of-treatment response, a virologic relapse occurs. Rates of virologic relapse are even higher for IFN-α monotherapy than for combination therapy with IFN-α plus ribavirin. In patients treated with IFN-α alone, HCV RNA cannot be detected by reverse transcription-PCR (RT-PCR) at the end of treatment and the end of follow-up in approximately 30 and 15% of cases, respectively (4, 6, 7, 11, 12). Thus, ribavirin in the current standard combination treatment enhances virologic end-of-treatment response and reduces relapse rates (13, 16). Recently, treatment of patients with chronic hepatitis C with long-acting polyethylene glycol-modified IFN (peginterferon) resulted in an increase in virologic response rates (24, 25). In patients treated for 48 weeks with 180 μg of peginterferon α-2a, HCV RNA was undetectable by RT-PCR in 69 and 39% of patients at the end of treatment and the end of a 24-week follow-up period, respectively (25).
Determination of virologic response is currently based on RT-PCR methods for measurement of HCV RNA in serum or plasma with a lower detection limit of about 100 HCV RNA copies/ml (∼50 IU/ml). Transcription-mediated amplification (TMA) is an isothermal, autocatalytic target amplification method, which has the potential to detect less than 50 HCV RNA copies/ml (<10 IU/ml) (21). The high sensitivity of the TMA-based assay may allow for the detection of low HCV RNA levels in end-of-treatment specimens from patients with subsequent virologic relapse. In a recent pilot study, residual HCV RNA was detected by TMA at the end of treatment in 36% of patients with virologic relapse after standard therapy using IFN-α with or without ribavirin (20). In the present study, end-of-treatment and end-of-follow-up plasma samples were investigated by the TMA-based assay to test for residual HCV RNA in a large cohort of patients who were chronically infected with HCV and were treated with standard IFN α-2a or peginterferon α-2a.
(Part of this work was presented at the Annual Meeting of the European Association for the Study of the Liver (EASL), Prague, Czech Republic, 18 to 22 April 2001.)
MATERIALS AND METHODS
Patients.
Between December 1997 and November 1999, 531 IFN-naïve patients with chronic HCV infection were prospectively enrolled in a global phase III open-label, parallel-dose, randomized, multinational trial. Randomization for subcutaneous treatment either with 180 μg of peginterferon α-2a once per week (PEGASYS; F. Hoffmann-La Roche, Ltd., Basel, Switzerland) for 48 weeks or with IFN α-2a (Roferon-A; F. Hoffmann-La Roche, Ltd.) at a dose of 6 million units thrice weekly for 12 weeks followed by 3 million units thrice weekly for 36 weeks was performed. All patients were monitored until week 72 for determination of sustained virologic response. Written informed consent was obtained from each patient, and the ethics committee at each site approved the study. Diagnosis of chronic HCV infection was based on two elevated serum aminotransferase levels within 6 months of treatment initiation, a positive anti-HCV antibody test, an HCV RNA level above 2,000 copies/ml as determined by a quantitative PCR-based assay, and a pretreatment liver biopsy result consistent with a diagnosis of chronic hepatitis C. All patients were negative for hepatitis B surface antigen and antibodies to human immunodeficiency virus. Further details of study design and assessments have been reported elsewhere (25).
According to the protocol, end-of-treatment samples were obtained 7 days after the last dosing of peginterferon α-2a and 1 to 2 days after the last dosing of standard IFN α-2a. Blood samples drawn from patients for HCV RNA tests were routinely centrifuged within 2 h at the site and were sent on dry ice to the central laboratory. Virologic response was defined as HCV RNA undetectable by RT-PCR at the central laboratory. Additional plasma samples were stored at −80°C.
Week 48 (end of treatment) and week 72 (24 weeks after discontinuation of therapy) plasma samples from all patients who received peginterferon α-2a (n = 267) were selected for retesting by the TMA-based method. Plasma samples of patients who were treated with standard IFN α-2a in the same study and had a virologic relapse after the end of treatment were retested by the TMA-based assay as controls.
PCR-based measurements of HCV RNA.
Qualitative HCV RNA testing within the phase III trial was performed using the Cobas Amplicor HCV version 2.0 (Roche Diagnostics Inc., Mannheim, Germany) assay. In this test, cDNA is made from HCV RNA by RT and is then amplified by PCR under a single set of conditions using the DNA polymerase of Thermus thermophilus (a single-tube, single-enzyme, and single-primer-set process). Details of the assay have been described elsewhere (20). Cobas Amplicor HCV version 2.0 uses 200 μl of plasma for RNA extraction and achieves a sensitivity of 100 HCV RNA copies/ml (50 IU/ml). Compared with version 1.0, Amplicor HCV version 2.0 is independent of HCV genotypes (5, 10). Quantification of HCV RNA was performed by RT-PCR with an internal RNA standard derived from the 5′ noncoding region of HCV (Amplicor HCV Monitor 2.0; Roche Diagnostics). All procedures were performed according to the manufacturer's instructions.
Analyses of plasma samples by TMA.
Stored plasma samples from the end of treatment (week 48) and end of follow-up (week 72) were available from 219 of 267 patients who received 180 μg of peginterferon α-2a once weekly. Plasma samples were not available from patients who prematurely discontinued treatment or when the plasma sample volume was insufficient after RT-PCR testing. In addition, end-of-treatment and end-of-follow-up plasma samples from 18 of 23 virologic relapsers treated with standard IFN α-2a in the same study were available for TMA retesting. All samples were blinded and shipped on dry ice from the study central laboratory for retesting by TMA.
The TMA-based assay (VERSANT HCV RNA Qualitative Assay; Bayer Diagnostics, Emeryville, Calif.) consists of three steps that are performed in a single tube: target capture, target amplification, and specific detection of target amplicons by hybridization protection assay (21). An internal control is added to each sample before the extraction step and is processed through the assay. The appropriate signal generated from the internal control is used as an indication that the result is a valid one. The assay employs 500 μl of plasma. The test system has been previously described in detail (20).
The analytical sensitivity of the VERSANT HCV RNA Qualitative Assay was described by Sawyer et al. (21). In that study the assay had sensitivities of 93 and 100% detection at 25 and 50 HCV RNA copies/ml, respectively (21). The assay detected HCV RNA in dilutions of the World Health Organization HCV standard in 96 and 100% of the replicates tested that contained 5 and 10 IU/ml, respectively. The sensitivity of the assay is reported to be independent of HCV genotypes (21). The clinical specificity is reported to be above 99.5% (21).
Data analysis.
Clinical and biochemical characteristics of patients are expressed as mean, median, and standard deviation as appropriate. Distribution of continuous, independent, nonparametric variables were analyzed by the Mann-Whitney U test (two-tailed). P values of <0.05 were considered significant.
RESULTS
Matched plasma samples from the end of treatment (week 48) and end of follow-up (week 72) were available from 219 of 267 patients who received peginterferon α-2a treatment. Retesting by the TMA-based assay (VERSANT HCV RNA Qualitative Assay) was not possible due to insufficient plasma volumes (<500 μl) in 21 week-48 and 20 week-72 samples. In addition, retesting by the TMA-based assay was determined to be invalid due to unknown errors during the testing procedure in three week-48 and three week-72 samples. In five week-48 samples with insufficient volume for TMA retesting, an invalid test result was obtained in the corresponding week-72 sample or vice versa. Thus, altogether 42 patients had to be excluded from further analyses, whereas for 177 of 219 patients, combined results from end-of-treatment and end-of-follow-up plasma samples were available.
Virologic response was defined by PCR-based HCV RNA testing using the Cobas Amplicor HCV version 2.0 assay. A sustained virologic response with undetectable HCV RNA by RT-PCR at the end of treatment (week 48) and at the end of follow-up 24 weeks after termination of therapy (week 72) was achieved in 78 of 177 patients (44%) treated with peginterferon α-2a. In 60 of 177 patients (34%), HCV RNA was not detectable at the end of treatment, but a virologic relapse occurred after discontinuation of therapy and 39 of 177 patients (22%) were virologic nonresponders. Pretreatment clinical, biochemical, virologic, and histological characteristics of the three groups are summarized in Table 1.
TABLE 1.
Pretreatment clinical, virologic, biochemical, and histological characteristics of patientsa
| Category and no. of patientsb | Sex (male/ female) | Age (yr) | Wt (kg) | Body surface area (m2) | No. (%) of patients belonging to genotypec:
|
Amt of pretreatment HCV RNA (copies/ml) | Amt of ALTd (U/liter) at:
|
Total HAIe score | No. (%) of patients at histological stage of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a/b | 2 | 3 | 4 | Other/ untypeable | Baseline | End of treatment (week 48) | End of follow-up (week 72) | Non-cirrhosis | Bridging fibrosis | Cirrhosis | |||||||
| Virologic sustained responders (78) | 46/32 | 39 ± 9.8 | 71 ± 14.2 | 1.8 ± 0.2 | 35 (45) | 13 (17) | 27 (35) | 2 (2) | 1 (1) | (4.2 ± 6.9) × 106 | 156 ± 137 | 44 ± 22 | 22 ± 16 | 9.1 ± 3.2 | 65 (83) | 9 (12) | 4 (5) |
| Virologic relapse patients (60) | 43/17 | 43 ± 9.7 | 77 ± 12.7 | 1.9 ± 0.2 | 44 (73) | 3 (5) | 10 (17) | 2 (3) | 1 (2) | (9.2 ± 9.1) × 106 | 107 ± 73 | 38 ± 19 | 94 ± 70 | 9.1 ± 3.0 | 51 (85) | 5 (8) | 4 (7) |
| Virologic nonresponders (39) | 22/17 | 44 ± 11 | 79 ± 18.3 | 1.9 ± 0.2 | 31 (79) | 1 (3) | 7 (18) | 0 | 0 | (11.5 ± 17.2) × 106 | 97 ± 59 | 77 ± 48 | 98 ± 80 | 8.0 ± 3.3 | 36 (92) | 2 (5) | 1 (3) |
All ± values represent means ± standard deviations.
Sustained responders, HCV RNA undetectable on week 72 (24 weeks after discontinuation of therapy; relapsers (end of treatment responders), HCV RNA undetectable on week 48 (end of treatment) but with virologic relapse thereafter; nonresponders, HCV RNA positive at end of treatment and thereafter. Classification of the three groups was based on results of Cobas Amplicor HCV version 2.0.
Genotyping was performed with a reverse hybridization assay (INNO LiPA HCV-II; Innogenetics, Ghent, Belgium).
Normal ranges for ALT: male patients, 6 to 43 U/liter; female patients, 6 to 34 U/liter.
HAI, histology activating index.
Among patients with sustained virologic response to treatment with peginterferon α-2a, residual HCV RNA was detected by the TMA-based assay in 3 of 78 (4%) end-of-treatment samples and none of the 78 end-of-follow-up samples (Table 2). A complete concordance between PCR-based results and TMA-based results was observed in all virologic nonresponders, as HCV RNA was detectable in all plasma samples from the 39 nonresponder patients at the end of treatment and the end of follow-up respectively (Table 2). In relapse patients, according to PCR results, HCV RNA was detectable by the TMA-based assay in 4 of 60 end-of-treatment plasma samples (7%) and in all end-of-follow-up samples (Table 2). In patients with virologic relapse after the end of treatment who were treated with standard IFN, HCV RNA was detectable by TMA in 6 of 18 end-of-treatment samples (33%) and in all end-of-follow-up plasma samples.
TABLE 2.
Results from retesting of end-of-treatment and end-of-follow-up plasma samples by TMA (VERSANT HCV RNA Qualitative Assay)
| Patient category (n)a | No. of patients who were
|
|||
|---|---|---|---|---|
| HCV RNA positive by TMA at:
|
HCV RNA negative by TMA
|
|||
| ETRb | EFUb | ETR | EFU | |
| Sustained responders (78) | 3 | 0 | 75 | 78 |
| Relapse patients (60) | 4 | 60 | 56 | 0 |
| Nonresponders (39) | 39 | 39 | 0 | 0 |
Virologic response of patients is defined according to the results of RT-PCR (Cobas Amplicor HCV version 2.0).
ETR, end of treatment (week 48); EFU, end of follow-up (week 72).
All four TMA-positive peginterferon α-2a-treated relapse patients were infected with HCV genotype 1 (HCV-1). However, the majority of peginterferon α-2a-treated patients (44 of 60) (73%) were infected with HCV-1 a/b subtypes, while only 16 of 60 peginterferon α-2a-treated relapse patients (27%) were infected with non-subtype 1 genotypes. The six TMA-positive standard IFN-treated relapse patients were infected with genotypes 1a (n = 3), 2c (n = 1), and 3a (n = 2).
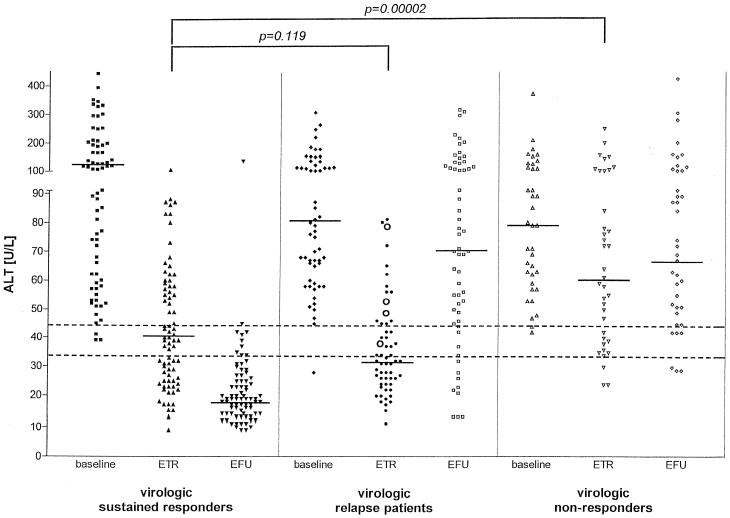
The differences in baseline alanine aminotransferase (ALT) levels (mean ± standard deviation) among sustained virologic responders (156 ± 137 U/liter), relapsers (107 ± 73 U/liter) and nonresponders (97 ± 59 U/liter) treated with peginterferon α-2a were not significant (Fig. 1). Mean end-of-treatment ALT levels were similar for sustained responders (44 ± 22 U/liter) and virologic relapsers (38 ± 19 U/liter), whereas mean ALT levels in virologic nonresponders (77 ± 48 U/liter) were significantly higher at the end of treatment than in sustained responders (44 ± 22 U/liter; P < 0.0001) (Fig. 1). Mean (± standard deviation) ALT levels of the four TMA-positive virologic relapse patients at the end of treatment (55 ± 16 U/liter) were significantly higher than for TMA-negative virologic relapse patients (37 ± 19 U/liter; P = 0.034) (Fig. 1). Interestingly, in 41 of 78 patients (53%) treated with peginterferon α-2a who achieved a sustained virologic response, elevated ALT levels were observed at the end of treatment (Fig. 1). At the end of follow-up, only 4 of 78 sustained virologic responders (4%) showed ALT levels above the upper limit of normal, whereas elevated ALT levels were detected in the majority of virologic relapse (85%) and nonresponder (87%) patients (Fig. 1).
FIG. 1.
ALT levels of sustained virologic responders, virologic relapse patients, and nonresponders measured during baseline (before initiation of therapy), end-of-treatment (ETR, week 48), and end-of-follow-up (EFU, week 72) visits. Horizontal bars indicate median values of the cohorts. Dotted lines indicate the upper limit of normal of ALT levels for men (43 U/liter) and women (34 U/liter). Open circles in the panel of virologic relapse patients at the end of treatment (ETR) indicate patients who had HCV RNA detectable by the TMA-based assay (VERSANT HCV RNA Qualitative Assay).
DISCUSSION
The underlying biological mechanisms for virologic relapse after antiviral therapy of patients with HCV infection are unknown. Generally two possibilities may be considered. Treatment leads to a complete replication arrest, but nonreplicating virions remain in hepatic or extrahepatic sites and resume replication after discontinuation of antiviral therapy. Alternatively, treatment does not completely suppress virus replication, and very low replication rates remain in hepatic or extrahepatic sites (1, 8, 9, 14, 15, 19, 22, 23). Testing the latter hypothesis requires the use of a highly sensitive assay to measure the residual viremia undetectable by currently used PCR-based techniques. TMA is an isothermal, autocatalytic method for detection of targets such as viral RNA. When different HCV RNA reference panels (World Health Organization, U.S. Food and Drug Administration, Paul Ehrlich Institute, and Pelypsy) are used, the TMA-based assay (VERSANT HCV RNA Qualitative Assay) has been shown to detect as few as 25 to 50 HCV RNA copies/ml (5 to 10 IU/ml) (21), compared with 100 copies/ml (50 IU/ml) by PCR-based methods (5).
In the present study, patients treated with a long-acting IFN (peginterferon α-2a) were investigated. The attachment of a 40-kDa polyethylene glycol moiety to IFN α-2a leads to generation of a drug which has substantially more sustained absorption, lower clearance, and a longer half-life than unmodified IFN-α. As reported previously, following therapy with peginterferon α-2a, virologic end-of-treatment and sustained responses were 69 and 39%, respectively (25). Thus, in a considerable proportion of patients (43%), a virologic relapse occurred after discontinuation of therapy. Retesting of stored end-of-follow-up plasma samples by the TMA-based assay confirmed PCR-based results for virologic response at week 72 (end of follow-up). All end-of-follow-up plasma samples from patients with sustained virologic response had no detectable HCV RNA, and all end-of-follow-up plasma samples from patients with virologic relapse or no response were HCV RNA positive by the TMA-based assay. Thus, the sensitivity of currently available PCR-based assays is sufficient for assessment of sustained virologic response 24 weeks after termination of therapy.
HCV RNA was detected by TMA at the end of treatment in 3 of 78 (4%) plasma samples from patients with subsequent, sustained, virologic response. The results of retesting these samples, which scored HCV RNA positive by the TMA-based assay but HCV RNA negative by PCR, might be considered false positive. However, it is possible that the TMA results reflect low levels of viremia not detected by PCR-based techniques at the end of treatment. This residual viremia might reflect late responders or, possibly, replication-incompetent virions which cleared completely after discontinuation of therapy. Residual HCV RNA detected by PCR in end-of-treatment samples of patients with subsequent, sustained, virologic response has also been described previously (16). For virologic nonresponders a complete concordance between PCR-based results and TMA was observed at the end of treatment.
In 4 of 60 (7%) patients with virologic relapse, HCV RNA was detected by TMA in end-of-treatment samples. In comparison, in patients who exhibit virologic relapse after discontinuation of standard IFN α-2a therapy, 6 of 18 (33%) end-of-treatment plasma samples tested HCV RNA positive by TMA. The latter data are in accordance with a previous study showing that 36% of end-of-treatment plasma samples from virologic relapse patients treated with standard IFN with or without ribavirin tested HCV RNA positive by the TMA-based assay (20).
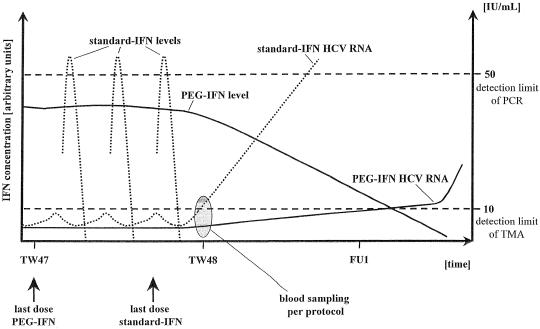
The PCR testing in the previous (20) study and in the present study was performed using Amplicor HCV version 2.0 and the automated Cobas Amplicor HCV version 2.0, respectively. Both test procedures achieve a similar sensitivity. Thus, the HCV RNA detection rate of the TMA-based assay in end-of-treatment plasma from virologic relapsers is apparently dependent on the applied IFN-α preparation (7% versus 33 to 36% in patients treated with peginterferon α-2a and standard IFN-α, respectively). End-of-treatment plasma samples were obtained according to the study protocol 1 to 2 days after the last standard IFN-α injection and 7 days after the last peginterferon α-2a dosing. The difference in half-life in serum between unmodified IFN-α and peginterferon α-2a is considerable (8 h versus 60 to 80 h) (2). Standard serum IFN-α levels are generally undetectable 24 to 36 h after the last injection, whereas peginterferon α-2a is still detectable in serum after a 48-week treatment period for up to 2 to 4 additional weeks of the follow-up period. Thus, most likely the different pharmacokinetic properties of the two drugs explain the differences in the detection rate of the TMA-based assay in virologic relapsers in end-of-treatment plasma samples (Fig. 2). In relapse patients treated with standard IFN-α, HCV RNA may be detectable by TMA but not yet by PCR 1 to 2 days after treatment discontinuation. In patients treated with peginterferon α-2a, detection of a relapse will require a longer posttreatment period before HCV RNA can be detected, as peginterferon α-2a will suppress viral replication as long as it is circulating in the body (Fig. 2). Future studies should prospectively investigate the pharmacokinetics of peginterferon α-2a after treatment discontinuation, and they should include serial HCV RNA measurements by TMA and/or PCR techniques to define the time points of possible HCV RNA detection in patients with relapse. According to the terminal half-life of peginterferon α-2a, detection of virologic relapse could be delayed by several weeks compared with the result for patients treated with standard IFN-α.
FIG. 2.
Proposed model of virologic relapse after the end of treatment in a representative patient treated with standard IFN α-2a (standard IFN) and peginterferon α-2a (PEG-IFN). TW, treatment week. FU, follow-up week.
Interestingly, a discordance between the virologic and biochemical response at the end of treatment was observed in many patients treated with peginterferon α-2a. In 53% (41 of 78) of patients with undetectable HCV RNA at the end of treatment and subsequent, sustained, virologic response, ALT levels above the upper normal limit were observed at the end of treatment, compared with only 5% (4 of 78) of patients at the end of follow-up (Fig. 1). However, patients who had a virologic response but not a biochemical response at the end of treatment had better overall virologic and biochemical responses at the end of follow-up than did patients who had both virologic and biochemical responses at the end of treatment (25).
In conclusion, residual HCV RNA can be detected by the TMA-based assay (VERSANT HCV RNA Qualitative Assay) in 7 and 33% of end-of-treatment samples from patients who exhibit a virologic relapse following therapy with peginterferon α-2a and standard IFN-α, respectively. The lower rate of detection of residual HCV RNA in patients treated with peginterferon α-2a may be due to the pharmacokinetics of this drug leading to a maintained antiviral pressure on HCV replication beyond the time point of treatment discontinuation. Future investigations have to elucidate whether patients with PCR-negative and TMA-positive results for HCV RNA at the end of treatment benefit from prolonged treatment periods.
REFERENCES
- 1.Afonso A M R, Jiang J, Penin F, Tareau C, Samuel D, Petit M A, Bismuth H, Dussaix E, Féray C. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J Virol. 1999;73:9213–9221. doi: 10.1128/jvi.73.11.9213-9221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Algranati N E, Sy S, Modi M. A branched methoxy 40 kDa polyethylene glycol (PEG) moiety optimizes the pharmacokinetics (PK) of peginterferon alfa-2a (PEG-IFN) and may explain its enhanced efficacy in chronic hepatitis C (CHC) Hepatology. 1999;30(Suppl. 4):190A. [Google Scholar]
- 3.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, et al. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 4.Carithers R L, Jr, Emerson S S. Therapy of hepatitis C: meta-analysis of interferon alfa-2b trials. Hepatology. 1997;26(Suppl. 1):83S–88S. doi: 10.1002/hep.510260715. [DOI] [PubMed] [Google Scholar]
- 5.Doglio A, Laffont C, Caroli-Bosc F X, Rochet P, Lefebvre J. Second generation of the automated Cobas Amplicor HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J Clin Microbiol. 1999;37:1567–1569. doi: 10.1128/jcm.37.5.1567-1569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell G C. Therapy of hepatitis C: interferon alfa-n1 trials. Hepatology. 1997;26(Suppl. 1):96S–100S. doi: 10.1002/hep.510260717. [DOI] [PubMed] [Google Scholar]
- 7.Heathcote J. Consensus interferon: a novel interferon for the treatment of hepatitis C. J Viral Hepat. 1998;5(Suppl. 1):13–18. doi: 10.1046/j.1365-2893.1998.0050s1013.x. [DOI] [PubMed] [Google Scholar]
- 8.Laskus T, Radkowski M, Piasek A, Nowicki M, Horban A, Cianciara J, Rakela J. Hepatitis C virus in lymphoid cells of patients coinfected with human immunodeficiency virus type 1: evidence of active replication in monocytes/macrophages and lymphocytes. J Infect Dis. 2000;181:442–448. doi: 10.1086/315283. [DOI] [PubMed] [Google Scholar]
- 9.Lau G K, Davis G L, Wu S P, Gish R G, Balart L A, Lau J Y. Hepatic expression of hepatitis C virus RNA in chronic hepatitis C: a study by in situ reverse-transcription polymerase chain reaction. Hepatology. 1996;23:1318–1323. doi: 10.1002/hep.510230604. [DOI] [PubMed] [Google Scholar]
- 10.Lee S C, Antony A, Lee N, Leibow J, Yang J Q, Soviero S, Gutekunst K, Rosenstraus M. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J Clin Microbiol. 2000;38:4171–4179. doi: 10.1128/jcm.38.11.4171-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W M. Therapy of hepatitis C: interferon alfa-2a trials. Hepatology. 1997;26(Suppl. 1):89S–95S. doi: 10.1002/hep.510260716. [DOI] [PubMed] [Google Scholar]
- 12.Lindsay K. Treatment of chronic hepatitis C: comparative virologic response rates among the different interferons. J Hepatol. 1999;31(Suppl. 1):232–236. doi: 10.1016/s0168-8278(99)80408-5. [DOI] [PubMed] [Google Scholar]
- 13.McHutchison J G, Poynard T. Combination therapy with interferon plus ribavirin for the initial treatment of chronic hepatitis C. Semin Liver Dis. 1999;19(Suppl. 1):57–65. [PubMed] [Google Scholar]
- 14.McHutchison J G, Poynard T, Davis G L, Esteban-Mur R, Harvey J, Ling M, Cort S, Fraud J J, Albrecht J, Dienstag J. Evaluation of hepatic HCV RNA before and after treatment with interferon alfa 2b or combined with ribavirin in chronic hepatitis. Hepatology. 1999;30(Suppl. 2):363A. [Google Scholar]
- 15.Morsica G, Tambussi G, Sitia G, Novati R, Lazzarin A, Lopalco L, Mukenge S. Replication of hepatitis C virus in B lymphocytes (CD19+) Blood. 1999;94:1138–1139. [PubMed] [Google Scholar]
- 16.Reichard O, Norkrans G, Frydén A, Braconier J H, Sönnerborg A, Weiland O. Randomised, double-blind, placebo-controlled trial of interferon a-2b with and without ribavirin for chronic hepatitis C. Lancet. 1998;351:83–87. doi: 10.1016/s0140-6736(97)06088-1. [DOI] [PubMed] [Google Scholar]
- 17.Reichard O, Schvarcz R, Weiland O. Therapy of hepatitis C: alpha interferon and ribavirin. Hepatology. 1997;26(Suppl. 1):108S–111S. doi: 10.1002/hep.510260719. [DOI] [PubMed] [Google Scholar]
- 18.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo Q L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleh M G, Tibbs C J, Koskinas J, Pereira L M, Bomford A B, Portmann B C, McFarlane I G, Williams R. Hepatic and extrahepatic hepatitis C virus replication in relation to response to interferon therapy. Hepatology. 1994;20:1399–1404. doi: 10.1002/hep.1840200604. . (Erratum, 21:900, 1995.) [DOI] [PubMed] [Google Scholar]
- 20.Sarrazin C, Teuber G, Kokka R, Rabenau H, Zeuzem S. Detection of residual hepatitis C virus RNA by transcription-mediated amplification in patients with complete virologic response according to polymerase chain reaction-based assays. Hepatology. 2000;32:818–823. doi: 10.1053/jhep.2000.17709. [DOI] [PubMed] [Google Scholar]
- 21.Sawyer L, Leung K, Friesenhahn M, Duey D, McMorrow M, Eguchi B. Clinical laboratory evaluation of a new sensitive and specific assay for qualitative detection of hepatitis C virus RNA in clinical specimens. J Hepatol. 2000;32(Suppl. 2):116A. [Google Scholar]
- 22.Sugano M, Hayashi Y, Yoon S, Kinoshita M, Ninomiya T, Ohta K, Itoh H, Kasuga M. Quantitation of hepatitis C viral RNA in liver and serum samples using competitive polymerase chain reaction. J Clin Pathol. 1995;48:820–825. doi: 10.1136/jcp.48.9.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas H C, Torok M E, Forton D M, Taylor-Robinson S D. Possible mechanisms of action and reasons for failure of antiviral therapy in chronic hepatitis C. J Hepatol. 1999;31(Suppl. 1):152–159. doi: 10.1016/s0168-8278(99)80393-6. [DOI] [PubMed] [Google Scholar]
- 24.Trepo C, Lindsay K, Niederau C, Shiffman M, Gordon S, Hoefs J, Schiff E, Marcellin P, Bacon B, Fang J, Garaud J, Albrecht J. Pegylated interferon alfa-2b (PEG-Intron) monotherapy is superior to interferon alfa-2b (Intron A) for the treatment of chronic hepatitis C. J Hepatol. 2000;32(Suppl. 2):29A. [Google Scholar]
- 25.Zeuzem S, Feinman S V, Rasenack J, Heathcote E J, Lai M Y, Gane E, O'Grady J, Reichen J, Diago M, Lin A, Hoffman J, Brunda M J. Peginterferon alfa-2a in patients with chronic hepatitis C. N Engl J Med. 2000;343:1666–1672. doi: 10.1056/NEJM200012073432301. [DOI] [PubMed] [Google Scholar]