Abstract
G-proteins are molecular on–off switches that are involved in transmitting a variety of extracellular signals to their intracellular targets. In animal and yeast systems, the switch property is encoded through nucleotides: a GDP-bound state is the “off-state” and the GTP-bound state is the “on-state”. The G-protein cycle consists of the switch turning on through nucleotide exchange facilitated by a G-protein coupled receptor and the switch turning off through hydrolysis of GTP back to GDP, facilitated by a protein designated REGULATOR OF G SIGNALING 1 (RGS). In plants, G-protein signaling dramatically differs from that in animals and yeast. Despite stringent conservation of the nucleotide binding and catalytic structures over the 1.6 billion years that separate the evolution of plants and animals, genetic and biochemical data indicate that nucleotide exchange is less critical for this switch to operate in plants. Also, the loss of the single RGS protein in Arabidopsis (Arabidopsis thaliana) confers unexpectedly weaker phenotypes consistent with a diminished role for the G cycle, at least under static conditions. However, under dynamic conditions, genetic ablation of RGS in Arabidopsis results in a strong phenotype. We explore explanations to this conundrum by formulating a mathematical model that takes into account the accruing evidence for the indispensable role of phosphorylation in G-protein signaling in plants and that the G-protein cycle is needed to process dynamic signal inputs. We speculate that the plant G-protein cycle and its attendant components evolved to process dynamic signals through signaling modulation rather than through on–off, switch-like regulation of signaling. This so-called change detection may impart greater fitness for plants due to their sessility in a dynamic light, temperature, and pest environment.
A mathematical model of the plant G-signaling switch agrees with recent data and suggests a role of 7-transmembrane regulator of G signaling in processing dynamical signals.
Introduction
Extracellular and intracellular signals operate on switches that turn on and off cellular processes that ultimately affect behavior. The properties of a switch are encoded in its architecture (Ghusinga et al., 2021a). For example, speed, sensitivity, and susceptibility to molecular noise are a switch’s properties that emerge from the way its components interact. Many extracellular signals are perceived in animals by 7-transmembrane (7-TM) receptors on the plasma membrane, known as a G-protein coupled receptors (GPCRs) having a catalytic guanine exchange factor (GEF) property. GPCRs may be viewed as 7-TM GEFs. Associated with a GPCR at the cytoplasmic side is a heterotrimeric complex (simply G complex here) comprising a guanine-nucleotide binding subunit Gα that has GTPase activity and its obligate Gβγ dimer. This heterotrimer is stable when the form of the bound nucleotide is GDP representing the “switch off” state and unstable when it is GTP, representing the “switch on” state (Sprang, 2016). Binding of the agonist to the GPCR promotes a conformation change that propagates to the associated G complex catalyzing the release of the bound GDP on the G subunit thus promoting binding of GTP. The GTP-bound Gα subunit dissociates at least in part from the Gβγ dimer, enabling each of these to interact with other partners to effect cellular behavior (hereafter, G signaling). The switch turns off upon hydrolysis of the bound GTP to GDP by an intrinsic rate of catalysis. This intrinsic hydrolysis is accelerated by as much as three orders of magnitude by a regulator of G signaling (RGS) protein having GTPase activating protein (GAP) activity (Ross and Wilkie, 2000; Hollinger and Hepler, 2002). The switching from off to on to off will be simply referred here as the G cycle.
Much of the G protein research outside of animal cells and yeast has been performed using the genetic model Arabidopsis (Arabidopsis thaliana; Urano and Jones, 2014). Despite a similar list of components comprising the switch, the chemical flux is reversed in plant cells and probably in protists too (Urano et al., 2013a). Plant cells do not require a 7-TM GEF (such as the GPCR in animals) to catalyze guanine nucleotide exchange because this occurs spontaneously and rapidly in vitro (Jones et al., 2011b) and is presumed to occur in vivo (Johnston et al., 2007). Instead, the GDP-bound off state is maintained, at least in part, by a 7-TM RGS protein (Chen et al., 2003). Genetic ablation of this 7-TM RGS does not abolish perception of any of the stimuli; therefore, the 7-TM RGS is not a receptor per se, rather a modulator of the flux as described above for the animal cell G cycle. The discrimination between different stimuli is likely done by kinases (Urano et al., 2012b; Fu et al., 2014; Tunc-Ozdemir et al., 2016). These phosphorylate both the 7TM-RGS (Tunc-Ozdemir et al., 2016; Hackenberg et al., 2017; Tunc-Ozdemir and Jones, 2017) and the Gα subunit (Li et al., 2018; Jia et al., 2019). Phosphorylated 7TM-RGS leads to de-repression of the Gα subunit allowing the Gα subunit to self-activate by exchanging GDP for GTP. It has been shown genetically that this GTP-bound Gα subunit is an output like in animal cells (Chen et al., 2003). Diploid plant genomes encode just one Gα subunit that is canonical in structure to the many Gα subunit types found in animal cells (Jones et al., 2011a).
RGS proteins operate upon the G cycle. Therefore, by deduction, organisms that have RGS proteins must use a G cycle. The prototype for the 7-TM RGS protein is the A. thaliana REGULATOR OF G SIGNALING 1 (AtRGS1; Chen et al., 2003). Except for some grasses (Hackenberg et al., 2017), homologs of AtRGS1 are found throughout the plant kingdom and even in the basal group of algae that gave rise to land plants. Moreover, nonplant genomes such as protists and fungal genomes encode 7-TM RGS proteins, some with multiple paralogs; for instance, the protist Trichomonas vaginalis has at least four 7-TM RGS proteins despite lacking the Gβγ dimer (Urano et al., 2013b) and the fungal pathogen Magnaportha oryza has eight (Zhang et al., 2011). This indicates that 7-TM RGS proteins are ancient and essential. If essential, then why do null mutations in AtRGS1 confer weak phenotypes under laboratory conditions (Urano et al., 2016b)? However, under a more natural condition when the light flux is high and/or dynamic, the phenotype of rgs1 null mutants is the stark inability to optimize photosynthesis efficiency (Liao et al., 2017). This suggests that the reason 7-TM RGS proteins are so highly conserved is that they provide fitness under naturally dynamic conditions and those few grass species that lack 7-TM proteins either do not experience these specific conditions in nature or have evolved another means to provide this fitness. It should be noted that cereals lacking the 7-TM RGS protein grow in open light and not under a forest canopy where light is dynamic due to shadows.
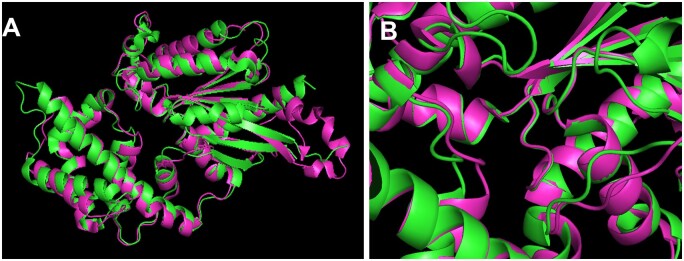
The Gα subunits of plant, animal, and protists have had the time to structurally diverge over 1.6 billion years of potential evolution, but they did not (Jones et al., 2011a). This structural conservation is shown in Figure 1A as an overlay of two Gα subunit crystals structures, one from a plant (green) and one from an animal (magenta). The root mean square deviation (RMSD) between the two entire structures is only 1.3 Å, less than the length of a carbon–carbon bond, while the structure that interacts with the guanine nucleotide (Figure 1B) has an RMSD that is nearly zero, meaning they are essentially identical in the active site. This indicates that nucleotide binding and hydrolysis, i.e. the structure important for the G cycle itself, has had major constraints on its structure evolution. It is therefore clear that nucleotide binding and hydrolysis must be as critical for the plant G cycle as it is for the animal G cycle.
Figure 1.
Plant and animal G subunits have not changed in structure over the 1.6 billion years of potential evolution and show particular conservation in the structure that is important for G cycling. A, Overlay of crystal structure of the plant G subunit (AtGPA1, PDB = 2XTZ; Jones et al., 2011a) and animal (rat Gi1, PDB = 3FFB; Kapoor et al., 2009). Structure 2xTZ is shown in green and structure 3FFB is shown in magenta. The nucleotide binding and hydrolysis pocket is in the center. The overall RMSD is 1.3 Å. B, The nucleotide pocket indicated in (A) is shown in detail with a slight tilting for better view.
However, there are observations that are inconsistent with the conclusion that plant G cycling is important for G signaling. The first came early in the history of plant G protein research: Adjobo-Hermans and coworkers (2006) were unable to detect nucleotide-dependent structural differences in the heterotrimeric complex. They used high-resolution, förster resonance energy transfer in combination with flourescence lifetime imaging microscopy to measure the distance (resolution <10 Å, Gadella, 1999) between subunits of the complex with nanosecond resolution; none was detected. This observation was inconsistent with the animal G cycle paradigm, but, unfortunately, little attention was paid to this paradox. Just a year before, Iwasaki’s group showed that many of the phenotypes of the loss-of-function mutation in the rice G subunit gene such as dwarfness were rescued to wild-type by a Gα subunit that is locked in the GTP-bound state (Oki et al., 2005). This indicates that cycling between GDP and GTP is not critical for function, at least not for the developmental traits that were scored. Another observation challenging a role for G cycling in plants was the discovery by the Assmann group of a set of plant Gα subunit homologs called extra-large G-proteins (XLGs; Lee and Assmann, 1999; Ding et al., 2008; Heo et al., 2012) that lacked the known critical structure (determined in animals) for guanine nucleotide binding and hydrolysis (Temple and Jones, 2007; Chakravorty et al., 2015). For some time, XLGs were dismissed from consideration because, other than homology, there was no strong evidence that XLGs operated in plant G signaling. However, that changed when it was showed that combining genetic ablation of all the XLGs together with the single canonical Gα subunit recapitulated the loss of the Gβγ dimer (Urano et al., 2016a), a mutation that seems to completely eliminate G signaling in plants (Ullah et al., 2003). These strange XLG proteins were thought to work somewhere in plant G signaling; only after discovery of their physical interactions with the Gβγ dimer and with its cognate RGS protein in a nucleotide-independent manner (Lou et al., 2020) did it become clear that this noncanonical Gα subunit that lacks nucleotide binding and hydrolysis, nonetheless plays a role in the plant G cycle (Liang et al., 2017). Another observation was that altering the G cycle by genetic ablation of the RGS1 gene had little effect on plant G signaling (Chen et al., 2003, 2006; Urano et al., 2016b), that is until the response to dynamic signal was measured (discussed below). The most recent observation challenging a role for the G cycle in plant G signaling is the observation that a point mutation in a plant G subunit that eliminates GTP binding nonetheless is able to rescue the Gα subunit null mutant (Maruta et al., 2019). After all, if an empty G subunit acts almost as well as the wild-type Gα subunit, then it seems reasonable to challenge a role for G cycling in plant G signaling. Can a new model reconcile this paradox?
As mentioned above, genetic loss of the RGS protein, clearly shown to control the active state of the G complex in vitro, has many morphological phenotypes weaker than expected for a master regulator of the G cycle. However, there is one phenotype of the rgs1 null mutant that is rather profound. Under dynamic and/or high light irradiation, rgs1 mutants lack the ability to adjust photosynthetic efficiency, whereas these mutants behave like the wild-type when the light is constant (Liao et al., 2017)). Something like this time-dependent signal presentation was seen before with plant G protein mutants. Roots respond to touch by secreting ATP (Weerasinghe et al., 2009), a signal that regulates calcium-based responses. When a second touch is applied to the wild-type root, there is no ATP secretion for another 540 s; however, G protein mutants respond to this second mechano-stimulation right away. This raises the possibility that plant G cycling plays a role in time-dependent signaling. If that is the case, then what is the mechanism for activation of plant G signaling and how are signals discriminated?
It is possible that this mechanism for activation and the discrimination of signals in plant G signaling is accomplished by receptor kinases (Tunc-Ozdemir et al., 2016, 2017; Tunc-Ozdemir and Jones, 2017; Chakravorty and Assmann, 2018; Liang et al., 2018). Plant genomes have had an expansive amplification of receptor-like kinases (RLKs), analogous to the amplification of GPCRs in animals and while animal G signaling does involve kinases, those kinases modulate signal transduction and are not presently known to be involved in signal perception and G activation. RLKs phosphorylate AtRGS1 and A. thaliana G PROTEIN ALPHA SUBUNIT 1 (AtGPA1) to alter their function in a way anticipated to affect switching, raising the possibility that RLKs are the discriminators in this switch. Thus, what appears to be unfolding currently is that a signal-dependent phosphorylation–dephosphorylation cycle is the mechanism underlying the plant G signaling on/off switch.
We are left with several questions: Does phosphorylation underlie the switch mechanism? What is the role of nucleotide binding? What is the role of AtRGS1? We explore these questions by formulating a mathematical model that is consistent with the evidence thus far. Our model provides a simple resolution of the paradox about nucleotide binding as well as the role of AtRGS1 by proposing that the phosphorylation status governs the switch activity. The steady-state behavior of the model for a constant signal strength can be explained as follows. The switch now takes four states, two of which are phosphorylated and the other two are not. In the absence of any signal, the switch will primarily live in the GDP-bound off state in wild-type and GTP-bound off state in the rgs1 mutant, both of which do not have any strong phenotypes under laboratory conditions. When a signal arrives, phosphorylation of AtGPA1 through RLK occurs, turning the switch on. Because AtRGS1 is also phosphorylated, decoupling its GAP activity from the Gα subunit, the amount of the signal in either of the phosphorylated state would be governed by the amount of AtRGS protein left on the plasma membrane. In case of the rgs1 mutant, the switch will primarily live in the GTP bound on state. Except for some weak signal strengths (for constant signals), it would be difficult to distinguish the wild-type and rgs1 mutant if the phosphorylated GTP bound state is the only signaling competent state.
Results
Revising the model of G-signaling
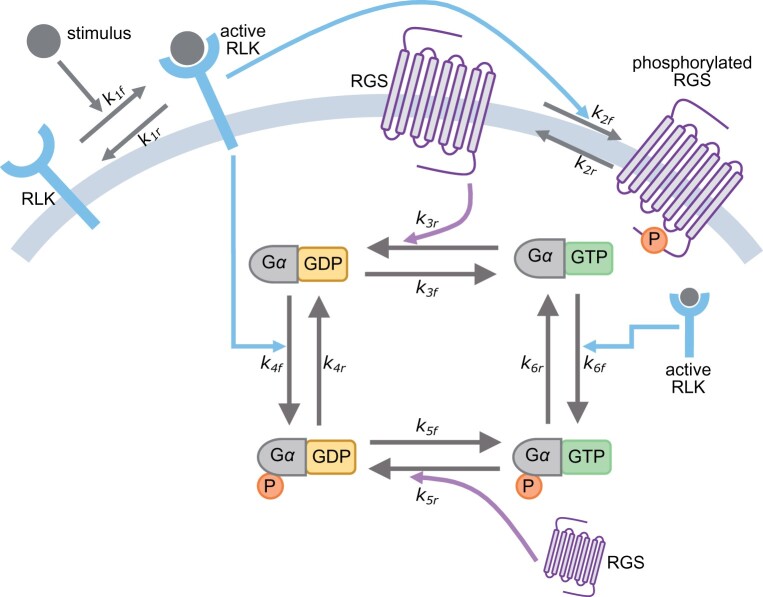
A simple model comprising spontaneous nucleotide exchange and de-repression of the switch through the hydrolysis rate is the core of G-signaling paradigm in plants (Urano et al., 2012a; Liang et al., 2018; Ghusinga et al., 2021a). In this model, there is no G-protein switch without nucleotide exchange. Furthermore, it predicts that the rgs1 null mutant would be constitutively “on” because the basal rate of hydrolysis is much smaller than the rate of nucleotide exchange (Johnston et al., 2007; Jones et al., 2011a, 2012; Urano et al., 2012a; Bradford et al., 2013). However, the recent evidence discussed above suggests that neither of these predictions hold, necessitating the revision of this model. We propose a revised model in Figure 2 with the following essential elements:
Figure 2.
Proposed model of G-protein signaling in plants. A RLK is activated upon presentation of its cognate stimulus. The active RLK phosphorylates the RGS, which consequently abolishes its ability to accelerate the hydrolysis of the GTPase cycle. Moreover, the active RLK also phosphorylates the Gα, resulting in four states of the switch.
The switch consists of four states, determined by the phosphorylation status and the bound nucleotide (GDP or GTP). The signaling response to a stimulus is driven by the state that is phosphorylated and is bound to GTP; the remaining three states of the switch are not signaling competent.
A stimulus is recognized by its cognate RLK, resulting in the RLK becoming active.
The active RLK phosphorylates RGS. The phosphorylated RGS loses the ability to accelerate the hydrolysis of the GTP-bound switch to the GDP-bound one.
The active RLK also phosphorylates both GDP-bound and GTP-bound forms of the switch.
We further assume that the signaling occurs at a much faster rate than the turnover of the components in the signaling pathway. Thus, the total amount of each of the components in the model is constant.
How does this model reconcile the paradox of plant G-signaling? That nucleotide exchange may not be important in plant G-signaling arises from the observation that AtGPA1Q222L, a mutant deficient in GTP hydrolysis, complements some gpa1 phenotypes (Maruta et al., 2019). In our proposed model, the lack of GTP hydrolysis implies that the four states of the switch effectively reduce to two states: unphosphorylated GTP bound state and phosphorylated GTP bound state. Among these two states, if the off state is the unphosphorylated GTP-bound state and the on state is the phosphorylated GTP-bound state, then the model is consistent with the AtGPA1Q222L data. Furthermore, the mild phenotypes shown by the rgs1 null mutant under the laboratory environment are also in agreement with the assumption that the only signaling competent state of the switch is the one with both phosphorylation and GTP. Similar to the AtGPA1Q222L data, the lack of AtRGS1 implies that the switch primarily operates through the GTP bound state. What remains to be resolved is the role of RGS1 and whether the model presented herein is consistent with the observation that although the rgs1 mutant has mild phenotypes, the mutant does show a strong phenotype when subjected to pulsating input (Liao et al., 2017). We investigate this by analyzing the mathematical description of the model, a set of ordinary differential equations (ODEs) parameterized by the rate constants depicted in Figure 2. The ODEs for the wild-type and the rgs1 mutant are given by Equations 2 and 3, respectively, in the “Materials and methods” section. Using these equations, we explore whether our model can generate signaling responses such that both wild-type and rgs1 mutant respond as similarly as possible for a step stimulus with constant amplitude and as differently as possible for a sinusoidal stimulus.
We translate the above objective to mathematical terms by searching for the values of rate constants that minimize the following cost function:
where and are the signaling outputs (fraction of Gα that is phosphorylated and bound to GTP) for wild-type and rgs1 mutant, respectively. The term Tmax is the time that is large enough for all the transient responses to approximately reach their steady states, assuming that the stimulus arrives at time t = 0. The first part of the cost function captures the time-average of the difference (squared) between the response of a wild-type system and the rgs1 mutant for a constant stimulus. The second part of the cost function captures the time-average of the difference (squared) for a sinusoidal stimulus. The negative sign of the second part signifies the maximization of the difference (squared) for the sinusoid input. It is also worth noting that for a given set of parameters, the choice of Tmax has very little effect on the value of the cost function as the system approaches the steady state. To search the parameter space, we put physiological constraints to inform relationships between some of the rate constants and for the remaining parameters we searched the parameter space spanning several orders of magnitude in each parameter using an evolutionary algorithm (“Materials and methods” section).
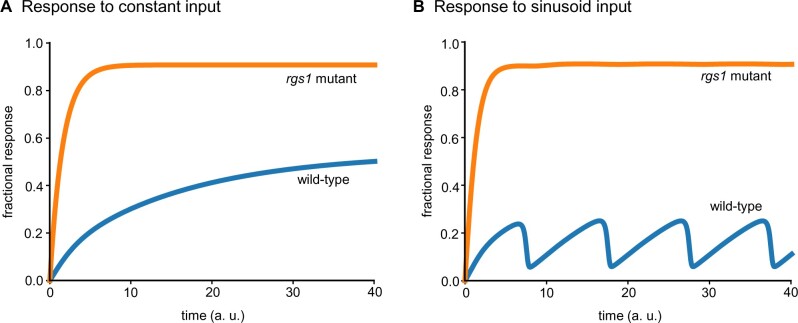
The violin plots for the top 50 parameter sets that minimize the cost function in Equation (1) are shown in Supplemental Figure S1 and the corresponding average signaling responses of the models for wild-type and rgs1 mutant are shown in Figure 3. Notably, the signaling responses for the constant input monotonically increase to their respective steady states (Figure 3A). The response for the rgs1 mutant is more than that of the wild-type, congruent with a higher reactive oxygen species production in the rgs1 mutant (Liang et al., 2018; Ghusinga et al., 2021b) and increased growth (Chen et al., 2003). It is possible for this difference to result in a mild phenotype provided whereby the fractional signaling response affecting the phenotype is lower than the response of the wild-type system. As encoded in the cost function in Equation (1), a more dramatic difference between the responses of wild-type and rgs1 mutant is seen for a sinusoid input (Figure 3B). Here, the wild-type system generates a response that resembles the sinusoid input whereas the rgs1 mutant is unable to follow the dynamics of the input. Our results thus demonstrate that the model in Figure 2 can recapitulate the observation that wild-type and rgs1 mutant phenotypes are similar under constant environment but different under dynamic environment. To understand the reasons behind this behavior of the model, we examine the top 50 parameter sets in Supplemental Figure S1. We find that:
Figure 3.
Mathematical model can recapitulate experimental observations that the rgs1 mutant shows mild phenotype for constant inputs and shows strong phenotype for dynamic inputs. A, In response to a step input with constant amplitude, the signaling responses of both wild-type and rgs1 mutant increase over time to their respective steady states. The response of rgs1 mutant is higher than the wild-type, consistent with a role of negative regulator for RGS in G signaling. B, Model behavior for parameters that minimize the difference between the wild-type response and the rgs1 mutant’s response under a constant stimulus and maximize those differences under a sinusoidal input. For both (A) and (B), the best fit parameter set obtained through the evolutionary algorithm is used.
The phosphorylation rate of RGS is a couple of orders of magnitude higher than the phosphorylation rates of and (. Likewise, the dephosphorylation rate of RGS is approximately an order of magnitude higher than the dephosphorylation rates of and . This means that as the frequency of the stimulus increases, the phosphorylation states of Gα lose their ability to track the stimulus before that of the nucleotide states.
The GAP activity of RGS for () is much higher than that for . Therefore, the change in the dynamics of RGS affect before its unphosphorylated counterpart.
These observations suggest that the switch follows an AND logic. That is, to be active, the Gα must be GTP bound and phosphorylated. In the absence of RGS, Gα is always in the GTP-bound state. When exposed to a constant stimulus, Gα becomes phosphorylated and activated, and therefore behaves similar to the wild-type. However, when the stimulus is time dependent, the rgs1 mutant is not able to track signals with sufficiently high frequencies, because of the slow kinetics of phosphorylation and dephosphorylation relative to the RGS-mediated hydrolysis rate. These observations are indeed consistent with the dynamics of for wild-type and the rgs1 mutant, obtained by simulating the model for the best parameter set from the evolutionary algorithm, as shown in Supplemental Figure S2.
Discussion
Our goal was to present a mathematical model that reconciles evidence that conflicts with the previous understanding of the plant G signaling. We acknowledge that the model is theoretical although it fits and describes the entire body of observations on plant G signaling to date. This model brings to bear several points:
G cycling in plants does not regulate the on–off state; rather, a signal-dependent phosphorylation–dephosphorylation cycle regulates these states. As such, in the absence of phosphorylation induced by a signal, one expects that there is no conformational difference between the GDP state and the GTP-bound state of the heterotrimeric complex and this is what was observed (Adjobo-Hermans et al., 2006).
The phosphorylated GTP-bound state of AtGPA1 is the active molecule in G signaling. The observation that a mutation that locks AtGPA1 into the GTP-bound conformation rescues the gpa1 null mutant indicates that the GTP-bound subunit is in the active state and that, despite its in vitro property of spontaneous nucleotide exchange, is not at equilibrium in vivo.
G cycling is required to properly detect dynamic signals such as light (Liao et al., 2017) and possibly mechanostimulation (Weerasinghe et al., 2009) and perception of pathogen-associated molecular pattern molecules such as flg22 from bacteria (Liang et al., 2016; Watkins et al., 2021).
The view that this mathematical model affords solves the plant G cycle paradox introduced above with perhaps the exception of the observation that AtGPA1S52C, an AtGPA1 mutant lacking GTP binding (empty state), rescues the gpa1 null mutant to wild-type phenotypes (Maruta et al., 2019). However, the plant model proposes a solution wherein the empty state adopts a conformation that is similar to or exactly like the GTP-bound conformation. This is the case for an animal G subunit where the empty state was chemically stabilized and as such was revealed that it mimicked the GTP-bound state (Andhirka et al., 2017); so, this proposal is not without precedence. It should be noted that in both animal cells where the rate-limiting step is nucleotide exchange and in plant cells where the rate-limiting step is hydrolysis, this empty state is transient and likely inconsequential to the switch property. However, in the point mutant described by Maruta et al. (2019) where GTP binding was undetectable above background, the empty state therefore is not transient, rather it is stabilized. From a geneticist point of view, this is the equivalent of introducing a neomorphic trait through a gain-of-function mutation. In this case, the new trait is a biochemical property compared to a new phenotype like an ectopic limb.
Although the model presented herein is consistent with the evidence thus far, its rigorous validation requires further experiments. One set of such experiments would be to investigate the phenotypes of different mutants of AtGPA1 that are deficient in phosphorylation. We would expect that the G-protein switch in a phosphorylation-deficient mutant of AtGPA1 would be unable to operate and therefore unable to complement the gpa1 null mutant static signal phenotypes. The mutant AtGPA1 would have the wild-type phenotype for nucleotide binding and hydrolysis. Another set of experiments to challenge the model would be to investigate the responses to dynamic inputs for the AtGPA1Q222L mutant, which according to the model should be similar to the rgs1 null mutant. Ultimately, a high-resolution peek into the responses to dynamic stimuli, instead of simply looking at physiological phenotypes, would be the true litmus test of our model. We also note that to avoid overparameterization of the mathematical model, we chose to exclude the Gβγ dimer and any role that the XLG Gα subunits would have through competition with AtGPA1 interactions with its RGS protein and Gβγ dimer partners. While this is unnecessary for the present purpose to reconcile the paradoxes, it is, nonetheless, paramount to include in future studies to complete the picture of plant G signaling. Finally, it would also be worthwhile to investigate system-level properties of the switch architecture proposed herein, much like the analyses performed in (Ghusinga et al., 2021a).
Materials and methods
Ordinary differential equations governing the dynamics of the proposed model
We write the ODEs that govern the dynamics of the model schematic depicted in Figure 2, assuming mass-action kinetics. Let , , and respectively denote the levels of RLK, RGS, and Gα. We denote the active or phosphorylated forms with a superscript (e.g. is the phosphorylated RGS) and use subscript ( or ) to represent the nucleotide status of Gα (e.g. is the phosphorylated and GDP-bound form of Gα). Finally, we absorb the total amounts of each of these components in the rate constants, such that the ODEs represent the amount of the components divided by their respective total amount. With these, the ODEs for the wild-type model are:
In these equations, is the stimulus (possibly time-varying) that is turned on at . The initial conditions are assumed to be the pre-stimulus steady states.
For the rgs1 mutant, setting the total amount of RGS to zero yields the following ODEs:
Same as the ODEs for the wild-type, we set the initial conditions to be pre-stimulus steady states.
Parameter sets minimizing the cost function
Our goal is to test whether the proposed model is capable of qualitatively recapitulating the experimental observations that the rgs1 mutant shows mild phenotypes in laboratory conditions, except for when challenged with a dynamic input (Liao et al., 2017). To that end, we defined the cost function Equation (1) to minimize the difference between the wild-type and rgs1 mutant for a constant stimulus but maximize the difference for a sinusoid stimulus. To search for parameters (rate constants in Figure 2) that minimize the cost function in Equation (1), we used an evolutionary algorithm that was implemented using Distributed Evolutionary Algorithms in Python, a python-based framework to build and execute evolutionary algorithms (Fortin et al., 2012).
We supplement our parameter search using available experimental data about the rate constants. In particular, we set and . The first assumption implies that without RGS, the nucleotide exchange rate is 10 times the basal rate of hydrolysis; therefore, the fraction of is ∼90%. The second assumption means that RGS speeds up the hydrolysis by approximately 100 times. Consequently, the fraction of is ∼10%. We note that the GTP binding rate and the basal hydrolysis rate were reported to be 14.4 min−1 and 0.012 min−1 (Johnston et al., 2007), which corresponds to ∼99% Gα being GTP bound, but we chose to not use these precise values because the estimations of GTP binding rate for a plant Gα subunit has a range of ∼10-fold in Arabidopsis (A. thaliana; Johnston et al., 2007; Jones et al., 2011a, 2012; Urano et al., 2012a; Bradford et al., 2013) and ∼40-fold between Arabidopsis and rice (Iwasaki et al., 1997). Likewise, the GTP hydrolysis rate has a range of about three-fold in Arabidopsis (Johnston et al., 2007; Jones et al., 2011a, 2012; Urano et al., 2012a; Bradford et al., 2013). The two-fold difference between the basal hydrolysis rate and the acceleration in hydrolysis by RGS is within the range reported in the animal field (Ross and Wilkie, 2000; Hollinger and Hepler, 2002), although for Arabidopsis the same was reported to be approximately 35 times (Johnston et al., 2007,).
We further restrict our search space by setting and , both of which mean that the phosphorylation and de-phosphorylation rates of Gα do not depend upon the nucleotide (GDP or GTP). Finally, we also assume that the nucleotide exchange rate and the basal rate of hydrolysis of GTP-bound states are the same, notwithstanding the phosphorylation status, i.e. and Although these assumptions may not be exactly accurate in quantitative terms, they either capture the known physiological constraints or restrict our parameter search space considerably. This is acceptable because we are only investigating whether the proposed model can recapitulate the data. If the model exhibits the desired behaviors in the restricted search space, it would also do so if we relaxed these assumptions. For each of the remaining parameters, we set a range spanning six-orders of magnitude (10−3, 103). Each generation of the evolutionary algorithm had 1,000 initiation parameter sets uniformly sampled over the search space. These parameter sets then underwent selection, crossover, and mutation for 100 generations. We specifically consider the case where the constant and sinusoid input (stimuli) are, respectively, given by and . The top 50 parameter sets that minimized the cost function in Equation (1) are shown in Supplemental Figure S1. We chose the sinusoid input such that, on average, the system sees the same input as the constant input.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Top 50 parameter sets that minimize the cost function in Equation (1).
Supplemental Figure S2 . Dynamics of α*GDP for the best parameter set obtained from the evolutionary algorithm.
Supplementary Material
Acknowledgments
The authors thank members of the Jones and Elston labs for discussions.
Funding
A.M.J. is supported by grants from the National Institute of General Medical Sciences (NIGMS, GM065989) and National Science Foundation (MCB-1713880 and IOS-2034929). T.C.E. is supported by the grant R35GM127145 from the NIGMS.
Conflict of interest statement. The authors declare no conflict of interest.
A.M.J. conceived the project with specific input from K.R.G. and T.C.E. K.R.G. and T.C.E. designed the experiments. K.R.G. executed the experiments and analyzed the results. K.R.G. prepared the figures with specific input from T.C.E. K.R.G., T.C.E., and A.M.J. wrote and edited the manuscript.
Author responsibility
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Alan M. Jones (alan_jones@unc.edu).
References
- Adjobo-Hermans MJW, Goedhart J, Gadella TWJ Jr (2006) Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. J Cell Sci 119:5087–5097 [DOI] [PubMed] [Google Scholar]
- Andhirka SK, Vignesh R, Aradhyam GK (2017) The nucleotide-free state of heterotrimeric G proteins α-subunit adopts a highly stable conformation. FEBS J 284:2464–2481 [DOI] [PubMed] [Google Scholar]
- Bradford W, Buckholz A, Morton J, Price C, Jones AM, Urano D (2013) Eukaryotic G protein signaling evolved to require G protein–coupled receptors for activation. Sci Signal 6:ra37–ra37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Assmann SM (2018) G protein subunit phosphorylation as a regulatory mechanism in heterotrimeric G protein signaling in mammals, yeast, and plants. Biochem J 475:3331–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Gookin TE, Milner MJ, Yu Y, Assmann SM (2015) Extra-large G proteins expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiol 169:512–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Gao Y, Jones AM (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol 141:887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301:1728–1731 [DOI] [PubMed] [Google Scholar]
- Ding L, Pandey S, Assmann SM (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J 53:248–263 [DOI] [PubMed] [Google Scholar]
- Fortin F-A, Rainville F-MD, Gardner M-A, Parizeau M, Gagné C (2012) DEAP: Evolutionary algorithms made easy. J Machine Learn Res 13:2171–2175 [Google Scholar]
- Fu Y, Lim S, Urano D, Tunc-Ozdemir M, Phan NG, Elston TC, Jones AM (2014) Reciprocal encoding of signal intensity and duration in a glucose-sensing circuit. Cell 156:1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadella TWJ Jr (1999) Fluorescence lifetime imaging microscopy (FLIM): instrumentation and applications. InMason W, ed, Fluorescent and Luminescent Probes for Biological Activity, Ed 2 .Academic Press, London, pp 467–479 [Google Scholar]
- Ghusinga KR, Jones RD, Jones AM, Elston TC (2021a) Molecular switch architecture determines response properties of signaling pathways. Proc Natl Acad Sci USA 118:e2013401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghusinga KR, Paredes F, Jones AM, Colaneri A (2021b) Reported differences in the flg22 response of the null mutation of AtRGS1 correlates with fixed genetic variation in the background of Col-0 isolates. Plant Signal Behav 16:1878685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, McKain MR, Lee SG, Choudhury SR, McCann T, Schreier S, Harkess A, Pires JC, Wong GK-S, Jez JM, et al. (2017) Gα and regulator of G-protein signaling (RGS) protein pairs maintain functional compatibility and conserved interaction interfaces throughout evolution despite frequent loss of RGS proteins in plants. New Phytol 216:562–575 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S, Assmann SM (2012) Ca2+-dependent GTPase, Extra-large G Protein 2 (XLG2), promotes activation of DNA-binding protein related to vernalization 1 (RTV1), leading to activation of floral integrator genes and early flowering in Arabidopsis. J Biol Chem 287:8242–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev 54:527–559 [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, Kato T, Kaidoh T, Ishikawa A, Asahi T (1997) Characterization of the putative α subunit of a heterotrimeric G protein in rice. Plant Mol Biol 34: 563–572 [DOI] [PubMed] [Google Scholar]
- Jia H, Song G, Werth EG, Walley JW, Hicks LM, Jones AM (2019) Receptor-like kinase phosphorylation of Arabidopsis heterotrimeric G-protein Gα-subunit AtGPA1. Proteomics 19:1900265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen J-G, Siderovski DP, Jones AM, Willard FS (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. PNAS 104:17317–17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Duffy JW, Machius M, Temple BRS, Dohlman HG, Jones AM (2011a) The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Sci Signal 4: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Jones AM, Temple BRS, Dohlman HG (2012) Differences in intradomain and interdomain motion confer distinct activation properties to structurally similar Gα proteins. PNAS 109:7275–7279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Temple BRS, Jones AM, Dohlman HG (2011b) Functional reconstitution of an atypical G protein heterotrimer and Regulator of G Protein Signaling Protein (RGS1) from Arabidopsis thaliana. J Biol Chem 286:13143–13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N, Menon ST, Chauhan R, Sachdev P, Sakmar TP (2009) Structural evidence for a sequential release mechanism for activation of heterotrimeric G proteins. J Mol Biol 393:882–897 [DOI] [PubMed] [Google Scholar]
- Lee Y-RJ, Assmann SM (1999) Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): a new class of G-protein. Plant Mol Biol 40:55–64 [DOI] [PubMed] [Google Scholar]
- Li B, Tunc-Ozdemir M, Urano D, Jia H, Werth EG, Mowrey DD, Hicks LM, Dokholyan NV, Torres MP, Jones AM (2018) Tyrosine phosphorylation switching of a G protein. J Biol Chem 293:4752–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. (2016) Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5:e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, Bi G, Li L, Zhang X, Chai J, et al. (2018) Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res 28:529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Gao Y, Jones AM (2017) Extra Large G-Protein interactome reveals multiple stress response function and partner-dependent XLG subcellular localization. Front Plant Sci 8:1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao K-L, Jones RD, McCarter P, Tunc-Ozdemir M, Draper JA, Elston TC, Kramer D, Jones AM (2017) A shadow detector for photosynthesis efficiency. J Theor Biol 414:231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou F, Abramyan TM, Jia H, Tropsha A, Jones AM (2020) An atypical heterotrimeric Gα protein has substantially reduced nucleotide binding but retains nucleotide-independent interactions with its cognate RGS protein and Gβγ dimer. J Biomol Struct Dyn 38:5204–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Chakravorty D, Urano D, Assmann SM, Botella JR (2019) Nucleotide exchange–dependent and nucleotide exchange–independent functions of plant heterotrimeric GTP-binding proteins. Sci Signal 12: eaav9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki K, Fujisawa Y, Kato H, Iwasaki Y (2005) Study of the constitutively active form of the α subunit of rice heterotrimeric G proteins. Plant Cell Physiol 46:381–386 [DOI] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem 69:795–827 [DOI] [PubMed] [Google Scholar]
- Sprang SR (2016) Invited review: activation of G proteins by GTP and the mechanism of Gα-catalyzed GTP hydrolysis. Biopolymers 105:449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple BRS, Jones AM (2007) The plant heterotrimeric G-protein complex. Annu Rev Plant Biol 58:249–266 [DOI] [PubMed] [Google Scholar]
- Tunc-Ozdemir M, Jones AM (2017) Ligand-induced dynamics of heterotrimeric G protein-coupled receptor-like kinase complexes. PLoS One 12:e0171854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunc-Ozdemir M, Li B, Jaiswal DK, Urano D, Jones AM, Torres MP (2017) Predicted functional implications of phosphorylation of Regulator of G Protein Signaling Protein in plants. Front Plant Sci 8:1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunc-Ozdemir M, Urano D, Jaiswal DK, Clouse SD, Jones AM (2016) Direct modulation of heterotrimeric G protein-coupled signaling by a Receptor Kinase Complex. J Biol Chem 291:13918–13925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM (2003) The β-Subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes[W]. Plant Cell 15:393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Chen J-G, Botella JR, Jones AM (2013a) Heterotrimeric G protein signalling in the plant kingdom. Open Biol 3:120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Fu Y, Jones AM (2013b) Activation of an unusual G-protein in the simple protist Trichomonas vaginalis. Cell Cycle 12:3127–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones AM (2014) Heterotrimeric G protein–coupled signaling in plants. Annu Rev Plant Biol 65:365–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM (2012a) G protein activation without a GEF in the Plant Kingdom. PLoS Genetics 8:e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Maruta N, Trusov Y, Stoian R, Wu Q, Liang Y, Jaiswal DK, Thung L, Jackson D, Botella JR, et al. (2016a) Saltational evolution of the heterotrimeric G protein signaling mechanisms in the plant kingdom. Sci Signal 9: ra93 [DOI] [PubMed] [Google Scholar]
- Urano D, Miura K, Wu Q, Iwasaki Y, Jackson D, Jones AM (2016b) Plant morphology of heterotrimeric G protein mutants. Plant Cell Physiol 57:437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Taylor JP, Jones AM (2012b) Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol 14:1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JM, Ross-Elliott TJ, Shan X, Lou F, Dreyer B, Tunc-Ozdemir M, Jia H, Yang J, Oliveira CC, Wu L, et al. (2021) Differential regulation of G protein signaling in Arabidopsis through two distinct pathways that internalize AtRGS1. Sci Signal [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe RR, Swanson SJ, Okada SF, Garrett MB, Kim S-Y, Stacey G, Boucher RC, Gilroy S, Jones AM (2009) Touch induces ATP release in Arabidopsis roots that is modulated by the heterotrimeric G-protein complex. FEBS Lett 583:2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tang W, Liu K, Huang Q, Zhang X, Yan X, Chen Y, Wang J, Qi Z, Wang Z, et al. (2011) Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathogen 7:e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.