Summary
One of the impacts of the coronavirus disease 2019 (COVID-19) pandemic has been a push for researchers to better exploit synthetic data and accelerate the design, analysis, and modeling of clinical trials. The unprecedented clinical efforts caused by COVID-19’s emergence will certainly boost future robust and innovative approaches of statistical sciences applied to clinical fields. Here, we report the development of SASC, a simple but efficient approach to generate COVID-19-related synthetic clinical data through a web application. SASC takes basic summary statistics for each group of patients and attempts to generate single variables according to internal correlations. To assess the “reliability” of the results, statistical comparisons with Synthea, a known synthetic patient generator tool, and, more importantly, with clinical data of real COVID-19 patients are provided. The source code and web application are available on GitHub, Zenodo, and Mendeley Data.
Keywords: SASC, virtual cohort, VC, synthea, COVID-19, clinical trial, real-world data
Highlights
-
•
From summaries of grouped variables, it is possible to generate synthetic data
-
•
Similarity between real and virtual cohorts visualized with Kaplan-Meyer plots
-
•
Shiny app for tuning and downloading the synthetic cohort provided to users
-
•
SASC performances and result accuracies are similar to or better than Synthea results
The bigger picture
Virtual cohorts built on synthetic data have received attention lately as they can change the way we work with clinical data. Generating synthetic data very close to "real-world" data without having ethical restrictions can considerably help data analysts in several aspects such as didactic efforts, statistical modeling, and assessments of interventions. We developed SASC, a tool that uses summary statistics of real clinical data to produce synthetic data having the same or a very close summary to the reference variables. SASC displays a performance comparable with, or superior to, that of other virtual cohort tools like Synthea and allows users to optimize the synthetic dataset produced through a Shiny visualizer application interactively.
SASC shows that to generate virtual cohorts of patient-level data, no black-box tools are needed. More-than-acceptable datasets can be achieved without neural-network-based virtual cohort generators (e.g., GAN) where generation processes are difficult follow and understand. Starting from patient-level cohort data summaries of grouped variables, and using a modified random-number-generation routine in R, synthetic datasets can be generated possessing quantitative characteristics similar to the real cohort, allowing further analysis in an easy flow scheme.
Introduction
In recent years, there has been growing interest in the re-use of clinic data for analyzing patients' disease phenotypes and their relative treatment. The coronavirus disease 2019 (COVID-19) pandemic has fueled this interest due to the need for accelerated design and analysis of clinical trials. Developments that can rapidly analyze clinical data in a reproducible manner and in a secure and privacy-preserving environment have become increasingly necessary. A virtual patient (VP) could be an answer to these needs. First described in 1971, VPs were able to mimic real clinical scenarios with the help of a computer program.1 Because of this, it then became possible to analyze physical exams, design diagnoses, and provide therapies based on a patient’s clinical history.2
A collection of VPs forms a virtual cohort (VC). In this regard, VCs can support several professionals: a novice can aid in simulating different clinical scenarios, a medical doctor could find suggestions for therapies for special patients, and, in general, clinical statisticians can use VPs and VCs as the stimuli for problem-based learning approaches.3 Indeed, the primary use of VCs began mainly for educational purposes.4
To create synthetic data, there are at least two broad methodological categories to choose from, each with its own benefits and drawbacks. The first includes drawing numbers from distributions based on certain statistical methods, while the other uses agent-based or artificial intelligence (AI)-based modeling methods. The latter of these approaches aims to explain an observed behavior using a mathematical model for driving the data-creation step.5
Belonging to the latest approach, the AI-driven methodologies are based on deep learning models, like variational autoencoders (VAEs) and generative adversarial networks (GANs). Another interesting approach based on agent-based6 simulations has been published recently. All of these generation techniques are mainly devoted to capturing and preserving internal variable correlations. Even though these methods represent an improvement of the overall capability of synthetic data to mirror every aspect of the reality, their complexity sometimes makes them obscure and non-transparent.7
When talking about mathematical models, they become more accurate when they get closer to observations seen in real populations. For this to happen, the variability of real patients must be taken into account, thereby simulating diversity in VPs. In principle, a trivial method to generate VP is adding, for instance, random noise to existing, real data.8 However, this approach does not have much success. The reason for this is the preservation of inter- and intra-patients' correlations, which are of vital importance for a reliable VC generation.9
In addition to this, legal and ethical constraints are to be evaluated, which complicate the sharing and re-use of real patient-level data beyond summary statistics, even when anonymization is accomplished in Europe according to the General Data Protection Regulation (GDPR) (https://gdpr-info.eu/). This is true not only at the inter-organization level but also at the intra-organization level.10 Therefore, clinical data “silos” exist. This is increasingly becoming an issue, as medicine as a whole is driven by the availability of electronic health reports (EHRs) where opportunities for digitalized storage, access, retrieval, and analysis, including the emerging use AI, exist.11 A practical way to overcome these objectives and legal hindrances could be the generative approach to very “realistic” patient-level data (i.e., a VC) possessing the same statistical features and properties as the source clinical data. The real advantage of this approach is manifold: first, clinical data often have missing data, which have to be imputed by the analyst wherever possible, while VCs contain no missing data if not inserted intentionally. Secondly, patient-recruitment difficulties are obvious to overcome: practical recruitment restrictions or preferences, for example, gender, habit selection (e.g., only smokers), and geo-localization can be easily implemented, provided that suitable reference “real” demographic data are available. Finally, “clinical-like” data can be inexpensive while maintaining a high-quality level, and, thus, their modeling could improve clinical trial planning or analysis.
Scope of the paper
In the present work, we describe SASC, a straightforward approach to generate VCs starting either from real patient-level data or from a summary table of a clinical trial. The VC-generating script is the result of a multilateral effort that included the collection of literature data as well as finding real, anonymized, clinical patient-level data that is publicly available. The acquisition of anonymized clinical data, with which we compare the synthetic data, has been quite challenging work. Even though the quest for re-purposed and effective therapies against the COVID-19 pandemic has generated numerous clinical trials, very few of them are easily accessible to researchers. Following the approach of Tucker et al.12 and using the data contained in their supplementary materials, we designed the present study as a comparison between a real clinical dataset, results from Synthea, and the synthetic datasets coming from SASC.
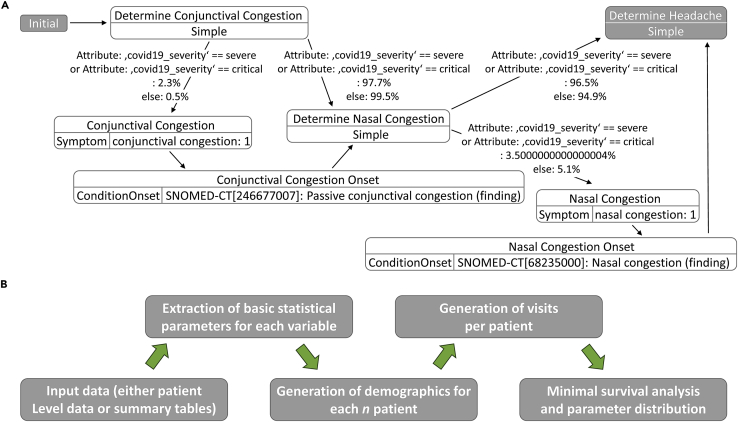
Fundamentally, numerical random generation under constraints is the core of SASC. This is true for both observational and interventional VCs, thereby assuring maintenance of intra-patient and cross-patient correlations that are important in achieving naturally “reliable” VC data, especially when there is only a qualitative assessment available for comparison. For a more quantitative assessment, several approaches have been offered.13,14 For simplicity, we did not generate metrics to compare cohorts similar to Tucker et al.,12 given the scope of this work, but rather focused on the similarity of single-parameter distributions and their relative correlations. Both VCs generated by SASC and Synthea share a common principle: the longitudinal component in terms of medical visits (called “encounters” in Synthea output) is generated patient-by-patient like a storybook in development.15 The major difference between the two is that Synthea generates a mixed population of sick versus healthy VPs, while SASC generates only VPs with a predefined ratio of outcomes (Figure 1).
Figure 1.
Different workflows for data generation
(A and B) Schema followed for the generation of data for (A) Synthea (from GitHub: https://synthetichealth.github.io/module-builder/#covid19/symptoms) and (B) SASC. Contrary to Synthea, SASC takes real clinical data at the patient level as the starting point, generates summary tables and relative grouped statistics according to demographics and outcomes, and, for each desired n virtual patient, recreates relative demographics along with the number of visits. In the end, it provides a survival analysis on the synthetically generated clinical parameters.
Due to potential ethical issues relating to the use of the algorithm, we focus on possible methodological concerns following the classification of Mittelstadt et al.16 For example, concerns implying causality can be directly excluded for SASC, as it does not generate therapeutic suggestions or causal dependencies between conditions and clinical parameters. Moreover, epistemic concerns (e.g., inconclusive or inscrutable evidence) should not affect SASC, as its code is transparent and public, and the risk of inconclusive or opaque decisions made on downstream machine-learning tasks (using the SASC results) is not within SASC’s responsibility, in our view. Biases in the generation of variable values are likewise rare, as they are strictly dependent on the reference clinical data summary values (Tables 1 and 2). SASC, therefore, has the ability to recreate the same features of the reference cohorts (Table 3), and we tried to demonstrate this using a reference COVID-19 dataset.
Table 1.
Real cohort statistics − one
| Parameter | Statistics |
|||||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Outcome | survived | survived | survived | survived | dead | dead | dead | dead |
| Gender | M | M | F | F | M | M | F | F |
| Age | 53.4 | 14.0 | 47.3 | 15.1 | 68.5 | 11.4 | 72.5 | 10.2 |
| Hospitalization days | 15.0 | 4.9 | 15.8 | 6.9 | 10.7 | 7.8 | 10.4 | 7.6 |
| Albumin | 35.6 | 4.5 | 36.8 | 4.0 | 27.7 | 4.8 | 27.6 | 4.8 |
| Neutrophil, % | 67.6 | 14.7 | 64.4 | 13.3 | 90.1 | 8.4 | 90.0 | 5.9 |
| Prothrombin activity | 94.3 | 11.6 | 95.7 | 12.7 | 64.6 | 20.7 | 69.0 | 15.5 |
| Neutrophil count | 4.6 | 2.7 | 3.9 | 2.7 | 11.7 | 6.6 | 11.4 | 5.4 |
| Lymphocyte, % | 22.3 | 11.1 | 26.3 | 11.1 | 5.6 | 5.7 | 6.0 | 4.0 |
| D-D dimer | 1.2 | 2.3 | 1.3 | 3.2 | 12.9 | 9.0 | 13.5 | 9.5 |
| Lactate dehydrogenase | 265.6 | 115.8 | 230.6 | 67.8 | 718.8 | 416.3 | 673.3 | 346.9 |
| Fibrin degradation products | 8.3 | 18.1 | 4.6 | 1.8 | 93.8 | 66.2 | 80.2 | 56.7 |
| High sensitivity C-reactive protein | 30.6 | 41.7 | 19.4 | 33.5 | 138.6 | 79.8 | 109.8 | 73.4 |
Summary table of relevant statistical parameters from the real COVID-19 cohort used in the SASC VC-generating script.
Table 2.
Real cohort statistics – two
| Category | Characteristics | Female |
Male |
p value | ||
|---|---|---|---|---|---|---|
| Survived (N = 103) (%) | Dead (N = 48) (%) | Survived (N = 98) (%) | Dead (N = 126) (%) | |||
| Age | – | 48 (15.4) | 70 (11.8) | 53 (14.3) | 68 (11.8) | |
| Gender | F | 103 (100) | 48 (100) | 0 (0) | 0 (0) | <0.001 |
| Gender | M | 0 (0) | 0 (0) | 98 (100) | 126 (100) | |
| Outcome | alive | 103 (100) | 0 (0) | 98 (100) | 0 (0) | <0.001 |
| Outcome | dead | 0 (0) | 48 (100) | 0 (0) | 126 (100) | |
| Age | young | 25 (24) | 0 (0) | 16 (16) | 2 (1.6) | <0.001 |
| Age | middle age | 28 (27) | 41 (85) | 33 (34) | 99 (79) | |
| Age | old | 50 (49) | 7 (15) | 49 (50) | 35 (20) | |
Group percentages extracted from COVID-19 real data. These values have been used in the generation step of independent values (demographic and outcome). Characteristics for age: middle age, people aged between 36 and 60 years; older, people aged >60 years; and younger, people aged <35 years.
Table 3.
Virtual cohort statistics
| Parameter | Statistics |
|||||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Outcome | survived | survived | survived | survived | dead | dead | dead | dead |
| Gender | M | M | F | F | M | M | F | F |
| Age | 54.9 | 16.7 | 51.4 | 22.6 | 67.3 | 10.7 | 55.5 | 14.6 |
| Albumin | 36.2 | 9.3 | 39.4 | 10.5 | 28.0 | 8.0 | 31.1 | 8.5 |
| Neutrophil, % | 39.2 | 10.1 | 34.9 | 9.5 | 76.0 | 19.1 | 68.2 | 18.5 |
| Prothrombin activity | 89.2 | 29.4 | 102.2 | 29.0 | 71.4 | 23.4 | 84.3 | 22.2 |
| Neutrophil count | 3.7 | 2.6 | 2.8 | 2.4 | 7.9 | 5.3 | 5.3 | 4.5 |
| Lymphocyte, % | 19.6 | 15.3 | 28.5 | 18.0 | 13.6 | 10.5 | 18.6 | 11.2 |
| D-D dimer | 4.6 | 3.6 | 3.5 | 3.1 | 9.4 | 7.1 | 7.5 | 6.2 |
| Lactate dehydrogenase | 278.8 | 160.5 | 196.8 | 135.3 | 528.1 | 319.2 | 434.7 | 310.1 |
| Fibrin degradation products | 38.2 | 25.9 | 29.7 | 20.7 | 75.6 | 53.0 | 60.3 | 44.8 |
| High sensitivity C-reactive protein |
49.3 | 32.4 | 35.0 | 27.0 | 94.8 | 63.7 | 71.9 | 53.6 |
Summary table for relevant variables generated by values in SASC VC showing results comparable to Table 1.
Results and discussion
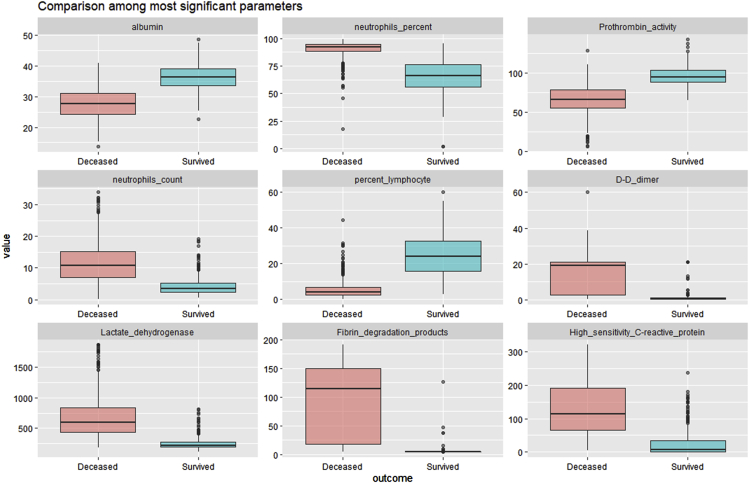
Our study started with a public longitudinal observational COVID-19 cohort of 375 participants with limited demographic parameters and pharmacological variables. The participants' outcomes were either deceased or survived, and, thus, the first analysis was concerned with the identification and characterization of these pharmacological variables that showed the highest positive and negative correlation with the outcome (Figure 2).
Figure 2.
Boxplot of variables correlating with outcome
Nine representative pharmacological variables from a public longitudinal observational COVID-19 cohort dataset with highest positive and negative correlations (absolute Pearson’s correlation coefficient >0.6) with regard to outcome (0 = survived, 1 = deceased). The selection of the above variable intends to highlight the most outcome-correlated variables and, as such, the most relevant for downstream modeling attempts using the synthetic data generated by SASC. All variables, however, have been generated using the same methodology.
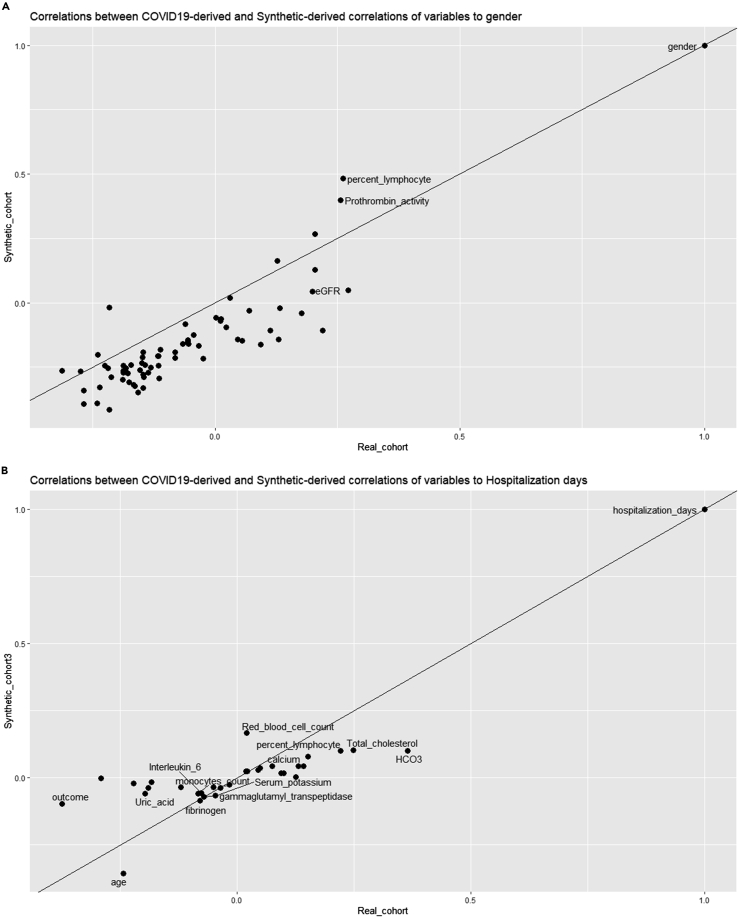
To demonstrate the validity of the process beyond the distribution shown above, we generated a correlation plot (Figure 3) between the correlations of variables from the real COVID-19 cohort and the SASC-generated synthetic cohort to gender and hospitalization time. This was an exemplary choice as it was not practical to generate extended heatmaps or correlation plots for all variables measured. Even though the distribution limits are maintained during the SASC variable-generation process, it is not certain that the correlation is strictly maintained, as the correlation also depends on the number of visits per patient and can differ after multiple SASC runs due to the random generator. To simplify the complexity of the quantitative comparison needed, we decide to track comparisons between real and generated synthetic variables by sampling their correlations to a common reference.
Figure 3.
Comparison correlation plots
(A and B) Here, correlation between the SASC-generated VC against the real COVID-19 cohort toward gender (A) and hospitalization days (B) are shown. Due to density, only some points are labeled to show their provenance. Each point coordinate represents the correlation obtained toward a third variable taken as reference (in this case, gender and hospitalization days) for the two cohorts (x axis = real cohort; y axis = VC).
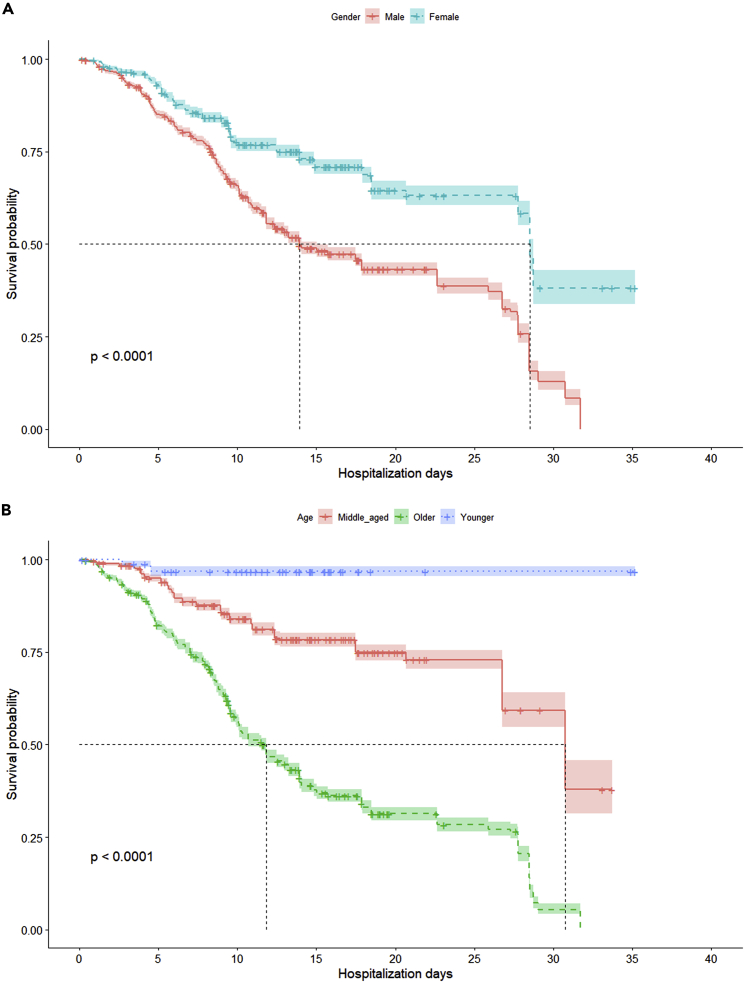
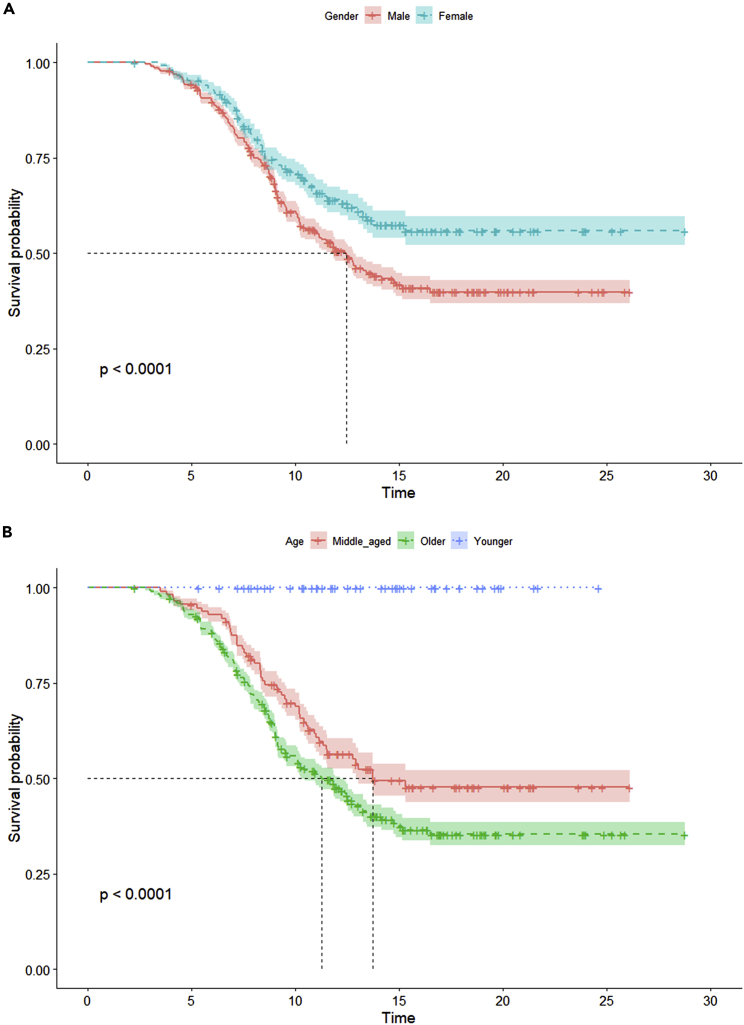
We also performed survival analysis on both the gender and age groups of participants. We noted well-known trends (Figure 4), including women showing higher hospitalization days with median survival probability and older patients showing less than half the hospitalization days at median survival probability than the other age groups. Kaplan-Meier analysis17 and single-parameter distribution comparison constitutes the minimal number of comparisons that our VC generation approach should satisfy in order to be assessed as a “realistic” VC and, thus, acceptable as an output (Figure 5).
Figure 4.
Survival analysis for real cohort
(A and B) Survival analysis (with Kaplan-Meier plot) of the public longitudinal observational COVID-19 cohort for (A) gender and (B) age groups. The age group has been defined as “young” with ages <35, “old” with ages >60, and “middle aged” for all remaining ages.
Figure 5.
Survival analysis for virtual cohort
(A and B) Kaplan-Meier survival plots related to a SASC VC generated for (A) gender and (B) age groups. The parameters were restricted to a maximum of seven visits to a medical doctor within a time interval of 10 days for all virtual patients. Characteristics for age: middle age, people aged between 36 and 60 years; older, people aged >60 years; younger, people aged <35 years.
Comparison of SASC cohort with real data
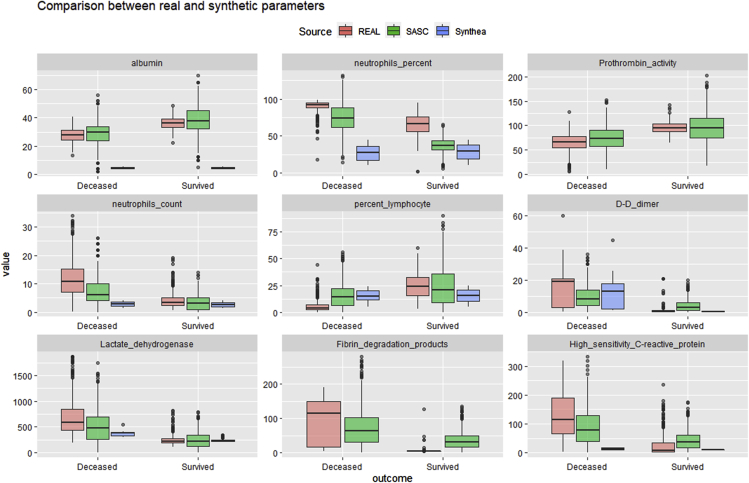
In order to further assess the reliability of the VC data generated, we conducted a comparison of the most relevant parameter distributions (absolute correlation with outcome >0.6) using the variables selected in Figure 2 (Figure 6).
Figure 6.
Summary boxplot comparison
Boxplot comparisons for each generated numerical parameter. Synthea parameter (blue) values seem to be, in general, more compressed than the ones of the real (orange) and SASC (green) datasets.
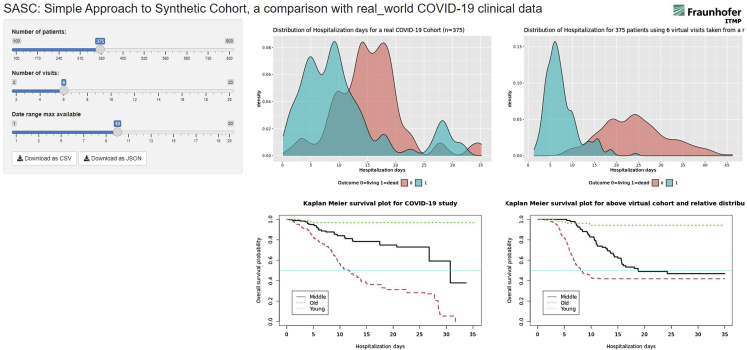
The SASC Shiny app (Figure 7), with its built-in survival analysis, can provide a similar display. When using specific parameters, users have dynamical visual evidence on how far the generated VC lays from the survival analysis of the real clinical data. Upon identification of discrepancies, users can improve the method to generate a novel VC.
Figure 7.
Shiny app display
Shiny app display page: the left cursors generates the right panels of visualization for a novel VC. The central fixed plots report the real cohort with the hospitalization day distributions for surviving and deceased patients in the upper central quadrant.
Comparison of SASC cohort with Synthea COVID-19 cohort
We have demonstrated that VCs generated using classical statistical parameters like mean and standard deviation can satisfy conditions for reliability. In Figure 6, the three sources of data (i.e., real, SASC, and Synthea cohorts) are compared in a boxplot for a selection of variables. Due to initial limitations of its novel COVID-19 module, Synthea did not generate two of the parameters, “Prothrombin activity” and “Fibrin degradation products”, and hence their corresponding blue boxes are not present in the boxplot analysis. Moreover, some clinical measurements play different roles whether they are generated in Europe or the US.
The Synthea module for COVID-19 is still in a beta version according to the authors; therefore, it is possible that it has not yet been optimized. Despite this, Synthea has been widely used in several other experiments, allowing users to design modules and add or modify parameters in case the cohort requires specific clinical design.18 Moreover, it also provides FHIR-formatted multi-output, which we did not use in this case.19
In our comparison reported herein, we have focused on clinically relevant parameters that provided a clear correlation with a real COVID-19 cohort outcome. All nine relevant parameters showed a positive or negative Pearson correlation >0.6. Besides this, comparison between the limited demographic variables was performed as shown in Tables 1 and 3. Synthea is US-centric and therefore uses US resident population data including demographic annotations, ethnicity, race, geo-localization within the US, marital status, smoking habits, etc. These parameters were absent in the real reference dataset and were thus excluded from the comparison. All numerical and ordinal data summarized in tabular format are derived from Figure 1A.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrea Zaliani (andrea.zaliani@itmp.fraunhofer.de).
Materials availability
The reference COVID-19 dataset was obtained from the Clinical Practice Research Datalink (CPRD).20 This synthetic dataset was based on real, anonymized, primary care patient data extracted from the CPRD Aurum database (https://www.cprd.com/primary-care).21 Patients were typically in primary care with symptoms of COVID-19 (confirmed/suspected) and control participants with a negative COVID-19 test result. For the purpose of this paper, we made use of the CRPD COVID-19 symptoms and risk factors synthetic dataset (v.2021.04.001). The dataset contains information on sociodemographic and clinical risk factors from December 3, 2019, to March 13, 2021. The dataset has also been made public here: Mendeley Data: https://doi.org/10.17632/ptz6zhknyp.1.
Generation of VC
Analogous to real cohort analysis, we generated a summary table from the public longitudinal observational COVID-19 cohort with the most important demographic and medical parameters. Although there are several methods to generate VCs from such a table, we chose to extract a set of probabilities for grouped demographics (e.g., age and gender) from real data and used these probabilities to drive the generation of independent, normally distributed random values. Using this strategy, we generated participant outcomes (i.e., death and survival) too.
Taking advantage of a table of grouped probabilities (Table 1), we generated random variables normally distributed for each group of VPs for what concerns their outcome, age, gender, and visit numbers (see outcome, gender, and age in Table 2). All other numerical variables were generated on the basis of their grouped averages and standard deviations (as depicted in Table 1) under the given constraint of their correlations with outcome and/or gender, following a general principle described in the following link (https://stats.stackexchange.com/questions/15011/generate-a-random-variable-with-a-defined-correlation-to-an-existing-variables/15035#15035). However, this generative step is only meaningfully applied for existing correlations with Pearson’s index higher than 0.6 in absolute value and is not warranted for all. This is an aspect clearly visible in Figure 4.
As the “hospitalization day” variable was used to analyze the events (death or survival) in the real clinical cohort, the optimal manner to mimic this variable was the number of visits performed within the same overall time span as that of the reference cohort. As this time-range variable seemed to be important in the real cohort analysis, we decided to implement its variation within the SASC Shiny app, where two control sliders were provided to users.
The overall workflow is depicted in Figure 1B.
For what concerns the generation of visiting time for all patients, we used the hospitalization days reference as a driving variable. From the hospitalization days distribution plot (Figure 7; Table 1), it is clear that patients who did not survive had shorter hospitalization times, so we used a maximal number of visits capped at seven and a time span of 10 days, and we defined the longitudinal variable hospitalization days as the difference in time between the start of the study (fixed for all) and the last medical doctor’s visit performed. This analysis generated the Kaplan-Meier plots reported in Figure 4, where it is evident that the age, class, and gender survival patterns are retained qualitatively and quantitatively, both in terms of ranking and significance compared with the real cohort.
As illustrated in Table 3, the variable summary of the VC is close to that of the reference cohort shown in Table 1. The main difference between the SASC-generated and reference-grouped summary tables is the reduction in difference between genders based on mortality. The lowest difference is evident between the older and middle-aged individuals. To display these results, we programmed a so-called Shiny app where we generated different VCs dynamically by modifying the two main parameters used (i.e., number of visits and time span of visits). Thus, this Shiny app supports users by generating diverse VCs according to the parameter chosen and provides an impression of the relative “reliability” of the VC synthetized.
Advantages and limitations of SASC approach
SASC, differently from Synthea, does not need any local installation and runs on the web, allowing users to directly download the results. Although the SASC Shiny app has been dockerized and can run on cloud environments (such as Binderhub), in principle, any non-public cloud environment could be able to host this application. Synthetic data generated by SASC require pre-existing, ethically compliant clinical datasets. In this sense, SASC-synthetized data are aimed toward educational purposes or for the validation of imputation techniques. Moreover, the COVID-19 observational reference studies for this analysis are typically of a few months' duration, and correlations of parameters with gender or age can differ on longer timescales. SASC also implies normal distributions for the variables it generates, which may not always be the case, and this is a limitation of the method. An initial survey, however, elucidated that the variables were predominant in either normal or lognormal distributions. In this first version of SASC, we focused on the basic structure of the data generative model. Comparisons provided here are limited, and we are actively working on defining novel but simple ways to compare real cohorts and VCs along the line of the work of Tucker et al.12 The different longitudinal dimension of the cohorts implies, in our eyes, the need to use robust and convenient summaries of clinical variables per patient as comparative parameters. Comparisons among internal correlations, though, is a tough task and needs a study of its own.
Conclusions
When legal and ethical constraints influence sharing and re-use of health data, the generation of synthetic reliable participant data can be a viable solution if ethically compliant algorithms have generated them. As SASC only uses classical statistical distribution, the use of synthetic data is not only “safer” for participant data privacy but also represents a valuable source for exploratory data analysis (EDA), clustering approaches, testing of novel modeling imputation technologies, and development of analytical workflows. We report herein a simple and efficient R script tool aimed at generating VCs starting from a reference clinical dataset. The comparison with a real COVID-19 clinical cohort resulted in similar conclusions from Kaplan-Meier plots, which validates our approach.
Acknowledgments
We would like to thank Sarah Mubeen and Dr. Sheraz Gul for reviewing the manuscript and providing us with valuable suggestions. We are indebted to Dr. Jean-Marie Burel at the University of Dundee for their support in the dockerization of the app. This research was done in the “COPERIMOplus” initiative and is supported by the Fraunhofer “Internal Programs” under grant no. Anti-Corona 840266.
Author contributions
Conceptualization, T.K., A.Z., C.C., G.W., and D.L.; writing – review & editing, A.Z., Y.G., and C.C.; writing – original draft, A.Z., T.K., G.W., and D.L.; supervision, A.Z. and Y.G.; methodology, T.K. and A.Z.; coding and reviewing the application, Y.G. and A.Z. All authors have read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 9, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2022.100453.
Contributor Information
Takoua Khorchani, Email: takoua.khorchani@etu.univ-lyon1.fr.
Andrea Zaliani, Email: andrea.zaliani@itmp.fraunhofer.de.
Supplemental information
Data and code availability
For generating VCs from Synthea, we used the newly added COVID-19 module (Figure 1A).18 We generated a 375-population cohort, whose data are available in the supplemental information (GitHub: https://github.com/synthetichealth/synthea). From those data, some comparison variable values have been used to generate a boxplot figure (Figure 6).
The original dataset, the code for the Shiny app, and the dockerized form of it are also publicly deposited on Zenodo at Zenodo: https://zenodo.org/record/5896935#.Ye6cpnrMJgC and at Mendeley Data: https://doi.org/10.17632/ptz6zhknyp.1. The SASC viewer (Figure 7) is accessible on GitHub at GitHub: https://github.com/Fraunhofer-ITMP/SASC/tree/v1.0. The Shiny was built with RStudio v.1.2.1335 (https://www.rstudio.com).
References
- 1.Harless W.G., Drennon G.G., Marxer J.J., Root J.A., Miller G.E. CASE: a computer-aided simulation of the clinical encounter. Acad. Med. 1971;46:443–448. [PubMed] [Google Scholar]
- 2.Cook D.A., Triola M.M. Virtual patients: a critical literature review and proposed next steps. Med. Educ. 2009;43:303–311. doi: 10.1111/j.1365-2923.2008.03286.x. [DOI] [PubMed] [Google Scholar]
- 3.Bergin R.A., Fors U.G. Interactive simulated patient—an advanced tool for student-activated learning in medicine and healthcare. Comput. Educ. 2003;40:361–376. doi: 10.1016/S0360-1315(02)00167-7. [DOI] [Google Scholar]
- 4.Ellaway R., Masters K. AMEE Guide 32: e-Learning in medical education Part 1: learning, teaching and assessment. Med. Teach. 2008;30:455–473. doi: 10.1080/01421590802108331. [DOI] [PubMed] [Google Scholar]
- 5.Macal C.M., North M.J. Proceedings of the Winter Simulation Conference. IEEE; 2005. Tutorial on agent-based modeling and simulation; p. 14. [DOI] [Google Scholar]
- 6.Popper N., Zechmeister M., Brunmeir D., Rippinger C., Weibrecht N., Urach C., Bicher M., Schneckenreither G., Rauber A. Synthetic reproduction and augmentation of COVID-19 case reporting data by agent-based simulation. Data Sci. J. 2021;20:16. doi: 10.5334/dsj-2021-016. [DOI] [Google Scholar]
- 7.Kornish D., Ezekiel S., Cornacchia M. 2018 IEEE Applied Imagery Pattern Recognition Workshop (AIPR) IEEE; 2018. Dcnn augmentation via synthetic data from variational autoencoders and generative adversarial networks; pp. 1–6. [DOI] [Google Scholar]
- 8.Eymard N., Volpert V., Kurbatova P., Volpert V., Bessonov N., Ogungbenro K., Aarons L., Janiaud P., Nony P., Bajard A., et al. Mathematical model of T-cell lymphoblastic lymphoma: disease, treatment, cure or relapse of a virtual cohort of patients. Math. Med. Biol. a J. IMA. 2018;35:25–47. doi: 10.1093/imammb/dqw019. [DOI] [PubMed] [Google Scholar]
- 9.Chase J.G., Preiser J.C., Dickson J.L., Pironet A., Chiew Y.S., Pretty C.G., Shaw G.M., Benyo B., Moeller K., Safaei S., et al. Next-generation, personalised, model-based critical care medicine: a state-of-the art review of in silico virtual patient models, methods, and cohorts, and how to validation them. Biomed. Eng. Online. 2018;17:1–29. doi: 10.1186/s12938-018-0455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gootjes-Dreesbach L., Sood M., Sahay A., Hofmann-Apitius M., Fröhlich H. Variational autoencoder modular bayesian networks for simulation of heterogeneous clinical study data. Front. Big Data. 2020;3:16. doi: 10.3389/fdata.2020.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fröhlich H., Balling R., Beerenwinkel N., Kohlbacher O., Kumar S., Lengauer T., Maathuis M.H., Moreau Y., Murphy S.A., Przytycka T.M., et al. From hype to reality: data science enabling personalized medicine. BMC Med. 2018;16:1–15. doi: 10.1186/s12916-018-1122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tucker A., Wang Z., Rotalinti Y., Myles P. Generating high-fidelity synthetic patient data for assessing machine learning healthcare software. NPJ Digit. Med. 2020;3:1–13. doi: 10.1038/s41746-020-00353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fultz S.L., Skanderson M., Mole L.A., Gandhi N., Bryant K., Crystal S., Justice A.C. Development and verification of a" virtual" cohort using the national VA health information system. Med. Care. 2006:S25–S30. doi: 10.1097/01.mlr.0000223670.00890.74. [DOI] [PubMed] [Google Scholar]
- 14.Li Z., Mirams G.R., Yoshinaga T., Ridder B.J., Han X., Chen J.E., Stockbridge N.L., Wisialowski T.A., Damiano B., Severi S., et al. General principles for the validation of proarrhythmia risk prediction models: an extension of the CiPA in silico strategy. Clin. Pharmacol. Ther. 2020;107:102–111. doi: 10.1002/cpt.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walonoski J., Kramer M., Nichols J., Quina A., Moesel C., Hall D., Duffett C., Dube K., Gallagher T., McLachlan S. Synthea: an approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J. Am. Med. Inform. Assoc. 2018;25:230–238. doi: 10.1093/jamia/ocx079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittelstadt B.D., Allo P., Taddeo M., Wachter S., Floridi L. The ethics of algorithms: mapping the debate. Big Data Soc. 2016;3:1–21. doi: 10.1177/2053951716679679. [DOI] [Google Scholar]
- 17.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 18.Walonoski J., Klaus S., Granger E., Hall D., Gregorowicz A., Neyarapally G., Watson A., Eastman J. Synthea™ Novel coronavirus (COVID-19) model and synthetic data set. Intell-Based Med. 2020;1:100007. doi: 10.1016/j.ibmed.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxhelaku S., Kika A. Proceedings of International Academic Conferences (No. 9211566) International Institute of Social and Economic Sciences; 2019. Improving interoperability in healthcare using Hl7 Fhir. [Google Scholar]
- 20.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jin L., Zhang M., et al. A machine learning-based model for survival prediction in patients with severe COVID-19 infection. medRxiv. 2020;27:2002. doi: 10.1101/2020.02.27.20028027. Preprint at. [DOI] [Google Scholar]
- 21.Herrett E., Gallagher A.M., Bhaskaran K., Forbes H., Mathur R., Van Staa T., Smeeth L. Data resource profile: clinical practice research datalink (CPRD) Int. J. Epidemiol. 2015;44:827–836. doi: 10.1093/ije/dyv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For generating VCs from Synthea, we used the newly added COVID-19 module (Figure 1A).18 We generated a 375-population cohort, whose data are available in the supplemental information (GitHub: https://github.com/synthetichealth/synthea). From those data, some comparison variable values have been used to generate a boxplot figure (Figure 6).
The original dataset, the code for the Shiny app, and the dockerized form of it are also publicly deposited on Zenodo at Zenodo: https://zenodo.org/record/5896935#.Ye6cpnrMJgC and at Mendeley Data: https://doi.org/10.17632/ptz6zhknyp.1. The SASC viewer (Figure 7) is accessible on GitHub at GitHub: https://github.com/Fraunhofer-ITMP/SASC/tree/v1.0. The Shiny was built with RStudio v.1.2.1335 (https://www.rstudio.com).