Abstract
Somatic embryogenesis is a type of plant cell totipotency where embryos develop from nonreproductive (vegetative) cells without fertilization. Somatic embryogenesis can be induced in vitro by auxins, and by ectopic expression of embryo-expressed transcription factors like the BABY BOOM (BBM) AINTEGUMENTA-LIKE APETALA2/ETHYLENE RESPONSE FACTOR domain protein. These different pathways are thought to converge to promote auxin response and biosynthesis, but the specific roles of the endogenous auxin pathway in somatic embryogenesis induction have not been well-characterized. Here we show that BBM transcriptionally regulates the YUCCA3 (YUC3) and YUC8 auxin biosynthesis genes during BBM-mediated somatic embryogenesis in Arabidopsis (Arabidopsis thaliana) seedlings. BBM induced local and ectopic YUC3 and YUC8 expression in seedlings, which coincided with increased DR5 auxin response and indole-3-acetic acid (IAA) biosynthesis and with ectopic expression of the WOX2 embryo reporter. YUC-driven auxin biosynthesis was required for BBM-mediated somatic embryogenesis, as the number of embryogenic explants was reduced by ca. 50% in yuc3 yuc8 mutants and abolished after chemical inhibition of YUC enzyme activity. However, a detailed YUC inhibitor time-course study revealed that YUC-dependent IAA biosynthesis is not required for the re-initiation of totipotent cell identity in seedlings. Rather, YUC enzymes are required later in somatic embryo development for the maintenance of embryo identity and growth. This study resolves a long-standing question about the role of endogenous auxin biosynthesis in transcription factor-mediated somatic embryogenesis and also provides an experimental framework for understanding the role of endogenous auxin biosynthesis in other in planta and in vitro embryogenesis systems.
The BABY BOOM transcription factor induces YUCCA-dependent auxin biosynthesis during somatic embryogenesis to maintain embryo identity and development.
Introduction
Totipotency is the capacity of a single cell to regenerate into a complete organism (Condic, 2014). Totipotency is restricted to the zygote in sexually reproducing plants, but some asexually reproducing plants also produce embryos from vegetative cells and from unfertilized gametes (Pichot et al., 2001; Garcês et al., 2007; Schmidt, 2020). Induced totipotency refers to the ability of cells to develop into embryos when cultured in vitro (Fehér, 2019). Somatic embryogenesis is a type of totipotency in which vegetative (nongametophytic) cells are induced to develop into embryos after exposure to exogenous growth regulators, in particular the synthetic auxin 2,4-dichlorophenoxy acetic acid (2,4-D), or by ectopic expression of embryo or meristem identity transcription factors (Horstman et al., 2017a; Fehér, 2019; Karami et al., 2021b). Both inducer treatments promote cell division and also reprogram cells in a multicellular explant toward somatic embryogenesis or toward pluripotent pathways resulting in callus formation and organogenesis. How both 2,4-D and transcription factors induce a subset of cells in an explant to develop specifically into somatic embryos is not known, but roles for chromatin modifications as well as for changes in expression of embryo identity genes and plant growth regulator pathway genes have been proposed (De-la-Peña et al., 2015; Horstman et al., 2017a; Wang et al., 2020; Wójcik et al., 2020).
2,4-D efficiently induces somatic embryogenesis in a wide range of explants in the model plant Arabidopsis (Arabidopsis thaliana). As in other plants, Arabidopsis somatic embryos either develop directly from the explant (Luo and Koop, 1997; Gaj, 2001; Kobayashi et al., 2010) or indirectly from embryogenic callus (Ikeda-Iwai et al., 2003; Su et al., 2009). In the direct system, fully differentiated embryos with root and shoot meristems and cotyledons develop in the presence of 2,4-D, while in the indirect system removal of 2,4-D from the culture medium is usually required to promote differentiation (patterning) of pro-embryogenic masses, which are multicellular embryos lacking radial and apical–basal patterning (Halperin and Jensen, 1967; Gaj, 2011). Ectopic expression of specific embryo or meristem identity transcription factors also induces somatic embryo formation, but can do so in the absence of exogenous plant growth regulators (Horstman et al., 2017a). Among these are the LEAFY COTYLEDON 1 (LEC1) HAP3/CCAAT binding protein, the LEC2 B3-domain protein, and the BABY BOOM (BBM) clade of AINTEGUMENTA-LIKE (AIL) APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factors, which also includes the PLETHORA (PLT) proteins (Lotan et al., 1998; Stone et al., 2001; Gaj et al., 2005; Horstman et al., 2017b). Ectopic over-expression of these transcription factors in germinating seeds induces direct somatic embryo formation on above-ground organs of seedlings, including the cotyledon petioles, tip and margin and the shoot apical meristem. The mechanisms driving transcription factor-induced somatic embryogenesis have not been well-studied, but like 2,4-D-induced somatic embryogenesis, are thought to require chromatin-level changes as well as deregulation of embryo/meristem identity transcription factor and auxin pathway genes (Horstman et al., 2017a; Tian et al., 2020; Wójcik et al., 2020).
Transcriptional activation of auxin biosynthesis genes is one of the common regulatory points downstream of 2,4-D and transcription factor-induced somatic embryogenesis. Plants synthesize auxin by different pathways (Normanly, 2010; Zhao, 2014). The major auxin in Arabidopsis is indole-3-acetic acid (IAA), which is mainly synthesized through the TRYPTOPHAN AMINOTRANSFERASE ARABIDOPSIS (TAA)/YUCCA (YUC) pathway (Zhao, 2014). Enzymatic activity of (TAA1 and TAA1-RELATED PROTEINS (TARs) convert TRP into the intermediate product indole-3-pyruvic acid (IPyA), which is then converted into IAA by the YUC flavin-dependent monooxygenases (Stepanova et al., 2011). The Arabidopsis genome contains three TAA1/TAR genes and 11 YUC monooxygenase genes that are differentially expressed during plant development (Cheng et al., 2006, 2007; Wang et al., 2011; Hentrich et al., 2013; Robert et al., 2013). Arabidopsis TAA/TARs and YUC proteins each function in a redundant manner, such that many of their functions only become evident in higher-order mutant combinations (Cheng et al., 2006, 2007; Wang et al., 2011; Robert et al., 2013).
Endogenous auxin, mainly IAA, is often elevated in cells or tissues undergoing 2,4-D-induced somatic embryogenesis (Michalczuk et al., 1992; Charrière et al., 1999; Pasternak et al., 2002). In the Arabidopsis direct somatic embryogenesis system, exposure of immature zygotic embryo explants to 2,4-D induces expression of YUC1 and YUC4 early in somatic embryogenesis, followed later by TAA1 and YUC10 expression (Wójcikowska et al., 2013). Single yuc mutants have no obvious phenotype under normal growth conditions, except the yuc8-1 mutant, which shows reduced seed set (Cheng et al., 2006, 2007; Ståldal et al., 2012). However, in 2,4-D-induced somatic embryo cultures, single yuc2 and yuc4 mutants produce fewer embryogenic explants and fewer somatic embryos per explant compared to wild-type (WT) explants (Wójcikowska et al., 2013). In the indirect somatic embryogenesis system, where embryos develop after an initial callus phase, YUC gene expression (YUC1, YUC2, YUC4, and YUC6) is detected late in the development of embryogenic callus and then increases after transfer of the callus to 2,4-D-free medium (Bai et al., 2013). In this system, the quadruple yuc1 yuc2 yuc4 yuc6 mutant shows a normal progression of somatic embryogenesis, while the yuc1 yuc4 yuc10 yuc11 mutant produces only a few malformed somatic embryos (Bai et al., 2013). Treatment with the YUC enzyme inhibitor yucasin drastically reduces somatic embryo formation from Coffea canephora explants (Uc-Chuc et al., 2020). It is clear that endogenous auxin biosynthesis has a role in 2,4-D-induced somatic embryo induction, but when and how auxin biosynthesis specifically promotes somatic embryogenesis is not known.
LEC and BBM/PLT transcription factors have also been shown to bind to and/or transcriptionally regulate auxin biosynthesis genes during normal plant development and under conditions that promote somatic embryo development. Ectopic LEC2 expression induces YUC2 and YUC4 expression early during somatic embryo development from seedlings (Stone et al., 2008), and ectopic LEC1 expression induces YUC gene expression during 2,4-D-induced somatic embryogenesis from immature zygotic embryos (YUC1, YUC4, and YUC10) and from seedlings (YUC10; Junker et al., 2012; Wójcikowska et al., 2013). CHOTTO1/EMBRYOMAKER/PLT5/AIL5 binds to and transcriptionally regulates YUC4 in the shoot apex (Pinon et al., 2013), while PLT2/AIL4 binds to and transcriptionally regulates YUC3 and YUC8 in the root tip (Santuari et al., 2016). BBM/AIL2 also binds to YUC3 and YUC8 during 2,4-D- and BBM-induced somatic embryogenesis, but it is not known if BBM also transcriptionally regulates these genes (Horstman et al., 2017b). Although auxin biosynthesis genes are downstream targets of embryo identity transcription factors during somatic embryogenesis, it is not known whether auxin biosynthesis is required to promote transcription factor-driven somatic embryogenesis.
Here we examined the role of YUC-dependent IAA biosynthesis in BBM-induced somatic embryogenesis from Arabidopsis seedling cotyledons. Using a combination of genetic analysis, pharmacological inhibition and cell fate analysis we show that YUC-dependent IAA biosynthesis is essential for BBM-mediated somatic embryogenesis, but that this pathway is only required after the initiation of totipotency, for the subsequent proliferation and differentiation of embryogenic cells.
Results
Developmental steps in BBM-induced somatic embryogenesis
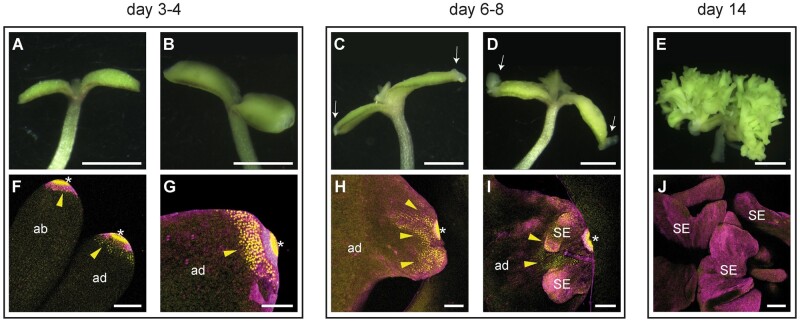
The normal course of somatic embryogenesis in seedlings from dexamethasone (DEX)-treated 35S:BBM-GR seeds has been described previously (Horstman et al., 2017b; Godel-Jedrychowska et al., 2020) and is summarized in Figure 1. DEX treatment induces posttranslational nuclear localization of the BBM-GR fusion protein (Horstman et al., 2017b), allowing comparison of samples with and without ectopic BBM activity. Embryogenic cell divisions are observed in the cotyledons of DEX-treated 35S:BBM-GR seedlings around Days 3–4 of culture (Figure 1, A and B). These divisions begin at the cotyledon tip, followed by the cotyledon margin and shoot apex and are visualized as thickened, smooth, and light green tissue. By Days 6–8 of culture small embryogenic protrusions can be observed on the dividing tip (Figure 1, C and D) and by Day 14 a mass of primary and secondary somatic embryos develops on the seedling cotyledon (Figure 1E).
Figure 1.
Overview of BBM-induced somatic embryogenesis. A–E, Light micrographs of representative DEX-treated 35S:BBM-GR explants. F–J, Confocal laser scanning micrographs of WOX2:YFP expression at the cotyledon tip of DEX-treated 35S:BBM-GR explants. The day of culture is indicated above the images. Arrowheads, WOX2-YFP expression. Arrows, growth protrusions. Asterisks, autofluorescence. ad, adaxial side. ab, abaxial side. SE, somatic embryo. Scale bars: A–E, 1 mm; F–J, 100 µm.
Previously we showed that the embryo identity and BBM direct target gene LEC1 is expressed on the cotyledon tip of DEX-treated 35S:BBM-GR seeds as early as 1 d after DEX treatment and becomes more highly expressed at the cotyledon tip and margin when these tissues begin to proliferate (Horstman et al., 2017b). We followed the expression of the WOX2:NLS-3xYFP embryo marker to determine whether embryo identity genes that are not direct BBM targets are expressed in the same way. During the first 2 d of culture WOX2:NLS-3xYFP expression was detected in both control (mock-treated) and DEX-treated seedlings throughout the seedling, and in the cotyledon on the abaxial and adaxial surface (Supplemental Figure S1, A and B; Figure 8). The nuclear WOX2-YFP signal could no longer be detected in the control seedling cotyledons from Day 3 onward (Supplemental Figure S1C), but was maintained and became restricted to the tip of the cotyledon in the DEX-treated seedlings (Figure 1, F and G). During Days 6–8 of culture, WOX2-YFP expression was observed on the explant in the region where embryos develop and in the embryogenic growths of most DEX-treated control seedlings (Figure 1, H and I). In the 35S: BBM-GR line used in this study, 10%–15% of the seedlings do not form somatic embryos and the same proportion of seedlings lacked WOX2-YFP expression in the cotyledon (Supplemental Figure 1D). By Day 14 of culture WOX2-YFP expression could only be detected in ca. 20% of these embryos (Supplemental Table S1).
Figure 8.
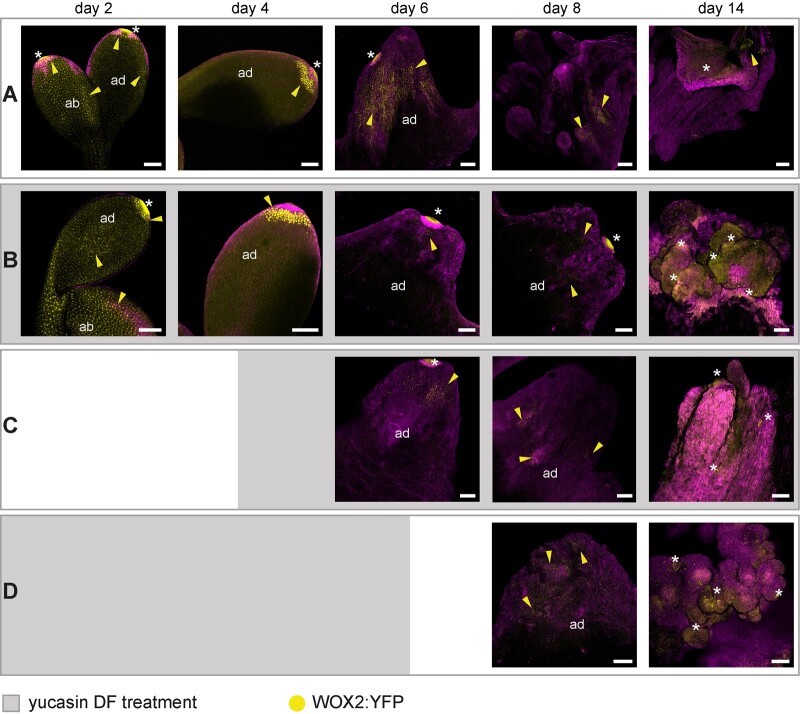
Auxin biosynthesis is required to maintain BBM-induced totipotency. Confocal laser scanning micrographs of cotyledon/cotyledon tips of 35S:BBM-GR explants grown with DEX, with or without the YUC enzyme inhibitor YDF (100 µM). A, Control DEX-treated explants. B–D, Explants treated with DEX and YDF, which was added or removed on the days indicated by gray blocks in each row. Samples were counter stained with SR2200 (magenta). The day of culture is indicated in the panels. Yellow arrowheads, WOX2 expression (yellow signal). Asterisks, autofluorescence. Scale bars, 100 μm.
The above data indicate that expression of the BBM direct target gene LEC1 precedes expression of the nontarget gene WOX2. Both LEC1 and WOX2 are initially expressed on the cotyledon tip, the site where somatic embryo formation is first initiated. LEC1 is a major regulator of early and late embryo development pathways and overexpression of LEC1 induces spontaneous somatic embryogenesis. LEC1 also acts a pioneer factor at the FLOWERING LOCUS C gene by promoting an active chromatin state (Tao et al., 2017). Activation of LEC1 expression by BBM might therefore be required for promoting chromatin accessibility at BBM target loci and/or for parallel activation of early embryo development genes.
BBM regulates auxin pathway genes
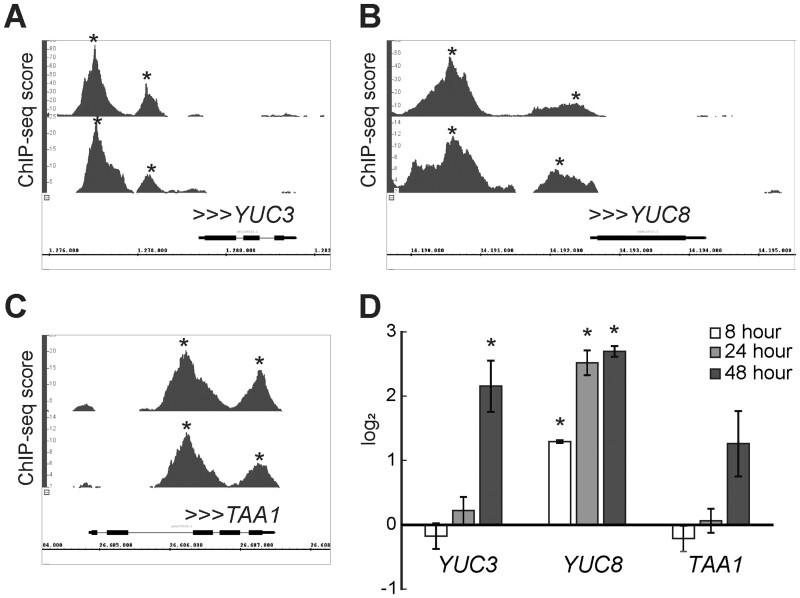
The BBM transcription factor binds a number of key regulatory genes during 2,4-D and BBM-induced somatic embryogenesis, including genes that promote in vitro regeneration and meristem identity and proliferation (Supplemental Data Set S1; Horstman et al., 2015; Horstman et al., 2017b). Among the direct BBM gene targets are also a number of auxin pathway genes, including the YUC3, YUC8, and TAA1 auxin biosynthesis genes. The BBM-binding sites at these loci are shown in Figure 2, A–C. To determine whether BBM also transcriptionally regulates these genes, we analyzed their expression using reverse transcription quantitative PCR (RT-qPCR) in DEX-treated 35S:BBM-GR seeds at 8, 24, and 48 h after imbibition (pregermination). YUC3 and YUC8 expression was significantly upregulated in DEX-treated 35S:BBM-GR seeds compared to DEX-treated WT seeds, with YUC3 expression (48 h) lagging behind that of YUC8 (8 h), while TAA1 expression was not significantly regulated (Figure 2D). We therefore focused our efforts on YUC3 and YUC8 as candidate early auxin biosynthesis target genes.
Figure 2.
BBM binds and regulates the expression of auxin biosynthesis genes. A–C, ChIP-seq BBM binding profiles for auxin biosynthesis genes in somatic embryo tissue. The binding profiles for 35S::BBM-GFP (upper profile) and BBM::BBM-YFP (lower profile) are shown. The x-axis shows the nucleotide position of DNA binding in the selected genes (TAIR 10 annotation), the y-axis shows the ChIP-seq score, and the arrowheads indicate the direction of gene transcription. Peaks with scores above 1.76 for 35S::BBM-GFP and 3.96 for pBBM::BBM-YFP were considered statistically significant (*, false discovery rate < 0.05). The ChIP-seq data were generated in Horstman et al. (2015). The ChIP-seq data and data analysis can be downloaded from GEO (GSE52400). The plots were generated using Integrated Genome Browser. D, The relative expression of auxin biosynthesis genes during seed germination was determined by qPCR for DEX-treated 35S::BBM-GR seedlings using mock-treated Col-0 seeds as the calibrator and the SAND gene (Czechowski et al., 2005) as the reference. Error bars indicate standard errors of the three biological replicates in the same genetic background. Asterisk, statistically significant change in gene expression levels, determined using Student’s t-test (P < 0.05).
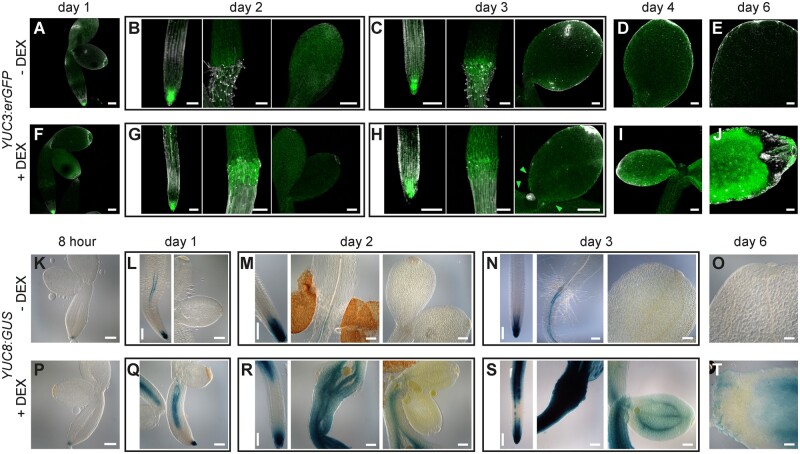
Next, we examined the spatial and temporal regulation of YUC3/YUC8 expression in 35S:BBM-GR seeds carrying the YUC3:erGFP or the YUC8:β-glucuronidase (GUS) reporters. Seeds were imbibed and then cultured with or without 10 µM DEX. In WT Arabidopsis seedlings, YUC3 is expressed in the root meristem and root–hypocotyl transition zone and YUC8 is expressed in the root vascular tissue and meristem (Ståldal et al., 2012; Chen et al., 2014; Santuari et al., 2016; Figure 3) BBM-enhanced YUC3 expression was observed in the root–hypocotyl transition zone from Day 2 of culture (Figure 3, B, C, G, and H), followed by weak, but consistent ectopic expression on the proximal cotyledon margin on Day 3 (Figure 3, C and H) and the entire cotyledon surface by Day 4 (Figure 3, D, E, I, and J). Enhanced YUC8 expression in the hypocotyl vascular tissue was observed after 1 d of culture (Figure 3, L and Q), and 35S:BBM-induced changes in hypocotyl morphology were already visible after 2 d of culture (Figure 3, M and R). Ectopic expression of YUC8 was observed in the cotyledons starting from Day 3 of culture (Figure 3, N and S). As in the root–hypocotyl, YUC8 was also expressed in the cotyledon vascular tissue. After 6 d of culture, areas lacking YUC3 and YUC8 expression were observed in a region close to the cotyledon tip (Figure 3, J and T), corresponding to the first sites of somatic embryo induction in DEX-treated 35S:BBM-GR lines (Figure 1, B and C). Notably, expression of a YUC3:GUS reporter that lacks the BBM binding site motif and that is not expressed in the root meristem (Chen et al., 2014) did not show altered expression in DEX-treated 35S:BBM-GR seedlings (Supplemental Figure S2, B and C).
Figure 3.
BBM overexpression induces ectopic expression of YUC3 and YUC8. Images of roots, hypocotyls and cotyledons from YUC reporter lines in a 35S:BBM-GR background with (solid gray line) or without (dashed gray line) DEX treatment. The day of culture is shown above the images. A–J, Confocal light scanning micrographs of YUC3:erGFP expression. K–T, Light micrographs of YUC8:GUS expression. Scale bars, 100 μm.
Together these analyses show that BBM transcriptionally regulates YUC3 and YUC8 expression early during somatic embryo induction, both in their native expression domain in the root/hypocotyl, as well as ectopically in the cotyledon. Ectopic YUC expression in cotyledons also coincided with the onset of ectopic WOX2 expression (Figure 1G), suggesting a major change in cotyledon cell fate at this time point. BBM-induced YUC3/YUC8 expression in cotyledons lagged behind YUC3/YUC8 expression in the root/transition zone. Germination relies mainly on translation of stored mRNAs (Sano et al., 2020), and postgermination light-grown cotyledons only undergo a few cell divisions (Sano et al., 2020), thus de novo BBM-induced transcription in cotyledons might require activation of cell division and/or reprogramming of chromatin to a transcriptionally active state, processes that are already active in the root and hypocotyl.
BBM enhances auxin response and biosynthesis
The above results indicate that YUC3 and YUC8 are transcriptionally regulated by BBM early during somatic embryo induction. We therefore investigated whether these changes are reflected in increased auxin response and IAA levels in seedlings.
We used DR5 reporters to follow the temporal and spatial dynamics of auxin response during BBM-mediated somatic embryogenesis. 35S:BBM-GR DR5 seeds were germinated with or without 10 μM DEX and DR5 expression followed in the explants for 7 d (Figure 4). Weak DR5 expression was observed on the adaxial and abaxial surfaces of cotyledons (Figure 4, A and D) of both DEX-treated and control seedlings after 1 d of culture. From Day 3 of culture onward, DR5 expression in the vascular tissue extended further into the root elongation zone in DEX-treated seedlings than in control seedlings (Figure 4, B and E). At this time, DR5 expression was no longer visible in control cotyledons, but broadened and increased in intensity on the adaxial surface of cotyledons from DEX-treated samples (Figure 4, C and F), where it localized to the adaxial epidermal/subepidermal layers and the vascular bundles (Figure 4G). In the following days, DR5 expression continued to increase in DEX-treated seedlings, especially along the cotyledon margin (Figure 4, H and I). Starting around Day 4, an auxin minimum as visualized by low DR5 expression (Figure 4, H–K) could be seen next to the cotyledon tip where embryogenic protrusions develop.
Figure 4.
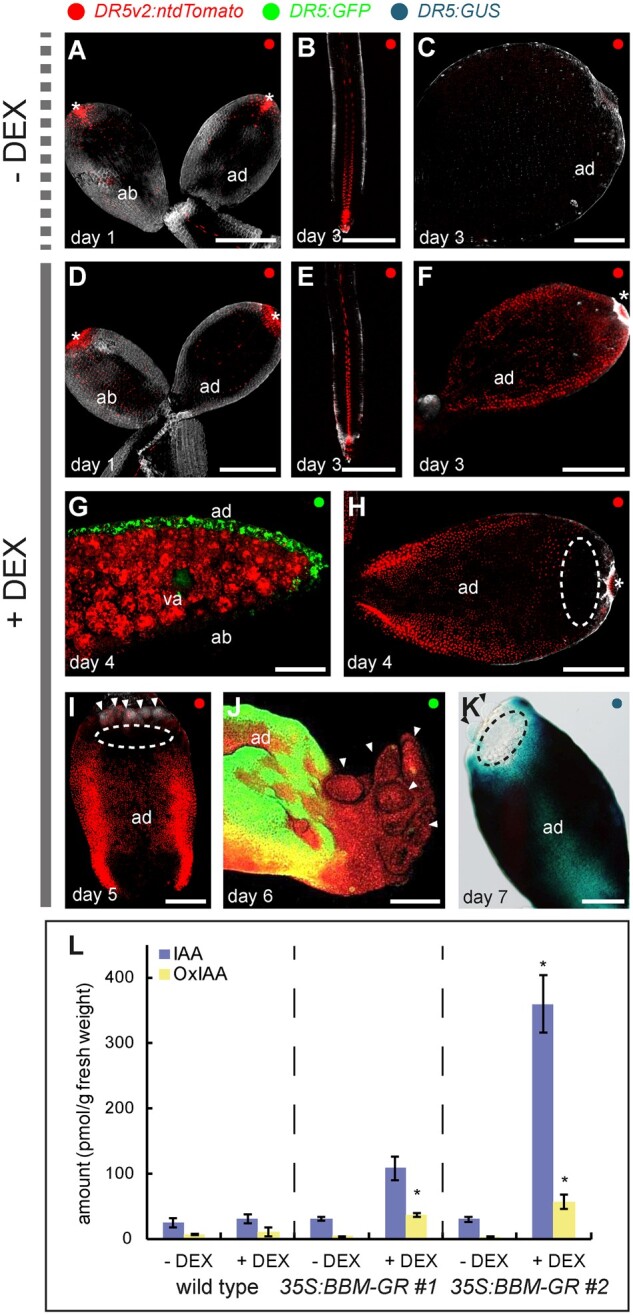
BBM expression enhances DR5 auxin response and IAA biosynthesis. Confocal laser scanning micrographs of cotyledons or roots from 35S:BBM-GR DR5 seedlings grown without (A–C) and with (D–J) DEX. D–F, H and I are images of DR5v2:ntdTomato cotyledons or roots. G and J, These are images of DR5:GFP cotyledons. The images in (G) and (J) are counterstained with FM4-64. K, Light image of DR5:GUS expression in the cotyledon of a DEX-treated 35S:BBM-GR seedling. Samples were counter stained with SR2200 (gray, A–F, H and I) or outlined using red autofluorescence (G and J). The dashed ellipses in (H), (I), and (K) indicate the DR5 minimum. Small embryogenic protrusions are indicated with arrowheads in (I) and (J). va, vascular tissue; asterisks autofluorescence. Scale bars, 200 μm. L, IAA and oxIAA concentrations in seedlings of WT Col-0 and two 35S:BBM-GR lines grown in the absence or presence of DEX (three technical replicates, each 200 mg). *, samples that showed statistically significant differences in IAA or oxIAA concentrations compared to the non-DEX treated 35S:BBM-GR control (Student’s t test, p < 0.05). Error bars represent the standard deviation of the replicates.
Auxin response reporters measure the sum of auxin signaling processes, and since BBM binds different types of auxin-pathway genes (Horstman et al., 2017b), we determined whether the enhanced DR5 response observed in BBM overexpression lines can be explained by changes in IAA levels. WT seeds and seeds from two independent 35S:BBM-GR lines differing in somatic embryo production rate were cultured with or without DEX for 3 d before measuring IAA and the IAA catabolite oxindole-3-acetic acid (oxIAA). Oxidation of IAA to oxIAA reduces auxin activity and plays an important role in maintaining auxin homeostasis (Stepanova and Alonso, 2016). Seedlings of both 35S:BBM-GR lines treated with DEX showed higher IAA levels than the WT seedlings and 35S:BBM-GR seedlings without DEX treatment (Figure 4L), but only the increase of IAA content in line 2 was significant compared to the WT control. The different IAA levels in these two lines might reflect the differences in penetrance of their somatic embryogenesis phenotypes (50% in line 1 and 100% in line 2).
The above data indicate that BBM overexpression induces a de novo auxin response on the adaxial cotyledon surface. The spatial localization of the DR5 auxin response in DEX-treated 35S:BBM-GR and WT seedlings started to diverge around the third day of culture, the time point at which YUC3/YUC8 gene expression and IAA levels also increased in DEX-treated 35S:BBM-GR cotyledons. This suggests that the enhanced auxin response observed in 35S:BBM-GR seedlings is due, at least in part, to increased IAA biosynthesis. This increase in YUC3/YU8 and DR5 expression was followed a few days later by DR5 and YUC3/YUC8 expression minima at the site of multicellular somatic embryo formation on the cotyledon tip. Together this data suggest that enhanced/ectopic YUC expression and IAA biosynthesis coincides with the establishment of totipotent cell fate, but that multicellular somatic embryo development takes place in a low auxin response field.
YUC3 and YUC8 are required for efficient BBM-mediated somatic embryogenesis
To determine the roles of YUC3 and YUC8 in BBM-induced somatic embryogenesis, we generated two independent yuc3 yuc8 double mutant lines in a 35S:BBM-GR background using CRISPR-Cas9 mutagenesis (Supplemental Figure S3). Both independent yuc3 yuc8 mutants contained the same yuc3CR1 mutation, an 848 bp deletion plus a 38 bp insertion that removed part of the promoter and first exon (Supplemental Figure S3, A and B). The yuc8CR1 mutation has a 1 bp insertion downstream of and close to the translational start site, resulting in a premature stop codon (Supplemental Figure S3, A and B). The yuc8CR2 mutant line has a 3 bp deletion at the same position as the yuc8CR1 mutation resulting in loss of one amino acid (Supplemental Figure S3, A and B). This amino acid is not located in previously described functional domains (Supplemental Figure S3C) and might not affect the protein’s function. However, both the yuc3CR1 yuc8CR1 and yuc3CR1 yuc8CR2 mutants showed the reduced seed set phenotype that was previously described for the yuc8-1 allele (Supplemental Figure S3D; Ståldal et al., 2012). This suggests that the single amino acid deletion in the yuc8CR2 allele disrupts YUC8 function. Other than the reduced seed set phenotype, none of the two independent yuc3CR yuc8CR double mutant lines showed obvious phenotypic differences from WT seedlings under standard growth conditions.
To evaluate the effect of the yuc3CR yuc8CR double mutants on BBM-induced somatic embryogenesis, we cultured control 35S:BBM-GR seeds and seeds from the two 35S:BBM-GR yuc3CR yuc8CR lines for 14 d with 10 µM DEX and categorized the explants into three groups: explants with somatic embryos, explants with ectopic shoots but no somatic embryos, and explants without any ectopic structures (Figure 5). DEX-treated explants from both 35S:BBM-GR yuc3CR yuc8CR lines showed a statistically significant reduction in the capacity for somatic embryogenesis (ca. 50%) compared to the DEX-treated 35S:BBM-GR control explants (ca. 90%). Ectopic shoot formation was not affected in the DEX-treated 35S:BBM-GR yuc3CR yuc8CR lines compared to the control. These results are in line with observations in 2,4-D-induced direct and indirect somatic embryo cultures, where mutation of different YUC genes was shown to be detrimental for somatic embryogenesis (Bai et al., 2013; Wójcikowska et al., 2013).
Figure 5.
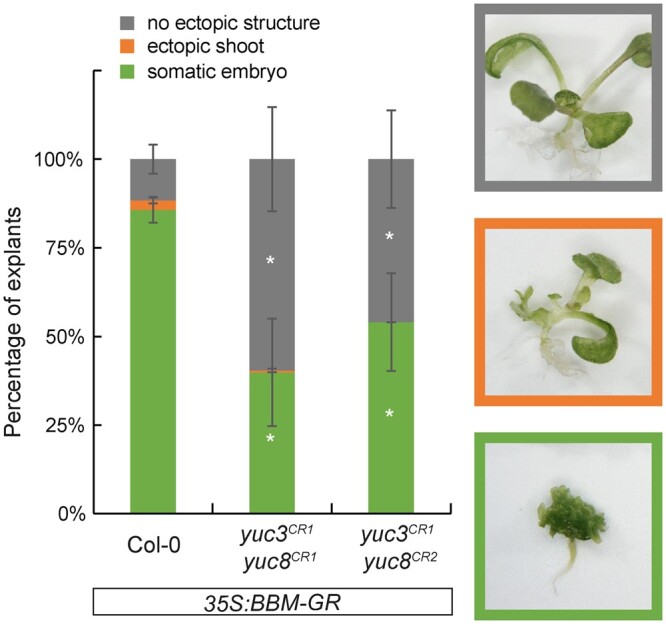
YUC-dependent auxin biosynthesis is required for efficient BBM-induced somatic embryogenesis. Regeneration phenotypes of 14-d-old explants from the indicated lines. The explants were categorized in three groups: explants with somatic embryos, explants with ectopic shoots and explants without any ectopic structures. Representative images are shown on the right. All seedlings were treated continuously with 10 μM DEX. Statistically significant differences in each category between the mutant lines and the 35S:BBM-GR control line were determined using Student’s t test (p < 0.05) and indicated with asterisks. Error bars represent standard deviation of at least two biological replicates (n > 227).
Auxin biosynthesis is required in a narrow developmental window for efficient BBM-induced somatic embryogenesis
Auxin biosynthesis genes are direct targets of embryo identity transcription factors like BBM, LEC1, and LEC2 and these proteins also control each other’s expression through complex transcriptional feedback loops (Tian et al., 2020; Wójcik et al., 2020). Given the possibility that additional YUC genes might be directly or indirectly regulated during BBM-induced somatic embryogenesis, we used a pharmacological approach to inhibit overall YUC activity. This approach also allowed us to dissect the role of YUC-dependent IAA biosynthesis in time by performing time course inhibitor addition-removal experiments.
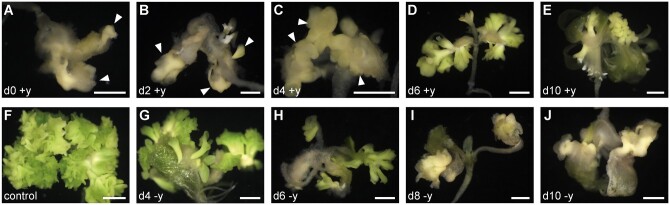
35S:BBM-GR seeds were cultured for 14 d in liquid medium with 10 µM DEX to activate the BBM protein. The YUC enzyme inhibitor yucasin (Nishimura et al., 2014) or the more stable analog yucasin difluorinated analog (yucasin DF [YDF];100 µM; Tsugafune et al., 2017) were added to or removed from the cultures at different time points to determine when YUC-mediated IAA biosynthesis plays a role in BBM-induced somatic embryogenesis. After three to 4 d of culture, the cotyledon margins of DEX-treated 35S:BBM-GR seedlings thicken due to increased cell division (Figure 1B). Multiple embryogenic protrusions develop from the adaxial surface of the cotyledon tip and margin around Day 6 of culture, followed by formation of histodifferentiated somatic embryos by 10 d of culture (Figures 1 and 6F). In contrast, the cotyledons of DEX-treated 35S:BBM-GR seedlings treated with 100 µM YUC enzyme inhibitor from Days 0, 2, and 4 onward developed into white callus-like structures, with or without white, dense amorphous structures (Figure 6, A–C and F; Supplemental Figure S4, A–C and F). In contrast, seedlings from cultures treated with YUC enzyme inhibitor from Day 6 onward formed somatic embryos were similar to the control samples, except that the number of somatic embryos was greatly reduced compared to control cultures (Figure 6, D and E; Supplemental Figure S4, D and E). Continuous treatment of DEX in combination with lower YUC enzyme inhibitor concentrations also reduced somatic embryo formation in 35S:BBM-GR seedlings, but to a lesser extent than with the 100 µM treatment (Supplemental Figure S5, A–E). The enhanced DR5:GFP expression in cotyledons of 4-d-old seedlings treated continuously with DEX was abolished after YUC enzyme inhibitor treatment (Supplemental Figure S5, K, L, and O), suggesting that YUC enzyme inhibitor treatment reduced BBM-induced IAA biosynthesis in the cotyledon.
Figure 6.
Auxin biosynthesis is required for BBM-mediated somatic embryogenesis. 35S:BBM-GR seeds were grown for 14 d in the presence of DEX and imaged at the indicated time points. The YUC enzyme inhibitor yucasin (100 µM) was added or removed during the culture period as indicated (day +y or day −y). A–E, DEX-treated samples to which YUC inhibitor was added on Day 0, 2, 4, 6, 8, or 10. F, DEX-treated control sample. G–J, DEX-treated samples in which YUC inhibitor was added on Day 0 and then removed on Day 4, 6, 8, or 10. Scale bars, 1 mm.
Next, we performed YUC inhibitor removal experiments to more accurately define the time point at which inhibition of auxin biosynthesis affects the progression of somatic embryogenesis. DEX and YUC enzyme inhibitor were added on Day 0 of culture and then the inhibitor was removed on Days 4, 6, 8, or 10 of culture (Figure 6, G–J; Supplemental Figure S4, G–J). Somatic embryos developed on the cotyledons of DEX-treated 35S:BBM-GR seedlings when YUC inhibitors were removed on or before Day 6, but the number of somatic embryos was reduced compared to nontreated control samples (Figure 6, G and H; Supplemental Figure S4, G and H). Somatic embryo formation could not be rescued when YUC enzyme inhibitor was removed after 6 d of treatment (Figure 6, I and J; Supplemental Figure S4, I and J).
Together these results suggest that YUC activity is essential for the normal progression of BBM-mediated somatic embryogenesis between the fourth and sixth day of culture. The YUC inhibitor concentrations that affect somatic embryo formation (25–100 µM; Supplemental Figure S5, A–E) are higher than those that affect root development in WT plants (1–10 µM; He et al., 2011), but similar to the concentration range (20–100 µM) that complemented the YUC1 overexpression phenotype (Nishimura et al., 2014). This suggests that BBM induces relatively high IAA levels in cotyledons or that cotyledons and developing somatic embryos are less sensitive to YUC enzyme inhibition than other tissues.
TAA/TAR proteins convert TRP to IPyA, which is then converted to IAA by YUC proteins. The TAA1 gene is also bound by BBM during BBM- and 2,4-D-induced somatic embryogenesis but was not transcriptionally-regulated by BBM during the first 2 d of culture (Figure 2, C and D). However, blocking TAA1/TAR enzyme activity in 35S:BBM-GR seedlings with kynurenine (kyn), a chemical inhibitor of TAA1/TAR activity (He et al., 2011) severely impaired somatic embryo formation (Supplemental Figure S5, F–J) and also abolished the BBM-induced DR5 response (Supplemental Figure S5, M–O). This inhibitory effect was not observed when kyn was added to the medium on Day 6 of culture (Supplemental Figure S6, D and E) or when kyn was removed by Day 8 of culture (Supplemental Figure S6, F–I), although fewer embryos developed than in the control samples. Thus TAA1/TAR-mediated auxin biosynthesis is also required for BBM-induced somatic embryogenesis, although the window in which TAA1/TAR enzymes are required is slightly broader than for YUC enzymes.
Auxin biosynthesis is required for the maintenance of BBM-induced totipotency
To determine how reduced IAA levels affect the progression of BBM-mediated somatic embryogenesis, we examined the development of auxin inhibitor-treated explants using thin sections and embryo identity reporters.
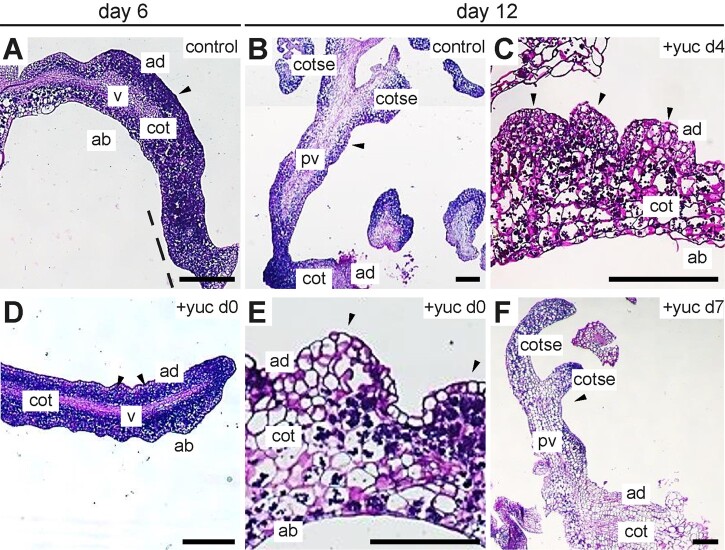
35S:BBM-GR seeds were germinated in medium containing DEX (control) with or without YUC enzyme inhibitor, which was added to the cultures during (Days 0 and 4) or after (Day 7) the critical time point for somatic embryo development. Thin sections were made six and 12 d after the start of culture. Thin sections of DEX-treated seedling cotyledons showed that the mesophyll and vascular cells had divided prolifically during the first 6 d of culture (Figure 7A). The proliferating adaxial mesophyll cells and cotyledon tip formed a continuous mass of cytoplasm-rich cells, which are characteristic for totipotent/meristematic cells (Huang and Yeoman, 1984; Prime et al., 2000; Kurczyńska et al., 2007; Verdeil et al., 2007; Godel-Jedrychowska et al., 2020). Callus-like cells, characterized by their reduced cytoplasmic staining, were visible on the adaxial surface of the cotyledon in the same explants (Figure 7A). By Day 12 of culture, the DEX-treated seedlings had formed (secondary) somatic embryos with defined apical–basal polarity (Figure 7B). When YUC enzyme inhibitor was added with DEX at the start of culture, the seedlings still produced cytoplasm-rich cells on the cotyledon surface, but with less overall cell proliferation compared to DEX-treated samples (Figure 7D). In addition, interspaced cell clusters formed along the adaxial surface of the cotyledon instead of the continuous band of proliferating cells observed in DEX-treated seedlings. These cell clusters became more callus-like by the 12th day of culture (Figure 7E). The cells in these callus-like clusters were covered by loosely connected epidermal cells, rather than densely packed cells in the control samples, indicating that they lost their capacity for meristematic/totipotent cell proliferation. The cotyledons of seedlings treated with YUC inhibitor on Day 4 resembled cotyledons from seedlings treated with inhibitor from Day 0 onward (Figure 7C). When YUC inhibitor was added on Day 7 of culture, somatic embryos with visible apical–basal polarity were formed on the cotyledons (Figure 7F), but the number of somatic embryos was reduced compared to the DEX-treated control. These data indicate that auxin biosynthesis is not absolutely required for the de novo induction of meristematic/totipotent cell proliferation, but rather is required to sustain these meristematic/totipotent cell divisions. These results also support the idea that auxin biosynthesis is also required after Day 6 of culture for efficient differentiated somatic embryo formation.
Figure 7.
YUC-dependent auxin biosynthesis is required for the formation of histodifferentiated somatic embryos. Light micrographs of thin cross sections of the cotyledons of DEX and YUC inhibitor (yucasin)-treated 35S:BBM-GR explants fixed on the days indicated above the images. The day of culture and the yucasin treatment (100 µM) is shown above and in the image panels, respectively. A and B, Explants from control samples treated with DEX from Day 0 until the end of the culture on Day 14. Panel B is a composite of different images from the same section. C–F, Explants from samples treated with DEX from Day 0 to Day 14, to which YUC enzyme inhibitor was added on Day 0 (D and E), Day 4 (C), or Day 7 (F). Black arrowhead, growth protrusions (A, C–E) and somatic embryos (B and F); cot, cotyledon; cotse, cotyledons of somatic embryos; v, vascular (A and D); pv, provascular tissue (B and F); dotted line, proliferating cotyledon tip. Scale bars, 200 μm.
To determine how reduced IAA levels alter embryo fate during BBM-induced somatic embryogenesis, we followed the expression of the WOX2:NLS-3xYFP embryo identity reporter in DEX-treated 35S:BBM-GR seedlings that were cultured in the presence of absence of YUC enzyme inhibitors. WOX2-YFP expression in seedlings treated continuously from Day 0 with 100 µM YUC enzyme inhibitor was similar to that of the control seedlings until Day 4 of culture (Figures 1F and 8B). The number of WOX2-YFP-positive seedlings decreased to half that of the control by Day 8 of culture and to zero by Day 14 (Supplemental Table S1; Figure 8B). When YDF was added on Day 4 of culture, the initial proportion of WOX2-YFP-expressing seedlings on Days 6 and 8 was similar to that of the DEX-treated control, but then decreased to zero on Day 14 (Supplemental Table S1; Figure 8C). Likewise, when YDF was added on Day 0 and then removed on Day 6 of culture, the number of seedlings initially showing WOX2-YFP expression was similar to the control, but then decreased to zero by Day 14 of culture (Supplemental Table S1; Figure 8D).
Taken together, these histology and cell fate experiments confirmed our observations on whole mount samples that is, that YUC-dependent IAA biosynthesis is not required for the initiation of embryo identity at the cotyledon tip in BBM overexpression lines, but is required later, in a narrow developmental window between Days 4 and 6 of culture, to maintain embryo identity and promote the development of embryogenic cell protrusions into histodifferentiated embryos. In the absence of YUC activity these embryogenic cells develop into callus-like structures.
Discussion
Ectopic expression of the AIL transcription factor BBM induces spontaneous adventitious organ formation (pluripotency) and embryogenesis (totipotency; Gordon-Kamm et al., 2019; Vijverberg et al., 2019). In WT plants, in vitro adventitious organ formation and somatic embryogenesis usually rely on exogenous auxin application, either alone or in combination with other hormones or abiotic stress treatments. A genetic relationship between BBM-like AILs and auxin in shoot and root meristem development, as well as binding and/or direct transcriptional regulation of YUC genes by AIL-family members has been shown (Pinon et al., 2013; Santuari et al., 2016), but neither has been described in the context of induced pluripotent or totipotent growth. Here we show that BBM regulates YUC gene expression and that YUC-dependent auxin biosynthesis has essential, but relatively late functions in BBM-mediated somatic embryogenesis. Our data suggest a two-step model in which BBM-induces expression of embryo identity genes like LEC1, LEC2, and FUSCA3 (FUS3) to establish cell totipotency (Horstman et al., 2017b), followed by induction of auxin biosynthesis to maintain embryo division and growth.
Multiple roles for auxin biosynthesis
Here we show that ectopic BBM expression induces expression of the canonical auxin biosynthesis pathway genes YUC3 and YUC8 (Figure 2D). Both of these genes are direct BBM targets in 2,4-D and BBM-induced somatic embryo cultures (Figure 2, A and B). BBM is expressed in the seedling root tip and throughout the zygotic embryo as early as the four-cell stage and becomes basally localized from the heart stage onward (Galinha et al., 2007; Horstman et al., 2015). Both YUC3 and YUC8 are expressed in the seedling root tip (Chen et al., 2014; Santuari et al., 2016; Figure 3), and in the zygotic embryo YUC3 is expressed in the suspensor and YUC8 in the basal region of the embryo proper (Robert et al., 2013). This overlap in BBM and YUC3/YUC8 expression suggests that BBM also regulates YUC3 and YUC8 expression during zygotic embryogenesis and root development in planta.
Reporter analysis showed that 35S:BBM-GR overexpression induces YUC3 and YUC8 expression in the root and hypocotyl, followed by expression in the cotyledons (Figure 3). The expansion of BBM-induced ectopic YUC3/YUC8 reporter expression from the below ground to the above-ground organs reflects the gradual increase in transcript levels detected by qPCR (Figure 2D). The increase in YUC expression in roots and cotyledons was also mirrored by increased DR5 expression in the same organs and by increased IAA biosynthesis (Figure 4). Together these results suggest that BBM induces enhanced and ectopic auxin biosynthesis gene expression and a concomitant increase in auxin levels.
We also observed that embryogenic protrusions develop in areas of low (DR5) auxin response (Figure 4) and low YUC3/YUC8 expression (Figure 3). In Arabidopsis, DR5 expression and auxin accumulation (as measured by the R2D2 [Ratiometric version of 2D2’s] reporter; Liao et al., 2015) are only reliably detected starting at the eight-cell embryo stage. This initial auxin response in the embryo proper is largely due to PIN-mediated auxin transport from the suspensor and from the surrounding maternal tissues (Friml et al., 2003; Robert et al., 2013). YUC and TAA1/TAR auxin biosynthesis genes are expressed later in zygotic embryos, in the embryo proper and suspensor from the 16-cell embryo stage onward (Stepanova et al., 2008; Robert et al., 2013). In 35S:BBM-GR explants, DR5 and YUC3 are initially expressed throughout the cotyledon and YUC8 in the cotyledon vasculature. Later, DR5 and YUC3/YUC8 expression is absent at the sites where WOX2-YFP expression is ectopically induced and where multicellular embryos emerge on the cotyledon tip and margin (Godel-Jedrychowska et al., 2020; Figures 1, F–I, 3, J and T, and 4, H–K). Reduced DR5 and YUC expression might simply reflect a switch in development from single or few-celled embryogenic structures to larger embryogenic clusters, analogous to early preglobular stage zygotic embryos, where neither DR5 nor characterized YUC genes are expressed. Alternatively, we have shown previously that this decrease in DR5 expression is accompanied by and requires increased callose production in plasmodesmata adjacent to sites of WOX2-YFP expression (Godel-Jedrychowska et al., 2020). Blocking callose production in DEX-treated 35S:BBM-GR seedlings prevents the formation of an auxin response minimum and completely blocks somatic embryo development. We hypothesized that auxin accumulation must be reduced locally to allow organized embryo growth and that callose deposition in surrounding plasmodesmata prevents passive auxin re-entry into these cells. Thus, a combined action of reduced auxin accumulation, reduced local auxin biosynthesis and reduction of the size of molecules that can pass through plasmodesmata might create a low auxin field that promotes the growth of multicellular embryogenic growth protrusions. Auxin biosynthesis inhibitor experiments showed that auxin is required later for further growth of these embryogenic protrusions into differentiated embryos; blocking YUC-dependent auxin biosynthesis results in conversion of embryogenic cells to callus-like structures rather than somatic embryos. At this point, callose deposition and WOX2-YFP expression colocalize in the same cells, as embryogenic protrusions increase in size and differentiate into somatic embryos (Godel-Jedrychowska et al., 2020). Together these observations suggest a two-step dynamic and local regulation of auxin to allow 1) development of multicellular embryogenic cell clusters in a low auxin/auxin response area, followed by 2) development of these structures into histodifferentiated embryos with zygotic embryo-like auxin responses.
A positive transcriptional feedback loop for somatic embryogenesis
Somatic embryo formation was completely abolished when DEX-treated 35S:BBM-GR explants were treated continuously or before the sixth day of culture with YUC enzyme inhibitors, but the somatic embryogenesis rate in the 35S:BBM-GR yuc3CR yuc8CR lines was only reduced to about half of the control 35S:BBM-GR line (Figure 5). This result suggests that YUC3 and YUC8 are not the only YUC enzymes required for BBM-induced somatic embryogenesis. Previously we found that BBM also binds the LEC1, LEC2, and FUS3 transcription factor genes (Horstman et al., 2017b). Ectopic LEC1 expression was also induced in DEX-treated 35S:BBM-GR seedlings during the first day of culture. LEC1 and LEC2 expression in seedlings induces, respectively, YUC8 and YUC10 (Junker et al., 2012; Huang et al., 2015) and YUC1, YUC4, and YUC10 expression (Wójcikowska et al., 2013). LEC2 and FUS3 also cooperatively promote YUC4 expression during lateral root formation (Wójcikowska et al., 2013; Tang et al., 2017). The LEC transcription factors might partly compensate for the reduced auxin biosynthesis in yuc3CR yuc8CR mutant lines by inducing expression of other YUC genes. The known positive transcriptional interactions between the BBM and LEC transcription factors and their respective target genes (Horstman et al., 2017a; Tian et al., 2020) suggest that a positive feedback loop is established that maintains both embryo identity and auxin biosynthesis during BBM-induced somatic embryogenesis.
Auxin requirement during embryogenesis
In Arabidopsis, YUC gene expression is activated during 2,4-D-induced somatic embryogenesis in explants undergoing direct and indirect somatic embryogenesis (Bai et al., 2013; Wójcikowska et al., 2013). During 2,4-D-induced direct somatic embryogenesis from Arabidopsis immature zygotic embryo explants, overexpression of LEC2 can compensate for treatment with a suboptimal 2,4-D concentration or for treatment with auxins that are poor inducers of somatic embryogenesis, like IAA or 1-naphthaleneacetic acid (Wójcikowska et al., 2013). The lec1 and lec2 loss-of-function mutants show a severe reduction of the number of embryogenic explants in the presence of 2,4-D, as well as a shift from direct to indirect (callus-derived) somatic embryogenesis (Gaj et al., 2005). Conversely, ectopic expression of LEC2 in the presence of an optimal concentration of 2,4-D negatively affects somatic embryo formation, as it delays and reduces embryo induction and induces callus and shoot-like structures instead of somatic embryos (Ledwoń and Gaj, 2009). Although IAA levels were not measured directly in these studies, these results suggest that tight regulation of auxin levels is required to promote somatic embryogenesis: both too little and too much endogenous or exogenous auxin can inhibit somatic embryo formation, absolutely and/or in favor of shoot or callus production (Ledwoń and Gaj, 2009).
The above studies on 2,4-D-induced somatic embryogenesis in WT and different LEC backgrounds demonstrate a role for YUC-dependent auxin biosynthesis in promoting efficient somatic embryogenesis. However, these studies did not determine when and for which aspect of somatic embryogenesis YUC-dependent IAA biosynthesis was required. Our analyses indicated that both YUC expression and IAA levels increase as early as 3 d after BBM activation (Figures 3 and 4L). These changes also correspond with onset of embryo marker gene expression, including WOX2-YFP (Figure 1). However, our pharmacological experiments using YUC enzyme inhibitors showed that YUC-TAA1/TAR-dependent IAA biosynthesis is not required at this time point for the re-initiation of totipotent growth (Figure 6). YUC-dependent IAA biosynthesis is required later, between Days 4 and 6 of culture, for the maintenance of embryo identity and for embryo growth and histodifferentiation. In explants treated continuously or up until the sixth day of culture with YUC enzyme inhibitors, cytoplasm-rich embryogenic protrusions do not progress to patterned embryos, but rather form callus-like structures (Figure 7). How does BBM-induced auxin biosynthesis maintain embryo growth and development? Recently, Karami et al. (2021a) showed that induction of cell totipotency during 2,4-D and 35S:AHL15-induced somatic embryogenesis does not require the auxin efflux and influx machinery. Rather, auxin transport is required later, for the proper transition of embryogenic cells to multicellular embryos and for correct embryo differentiation. Similarly, it is likely that endogenous auxin supplied by BBM signaling is also required to establish the auxin gradients needed for embryo outgrowth and patterning.
During zygotic embryogenesis, YUC and TAA1 genes are expressed relatively late, during the transition from the globular/heart stage to the torpedo stage, where they are required for correct embryo patterning (Stepanova et al., 2008; Robert et al., 2013). TAA1/TAR and YUC genes are expressed earlier in the surrounding maternal ovule and seed coat, but maternally supplied auxin only appears to be required for proper embryo patterning (Robert et al., 2018). Although a complete description of all YUC genes and other TRP-independent IAA synthesis genes during zygotic embryogenesis is currently not available, this data, together with our observations on BBM-induced totipotency suggest that YUC-dependent auxin biosynthesis is not required for the initiation of embryo identity per se. In contrast, TRP-independent IAA biosynthesis has been shown to be essential for early zygotic embryo viability and patterning (Wang et al., 2015). TRP-independent auxin biosynthesis genes have not been identified as direct BBM targets, but might act downstream of other BBM target genes. Recently, Li et al. (2021) described a developmental pathway in which MATERNAL EFFECT EMBRYO ARREST45 induces the AIL gene AINTEGUMENTA, which in turn regulates YUC expression in the ovule integument to control embryo size. These results are in line with our observations on the role of YUC-dependent auxin biosynthesis in maintaining embryogenic cell divisions in vitro and suggest that similar seed functions might be co-opted by embryo identity transcription factors like BBM in embryogenic explants.
Conclusion
The importance of auxin for in vitro somatic embryogenesis is apparent in its widespread use as an exogenous inducer and in the requirement for endogenous auxin for efficient somatic embryo production. “Totipotency” transcription factors are not only rapidly induced in response to 2,4-D, but also induce somatic embryogenesis in the absence of exogenous auxin (Ledwoń and Gaj, 2009, 2011; Horstman et al., 2017a; Tian et al., 2020). These transcription factors also bind to and/or transcriptionally regulate auxin biosynthesis genes, making them good candidates for direct regulators of auxin biosynthesis in different somatic embryogenesis systems. We show that YUC-dependent auxin biosynthesis is required to maintain somatic embryo identity and promote growth, but not for the cell fate transition to embryogenesis. De novo induction of both embryo identity transcription factors and auxin biosynthesis therefore ensures that embryogenic cells proliferate and develop into somatic embryos.
Materials and methods
Plant material and growth conditions
The 35S:BBM-GR, WOX8gΔ:NLS-venusYFP3 (referred to here as WOX2:NLS-3xYFP), YUC8:GUS, YUC3:GUS, YUC3:erGFP, DR5v2:ntdTomato, DR5:GUS, and DR5:GFP lines were described previously (Benková et al., 2003; Růžička et al., 2007; Breuninger et al., 2008; Passarinho et al., 2008; Chen et al., 2014; Liao et al., 2015; Santuari et al., 2016). Due to BBM silencing upon outcrossing (Horstman et al., 2017b), the majority of 35S:BBM-GR lines containing reporter constructs were made by either transforming the 35S:BBM-GR vector to the reporter line (YUC8:GUS and WOX2:NLS-3xYFP) and then selecting highly embryogenic lines, or transforming the reporter vectors (DR5v2, YUC3:erGFP) to an existing embryogenic 35S:BBM-GR line. In the latter case, the transgenic lines were selected based on reporter expression. For the DR5:GUS and DR5:GFP reporter lines, crosses were made with a homozygous 35S:BBM-GR line and the progeny selected over four generations until nonsilenced homozygous lines with at least 90% penetrance of embryogenic explants and 100% reporter gene expression were recovered.
Seeds were sterilized with liquid bleach as described previously (Horstman et al., 2017a, 2017b). For liquid cultures, sterilized seeds were dispensed in 190 mL containers (Greiner, Kremsmünster, Austria) with 30 mL liquid half-strength Murashige and Skoog (1/2 MS-10) medium (Murashige and Skoog, 1962) with 1× MS vitamins, pH 5.8, and 1% sucrose (w/v). The liquid cultures were stratified at 4°C in the dark for up to 48 h before transfer to a rotary shaker (60 rpm) at 25°C (16 h light/8 h dark cycle) for the indicated time. For solid medium cultures, sterilized seeds were cultured at 21°C (16-h light/8-h dark cycle) on 1/2 MS-10 medium with 0.8% (w/v) agar.
Chemical treatments
DEX (Sigma, St. Louis, MO, USA) was dissolved in 70% ethanol and used at a final concentration of 10 µM in all experiments. Yucasin (Sigma; Nishimura et al., 2014), YDF (Tsugafune et al., 2017; provided by Hayashi lab) and kyn (Sigma) were all dissolved in DMSO and were added to the solid and liquid culture medium as described in the text. Mock-treated samples contained the same volume of ethanol or DMSO. The liquid medium and chemicals were refreshed every 6–7 d. Analysis of somatic embryogenesis phenotypes was performed with >3 replicates with >100 explants per treatment. The phenotypes shown were observed in 100% of the explants.
CRISPR-Cas9 mutagenesis
To avoid BBM silencing upon outcrossing (Horstman et al., 2017b), yuc3 yuc8 double mutants were generated by CRISPR-Cas9 mutagenesis directly in the 35S:BBM-GR background rather than by crossing with T-DNA mutants. CRISPR-Cas9 mutagenesis of YUC3 and YUC8 was performed using the U6-26 promoter for the single-guide RNAs (sgRNAs), an RPS5A promoter-driven Arabidopsis codon-optimized Cas9 gene (Fauser et al., 2014), and FAST-Red selection (Castel et al., 2019), all in vector pICSL4723 (Weber et al., 2011; Wang et al., 2019). Two sgRNAs targeting YUC3 and two sgRNAs targeting YUC8 were assembled into one vector to obtain yuc3CR yuc8CR double mutant lines. The sgRNAs and mutant genotyping primers are listed in Supplemental Table S2. The CRISPR-Cas9 vectors were transformed to a highly embryogenic 35S:BBM-GR line. Two double yuc3 yuc8 mutant lines, each with the same yuc3 mutation and a different yuc8 mutation were obtained (Supplemental Figure S3). Homozygous T4 CAS9-free yuc3 and yuc8 mutants were used for the analysis. Analysis of somatic embryogenesis efficiency was performed with at least two technical replicates with >99 explants per mutant line.
Transformation
All constructs were transformed using the floral dip method (Clough and Bent, 1998). Transgenic T1 seeds from CRISPR transformants were selected based on FAST-Red expression (Castel et al., 2019). Transgenic T1 seedlings with reporter lines were selected as described above. Homozygous mutant lines were used in all analyses.
Quantitative real-time RT-PCR
RNA was isolated using the InviTrap Spin plant RNA mini kit (Invitek Molecular, Washington, DC, USA; # 1064100300) with the addition of 25 µL Plant RNA isolation Aid (Ambion, Austin, TX, USA), followed by a DNAse treatment (TURBO DNA-free kit; Invitrogen, Waltham, MA, USA). cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) following the manufacturer’s instructions. Quantitative real-time RT-PCR (RT-qPCR) was performed using a BioRad MyiQ PCR machine with the SYBR green mix as described in Horstman et al. (2015). Relative gene expression was calculated with the 2−ΔΔCT method (Livak and Schmittgen, 2001) using the nonDEX treated (mock) samples as calibrators and the SAND gene (Czechowski et al., 2005) as the reference. Three biological replicates comprising germinating seeds/seedlings were used for each treatment. Statistically significant changes in gene expression levels were determined using Student’s t test P < 0.05. The qPCR DNA primers are shown in Supplemental Table S3.
Histology
Fresh material for sectioning was fixed overnight at 4°C in 3:1 absolute ethanol:glacial acetic acid and then dehydrated stepwise from 70% to 100% ethanol. The fixed material was infiltrated in Steedman’s wax and then sectioned and stained with 0.05% Toluidine Blue (w/v) as previously described (Wrobel et al., 2011). Images were taken with a Nikon Eclipse Ni microscope with a Nikon DS-Fi1 camera and NIS Elements L software (Nikon). About 9–12 explants per treatment were observed.
Microscopy
Confocal laser scanning microscopy was performed as previously described (Soriano et al., 2014; Horstman et al., 2017b). Samples were fixed with 4% (w/v) para-formaldehyde, counterstained with 0.1% (v/v) SCRI Renaissance 2200 (SR2200; Musielak et al., 2016) and then stored at 4°C for up to 2 weeks before imaging. Fluorescence was observed using a Leica SPE DM5500 confocal microscope using the LAS AF software. SR2200 was exited with the 405-nm laser line and fluorescence emission detected between 415 and 476 nm. GFP was excited with the 488-nm laser line and light emission detected between 505 and 540 nm. YFP was excited with the 488-nm laser line and detected between 517 and 597 nm. tdTomato was excited with the 561-nm laser line and light emission detected between 571 and 630 nm. Brightness/contrast adjustment was done in the LAS AF software and image cropping was done in ImageJ. About 9–20 explants were analyzed for each treatment. The images represent the majority of the examined explants or as noted in Supplemental Table S1.
GUS assays were performed for up to 22 h at 37°C, as previously described (Sieburth and Meyerowitz, 1997) using 2.5 mM potassium ferri and ferrocyanide. GUS-stained tissues were cleared in HCG (water:chloral hydrate:glycerol, 25:55.7:8.3; w/w) and then observed using a Nikon Optiphot microscope with differential interference contrast optics. Images were recorded with a Nikon DS-Fi1 camera and processed using NIS-Elements D 3.2 software and ImageJ. Light microscopy was performed using a ZEISS Stemi SV 11 microscope. The GUS assay was repeated 2 times with at least 40 explants examined for each timepoint. The images represent the majority of the examined explants.
IAA measurements
Seeds from WT Col-0 and two independent 35S:BBM-GR lines (two replicates per line) were grown for 24 h in liquid 1/2 MS-10 medium and then grown for an additional 3 d in the presence or absence of 10 µM DEX. IAA extraction and measurements were performed as in Ruyter-Spira et al. (2011) using ca. 100–250 mg fresh weight per sample.
Accession numbers
The previously published chromatin immunoprecipitation sequencing (ChIP-seq) data and data analysis (Horstman et al., 2015) can be downloaded from the Gene Expression Omnibus (GEO; GSE52400).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Confocal images of control 35S:BBM-GR WOX2:YFP explants.
Supplemental Figure S2. The BBM DNA binding motif in the YUC3 promoter is required for YUC3 expression in root meristems and BBM-induced YUC3 ectopic expression.
Supplemental Figure S3. CRISPR-Cas9-induced yuc3 and yuc8 alleles.
Supplemental Figure S4. Magnified images of 14-d-old DEX and DEX+YUC inhibitor-treated 35S:BBM-GR explants.
Supplemental Figure S5. Auxin biosynthesis inhibitors block somatic embryo formation and auxin response.
Supplemental Figure S6. TAA1/TAR auxin biosynthesis is required for BBM-mediated somatic embryogenesis.
Supplemental Table S1. Percentage of 35S:BBM-GR WOX2:NLS-3xYFP seedlings with YFP signal in the cotyledon tip or growth protrusion.
Supplemental Table S2. Single-guide RNAs used for CRISPR-Cas9 mutagenesis and primers used for genotyping CRISPR mutants.
Supplemental Table S3. DNA primers used for RT-qPCR.
Supplemental Data Set S1. BBM direct target genes as determine by ChIP-seq.
Supplementary Material
Acknowledgments
We thank Ewa Benková for the DR5:GUS and DR5:GFP lines, Dolf Weijers for the DR5v2:ntd Tomato plasmid, Thomas Laux for the WOX2gΔ:NLS-venusYFP3 (WOX2:YFP) plasmid, Yunde Zhao for the YUC8:GUS and YUC3:GUS plasmids, and Renze Heidstra for the YUC3:erGFP plasmid. We are very grateful to the Hayashi lab for the generous gift of YDF.
Funding
This research was funded by the Dutch Research Council (NWO; NWO-Groen grant 3184300100) and by the Technology Top Institute-Green Genetics (grant 4CC060RP).
Conflict of interest statement. None declared.
M.L., J.W.-M., I.H., A.H., B.C., and R.R. performed experiments and analyzed the data. M.L., J.W.-M., I.H., A.H., B.C., R.R., G.C.A., and K.B. contributed to the experimental set-up and data interpretation. M.L. and K.B. wrote the manuscript with input from all authors. K.B. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Kim Boutilier (kim.boutilier@wur.nl).
References
- Bai B, Su YH, Yuan J, Zhang XS (2013) Induction of somatic embryos in Arabidopsis requires local YUCCA expression mediated by the down-regulation of ethylene biosynthesis. Mol Plant 6: 1247–1260 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Castel B, Tomlinson L, Locci F, Yang Y, Jones JDG (2019) Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PLoS One 14: e0204778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrière F, Sotta B, Miginiac É, Hahne G (1999) Induction of adventitious shoots or somatic embryos on in vitro cultured zygotic embryos of Helianthus annuus: variation of endogenous hormone levels. Plant Physiol Biochem 37: 751–757 [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y (2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Condic ML (2014) Totipotency: what it is and what it is not. Stem Cells Dev 23: 796–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Diger Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Peña C, Nic-Can GI, Galaz-Ávalos RM, Avilez-Montalvo R, Loyola-Vargas VM (2015) The role of chromatin modifications in somatic embryogenesis in plants. Front Plant Sci 6: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79: 348–359 [DOI] [PubMed] [Google Scholar]
- Fehér A (2019) Callus, dedifferentiation, totipotency, somatic embryogenesis: what these terms mean in the era of molecular plant biology? Front Plant Sci 10: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gaj MD (2001) Direct somatic embryogenesis as a rapid and efficient system for in vitro regeneration of Arabidopsis thaliana. Plant Cell Tissue Organ Cult 64: 39–46 [Google Scholar]
- Gaj MD (2011) Somatic embryogenesis and plant regeneration in the culture of Arabidopsis thaliana (L.) Heynh. immature zygotic embryos. Methods Mol Biol 710: 257–265 [DOI] [PubMed] [Google Scholar]
- Gaj MD, Zhang S, Harada JJ, Lemaux PG (2005) Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222: 977–988 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Garcês HMP, Champagne CEM, Townsley BT, Park S, Malhó R, Pedroso MC, Harada JJ, Sinha NR (2007) Evolution of asexual reproduction in leaves of the genus Kalanchoë. Proc Natl Acad Sci USA 104: 15578–15583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godel-Jedrychowska K, Kulinska-Lukaszek K, Horstman A, Soriano M, Li M, Malota K, Boutilier K, Kurczynska EU (2020) Symplasmic isolation marks cell fate changes during somatic embryogenesis. J Exp Bot 71: 2612–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Kamm B, Sardesai N, Arling M, Lowe K, Hoerster G, Betts S, Jones T, Gordon-Kamm B, Sardesai N, Arling M, et al. (2019) Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 8: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin W, Jensen WA (1967) Ultrastructural changes during growth and embryogenesis in carrot cell cultures. J Ultrasructure Res 18: 428–443 [DOI] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. (2011) A small-molecule screen identifies L-Kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M, Böttcher C, Düchting P, Cheng Y, Zhao Y, Berkowitz O, Masle J, Medina J, Pollmann S (2013) The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J 74: 626–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman A, Bemer M, Boutilier K (2017a) A transcriptional view on somatic embryogenesis. Regeneration 4: 201–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman A, Fukuoka H, Muino JM, Nitsch L, Guo C, Passarinho P, Sanchez-Perez G, Immink R, Angenent G, Boutilier K (2015) AIL and HDG proteins act antagonistically to control cell proliferation. Development 142: 454–464 [DOI] [PubMed] [Google Scholar]
- Horstman A, Li M, Heidmann I, Weemen M, Chen B, Muino JM, Angenent GC, Boutilier K (2017b) The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol 175: 848–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang BC, Yeoman MM (1984) Callus proliferation and morphogenesis in tissue cultures of Arabidopsis thaliana L. Plant Sci Lett 33: 353–363 [Google Scholar]
- Huang M, Hu Y, Liu X, Li Y, Hou X (2015) Arabidopsis LEAFY COTYLEDON1 mediates postembryonic development via interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell 27: 3099–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Umehara M, Satoh S, Kamada H (2003) Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J 34: 107–114 [DOI] [PubMed] [Google Scholar]
- Junker A, Mönke G, Rutten T, Keilwagen J, Seifert M, Thi TMN, Renou JP, Balzergue S, Viehöver P, Hähnel U, et al. (2012) Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J 71: 427–442 [DOI] [PubMed] [Google Scholar]
- Karami O, Philipsen C, Rahimi A, Nurillah AR, Boutilier K, Offringa R (2021a) Endogenous auxin directs development of embryonic stem cells into somatic proembryos in Arabidopsis. bioRxiv https://doi.org/10.1101/2021.08.06.455432
- Karami O, Rahimi A, Mak P, Horstman A, Boutilier K, Compier M, van der Zaal B, Offringa R (2021b) An Arabidopsis AT-hook motif nuclear protein mediates somatic embryogenesis and coinciding genome duplication. Nat Commun 12: 2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nagayama Y, Higashi K, Kobayashi M (2010) Establishment of a tissue culture system for somatic embryogenesis from germinating embryos of Arabidopsis thaliana. Plant Biotechnol 27: 359–364 [Google Scholar]
- Kurczyńska EU, Gaj MD, Ujczak A, Mazur E (2007) Histological analysis of direct somatic embryogenesis in Arabidopsis thaliana (L.) Heynh. Planta 226: 619–628 [DOI] [PubMed] [Google Scholar]
- Ledwoń A, Gaj MD (2011) LEAFY COTYLEDON1, FUSCA3 expression and auxin treatment in relation to somatic embryogenesis induction in Arabidopsis. Plant Growth Regul 65: 157–167 [Google Scholar]
- Ledwoń A, Gaj MD (2009) LEAFY COTYLEDON2 gene expression and auxin treatment in relation to embryogenic capacity of Arabidopsis somatic cells. Plant Cell Rep 28: 1677–1688 [DOI] [PubMed] [Google Scholar]
- Li YJ, Yu Y, Liu X, Zhang XS, Su YH (2021) The Arabidopsis MATERNAL EFFECT EMBRYO ARREST45 protein modulates maternal auxin biosynthesis and controls seed size by inducing AINTEGUMENTA. Plant Cell 33: 1907–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D (2015) Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real- time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto MA, Matsudaira Yee K, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luo Y, Koop HU (1997) Somatic embryogenesis in cultured immature zygotic embryos and leaf protoplasts of Arabidopsis thaliana ecotypes. Planta 202: 387–396 [DOI] [PubMed] [Google Scholar]
- Michalczuk L, Cooke TJ, Cohen JD (1992) Auxin levels at different stages of carrot somatic embryogenesis. Phytochemistry 31: 1097–1103 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Musielak T, Bürgel P, Kolb M, Bayer M (2016) Use of SCRI renaissance 2200 (SR2200) as a versatile dye for imaging of developing embryos, whole ovules, pollen tubes and roots. Bioprotocol 6: e1935 [Google Scholar]
- Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, et al. (2014) Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J 77: 352–366 [DOI] [PubMed] [Google Scholar]
- Normanly J (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarinho P, Ketelaar T, Meiqing A, Ae X, Van J, Ae A, Maliepaard C, Mieke A, Hendriks W, Joosen R, et al. (2008) BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol 68: 225–237 [DOI] [PubMed] [Google Scholar]
- Pasternak TP, Prinsen E, Ayaydin F, Miskolczi P, Potters G, Asard H, Van Onckelen HA, Dudits D, Fehér A (2002) The role of auxin, pH, and stress in the activation of embryogenic cell division in leaf protoplast-derived cells of alfalfa. Plant Physiol 129: 1807–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichot C, El Mâtaoui M, Raddi S, Raddi P (2001) Surrogate mother for endangered cupressus: a rare cypress tree increases its chances by using a clever reproductive strategy. Nature 412: 39. [DOI] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez-Perez GF, Scheres B (2013) Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci USA 110: 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prime TA, Sherrier DJ, Mahon P, Packman LC, Dupree P (2000) A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21: 3488–3499 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J (2013) Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512 [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Loreto Gutièrrez C, Wójcikowska B, Pěnčík A, Novák O, Chen J, Grunewald W, Dresselhaus T, Friml J, et al. (2018) Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants 4: 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano N, Rajjou L, North HM (2020) Lost in translation: physiological roles of stored mRNAs in seed germination. Plants 9: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L, Gorte M, Prasad K, Bao D, Timmermans-Hereijgers JLPM, et al. (2016) The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28: 2937–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A (2020) Controlling apomixis: shared features and distinct characteristics of gene regulation. Genes (Basel) 11: 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano M, Li H, Jacquard C, Angenent GC, Krochko J, Offringa R, Boutilier K (2014) Plasticity in cell division patterns and auxin transport dependency during in vitro embryogenesis in Brassica napus. Plant Cell 26: 2568–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståldal V, Cierlik I, Landberg K, Myrenås M, Sundström JF, Eklund DM, Chen S, Baylis T, Ljung K, Sundberg E (2012) The Arabidopsis thaliana transcriptional activator STYLISH1 regulates genes affecting stamen development, cell expansion and timing of flowering. Plant Mol Biol 78: 545–559 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM (2016) Auxin catabolism unplugged: role of IAA oxidation in auxin homeostasis. Proc Natl Acad Sci USA 113: 10742–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci USA 105: 3151–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Zhao XY, Liu YB, Zhang CL, O’Neill SD, Zhang XS (2009) Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J 59: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LP, Zhou C, Wang SS, Yuan J, Zhang XS, Su YH (2017) FUSCA 3 interacting with LEAFY COTYLEDON 2 controls lateral root formation through regulating YUCCA 4 gene expression in Arabidopsis thaliana. New Phytol 213: 1740–1754 [DOI] [PubMed] [Google Scholar]
- Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y (2017) Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551: 124–128 [DOI] [PubMed] [Google Scholar]
- Tian R, Paul P, Joshi S, Perry S (2020) Genetic activity during early plant embryogenesis. Biochem J 477: 3743–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugafune S, Mashiguchi K, Fukui K, Takebayashi Y, Nishimura T, Sakai T, Shimada Y, Kasahara H, Koshiba T, Hayashi KI (2017) Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci Rep 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uc-Chuc MA, Pérez-Hernández C, Galaz-Ávalos RM, Brito-Argaez L, Aguilar-Hernández V, Loyola-Vargas VM (2020) YUCCA-mediated biosynthesis of the auxin IAA is required during the somatic embryogenic induction process in Coffea canephora. Int J Mol Sci 21: 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdeil JL, Alemanno L, Niemenak N, Tranbarger TJ (2007) Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends Plant Sci 12: 245–252 [DOI] [PubMed] [Google Scholar]
- Vijverberg K, Ozias-Akins P, Schranz ME (2019) Identifying and engineering genes for parthenogenesis in plants. Front Plant Sci 10: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015) Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA 112: 4821–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FX, Shang GD, Wu LY, Xu ZG, Zhao XY, Wang JW (2020) Chromatin accessibility dynamics and a hierarchical transcriptional regulatory network structure for plant somatic embryogenesis. Dev Cell 54: 742–757.e8 [DOI] [PubMed] [Google Scholar]
- Wang R, Tavano EC da R, Lammers M, Martinelli AP, Angenent GC, de Maagd RA (2019) Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci Rep 9: 1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu B, Wang H, Li J, Huang H, Xu L (2011) YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol 157: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS One 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik AM, Wójcikowska B, Gaj MD (2020) Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. Int J Mol Sci 21: 1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcikowska B, Jaskóła K, Gąsiorek P, Meus M, Nowak K, Gaj MD (2013) LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 238: 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel J, Barlow PW, Gorka K, Nabialkowska D, Kurczynska EU (2011) Histology and symplasmic tracer distribution during development of barley androgenic embryos. Planta 233: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y (2014) Auxin biosynthesis. Arab B 12: e0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.