Abstract
Extended-spectrum β-lactamases (ESBLs) are enzymes found in gram-negative bacilli that mediate resistance to extended-spectrum cephalosporins and aztreonam. In 1999, the National Committee for Clinical Laboratory Standards (NCCLS) published methods for screening and confirming the presence of ESBLs in Klebsiella pneumoniae, Klebsiella oxytoca, and Escherichia coli. To evaluate the confirmation protocol, we tested 139 isolates of K. pneumoniae that were sent to Project ICARE (Intensive Care Antimicrobial Resistance Epidemiology) from 19 hospitals in 11 U.S. states. Each isolate met the NCCLS screening criteria for potential ESBL producers (ceftazidime [CAZ] or cefotaxime [CTX] MICs were ≥2 μg/ml for all isolates). Initially, 117 (84%) isolates demonstrated a clavulanic acid (CA) effect by disk diffusion (i.e., an increase in CAZ or CTX zone diameters of ≥5 mm in the presence of CA), and 114 (82%) demonstrated a CA effect by broth microdilution (reduction of CAZ or CTX MICs by ≥3 dilutions). For five isolates, a CA effect could not be determined initially by broth microdilution because of off-scale CAZ results. However, a CA effect was observed in two of these isolates by testing cefepime and cefepime plus CA. The cefoxitin MICs for 23 isolates that failed to show a CA effect by broth microdilution were ≥32 μg/ml, suggesting either the presence of an AmpC-type β-lactamase or porin changes that could mask a CA effect. By isoelectric focusing (IEF), 7 of the 23 isolates contained a β-lactamase with a pI of ≥8.3 suggestive of an AmpC-type β-lactamase; 6 of the 7 isolates were shown by PCR to contain both ampC-type and blaOXA genes. The IEF profiles of the remaining 16 isolates showed a variety of β-lactamase bands, all of which had pIs of ≤7.5. All 16 isolates were negative by PCR with multiple primer sets for ampC-type, blaOXA, and blaCTX-M genes. In summary, 83.5% of the K. pneumoniae isolates that were identified initially as presumptive ESBL producers were positive for a CA effect, while 5.0% contained β-lactamases that likely masked the CA effect. The remaining 11.5% of the isolates studied contained β-lactamases that did not demonstrate a CA effect. An algorithm based on phenotypic analyses is suggested for evaluation of such isolates.
Resistance to β-lactam antimicrobial agents in gram-negative bacilli is primarily mediated by β-lactamases. Although a variety of β-lactamases have been described, the TEM and SHV enzymes are those most frequently observed among members of the family Enterobacteriaceae (7, 21). Mutations in the genes encoding the TEM and SHV β-lactamases can extend the spectrum of enzyme activity to include penicillins, the extended-spectrum cephalosporins (ESCs) (e.g., ceftazidime [CAZ], cefotaxime [CTX], and ceftriaxone), and aztreonam. Such enzymes are called extended-spectrum β-lactamases (ESBLs). ESBLs are predominantly derivatives of TEM and SHV enzymes (7); however, some oxacillin-hydrolyzing (OXA) (http://www.lahey.org/studies/webt.htm) (26, 36) and CTX-M β-lactamases (42) also show activity against these antimicrobial agents. In addition, the AmpC-type β-lactamases, some of which are encoded on plasmids (7, 10, 15, 20), can also mediate high-level cephalosporin resistance.
Although the original definition of what constituted an ESBL was primarily based on the substrates hydrolyzed by the enzymes, more recently the term ESBL has been limited to those β-lactamases that are inhibited by clavulanic acid (CA), in addition to showing the enhanced spectrum of activity. Although most ESBLs described to date are derived from TEM-1, TEM-2, and SHV-1, CTX-M and some OXA β-lactamases are also inhibited by CA (26, 35, 42). These latter classes of β-lactamases are not mentioned in National Committee for Clinical Laboratory Standards (NCCLS) guidelines M2-A7 and M7-A5 (29, 30). Thus, there continues to be some confusion regarding which β-lactamases are correctly classified as ESBLs.
Confirmation of ESBL production by CA inhibition can be difficult in some strains, not only because the activity of the β-lactamase varies with different substrates, but also because organisms may contain additional resistance mechanisms that can mask the presence of ESBL activity (L. S. Tzouvelekis, A. C. Vatopoulos, G. Katsanis, and E. Tzelepi, Letter, J. Clin. Microbiol. 37:2388, 1999). These could include AmpC-type enzymes (7, 31), porin changes (2, 23, 24), and TEM and SHV β-lactamases that are no longer inhibited by CA due to mutations in the coding sequences (4, 9, 37). Nonetheless, identification of ESBLs is important, since the activity of the ESCs in vivo may not be accurately predicted by susceptibility tests using the traditional NCCLS breakpoints (29).
In 1999, the NCCLS published methods for screening and confirming the presence of ESBLs in Klebsiella pneumoniae, Klebsiella oxytoca, and Escherichia coli (28). This study was conducted to evaluate the NCCLS ESBL phenotypic confirmatory tests that use CAZ and CTX with and without the inhibitor CA to identify the presence of ESBLs in isolates of K. pneumoniae and to assess the contributions of other β-lactamases to the ESBL phenotype.
MATERIALS AND METHODS
Bacterial isolates.
The bacterial isolates selected for this study included 139 K. pneumoniae isolates from 139 patients from 19 of the hospital laboratories participating in phases 1 and 2 of Project ICARE (Intensive Care Antimicrobial Resistance Epidemiology) (1, 14). The laboratories were located in 11 different U.S. states. The organisms were among those isolated between July 1994 and January 1997 by participating laboratories. The number of isolates per submitting laboratory ranged from 1 to 41 (median, 3). Isolates from two laboratories made up 46.8% of the total isolates (24 and 41 isolates per laboratory, respectively); the organisms demonstrated a variety of cephalosporin-resistant phenotypes and isoelectric focusing (IEF) patterns. The study isolates were chosen based on CAZ and CTX MICs from broth microdilution antimicrobial susceptibility testing performed for Project ICARE at the Centers for Disease Control and Prevention by using MIC plates prepared in house with cation-adjusted Mueller-Hinton broth (Difco brand, BD BioSciences, Sparks, Md.) (27). Isolates included in this study were those for which the CAZ or CTX MICs were ≥2 μg/ml, as per the NCCLS screening criteria for ESBL-producing organisms (28). Cefoxitin MICs were also determined, since, for the purposes of this study, resistance to cefoxitin was considered a surrogate marker for either porin loss or the presence of an AmpC-type enzyme (2, 23, 24).
Antimicrobial susceptibility testing.
The 139 isolates were subcultured from −70°C storage onto Trypticase soy agar plates containing 5% defibrinated sheep blood (BD) and then subcultured again before testing. The NCCLS ESBL phenotypic confirmatory tests with CAZ and CTX (28) were performed with each organism with the same bacterial suspension for both broth microdilution and disk diffusion methods. For broth microdilution, MIC plates containing CAZ (Glaxo-Wellcome, Research Triangle Park, N.C.) and CTX (Sigma, St. Louis, Mo.) with and without CA (Smith-Kline Beecham, Collegeville, Pa.) were prepared in house with cation-adjusted Mueller-Hinton broth (Difco). CAZ and CTX (in concentrations of 0.25 to 128 μg/ml) were tested alone and in combination with 4 μg of CA per ml. A subset of isolates were further tested by broth microdilution against cefepime (in concentrations of 0.03 to 32 μg/ml) alone and in combination with 4 μg of CA per ml.
For disk diffusion, Mueller-Hinton agar plates (BD) and disks containing 30 μg of CAZ or CTX (BD), with and without 10 μg of CA, were used for testing. Disks containing CA were prepared by applying 10 μl of a 1,000-μg/ml CA stock solution to each disk (28). The prepared disks were allowed to dry for 30 min before use.
Susceptibility testing results were interpreted according to the criteria established by the NCCLS (28). A ≥3 twofold-concentration decrease in a MIC for either CAZ or CTX tested in combination with CA versus its MIC when tested alone, or a ≥5-mm increase in zone diameter for CAZ or CTX tested in combination with CA versus its zone when tested alone, was considered indicative of ESBL production (i.e., the presence of a CA effect). Broth microdilution and disk diffusion tests were repeated for strains showing discrepant results.
Quality control strains used in this study for antimicrobial susceptibility testing included E. coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and the ESBL control strain, K. pneumoniae ATCC 700603. No ranges have been published for testing cefepime plus CA; however, in-house data collected over 35 test days showed an average decrease of 2 dilutions between cefepime and cefepime plus CA when tested with K. pneumoniae ATCC 700603.
IEF and PCR methods.
IEF and PCR were used for preliminary characterization of the β-lactamases and β-lactamase genes present in the K. pneumoniae isolates. IEF (8, 25) was performed to identify the number and isoelectric points of β-lactamases present. Based on our previous experience, bands with pIs of 5.2 to 6.5 were suggestive of TEM, those with pIs of 7.0 to 8.2 were suggestive of SHV, and those with pIs of ≥8.3 were suggestive of AmpC-type enzymes (7) (http://www.lahey.org/studies/webt.htm). While it is recognized that the pIs of other β-lactamases, such as the CTX-M type (pIs of 7.5 to 8.9) and OXA type (pIs of 5.5 to 9.0), fall within the IEF ranges used in this study, and the pIs of some TEM and AmpC-type enzymes fall outside the ranges (http://www.lahey.org/studies/webt.htm) (5, 12, 16), the suggested IEF ranges, when used in conjunction with the PCR results, proved to be very effective tools for ESBL characterization.
PCR was used to determine the presence of blaTEM and blaSHV in each organism as previously described (22, 38). Testing was repeated when there were discrepancies between PCR and IEF results. A subset of isolates were further evaluated by PCR for the presence of blaOXA, blaCTX-M, and ampC-type genes. Oligonucleotide primers designed to amplify the genes encoding the most common subgroups within the family of OXA β-lactamases are shown in Table 1. Primers OXA-1F and OXA-1R amplify the genes encoding OXA-1 and the closely related OXA-4 and OXA-30 β-lactamases. Primers OXA-2F and OXA-2R amplify the genes encoding OXA-2 and closely related OXA-3, OXA-15, and OXA-21. Primers OXA-10F and OXA-10R amplify the genes encoding OXA-10 and the closely related OXA-7, OXA-11, OXA-13, OXA-14, OXA-16, OXA-17, OXA-19, and OXA-28 enzymes. Primers CTX-M-10F and CTX-M-10R (Table 1) were selected to amplify a 534-bp blaCTX-M fragment (33). Primers CTX-M-2F and CTX-M-2R amplify blaCTX-M-2 and related genes (M. Galas, A. Petroni, R. Melana, A. Corso, M. Rodriguez, M. L. Cacace, A. M. Bru and A. Rossi, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-174, p. 119, 1998). Consensus primers used for the detection of ampC-type genes were those previously described (6). Control organisms included strains of E. coli containing either the blaTEM-1, blaTEM-9, blaSHV-1, blaSHV-3, blaOXA-3, blaOXA-4, blaOXA-7, blaCTX-M-5, or blaCTX-M-9 gene. Additional controls included E. coli C600 (negative control), Enterobacter cloacae P99 (ampC), and Citrobacter freundii 1836 (ampC).
TABLE 1.
Oligonucleotides used for PCR amplification
| Primer | Nucleotide sequence (5′ to 3′) | Locationa | Reference |
|---|---|---|---|
| blaoxa | |||
| OXA-1F | ACA CAA TAC ATA TCA ACT TCG C | 793 | 34 |
| OXA-1R | AGT GTG TTT AGA ATG GTG ATC | 1606 | 34 |
| OXA-2F | TTC AAG CCA AAG GCA CGA TAG | 234 | 11 |
| OXA-2R | TCC GAG TTG ACT GCC GGG TTG | 936 | 11 |
| OXA-10F | CGT GCT TTG TAA AAG TAG CAG | 253 | 17 |
| OXA-10R | CAT GAT TTT GGT GGG AAT GG | 904 | 17 |
| blaCTX-M | |||
| CTX-M-2F | ATG ATG ACT CAG AGC ATT CG | 6 | 3; Galas et al., 38th ICAAC |
| CTX-M-2R | TTA TTG CAT CAG AAA CCG TG | 889 | 3; Galas et al., 38th ICAAC |
| CTX-M-10F | GCA GCA CCA GTA AAG TGA TGG | 215 | 33 |
| CTX-M-10R | GCG ATA TCG TTG GTG GTA CC | 749 | 33 |
| ampC | |||
| CF-Ab | ATT CCG GGT ATG GCC GT | 175 | 6, 20 |
| CF-Bb | GGG TTT ACC TCA ACG GC | 1010 | 6, 20 |
| EC-Ac | CCC TTT GCT GCG CCC TGC | 57 | 6, 15 |
| EC-Bc | TGC CGC CTC AAC GCG TGC | 1162 | 6, 15 |
| COL-Ad | ACG ACG CTC TGC GCC TTA | 69 | 6, 18 |
| COL-Bd | AAG AAT CTG CCA GGC GGC | 1178 | 6, 18 |
Position number corresponds to the location of the first 5′ base of the oligonucleotide within the β-lactamase gene cited.
Consensus sequences of ampC genes of Citrobacter freundii.
Consensus sequences of ampC genes of Enterobacter cloacae.
Primers are specific for the ampC gene of E. coli K-12.
Cycling parameters with primer pairs OXA-1F and OXA-1R or OXA-10F and OXA-10R included a 5-min denaturation at 96°C, followed by 35 cycles of denaturation (96°C for 1 min), annealing (61°C for 1 min), and extension (72°C for 2 min), ending in a final extension period of 72°C for 10 min. These parameters differed from amplification with the other oligonucleotide primers as follows. For OXA-2F plus OXA-2R, the annealing temperature was 65°C. For CTX-M-2F plus CTX-M-2R, denaturation was at 94°C and the annealing temperature was 58°C. For CTX-M-10F plus CTX-M-10R, the annealing temperature was 60°C and there was a cycling extension period of 1 min at 72°C. For CF-A plus CF-B, EC-A plus EC-B, and COL-A plus COL-B, there was a cycling extension period of 1 min at 72°C and there were annealing temperatures of 54, 64, and 61°C, respectively. The IEF and PCR data were used to predict the potential resistance mechanisms in the isolates that did not produce a CA effect.
DNA sequence analysis of blaTEM and blaSHV genes was performed with a subset of isolates. An 867-bp blaTEM amplification product (38) and a 1,017-bp PCR product amplified with oligonucleotides located outside of the blaSHV coding region (32) were sequenced following purification on QIAquick spin columns (Qiagen, Chatsworth, Calif.). Cycle sequencing reactions were performed in a GeneAmp PCR System 9600 thermal cycler with the ABI Prism dRhodamine Terminator Cycle Sequencing Ready Reaction kit according to instructions provided by the vendor (Perkin-Elmer, Applied Biosystems Division [PE-ABI], Foster City, Calif.). Products from sequencing reactions were purified on Centri-Sep spin columns (Princeton Separations, Adelphia, N.J.) before analysis on an ABI Prism 377 DNA Sequencer (PE-ABI). In order to eliminate errors that may have been introduced during amplification, the DNA sequences of leading and lagging strands were determined for independent PCR products. DNA sequencing data were analyzed with DNASIS for Windows (Hitachi Software Genetic Systems, San Francisco, Calif.).
Pulsed-field gel electrophoresis (PFGE) was performed in the Project ICARE laboratory on subsets of isolates from the same institution. After overnight incubation of cultures in Trypticase soy broth (Remel, Lenexa, Kans.), cells were suspended in 1 mM Tris-EDTA (TE) buffer (10 mM Tris-HCl [Sigma], 1 mM EDTA [Sigma]) at pH 7.5, centrifuged, and resuspended in 1 mM TE buffer before the addition of melted 2% SeaPlaque agarose (BioWhittaker Molecular Applications [BMA], Rockland, Maine) (final agarose concentration = 1%). Plugs were prepared in nondisposable plug molds (Bio-Rad Laboratories, Hercules, Calif.). Deproteination of the samples was performed by incubating the plugs in a solution of 0.1 mg of proteinase K per ml (Life Technologies, Inc., Rockville, Md.) in Sarkosyl-EDTA-Tris (SaET) buffer (10 mM Tris-HCI, 0.1 M EDTA, 1% Sarkosyl [Sigma]) at pH 7.5 overnight in a 55°C water bath. After deproteination, the plugs were washed for 15 min at least four times in 1 mM TE buffer, and portions of the plugs were cut and incubated with the restriction endonuclease XbaI (New England BioLabs, Inc., Beverly, Mass.) overnight in a 37°C water bath. The plugs were placed in a 1% SeaKem Gold agarose (BMA) gel. The gel was run on a CHEF DR-III (Bio-Rad Laboratories) under the following conditions: pulse time, 5 to 40 s; run time, 16 h; temperature, 14°C; voltage, 6 V/cm. Banding pattern interpretation was based on published criteria (41).
RESULTS
The NCCLS ESBL phenotypic confirmatory tests were performed by disk diffusion and broth microdilution with 139 clinical isolates of K. pneumoniae for which the CAZ or CTX MICs were ≥2 μg/ml.
Disk diffusion results.
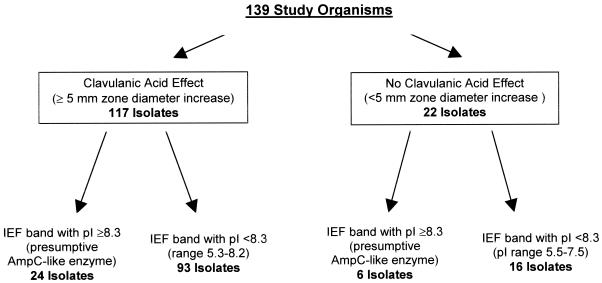
On initial testing by disk diffusion, a CA effect was observed for 117 (84.2%) isolates; i.e., the zone diameters for CAZ plus CA or CTX plus CA were at least 5 mm larger than the zone diameters for CAZ or CTX alone (Table 2). Of the 117 isolates, 104 (88.9%) showed a CA effect with both CAZ and CTX, 11 (9.4%) showed a CA effect with CAZ only, and 2 (1.7%) showed a CA effect with CTX only. The 22 isolates that failed to demonstrate a CA effect by disk diffusion testing had similar zone sizes for CAZ and CAZ plus CA and for CTX and CTX plus CA (Table 3); for these 22 isolates, the cefoxitin MICs were >32 μg/ml. Therefore, by disk diffusion testing, 117 isolates were classified as ESBL producers (Fig. 1).
TABLE 2.
Distribution of zone diameters among 117 isolates demonstrating a CA effect
| Zone size (mm) | CAZ
|
CAZ plus CA
|
Zone size (mm) | CTX
|
CTX plus CA
|
||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Cumulative % | No. of isolates | Cumulative % | No. of isolates | Cumulative % | No. of isolates | Cumulative % | ||
| 6–8 | 57a | 48.7 | 6–8 | 3 | 2.6 | ||||
| 9–11 | 21 | 66.7 | 9–11 | 6 | 7.7 | ||||
| 12–14 | 22 | 85.5 | 5 | 4.3 | 12–14 | 33c | 35.9 | ||
| 15–17 | 8 | 92.3 | 1 | 5.1 | 15–17 | 29 | 60.7 | 2 | 1.7 |
| 18–20 | 6 | 97.4 | 2 | 6.8 | 18–20 | 24 | 81.2 | 4 | 5.1 |
| 21–23 | 3 | 100 | 31 | 33.3 | 21–23 | 9 | 88.9 | 5.1 | |
| 24–26 | 52b | 77.8 | 24–26 | 8 | 95.7 | 8 | 12.0 | ||
| 27–29 | 25 | 99.1 | 27–29 | 3 | 98.3 | 42 | 47.9 | ||
| 30–32 | 1 | 100 | 30–32 | 2 | 100 | 55d | 94.9 | ||
| 33–35 | 33–35 | 6 | 100 | ||||||
Mode = 6 mm.
Mode = 25 mm.
Mode = 12 mm.
Mode = 31 mm.
TABLE 3.
Distribution of zone diameters among 22 isolates failing to demonstrate a CA effect
| Zone size (mm) | CAZ
|
CAZ plus CA
|
Zone size (mm) | CTX
|
CTX plus CA
|
||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | Cumulative % | No. of isolates | Cumulative % | No. of isolates | Cumulative % | No. of isolates | Cumulative % | ||
| 6–8 | 3 | 13.6 | 1 | 4.5 | 6–8 | ||||
| 9–11 | 5 | 36.4 | 5 | 27.3 | 9–11 | 1 | 4.5 | 1 | 4.5 |
| 12–14 | 7a | 68.2 | 7 | 59.1 | 12–14 | 2 | 13.6 | 2 | 13.6 |
| 15–17 | 5 | 90.9 | 7b | 90.9 | 15–17 | 4 | 31.8 | 4 | 31.8 |
| 18–20 | 90.9 | 90.9 | 18–20 | 9c | 72.7 | 11d | 81.8 | ||
| 21–23 | 1 | 95.5 | 1 | 95.5 | 21–23 | 4 | 90.9 | 2 | 90.9 |
| 24–26 | 1 | 100 | 95.5 | 24–26 | 1 | 95.5 | 1 | 95.5 | |
| 27–28 | 1 | 100 | 27–28 | 1 | 100 | 1 | 100 | ||
Mode = 12 mm.
Mode = 16 mm.
Mode = 19 mm.
Mode = 19 mm.
FIG. 1.
Flowchart of disk diffusion and IEF test results.
Broth microdilution results.
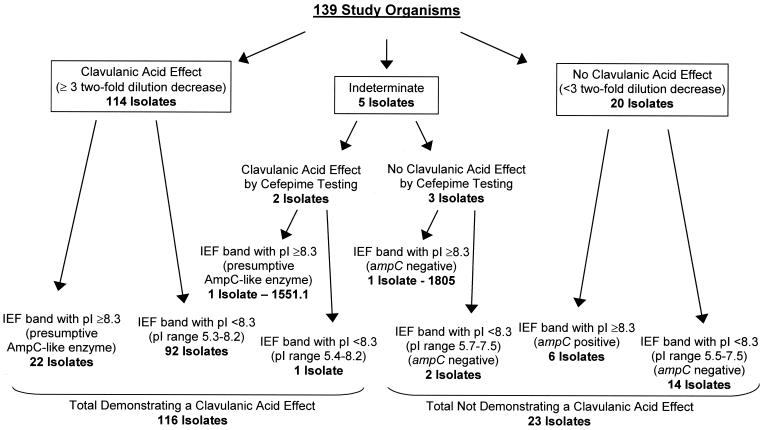
By broth microdilution, a CA effect was observed for 114 (82.0%) isolates (Fig. 2). For these isolates, the MICs for CAZ plus CA or CTX plus CA were decreased by ≥3 dilutions when compared to the MICs for CAZ or CTX alone (Table 4). Of the 114 isolates, 108 (94.7%) demonstrated a CA effect with both CAZ and CTX; the other 6 (5.3%) isolates showed a CA effect for CAZ only. The results for 5 of the 139 isolates were indeterminate, because the CAZ MICs were above the highest dilution tested (>128 μg/ml) and the CAZ plus CA MICs ranged from 64 (CAZ)/4 (CA) to >128/4 μg/ml (Fig. 2). The data for the remaining 20 isolates that failed to demonstrate a CA effect by broth microdilution testing are shown in Table 5.
FIG. 2.
Flowchart of broth microdilution and IEF test results. Only isolates that did not demonstrate a CA effect were tested by PCR for the presence of ampC-type genes.
TABLE 4.
Distribution of MICs among 114 isolates demonstrating a CA effect
| CAZ
|
CAZ plus CA
|
CTX
|
CTX plus CA
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ MIC (μg/ml) | No. of isolates | Cumulative % | CAZ/CA MIC (μg/ml) | No. of isolates | Cumulative % | CTX MIC (μg/ml) | No. of isolates | Cumulative % | CTX/CA MIC (μg/ml) | No. of isolates | Cumulative % |
| ≤0.25 | ≤0.25/4 | 41a | 36.0 | ≤0.25 | 1 | 0.9 | ≤0.25/4 | 107a | 93.0 | ||
| 0.5 | 0.5/4 | 34 | 65.8 | 0.5 | 1 | 1.8 | 0.5/4 | 4 | 97.4 | ||
| 1 | 1/4 | 31 | 93.0 | 1 | 4 | 5.3 | 1/4 | 97.4 | |||
| 2 | 1 | 0.9 | 2/4 | 5 | 97.4 | 2 | 7 | 11.4 | 2/4 | 97.4 | |
| 4 | 2 | 2.6 | 4/4 | 97.4 | 4 | 8 | 18.4 | 4/4 | 1 | 98.2 | |
| 8 | 4 | 6.1 | 8/4 | 97.4 | 8 | 13 | 29.8 | 8/4 | 1 | 99.1 | |
| 16 | 4 | 9.7 | 16/4 | 97.4 | 16 | 13 | 41.2 | 16/4 | 1 | 100 | |
| 32 | 4 | 13.2 | 32/4 | 3 | 100 | 32 | 26 | 64.0 | 32/4 | ||
| 64 | 16 | 27.2 | 64/4 | 64 | 27a | 87.7 | 64/4 | ||||
| 128 | 17 | 42.1 | 128/4 | 128 | 10 | 96.5 | 128/4 | ||||
| >128 | 66a | 100 | >128/4 | >128 | 4 | 100 | >128/4 | ||||
Mode.
TABLE 5.
Distribution of MICs among 20 isolates failing to demonstrate a CA effect
| CAZ
|
CAZ plus CA
|
CTX
|
CTX plus CA
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ MIC (μg/ml) | No. of isolates | Cumulative % | CAZ/CA MIC (μg/ml) | No. of isolates | Cumulative % | CTX MIC (μg/ml) | No. of isolates | Cumulative % | CTX/CA MIC (μg/ml) | No. of isolates | Cumulative % |
| ≤0.25 | ≤0.25/4 | ≤0.25 | ≤0.25/4 | ||||||||
| 0.5 | 0.5/4 | 1 | 5.0 | 0.5 | 1 | 5.0 | 0.5/4 | 1 | 5.0 | ||
| 1 | 1/4 | 5.0 | 1 | 1 | 10.0 | 1/4 | 5.0 | ||||
| 2 | 1 | 5.0 | 2/4 | 5.0 | 2 | 10.0 | 2/4 | 1 | 10.0 | ||
| 4 | 1 | 10.0 | 4/4 | 1 | 10.0 | 4 | 1 | 15.0 | 4/4 | 2 | 20.0 |
| 8 | 10.0 | 8/4 | 10.0 | 8 | 3 | 30.0 | 8/4 | 8a | 60.0 | ||
| 16 | 4 | 30.0 | 16/4 | 2 | 20.0 | 16 | 9a | 75.0 | 16/4 | 7 | 95.0 |
| 32 | 4 | 50.0 | 32/4 | 8a | 60.0 | 32 | 4 | 95.0 | 32/4 | 95.0 | |
| 64 | 3 | 65.0 | 64/4 | 4 | 80.0 | 64 | 95.0 | 64/4 | 95.0 | ||
| 128 | 7a | 100 | 128/4 | 4 | 100 | 128 | 1 | 100 | 128/4 | 1 | 100 |
| >128 | >128/4 | >128 | >128/4 | ||||||||
Mode.
The five isolates yielding indeterminate results were retested by broth microdilution with cefepime and cefepime plus CA in an attempt to obtain on-scale results. A ≥3 twofold-dilution decrease in cefepime MIC was demonstrated for two of the five isolates, 1402 and 1551.1 (Table 6), which were also shown by disk diffusion to demonstrate a CA effect. Therefore, by broth microdilution testing, 116 (83.5%) isolates were classified as ESBL producers.
TABLE 6.
Cefepime and cefepime plus CA results for isolates in which a ≥3-dilution difference between CAZ and CAZ plus CA could not be calculated
| Isolate | Broth microdilution MIC (μg/ml)a
|
Disk diffusion zone size (mm)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CAZ/CA | CTX | CTX/CA | Cefepime | Cefepime/CA | CAZ | CAZ plus CA | CTX | CTX plus CA | |
| 919 | >128 | 128/4 | 32 | 16/4 | 1 | 1/4 | 7 | 9 | 16 | 16 |
| 1400 | >128 | >128/4 | 64 | 32/4 | 2 | 1/4 | 6 | 8 | 14 | 14 |
| 1402 | >128 | 128/4 | 32 | 8/4 | 4 | 0.5/4 | 6 | 12 | 15 | 19 |
| 1551.1 | >128 | 64/4 | 64 | 32/4 | 8 | 0.25/4 | 6 | 14 | 14 | 16 |
| 1805 | >128 | 128/4 | 32 | 32/4 | 1 | 1/4 | 8 | 11 | 13 | 14 |
Boldface values indicate CA effect.
IEF and PCR testing.
All 139 isolates were examined by IEF to determine their β-lactamase profiles and by PCR to determine the presence of blaTEM and blaSHV. IEF analysis revealed that 136 of 139 isolates contained β-lactamases consistent with TEM or SHV as defined in this study. The three remaining isolates each demonstrated a single IEF band with a pI of 6.7, 6.85, or 8.5. A blaSHV gene product was detected in all three isolates by PCR, and each isolate demonstrated a CA effect. One of the 136 isolates demonstrated four bands by IEF (pIs of 5.7, 6.6, 7.1, and 7.5), consistent with the presence of both TEM and SHV β-lactamases; however, repeated PCR testing produced a product consistent only with blaSHV. No blaOXA gene was detected in this isolate by PCR, nor did it demonstrate a CA effect.
When tested by PCR, 138 of 139 isolates contained either blaSHV, blaTEM, or both. The single isolate that was negative repeatedly for blaSHV and blaTEM nonetheless produced one IEF band of pI 7.65 and demonstrated a CA effect with CAZ and CTX. The identity of this β-lactamase is under investigation.
IEF bands with pIs of ≥8.3 were noted in 30 of the 139 isolates, 23 of which showed a CA effect by broth microdilution, including strain 1551.1 which showed a CA effect by cefepime testing (Fig. 2). The cefoxitin MICs for these strains ranged from 4 to >32 μg/ml, suggesting that β-lactamases other than an AmpC type may be present. The seven isolates that failed to show a CA effect (including strain 1805) demonstrated cefoxitin MICs of >32 μg/ml (Fig. 2). Six of the seven isolates were shown by PCR to contain both an ampC-type gene and a blaOXA gene. The seventh isolate (strain 1805) was positive only with blaTEM primers. No blaCTX-M genes were identified in any of the seven isolates.
Sixteen of the 23 isolates that failed to demonstrate a CA effect by broth microdilution had β-lactamases with pIs of ≤7.5 and gave negative results by PCR for blaOXA, blaCTX-M, and ampC-type genes. The reasons for the lack of a CA effect in these isolates are unclear. Thirteen of the 16 isolates were from one laboratory; 11 contained both blaSHV and blaTEM, and 2 contained only blaSHV. PFGE analysis of the 13 isolates showed (i) one cluster of three related isolates (all with similar MICs and IEF profiles), (ii) another cluster of four related isolates (with different numbers of β-lactamases and various MICs), and (iii) six isolates with unique PFGE patterns (data not shown). Thus, the 13 isolates did not represent dissemination of a single clone. However, 12 of the 13 isolates contained an IEF band with a pI between 6.8 and 7.1, possibly indicating the presence of either a plasmid-borne β-lactamase gene other than blaTEM or blaSHV or a blaOXA or ampC variant that was undetected by the oligonucleotide primers used. The other three isolates that failed to show a CA effect were from three different laboratories. Although these isolates all contained blaSHV and blaTEM, they differed in the number and pIs of β-lactamases present in their IEF profiles.
DNA sequence analysis of β-lactamase genes from 5 of the 16 isolates (chosen on the basis of their unique patterns) failed to reveal the presence of a blaTEM or blaSHV with mutations known to confer resistance to β-lactamase inhibitors, such as CA or sulbactam (4, 9, 37) (http://www.lahey.org/studies/webt.htm), nor did the genes examined appear to encode mutations associated with ESBL phenotypes.
Discrepancies between broth microdilution and disk diffusion results.
Discrepant results were noted with three isolates, all of which produced borderline CA effects. Broth microdilution, but not disk diffusion, indicated a CA effect in isolate 976; however, the CAZ result was off-scale. On repeat testing, the CA effect was not observed (Table 7). By PCR, this isolate contained a blaTEM gene. Similarly, a CA effect was observed initially for isolate 1824 by disk diffusion, but not broth microdilution. However, the CA effect was not reproducible. The final isolate, 9632.1, gave borderline positive results by both broth microdilution and disk testing, but never simultaneously. The disk results decreased from 5 mm on initial testing (positive CA effect) to 3 to 4 mm on repeat testing (Table 7), while broth microdilution testing initially showed a 2-dilution difference, but gave a 3-dilution difference on repeat testing. Both isolates 1824 and 9632.1 contained blaTEM, blaSHV, blaOXA, and ampC-type genes by PCR.
TABLE 7.
Disk diffusion versus broth microdilution discrepancies
| Isolate | Band suggestive of AmpC on IEF | Broth microdilution MIC (μg/ml)a
|
Disk diffusion zone size (mm)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| CAZ | CAZ/CA | CTX | CTX/CA | CAZ | CAZ plus CA | CTX | CTX plus CA | ||
| 976 | Yes (pI 8.5) | >128 | 32/4 | 16 | 16/4 | 11 | 15 | 19 | 19 |
| Repeat | 128 | 64/4 | 8 | 16/4 | 10 | 14 | 19 | 16 | |
| Repeat | 128 | 64/4 | 8 | 16/4 | 10 | 14 | 19 | 16 | |
| 1824 | Yes (pI 8.4) | 128 | 32/4 | 16 | 16/4 | 11 | 16 | 16 | 17 |
| Repeat | 128 | 32/4 | 16 | 16/4 | 12 | 16 | 17 | 17 | |
| Repeat | 128 | 64/4 | 32 | 32/4 | 12 | 16 | 17 | 16 | |
| 9632.1 | Yes (pI 8.6) | 128 | 32/4 | 16 | 8/4 | 8 | 13 | 18 | 18 |
| Repeat | >128b | 64/4 | 16 | 16/4 | 8 | 11 | 14 | 15 | |
| Repeat | >128 | 32/4 | 16 | 16/4 | 7 | 11 | 15 | 14 | |
Boldface values indicate CA Effect.
Cefepime MIC = 1 μg/ml; cefepime/CA MIC = 0.25/4 μg/ml.
DISCUSSION
The NCCLS disk diffusion and broth microdilution ESBL confirmatory tests worked well for the 139 K. pneumoniae study isolates identified as potential ESBL producers by NCCLS screening criteria. By disk diffusion and broth microdilution testing, 117 (84.2%) and 114 (82.0%) of isolates tested, respectively, were confirmed as ESBL producers. Broth microdilution tests with cefepime and cefepime plus CA demonstrated a CA effect in two additional isolates, bringing the total to 116 confirmed ESBL producers by broth microdilution.
Broth microdilution testing failed to demonstrate a CA effect in 23 isolates even though they contained blaTEM and blaSHV genes. Seven of the 23 isolates demonstrated IEF bands with pIs of ≥8.3. Six of the seven also contained both AmpC and OXA β-lactamases (confirmed by ampC- and blaOXA-specific PCR assays), which, if not directly responsible for the ESC-resistant phenotype, likely masked any CA effect that may have been present. The remaining 16 isolates that failed to show a CA effect were negative by PCR for ampC-type, blaOXA, and blaCTX-M genes and contained β-lactamases with pIs outside the ranges observed for TEM and SHV in this study. This collection of isolates from four different hospitals probably represents multiple mechanisms of cephalosporin resistance that are not subject to CA inhibition. These may include genes encoding OXA enzymes (7, 21, 26) or AmpC-type enzymes (7, 31; Tzouvelekis et al., Letter) not covered by our primer sets, hyper β-lactamase production (13, 39, 43), porin changes (23), or novel inhibitor-resistant β-lactamases.
For most of the ESBL-producing isolates for which a CA effect was observed, the confirmatory test results comparing CAZ to CAZ plus CA and CTX to CTX plus CA were easily interpreted. Broth microdilution tests typically demonstrated MIC differences of ≥5 twofold dilutions, and disk diffusion tests showed zone diameter differences of ≥10 mm between the antimicrobial agent and its inhibitor combination. Relatively few of the results differed by only 3 to 4 dilutions or 5 to 7 mm, which made interpretation more difficult. One strain gave borderline results by both methods even on repeat testing.
Interpretation of NCCLS confirmatory test results produced by broth microdilution was not possible when the CAZ MICs were off-scale (>128 μg/ml). This occurred with five isolates for which CTX results were on-scale but did not show a CA effect. In these isolates, the results of testing cefepime and cefepime plus CA, a test not specified in NCCLS guidelines, were used to demonstrate a CA effect in two of the isolates. While both of these isolates, 1551.1 and 1402, were thought to produce an AmpC-type enzyme based on their IEF profile, an ampC fragment was amplified by PCR only from the former. However, there are additional ampC genes that are not detected by our three PCR primer sets, and isolate 1402 may contain such a gene. The MICs of cefepime, a cephalosporin with activity against Enterobacter, Serratia, and Pseudomonas species containing AmpC chromosomal β-lactamases, are often lower than those of CAZ and CTX for ESBL-producing Klebsiella species (19, 24, 40). Therefore, testing this antimicrobial agent in conjunction with CA can serve as a secondary indicator of ESBL production, particularly in organisms containing AmpC-type β-lactamases. Alternatively, the range of concentrations for CAZ could be extended beyond 128 μg/ml to improve performance of the broth microdilution test among isolates with higher MICs.
Difficulty in interpretation of CTX MICs also occurred due to off-scale results, but these were because the MICs were below the range tested rather than above it. For six ESBL producers, a ≥3-dilution difference between the CTX and CTX plus CA MICs could not be calculated because the CTX MIC range was ≤0.25 to 1.0 μg/ml and the CTX plus CA MICs were below the test range (≤0.25 μg/ml). However, in these cases, the CAZ and CAZ plus CA results were on-scale and confirmed ESBL production.
Most isolates contained either blaSHV, blaTEM, or both by PCR (138 of 139 [99%]) and contained bands suggestive of either TEM, SHV, or both by IEF testing (136 of 139 [98%]). Together, IEF and PCR make excellent screening tools for studying ESBL-containing strains of K. pneumoniae. The single isolate that was negative for blaSHV and blaTEM contains a β-lactamase that has yet to be identified. Three isolates were positive for blaSHV by PCR but contained only IEF bands outside the range defined for SHV enzymes (with pIs of 6.7, 6.85, and 8.5). This suggests that our IEF range for SHV enzymes may require adjustment for future screening studies. Given the ever increasing number of PCR primer sets required to identify β-lactamase genes (10 sets were used in this study, and several genes still remain unidentified), IEF remains a key tool for characterizing β-lactamase-producing isolates.
For this study, we chose K. pneumoniae isolates that were potential ESBL-producing organisms based on the NCCLS ESBL MIC screening criteria of ≥2 μg/ml for CAZ or CTX. Laboratories that screen for ESBL production by using the traditional intermediate or resistant breakpoints for CAZ and CTX may fail to detect potential ESBL producers. In this study, if a CTX MIC of >8 μg/ml had been used for screening for potential ESBL-producing strains, 34 confirmed ESBL producers would have been missed (Table 4). Similarly, if a CAZ MIC of >8 μg/ml had been used for screening, seven confirmed ESBL producers would have been missed. For disk diffusion, the traditional breakpoints of ≥22 mm for CTX and ≥17 mm for CAZ would have missed 13 and 9 ESBL-producing isolates, respectively (Table 2). In addition, disk diffusion testing of both CAZ and CTX with and without CA, as per NCCLS recommendations, confirmed more isolates as ESBL producers than did testing only CAZ and CAZ plus CA or CTX and CTX plus CA, alone.
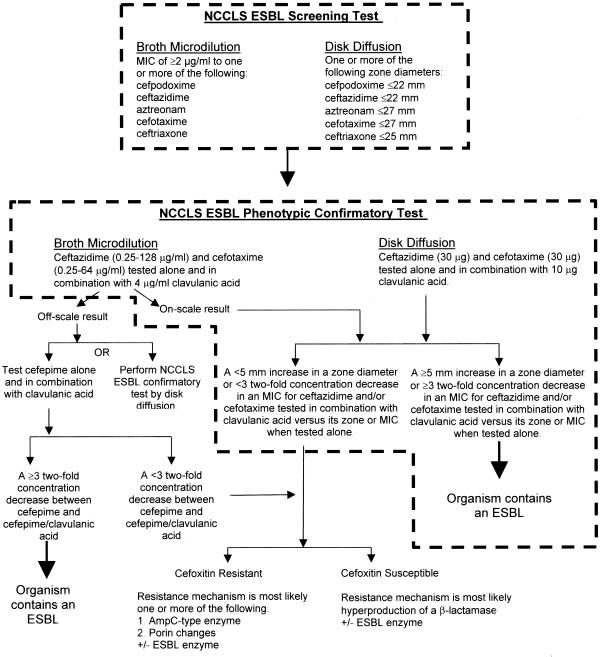
Similar to results published by Coudron et al. (10), we found cefoxitin resistance was a nonspecific indicator of AmpC β-lactamase production. Among the 23 isolates that failed to show a CA effect by broth microdilution, cefoxitin failed to differentiate among those with proven ampC resistance versus isolates with other mechanisms of resistance. Thus, cefoxitin did not prove useful in our study. To help guide laboratories in ESBL detection, we have developed an algorithm for ESBL testing based on phenotypic analysis. It is shown in Fig. 3.
FIG. 3.
Algorithm for ESBL testing by phenotypic methods.
ACKNOWLEDGMENTS
We thank Jana Swenson and Scott Fridkin for assistance in preparation of the manuscript and the microbiology personnel at Project ICARE hospitals for sending the isolates of K. pneumoniae.
Phase 3 of Project ICARE was supported in part by grants to the Rollins School of Public Health of Emory University by Astra-Zeneca Pharmaceuticals, Wilmington, Del. (full sponsor); Pfizer Incorporated, New York, N.Y. (full sponsor); Roche Laboratories, Nutley, N.J. (full sponsor); American Society for Health-System Pharmacists Research and Education Foundation, Bethesda, Md.; Bayer Corporation, Pharmaceuticals Division, West Haven, Conn.; Kimberly-Clark Corporation, Roswell, Ga.; National Foundation for Infectious Diseases, Bethesda, Md.; and Rhône-Poulenc Rorer (now Aventis Pharma), Collegeville, Pa.
REFERENCES
- 1.Archibald L, Phillips L, Monnet D, McGowan J E, Jr, Tenover F, Gaynes R. Antimicrobial resistance in isolates from inpatients and outpatients in the United States: the increasing importance of the intensive care unit. Clin Infect Dis. 1997;24:211–215. doi: 10.1093/clinids/24.2.211. [DOI] [PubMed] [Google Scholar]
- 2.Ardanuy C, Liñares J, Angeles Dominguez M, Hernández-Alléz S, Benedi V J, Martinez-Martinez L. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob Agents Chemother. 1998;42:1636–1640. doi: 10.1128/aac.42.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonomo R A, Rice L B. Inhibitor resistant class A beta-lactamases. Front Biosci. 1999;4:34–41. doi: 10.2741/A477. [DOI] [PubMed] [Google Scholar]
- 5.Bou G, Oliver A, Ojeda M, Monzón C, Martínez-Beltrán J. Molecular characterization of FOX-4, a new AmpC-type plasmid-mediated β-lactamase from an Escherichia coli strain isolated in Spain. Antimicrob Agents Chemother. 2000;44:2549–2553. doi: 10.1128/aac.44.9.2549-2553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bret L, Chanal-Claris C, Sirot D, Chaibi E B, Labia R, Sirot J. Chromosomally encoded AmpC-type β-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother. 1998;42:1110–1114. doi: 10.1128/aac.42.5.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush K, Singer S B. Effective cooling allows sonication to be used for liberation of beta-lactamases from gram-negative bacteria. J Antimicrob Chemother. 1989;24:82–84. doi: 10.1093/jac/24.1.82. [DOI] [PubMed] [Google Scholar]
- 9.Chaibi E B, Sirot D, Paul G, Labia R. Inhibitor-resistant TEM β-lactamases: phenotypic, genetic and biochemical characteristics. J Antimicrob Chemother. 1999;43:447–458. doi: 10.1093/jac/43.4.447. [DOI] [PubMed] [Google Scholar]
- 10.Coudron P E, Moland E S, Thomson K S. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38:1791–1796. doi: 10.1128/jcm.38.5.1791-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dale J W, Godwin D, Mossakowska D, Stephenson P, Wall S. Sequence of the OXA-2 β-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 1985;191:39–44. doi: 10.1016/0014-5793(85)80989-3. [DOI] [PubMed] [Google Scholar]
- 12.Fortineau N, Poirel L, Nordmann P. Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumoniae. J Antimicrob Chemother. 2001;47:207–210. doi: 10.1093/jac/47.2.207. [DOI] [PubMed] [Google Scholar]
- 13.French G L, Shannon K P, Simmons N. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and β-lactam–β-lactamase inhibitor combinations by hyperproduction of SHV-5 β-lactamase. J Clin Microbiol. 1996;34:358–363. doi: 10.1128/jcm.34.2.358-363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridkin S K, Steward C D, Edwards J R, Pryor E R, McGowan J E, Jr, Archibald L K, Gaynes R P, Tenover F C Project Intensive Care Antimicrobial Resistance Epidemiology Hospitals. Surveillance of antimicrobial use and antimicrobial resistance in U.S. hospitals: Project ICARE Phase 2. Clin Infect Dis. 1999;29:245–252. doi: 10.1086/520193. [DOI] [PubMed] [Google Scholar]
- 15.Galleni M, Lindberg F, Normark S, Cole S, Honore S, Joris B, Frère J. Sequence and comparative analysis of three Enterobacter cloacae ampC β-lactamase genes and their product. Biochem J. 1988;250:753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girlich D, Karim A, Spicq C, Nordmann P. Plasmid-mediated cephalosporinase ACC-1 in clinical isolates of Proteus mirabilis and Escherichia coli. Eur J Clin Microbiol Infect Dis. 2000;19:893–895. doi: 10.1007/s100960000386. [DOI] [PubMed] [Google Scholar]
- 17.Huovinen P, Huovinen S, Jacoby G A. Sequence of PSE-2 β-lactamase. Antimicrob Agents Chemother. 1988;32:134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaurin B, Grundström T. AmpC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaye K S, Fraimow H S, Abrutyn E. Pathogens resistant to antimicrobial agents — epidemiology, molecular mechanisms, and clinical management. Infect Dis Clin N Am. 2000;14:293–319. doi: 10.1016/s0891-5520(05)70249-x. [DOI] [PubMed] [Google Scholar]
- 20.Lindberg F, Normark S. Sequence of the Citrobacter freundii OS60 chromosomal AmpC β-lactamase gene. Eur J Biochem. 1986;156:441–445. doi: 10.1111/j.1432-1033.1986.tb09601.x. [DOI] [PubMed] [Google Scholar]
- 21.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mabilat C, Goussard S. PCR detection and identification of genes for extended-spectrum β-lactamases. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 553–559. [Google Scholar]
- 23.Martínez-Martínez L, Hernández-Alléz S, Albertí S, Tomás J M, Benedi V J, Jacoby G A. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1996;40:342–348. doi: 10.1128/aac.40.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Martínez L, Pascual A, Hernández-Allés S, Alvarez-Díaz D, Suárez A I, Tran J, Benedí V J, Jacoby G A. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother. 1999;43:1669–1673. doi: 10.1128/aac.43.7.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 26.Naas T, Nordmann P. OXA-type β-lactamases. Curr Pharm Des. 1999;5:865–879. [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Ninth information supplement, M100–S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7–A5. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M2–A7. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 31.Nordmann P. Trends in β-lactam resistance among Enterobacteriaceae. Clin Infect Dis. 1998;27(Suppl. 1):S100–S106. doi: 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- 32.Nüesch-Inderbinen M T, Kayser F H, Hächler H. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41:943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver A, Pérez-Díaz J C, Coque T M, Baquero F, Cantón R. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob Agents Chemother. 2001;45:616–620. doi: 10.1128/AAC.45.2.616-620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouellette M, Bissonnette L, Roy P H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase gene. Proc Natl Acad Sci USA. 1997;84:7378–7387. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philippon L N, Naas T, Bouthors A-T, Barakett V, Nordmann P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirel L, Girlich D, Naas T, Nordmann P. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid-and integron-located gene. Antimicrob Agents Chemother. 2001;45:447–453. doi: 10.1128/AAC.45.2.447-453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prinarakis E, Miriagou V, Tzelepi E, Gazouli M, Tzouvelekis L S. Emergence of an inhibitor-resistant β-lactamase (SHV-10) derived from an SHV-5 variant. Antimicrob Agents Chemother. 1997;41:838–840. doi: 10.1128/aac.41.4.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasheed J K, Jay C, Metchock B, Berkowitz F, Weigel L, Crellin J, Steward C, Hill B, Medeiros A A, Tenover F C. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother. 1997;41:647–653. doi: 10.1128/aac.41.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice L B, Carias L L, Hujer A M, Bonafede M, Hutton R, Hoyen C, Bonomo R A. High-level expression of chromosomally encoded SHV-1 β-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:362–367. doi: 10.1128/aac.44.2.362-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders C C. In vitro activity of fourth generation cephalosporins against Enterobacteriaceae producing extended-spectrum β-lactamases. J Chemother. 1996;8(Suppl. 2):57–62. [PubMed] [Google Scholar]
- 41.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzouvelekis L S, Tzelepi E, Tassios P T, Legakis N J. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int J Antimicrob Agents. 2000;14:137–142. doi: 10.1016/s0924-8579(99)00165-x. [DOI] [PubMed] [Google Scholar]
- 43.Xiang X, Shannon K, French G. Mechanism and stability of hyperproduction of the extended-spectrum β-lactamase SHV-5 in Klebsiella pneumoniae. J Antimicrob Chemother. 1997;40:525–532. doi: 10.1093/jac/40.4.525. [DOI] [PubMed] [Google Scholar]