Abstract
DNA methylation is an important epigenetic mark that regulates the expression of genes and transposons. RNA-directed DNA methylation (RdDM) is the main molecular pathway responsible for de novo DNA methylation in plants. Although the mechanism of RdDM has been well studied in Arabidopsis (Arabidopsis thaliana), most mutations in RdDM genes cause no remarkable developmental defects in Arabidopsis. Here, we isolated and cloned Five Elements Mountain 1 (FEM1), which encodes RNA-dependent RNA polymerase 2 (OsRDR2) in rice (Oryza sativa). Mutation in OsRDR2 abolished the accumulation of 24-nt small interfering RNAs, and consequently substantially decreased genome-wide CHH (H = A, C, or T) methylation. Moreover, male and female reproductive development was disturbed, which led to sterility in osrdr2 mutants. We discovered that OsRDR2-dependent DNA methylation may regulate the expression of multiple key genes involved in stamen development, meiosis, and pollen viability. In wild-type (WT) plants but not in osrdr2 mutants, genome-wide CHH methylation levels were greater in panicles, stamens, and pistils than in seedlings. The global increase of CHH methylation in reproductive organs of the WT was mainly explained by the enhancement of RdDM activity, which includes OsRDR2 activity. Our results, which revealed a global increase in CHH methylation through enhancement of RdDM activity in reproductive organs, suggest a crucial role for OsRDR2 in the sexual reproduction of rice.
The increase of genome-wide CHH methylation through RNA-directed DNA methylation in anther and pistil is required for proper expression of developmental genes and fertility in rice
Introduction
DNA methylation is a stable epigenetic mark that represses transposon expression and regulates gene expression in eukaryotes. In animals, DNA methylation undergoes reprogramming during development, and an error in DNA methylation or DNA demethylation can cause severe developmental defects (Smith and Meissner, 2013; Greenberg and Bourc His, 2019). Although DNA methylation in mammals mainly involves CG sequence contexts, DNA methylation in plants involves three sequence contexts: CG, CHG, and CHH (H = A, C, or T) (Law and Jacobsen, 2010; Du et al., 2015; Zhang et al., 2018). The ortholog of DNA methyltransferase1, METHYLTRANSFERASE1 (MET1), works together with three homologous proteins, VARIANT IN METHYLATION1 (VIM1), VIM2, and VIM3, to maintain CG methylation during DNA replication (Kankel et al., 2003; Saze et al., 2003; Woo et al., 2007; Stroud et al., 2013b). CHROMOMETHYLASE3 (CMT3) is responsible for the maintenance of CHG methylation by recognizing the dimethylated state of histone H3 lysine 9 (H3K9me2), which is established by KRYPTONITE/SUPPRESSOR OF VARIEGATION 3-9 HOMOLOGUE PROTEIN4 (KYP/SUVH4), SUVH5, and SUVH6 (Bartee et al., 2001; Lindroth et al., 2001; Du et al., 2012).
There are two molecular pathways for the methylation of CHH in plants. In one pathway, a chromatin-remodeling protein, DECREASE IN DNA METHYLATION1 (DDM1), releases Histone 1 (H1) to allow CMT2 to catalyze DNA methylation on long transposons (Zemach et al., 2013; Stroud et al., 2014). Another pathway is called RNA-directed DNA methylation (RdDM), which involves the biosynthesis of small noncoding RNAs and long noncoding RNAs (Zhang and Zhu, 2011; Matzke and Mosher, 2014; Matzke et al., 2015). Facilitated by four homologous chromatin-remodeling proteins (CLSY1–CLSY4) and a transcription factor-like protein DNA-BINDING TRANSCRIPTION FACTOR 1/SAWADEE HOMEODOMAIN HOMOLOG 1 (DTF1/SHH1) (Smith et al., 2007; Law et al., 2013; Zhang et al., 2013; Yang et al., 2018; Zhou et al., 2018), RNA polymerase IV (Pol IV) and RNA-dependent RNA polymerase 2 (RDR2) bind the targets and work together to synthesize Pol IV-dependent small RNAs (P4-RNAs) consisting of 25–50 nt (Blevins et al., 2015; Zhai et al., 2015; Yang et al., 2016; Ye et al., 2016). P4-RNAs are cleaved by DICER-LIKE3 (DCL3) to produce 24-nt small interfering RNAs (siRNAs) (Xie et al., 2004; Henderson et al., 2006), which are loaded into ARGONAUTE4 (AGO4) and ARGONAUTE6 (AGO6) (Mi et al., 2008; Duan et al., 2015). The long noncoding RNAs transcribed by polymerase V (Pol V) then pair with siRNAs in AGO4 (Wierzbicki et al., 2009). During this process, AGO4 recruits DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) to the DNA targets and catalyzes DNA methylation in three contexts (Zilberman et al., 2003; Zhong et al., 2014).
The mechanism of RdDM is well-understood in Arabidopsis, but because most Arabidopsis RdDM mutants lack visible developmental defects (Matzke and Mosher, 2014), and because only small numbers of transposons are released in Arabidopsis RdDM mutants (Zemach et al., 2013), the biological role of RdDM is unclear. Rice (Oryza sativa) might be a better model than Arabidopsis for studying the biological role of RdDM in plants because rice has a much larger genome (370 versus 125 Mb) and a much higher percentage of transposable elements (TEs, 39.5% versus 18.5%) (Ausin et al., 2016); the size of the genome and the percentage of TEs are more representative of plants in general in rice than in Arabidopsis. There is also evidence that de novo methylation affects development in rice. OsDCL3a knockdown rice plants, for example, exhibit dwarfism and a larger flag leaf angle (Wei et al., 2014). Mutation in NUCLEAR RNA POLYMERASE D1 (OsNRPD1) causes increased tillering via its regulation of OsMIR156 and DWARF 14 (D14) (Xu et al., 2020). OsDRM2 mutants caused pleiotropic developmental defects at vegetative and reproductive stages (Moritoh et al., 2012; Tan et al., 2016). In tomato (Solanum lycopersicum), which has a 900-Mb genome (Tomato Genome Consortium, 2012), mutations in NUCLEAR RNA POLYMERASE D1 or NUCLEAR RNA POLYMERASE E1 led to small fruits and abnormal leaves and flowers (Gouil and Baulcombe, 2016). A dominant mutation in MEDIATOR OF PARAMUTATION2 (MOP2), an NRPD/E2 gene, caused late flowering and sometimes a short stature and abnormal tassels in maize (Zea Mays) (Sidorenko et al., 2009). REQUIRED TO MAINTAIN REPRESSION 6 (RMR6), encoding the largest subunit of Pol IV, ensured sex determination and proper adaxial–abaxial polarity of the leaf sheath of maize (Parkinson et al., 2007; Erhard et al., 2009). In contrast, mutation in RMR7, another NRPD/E2 gene, did not affect maize development (Stonaker et al., 2009). Mutation in RMR1, encoding a SUCROSE NONFERMENTING2 protein that is required for siRNA production, caused no developmental defects but changed inbreeding behavior in maize (Hale et al., 2007, 2009). These pleiotropic developmental defects in rice, tomato, and maize mutants suggest that de novo methylation might be important in plant species with larger genomes than Arabidopsis. Recent research demonstrated that blocking RdDM caused mis-splicing of MULTIPOLAR SPINDLE1 (MPS1) and thereby affected meiosis in Arabidopsis (Walker et al., 2018).
The Arabidopsis rdr2 mutant eliminated 24-nt siRNA accumulation but had no obvious developmental defects (Xie et al., 2004). Rice RDR2 RNAi transgenic plants, in contrast, had a larger leaf angle and a greater tiller number than wild-type (WT) plants (Wei et al., 2014). Mutation in MOP1 caused late flowering in maize (Dorweiler et al., 2000; Alleman et al., 2006), and a recent study found that MOP1 affects the recombination rate (Zhao et al., 2021). These reports suggested that RDR2 proteins function in multiple biological processes.
In this study of rice, we found that CHH methylation levels were much higher in WT panicles, stamens, and pistils than in WT seedlings. This increase in the methylation levels in reproductive versus vegetative tissue, however, was largely absent in the osrdr2 mutants, which exhibited reproductive defects. We demonstrate that the increase in DNA methylation in WT panicles, stamens, and pistils depends on OsRDR2, and that the global increase in the DNA methylation in WT sexual organs may regulate the expression of adjacent genes in these organs. The results suggest that the upregulated expression of genes that encode RdDM components, including OsRDR2, is responsible for the increased CHH methylation in WT reproductive organs and is therefore responsible for the proper development of those organs.
Results
We used the cauliflower mosaic virus (CaMV) 35S promoter to ectopically express a gibberellin (GA) metabolic gene, OsGA2ox1 (Sakamoto et al., 2004), in the japonica rice variety TP309. As expected, most of the transgenic rice plants exhibited GA-deficient phenotypes including dwarfism, late flowering, and short and dark green leaves (Supplemental Figure S1, A and B; Zheng et al., 2021). A semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) assay showed that the expression level in one homozygous OsGA2ox1ectopic expression line (GAE) was substantially increased (Supplemental Figure S1C; Zheng et al., 2021). Interestingly, some offspring of the transgenic rice plants restored the developmental defects caused by GA deficiency (Supplemental Figure S1A) even if the offspring contained the 35S::OsGA2ox1 transgene (Supplemental Figure S1B). The expression level of OsGA2ox1 in the normal transgenic rice plants was reduced to the WT level (Supplemental Figure S1C), which suggests that gene silencing occurs in those plants. The gene silencing plants were, therefore, referred to as OsGA2ox1Silencing (GAS) plants. We planted GAS for more than 20 generations with at least 60 plants in each generation and did not find any revertants, suggesting that gene silencing of 35S::OsGA2ox1 in GAS is stable. Small RNA sequencing (sRNA-seq) showed that abundant 21-nt and 22-nt siRNAs and a few 23-nt, 24-nt, and 25-nt siRNAs are produced on the exons of OsGA2ox1 in GAE (Supplemental Figure S1D; Zheng et al., 2021). In contrast, there are no siRNAs on the exons of OsGA2ox1 in the TP309 WT (Supplemental Figure S1E; Zheng et al., 2021). Thus, the siRNAs in GAE were derived from the transgenic OsGA2ox1. Only CG methylation existed on the two 3′-exons of OsGA2ox1 in TP309 (Supplemental Figure S1E; Zheng et al., 2021). In GAE, however, methylation in CHG and CHH was evident (Supplemental Figure S1D; Zheng et al., 2021). The 35S promoter produced abundant 21-nt and 22-nt siRNAs and a few 24-nt siRNAs in GAE (Supplemental Figure S1F; Zheng et al., 2021). In addition, DNA methylation in all sequence contexts, but mainly in the CHH context, established in GAE (Supplemental Figure S1F; Zheng et al., 2021). These results suggested that noncanonical RdDM occurs for 35S::OsGA2ox1 in GAE.
The siRNAs disappeared and non-CG methylation was eliminated on OsGA2ox1 in GAS (Supplemental Figure S1D; Zheng et al., 2021). On the 35S promoter, in contrast, 21-nt and 22-nt siRNAs disappeared but 23-nt and 24-nt siRNAs increased in GAS (Supplemental Figure S1F; Zheng et al., 2021). Consequently, the DNA methylation in three contexts on the 35S promoter was substantially higher in GAS than in GAE (Supplemental Figure S1F; Zheng et al., 2021). We, therefore, suspected that canonical RdDM occurs for 35S::OsGA2ox1 in GAS.
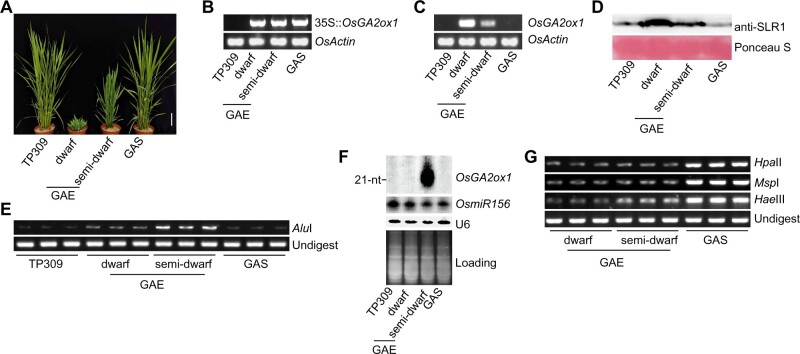
GAE plants displayed variations of GA-deficient phenotypes, including plant height, leaf length, and leaf color (Figure 1, A and B). The transcriptional levels of OsGA2ox1 in dwarf and semi-dwarf plants were positively corresponded with the GA-deficient phenotypes (Figure 1, A and C). The protein level of SLR1, the repressor protein in GA signaling (Ikeda et al., 2001), was positively corresponded with the GA-deficient phenotype (Figure 1D). The CHH methylation level in dwarf GAE was higher than in TP309 and GAS, and was much higher in semi-dwarf GAE than in dwarf GAE (Figure 1E). Northern blotting showed that abundant 21-nt and 22-nt siRNAs accumulated only in semi-dwarf GAE (Figure 1F), which is consistent with the high CHH methylation on OsGA2ox1 in semi-dwarf GAE. On the 35S promoter, CHH methylation progressively increased from dwarf GAE, to semi-dwarf GAE, and to GAS (Figure 1G). Methylation in CG and CHG contexts, in contrast, was established in GAS (Figure 1G). These results indicate that on the 35S::OsGA2ox1, noncanonical RdDM in semi-dwarf GAE switched into canonical RdDM in GAS, which resembles the de novo silencing of an active retrotransposon (Marí-Ordóñez et al., 2013).
Figure 1.
Gene silencing of 35S::OsGA2ox1. A, Morphology of 110-d-old TP309, dwarf GAE, semi-dwarf GAE, and GAS plants. Scale bar = 10 cm. B, Genotyping transgenic OsGA2ox1. OsActin served as the control. GAE, OsGA2ox1ectopic expression; GAS, OsGA2ox1silencing. C, The mRNA level of OsGA2ox1 as indicated by semi-quantitative RT-PCR. OsActin served as the control. D, Western blot detection of the protein level of SLR1. E, Chop-PCR assay on the gene body of OsGA2ox1 in the indicated genotypes. F, sRNA northern blot detection of the sRNA levels on the gene body of OsGA2ox1. G, Chop-PCR assay of the 35S promoter in the indicated genotypes.
Isolation and map-based cloning of Five Elements Mountain 1
To elucidate the molecular mechanism of epigenetic silencing of 35S::OsGA2ox1 in GAS plants, we conducted a genetic screen for mutations that suppress gene silencing of 35S::OsGA2ox1. More than 23,000 GAS plants that germinated from ethyl methyl sulfonate (EMS)-treated GAS seeds were planted and permitted to self-fertilize. The plants with GA-deficient phenotypes like dwarfism and short and dark green leaves were isolated from the M2 generation. The OsGA2ox1 expression levels in those plants were then assessed by RT-qPCR, and the plants with substantially increased expression levels (>20-fold) compared to GAS were selected and named five elements mountain (fem) plants (Figure 2, A–C; Zheng et al., 2021). In Journey to the West, a mythological Chinese story, the Monkey King disturbed the heavenly hierarchy. In response, Buddha imprisoned the Monkey King under Five Elements Mountain. Once the Five Elements Mountain was destroyed, the Monkey King was released. In our study, 35S::OsGA2ox1 as one transgene often threatens genome integrity. In GAE, 35S::OsGA2ox1 was ectopically expressed. In GAS, it was silenced. Thus 35S::OsGA2ox1 is analogous to the Monkey King; the gene silencing machinery responsible for suppressing 35S::OsGA2ox1 is analogous to the Five Elements Mountain. When the gene silencing pathway was disturbed, 35S::OsGA2ox1 was released.
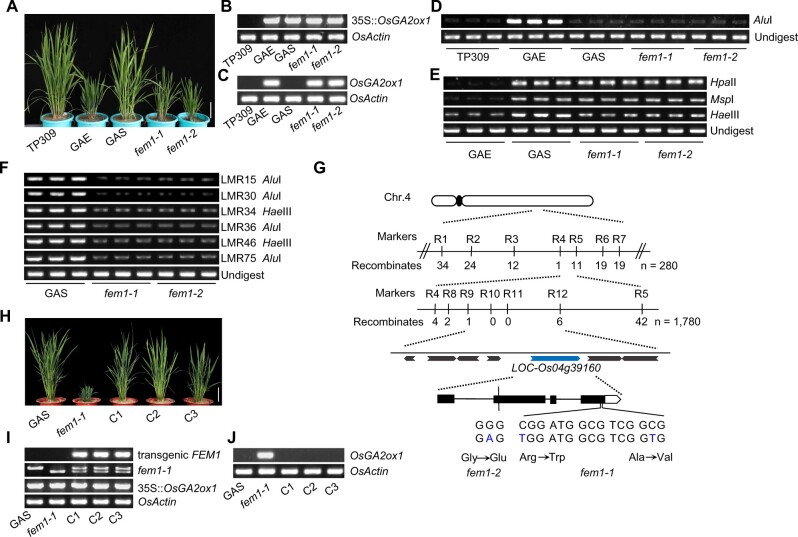
Figure 2.
Map-based cloning of FEM1. A, Morphology of 3-month-old TP309, GAE, GAS, fem1-1, and fem1-2 plants. Scale bar = 10 cm. B, Genotyping transgenic OsGA2ox1. OsActin served as the control. C, The mRNA level of OsGA2ox1 as indicated by semi-quantitative RT-PCR. OsActin served as the control. D, Chop-PCR assay of the gene body of OsGA2ox1. E, Chop-PCR assay of the 35S promoter. F, Chop-PCR assay of endogenous loci. LMR, Losing DNA methylation region. G, Diagram showing the map-based cloning of fem1-1. Mutations of fem1-1 and fem1-2 are indicated. H, Morphology of 3-month-old plants of GAS, fem1-1, and three complementary lines (C1–C3). Scale bar = 10 cm. I, Genotyping of complementary lines. J, The mRNA level of OsGA2ox1 in the indicated genotypes as determined by semi-quantitative RT-PCR.
We isolated about 200 recessive fem mutants, two of which (#45 and #30) were referred to as fem1-1 and fem1-2 because subsequent, separate map-based cloning and allelic tests demonstrated that they were allelic to each other (see below). A Chop-PCR assay indicated that the methylation level in CHH context on OsGA2ox1 in fem1 was as low as that in GAS (Figure 2D). On the 35S promoter, DNA methylation in CHH but not in CG and CHG was lower in fem1 mutants than in GAS (Figure 2E). On endogenous loci, the CHH methylation was also lower in fem1 mutants than in GAS (Figure 2F; Supplemental Figure S2). These results indicated that FEM1 probably regulates CHH methylation to control gene silencing.
Because both fem1-1 and fem1-2 with the transgene 35S::OsGA2ox1 were totally sterile, we crossed the heterozygous fem1-1/FEM1 (#45) with the indica variety Taichuang native 1 (TN1); the GA-deficient plants from the F2 population were selected and constituted a map-based cloning population. In total, 890 plants were used to locate the candidate gene in a 103-kb region (Figure 2G). Among seven genes in this region, only LOC_Os04g39160 contained two mutations (both were C to T), which caused two amino acid substitutions near its carboxylic terminal (Figure 2G). To confirm the positional cloning result, we conducted a genetic complementary assay using the native promoter to drive the genomic DNA of LOC_Os04g39160. Among 35 independently regenerated plants, 27 transgenic positive lines were similar to the WT in stature (Supplemental Figure S3, A and B). The mRNA levels of OsGA2ox1 were substantially lower in the complemented lines of T0 than in the fem1 mutant (Supplemental Figure S3C). In three representative lines in the T1 generation, the transgenic plants restored the GA-deficient phenotypes and reduced OsGA2ox1 expression (Figure 2, H–J). Chop-PCR assay showed that CHH methylation levels on the 35S promoter and six endogenous loci were recovered in the complementary lines (Supplemental Figure S3, D and E). The genetic complementation test demonstrated that mutation in LOC_Os04g39160 is responsible for suppressing the silencing of 35S::OsGA2ox1.
A phylogenetic tree showed that the protein encoded by LOC_Os04g39160 is homologous with the RDR2 protein in Arabidopsis and with the MOP1 protein in maize (Supplemental Figure S4A). Among the two amino acids that are mutated in fem1-1 (osrdr2-1), the amino acid alanine (A1080) is highly conserved in RDR2 proteins from 13 flowering plant species (Supplemental Figure S4B).
A subsequent independent map-based cloning of a different fem mutant (#30) also identified one mutation (G to A) in the same gene; this mutation caused a one-amino acid substitution from glycine to glutamic acid (Figure 2G). This glycine (G230) is highly conserved in RDR2 proteins in the tested plant species (Supplemental Figure S4C). Moreover, genetic analysis demonstrated that fem1-1 (osrdr2-1) and fem1-2 (osrdr2-2) were allelic for suppression of silencing of 35S::OsGA2ox1 (Supplemental Figure S4, D–F). Taken together, these results demonstrated that OsRDR2/FEM1 (LOC_Os04g39160) is required for silencing of 35S::OsGA2ox1.
Mutation in OsRDR2 causes sterility
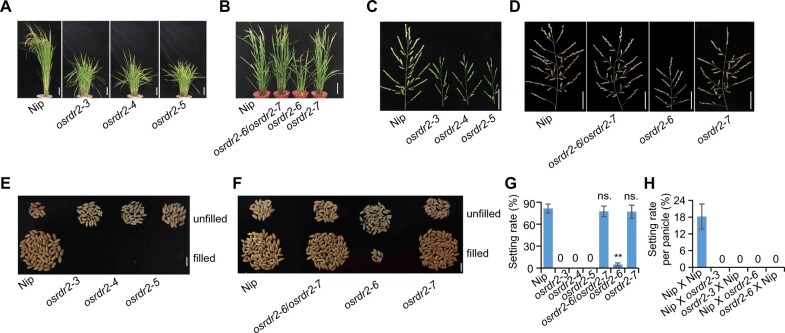
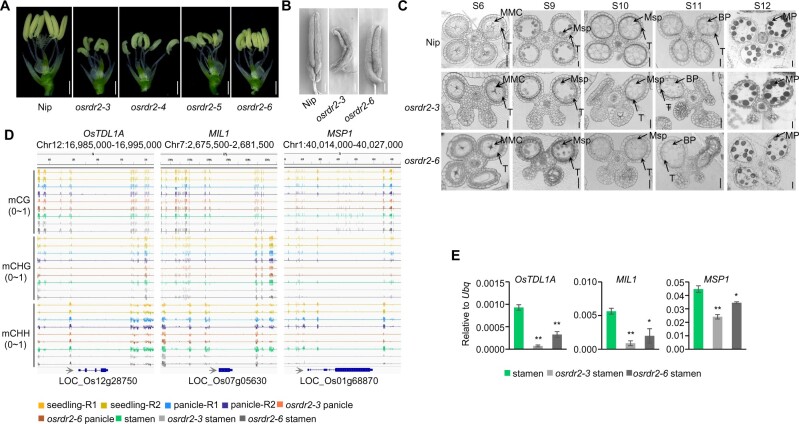
We knocked out OsRDR2 using CRISPR/Cas9 technology at two different single-guide RNA (sgRNA) sites in Nipponbare, which has a high-quality reference genome. sgRNA1 targeted the DNA sequence encoding the RNA-dependent RNA polymerase (RdRP) domain, and sgRNA2 targeted the C-terminal (Supplemental Figure S5, A and B). One-base insertion or deletion near the protospacer-adjacent motif in three osrdr2 alleles (osrdr2–3, osrdr2–4, and osrdr2–5) generated by sgRNA1 caused a premature stop codon and led to a truncated protein without an intact RdRP domain, indicating that the three osrdr2 alleles might be functional null mutants (Supplemental Figure S5B). The osrdr2–6 mutant generated by sgRNA2 contained 49 amino acid (aa) mutations with a 12-aa deletion on its C-terminal (Supplemental Figure S5B). The osrdr2–7 mutant contained a 2-aa deletion and a 1-aa substitution (ASR changed to G) (Supplemental Figure S5, A and B). Like the OsDCL3a and OsNRPD1 knockdown rice plants (Wei et al., 2014; Xu et al., 2020), osrdr2 mutant plants were shorter, and their flag leaf angle was much larger than that of WT plants with an empty vector (Figure 3, A and B). However, the statures of the osrdr2–6/osrdr2–7 and osrdr2–7 mutants were similar to that of WT (Figure 3B), and the three aa mutated in osrdr2–7 (ASR, 1074–1076) were not conserved (Supplemental Figure S4B). In addition, the panicles were smaller for osrdr2 than for the WT (Figure 3, C and D). Again, the panicle sizes of the osrdr2–6/osrdr2–7 and osrdr2–7 mutants were similar to that of the WT (Figure 3D). The osrdr2–3, osrdr2–4, and osrdr2–5 plants did not produce seeds; osrdr2–6 plants, however, produced a small number of seeds (Figure 3, C–G). The osrdr2–6/osrdr2–7 and osrdr2–7 plants produced a similar number of seeds as the WT (Figure 3, F and G). The reciprocal crosses of the WT with osrdr2–3 or osrdr2–6 failed to produce seeds (Figure 3H), suggesting that both the male and female organs of osrdr2 are defective.
Figure 3.
osrdr2 mutants are sterile. A, Morphology of the WT and various osrdr2 mutants. Scale bar = 10 cm. B, Morphology of the WT, osrdr2–6/osrdr2–7, osrdr2–6, and osrdr2–7 mutants. Scale bar = 10 cm. C, Panicle morphology of the WT, osrdr2–3, osrdr2–4, and osrdr2–5. Scale bar = 5 cm. D, Panicle morphology of the WT, osrdr2–6/osrdr2–7, osrdr2–6, and osrdr2–7. Scale bar = 5 cm. E, Unfilled (top) and filled (bottom) rice grains of the WT, osrdr2–3, osrdr2–4, and osrdr2–5. Scale bar = 1 cm. F, Unfilled (top) and filled (bottom) rice grains of the WT, osrdr2–6/osrdr2–7, osrdr2–6, and osrdr2–7. Scale bar = 1 cm. G, Seed setting rate of various genotypes (Data are means ± sd (Standard Deviation), n = 15) (**P < 0.01; ns, no significant difference by Student’s t test). H, Seed setting rate from reciprocal crosses between the WT and osrdr2 mutants (Data are means ± sd, n = 10).
OsRDR2 regulates the methylome and 24-nt siRNAs
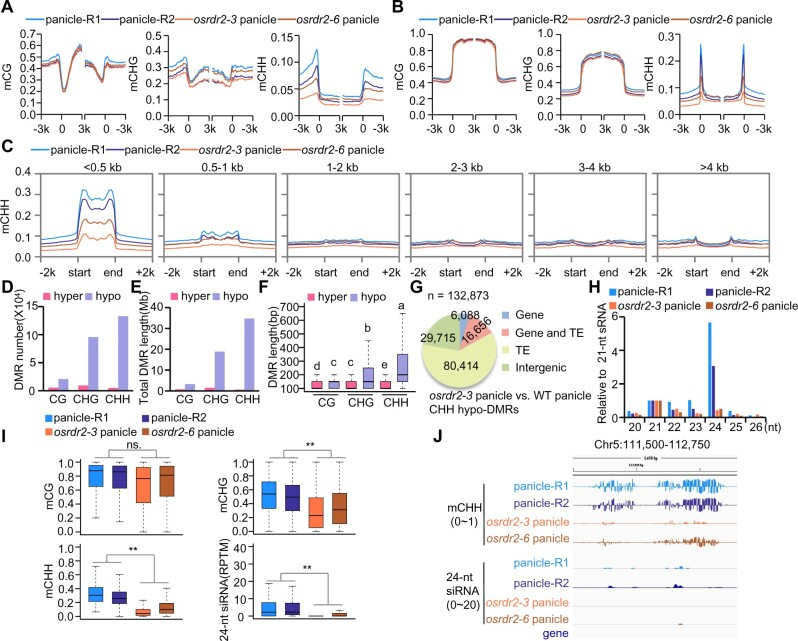
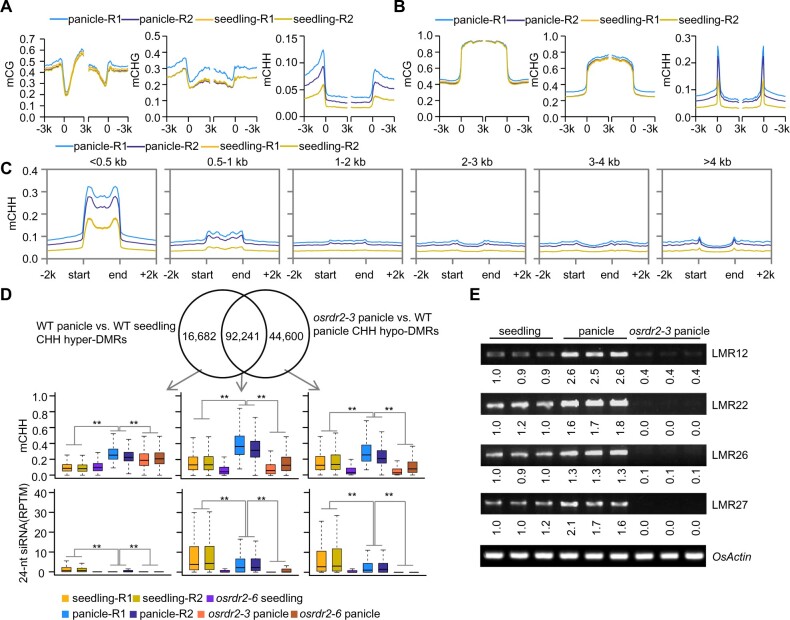
To elucidate the role of OsRDR2 in the methylome, we conducted whole-genome bisulfite sequencing (WGBS) for the panicles of osrdr2–3 and osrdr2–6 with the WT (panicle-R1) and the transgenic control (panicle-R2). Because tissue culture induces DNA hypomethylation (Stroud et al., 2013a), we used two kinds of WT controls in this study. The methylome with high quality (Supplemental Table S1) showed that mutation in OsRDR2 reduces the genome-wide methylation level (GML) of CHH, that is, the CHH methylation level dropped from 7.0% in WT replicate 1 to 2.9% in osrdr2–3, and from 5.1% in WT replicate 2 to 3.9% in osrdr2–6 (Supplemental Table S1). For genes, CHH methylation levels were higher for upstream and downstream regions in the WT than for the gene body (Figure 4A). The high CHH methylation levels on the borders of genes were substantially reduced in osrdr2–6 and nearly eliminated in osrdr2–3 (Figure 4A), suggesting that CHH methylation upstream and downstream of genes is dependent on OsRDR2. As was the case for genes, CHH methylation but not CG or CHG methylation on TEs was also dependent on OsRDR2 (Figure 4B). CHH methylation depended on OsRDR2 not only in panicles but also in seedlings (Supplemental Figure S6, A and B). To determine which kind of TEs were dominant targets of OsRDR2, we divided TEs into six subgroups based on length. On short TEs (<500 bp), the CHH methylation level was as high as 30% in the WT, but was 10% in the osrdr2–3 mutant (Figure 4C). On long TEs (>500 bp), however, the methylation level in the WT was very low, and the difference between the WT and osrdr2 was small (Figure 4C). That osrdr2 mainly regulated DNA methylation on short TEs was also apparent in seedlings (Supplemental Figure S6C).
Figure 4.
OsRDR2 regulates CHH methylation and 24-nt siRNA biosynthesis. A and B, Average CG, CHG, and CHH methylation levels on genes (A) and TEs (B) in the panicles of various genotypes (R1 represents WT-Replicate 1; R2 represents WT-Replicate 2). C, Average CHH methylation levels on TEs of different lengths in panicles of the indicated genotypes. D and E, The number (D) and length (E) of various DMRs in panicles of osrdr2–3. F, Box plots showing DMR lengths. Different letters indicate a significant difference according to Fisher’s least significant difference (LSD) (P < 0.05). Here and all subsequent box plots, the box indicates the upper and lower quartiles, the black lines are medians, and the whiskers corresponding to the minimum and maximum range. G, Genomic distribution of CHH hypo-DMRs in osrdr2–3 panicles. “Gene” indicates a DMR that overlaps with the gene body; “TE” indicates a DMR that overlaps with the TE body; “Gene and TE” indicates a DMR that overlaps with both the gene body and TE body; “Intergenic” indicates a DMR that does not overlap with either the gene body or the TE body. H, Abundance of small RNAs of different length relative to 21-nt in WT and osrdr2 panicles. I, Box plots indicating the methylation level of CG, CHG, and CHH, and 24-nt siRNA abundance on CHH hypo-DMR in osrdr2–3 panicles (**P < 0.01 by two-tailed z-test). J, Integrated Genome Browser view of CHH methylation levels, 24-nt siRNAs abundance on one representative CHH hypo-DMR in panicles of the indicated genotypes.
We further identified differentially methylated regions (DMRs) between osrdr2 panicles and WT panicles. Many hypo-DMRs but very few hyper-DMRs of CHG and CHH were identified in the osrdr2 panicles (Figure 4D; Supplemental Figure S6D; Supplemental Table S2), suggesting that OsRDR2 positively regulates DNA methylation. Among DMRs in panicles, CHH hypo-DMRs were the longest (Figure 4E; Supplemental Figure S6E). The total length of CHH hypo-DMRs covered 34.8 Mb in osrdr2–3 panicles and 23.4 Mb in osrdr2–6 panicles (Figure 4E; Supplemental Figure S6E). Furthermore, CHH hypo-DMRs were longer than other types of DMRs (Figure 4F; Supplemental Figure S6F). The majority of CHH hypo-DMRs of osrdr2 were located on TEs (Figure 4G; Supplemental Figure S6G). sRNA-seq (Supplemental Table S1) showed that 24-nt siRNAs were nearly eliminated in osrdr2 panicles (Figure 4H), which was consistent with the conclusion that RDR2 and MOP1 are essential for 24-nt siRNA biosynthesis (Xie et al., 2004; Lu et al., 2006; Nobuta et al., 2008). On the CHH hypo-DMRs of osrdr2–3 panicles, 23-nt and 24-nt siRNAs were significantly decreased in osrdr2–3 panicles (Figure 4, I and J; Supplemental Figure S7). In contrast, the abundance of 21-nt siRNAs in osrdr2–3 was not significantly different from that in the WT (Supplemental Figure S7). These results suggest that OsRDR2 activity is required for biosynthesis of siRNAs that subsequently affect DNA methylation.
Increased DNA methylation in panicles depends on OsRDR2
As was the case in panicles, the CHH hypo-DMRs were the main type of DMRs in osrdr2–6 seedlings (Supplemental Figure S8, A and B; Supplemental Table S2). The CHH hypo-DMRs were the longest among different DMRs for osrdr2–6 (Supplemental Figure S8C), and the CHH hypo-DMRs in seedlings were mainly located on TEs (Supplemental Figure S8D). The abundance of 24-nt siRNAs in osrdr2-6 was substantially decreased globally on the CHH hypo-DMRs in seedlings (Supplemental Figure S8, E–G), which was consistent with the results for panicles. However, the obvious difference between the number and length of hypo-DMRs in osrdr2–6 panicles versus osrdr2–6 seedlings indicated that the DNA methylation level might differ among organs. To understand this difference, we directly compared the GML in the WT panicles and seedlings, and found that CHH methylation levels in panicles were 7.0% in replicate 1 and 5.1% in replicate 2, which were substantially higher than the 3.1% in seedlings (Supplemental Table S1). On both genes and TEs, the CHH methylation level but not the CG or CHG methylation level was substantially higher in WT panicles than in WT seedlings (Figure 5, A and B). The increase in CHH methylation level on transposons mainly occurred on short TEs (Figure 5C). In contrast, the CG and CHG methylation levels in WT panicles and WT seedlings were similar on TEs of various lengths (Supplemental Figure S9, A and B). CHH hyper-DMRs were the main type of DMRs between panicles and seedlings (Supplemental Figure S9, C and D). The CHH hyper-DMRs were longer than other types of DMRs (Supplemental Figure S9E), and were also mainly located on TEs (Supplemental Figure S9F). On the CHH hyper-DMRs, the CG methylation level was similar in WT panicles and WT seedlings (Supplemental Figure S9G). The CHH level was significantly higher in WT panicles than in WT seedlings (Supplemental Figure S9G). However, the 24-nt siRNA abundance on the CHH hyper-DMRs was not increased but was significantly lower in WT panicles than in WT seedlings (Supplemental Figure S9G), which indicated that the increase in DNA methylation is not explained by an increase in siRNA abundance.
Figure 5.
Increase of CHH methylation in panicles depends on OsRDR2. A and B, Average CG, CHG, and CHH methylation levels on genes (A) and TEs (B) in panicles and seedlings. C, Average CHH methylation levels on TEs of different length in panicles and seedlings. D, Venn diagram of the overlap between CHH hyper-DMRs of panicles > seedlings and hypo-DMRs of osrdr2–3 < WT in panicles. Box plots indicate the methylation levels of CHH and 24-nt siRNA abundance on common and specific regions in panicles or seedlings of various genotypes (**P < 0.01 by two-tailed z-test). E, Chop-PCR assay of several panicle CHH hyper-DMRs. Values were calculated relative to the first sample.
The increased methylation on panicle CHH hyper-DMRs and on the OsRDR2-dependent DMRs in both panicles and seedlings occurred mainly on short TEs and in the CHH context, suggesting that the increased methylation in panicles may depend on OsRDR2. Among the 108,923 panicle CHH hyper-DMRs, ∼84.7% overlapped with OsRDR2-dependent DMRs in the panicle (Figure 5D), which suggested that the majority of panicle CHH hyper-DMRs is OsRDR2-dependent. In the overlapped regions, the high CHH methylation level in WT panicles was reduced in osrdr2 panicles (Figure 5D). Even on the specific panicle CHH hyper-DMRs versus seedling and the specific osrdr2–3 panicle CHH hypo-DMRs, the CHH methylation level was higher in WT panicles than in WT seedlings (Figure 5D), suggesting that the extent of dependence on OsRDR2 for increased methylation was greater in panicles than in seedlings. Consistent with that possibility, the CHH methylation level was higher in all OsRDR2-dependent CHH DMRs in panicles than in seedlings (Supplemental Figure S9H). A Chop-PCR assay showed that the increase in CHH methylation level in panicle CHH hyper-DMRs was dependent on OsRDR2 (Figure 5E; Supplemental Figure S10). However, the increase in CHH methylation was not accompanied by an upregulation of siRNA abundance (Figure 5D; Supplemental Figure S9, G and H).
Increased DNA methylation in stamens and pistils depends on OsRDR2
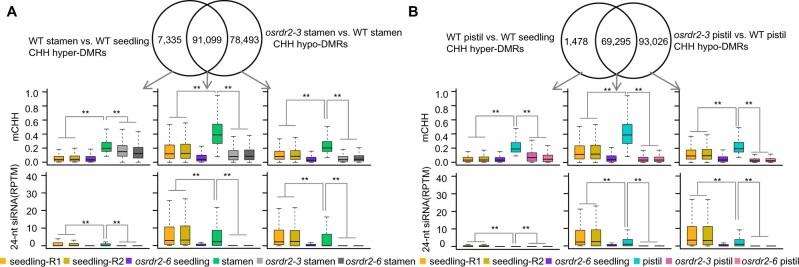
To determine whether the increased methylation in panicles involves an increase in methylation of the reproductive organs, we performed WGBS of stamens (Stage 12) and pistils (FG8) of the WT, osrdr2–3, and osrdr2–6 (Supplemental Table S1). On a genome-wide scale, the CHH methylation level was 5.5% in WT stamens and 4.7% in WT pistils, but was only 3.1% in WT seedlings (Supplemental Table S1). The CHH methylation levels were higher on both genes and TEs in stamens than in seedlings (Supplemental Figure S11, A and B). On TEs, CHH methylation in stamens mainly occurred on short TEs (Supplemental Figure S11C). There were 98,434 CHH hyper-DMRs between stamens and seedlings (Supplemental Figure S11D; Supplemental Table S3, hereafter these CHH hyper-DMRs were named “stamen hyper-DMRs”), which covered 17.8 Mb (Supplemental Figure S11E). Among the different types of DMRs, hyper-DMRs were the longest (Supplemental Figure S11F). Among the stamen hyper-DMRs, 59.4% were located on TEs (Supplemental Figure S11G). Mutation in OsRDR2 dramatically reduced GML in CHH (WT: 5.5%; osrdr2–3: 2.8%; osrdr2–6: 2.6%; Supplemental Table S1), and produced 162,809 CHH hypo-DMRs in stamens (Supplemental Figure S11H; Supplemental Table S3), which covered 45.5 Mb, that is, more than one-tenth of the rice genome (Supplemental Figure S11I). The CHH hypo-DMRs were the longest among the different DMRs (Supplemental Figure S11J), and 58.6% of the CHH hypo-DMRs were on TEs (Supplemental Figure S11K). About 92.5% of the stamen hyper-DMRs overlapped with OsRDR2-dependent CHH methylation regions in stamens (Figure 6A). In the overlapped regions, the CHH methylation level was higher in WT stamens than in WT seedlings, and mutation in OsRDR2 substantially reduced CHH methylation in both seedlings and stamens (Figure 6A). Like the overlapped DMRs, the CHH levels on the specific OsRDR2-dependent DMRs and on the specific stamen hyper-DMRs were higher in WT stamens than in WT seedlings (Figure 6A), suggesting that the increased methylation in stamens was largely OsRDR2-dependent. However, siRNA abundance was not higher in WT stamens (Stage 12) than in WT seedlings in those regions (Figure 6A), suggesting the absence of direct correlation between 24-nt siRNA abundance and CHH methylation levels.
Figure 6.
Increase of CHH methylation in stamens and pistils depends on OsRDR2. A, Venn diagram showing the overlap between stamen CHH hyper-DMRs (stamen > seedling) and osrdr2–3 CHH hypo-DMRs in stamens. Box plots indicate the methylation levels of CHH and 24-nt siRNAs abundance on the overlapped and specific regions in stamens or seedlings of the indicated genotypes (**P < 0.01 by two-tailed z-test). B, Venn diagram showing the overlap between pistil CHH hyper-DMRs (pistils > seedlings) and CHH hypo-DMRs of osrdr2–3 in pistils. Box plots indicate the methylation levels of CHH and 24-nt siRNA abundance on the common and specific regions in pistils or seedlings of the indicated genotypes (**P < 0.01 by two-tailed z-test).
As was the case for panicles and stamens, CHH levels on genes and TEs were higher in WT pistils than on the corresponding genes and TEs in WT seedlings (Supplemental Figure S12, A and B), and the increase in methylation in pistils occurred on short TEs (Supplemental Figure S12C). Mutation in OsRDR2 dramatically reduced GML in CHH in pistils (WT: 4.7%; osrdr2–3: 1.9%; osrdr2–6: 1.8%). We identified a total of 70,773 CHH hyper-DMRs of WT pistils versus WT seedlings (Supplemental Figure S12D; Supplemental Table S4; these CHH hyper-DMRs were named “pistil hyper-DMRs”) with a total length of 12.2 Mb (Supplemental Figure S12E). The pistil hyper-DMRs were the longest among the different DMRs (Supplemental Figure S12F). Of the pistil hyper-DMRs, 59.6% were located on TEs (Supplemental Figure S12G). When osrdr2–3 pistils were compared with WT pistils, 156,301 CHH hypo-DMRs were identified (Supplemental Figure S12H; Supplemental Table S4); these CHH hypo-DMRs covered 12.4% of the rice genome (46.3 Mb, Supplemental Figure S12I). The CHH hypo-DMRs were the longest among the different DMRs (Supplemental Figure S12J). Of the CHH hypo-DMRs, 58.6% were located on TEs (Supplemental Figure S12K). Among the pistil hyper-DMRs, 97.9% overlapped with hypo-DMRs in osrdr2–3 pistils (Figure 6B), suggesting that the increase in CHH methylation was largely OsRDR2-dependent. On the specific OsRDR2-dependent DMRs and the specific pistil hyper-DMRs, the CHH methylation levels were much higher in WT pistils than in WT seedlings (Figure 6B), suggesting that the increase in methylation might occur on all OsRDR2 targets in pistils. The 24-nt siRNA levels, however, were not increased on OsRDR2-dependent DMRs (Figure 6B). Again, this indicated that the increase in DNA methylation is not explained by an increase in siRNA abundance.
The CHH hyper-DMRs were longest in panicles (Figure 4E), shortest in pistils (Supplemental Figure S12E), and of intermediate length in stamens (Supplemental Figure S11E). However, the three types of DMRs largely overlapped with each other (Supplemental Figure S13A). On seven groups of DMRs, the CHH methylation levels were higher in panicles, stamens, and pistils than in seedlings, and the increase in CHH methylation was reversed in various organs of osrdr2 mutants (Supplemental Figure S13B), suggesting that methylation reinforcement largely occurred on common regions in panicles, stamens, and pistils, and that this reinforcement was OsRDR2-dependent.
Although the methylation levels in the same tissues were always lower in the osrdr2 mutant than in the WT, the methylation levels of osrdr2 differed among tissues (Supplemental Figure S13B), indicating that RdDM is not the only factor contributing to the enhancement of CHH methylation in reproductive organs. To examine this possibility, we compared CHH methylation levels of osrdr2–6 seedlings with those of osrdr2–6 panicles, osrdr2–6 stamens, and osrdr2–6 pistils. On OsRDR2-dependent loci in panicles, the CHH methylation level was significantly higher in osrdr2–6 panicles than in osrdr2–6 seedlings (Supplemental Figure S14A), indicating that another mechanism is also involved in the higher methylation in panicles than in seedlings. The higher methylation in osrdr2–6 stamens than in osrdr2–6 seedlings on OsRDR2-dependent loci was also significant (Supplemental Figure S14B). On OsRDR2-dependent loci, however, the CHH methylation level was significantly lower in osrdr2–6 pistils than in osrdr2–6 seedlings (Supplemental Figure S14C).
On both genes and TEs, the difference in CHH methylation in panicles and stamens versus seedlings was much greater for the WT than for osrdr2–6 (Supplemental Figure S14, D and E), suggesting that the RdDM pathway is largely responsible for the increase of DNA methylation in panicles and stamens. Although the CHH methylation on TEs and gene borders was higher in WT pistils than in WT seedlings, the CHH methylation on TEs and gene borders was lower in osrdr2–6 pistils than in osrdr2–6 seedlings (Supplemental Figure S14, D and E), suggesting that RdDM was responsible for the higher CHH methylation in WT pistils than in WT seedlings.
To investigate the effects of CHH methylation on gene expression, we performed RNA-seq for stamens (Stage 12) of the WT and osrdr2–3 (Supplemental Table S1). We identified 1,634 upregulated and 1,063 downregulated differentially expressed genes (DEGs, fold-change >2, false discovery rate (FDR) < 0.05) between osrdr2–3 stamens and WT stamens (Supplemental Table S5). Among the 1,634 upregulated DEGs, 1,461 genes (89.4%) overlapped with osrdr2–3 stamen CHH hypo-DMRs (If a DEG overlap with a DMR with the gene body, with the 1-kb promoter of one gene, or with the 1-kb terminator of one gene, we defined it as a hypo-DMR overlapped gene) (Supplemental Figure S15A). Among the 1,063 downregulated DEGs, 922 genes (86.7%) overlapped with osrdr2–3 stamen CHH hypo-DMRs (Supplemental Figure S15A). Both upregulated and downregulated DEGs overlapped with more osrdr2–3 stamen hypo-DMRs than all genes, random genes, or non-DEGs (Supplemental Figure S15B).
We identified 2,537 upregulated and 3,048 downregulated DEGs in comparisons of osrdr2–3 pistils with WT pistils (Supplemental Table S5; Supplemental Figure S15C). The osrdr2–3 CHH hypo-DMRs in pistils overlapped with 2,195 upregulated DEGs (86.5%) and with 2,624 downregulated DEGs (86.1%) (Supplemental Figure S15C). As was the case in stamens, DEGs in pistils overlapped with more osrdr2–3 CHH hypo-DMRs than all genes, random genes, or non-DEGs (Supplemental Figure S15D). The majority of DEGs overlapped with loci on which CHH methylation was dependent on OsRDR2 in both stamens and pistils, suggesting that OsRDR2-dependent DNA methylation likely regulates their transcription.
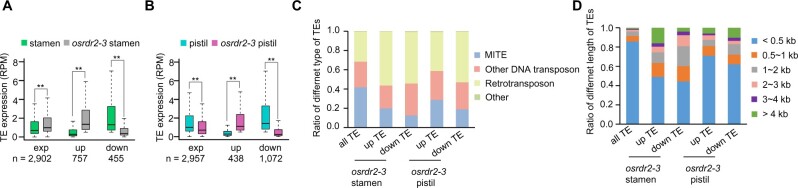
In stamens, 2,902 TEs were expressed in the WT or the osrdr2–3 mutant; among them, 757 and 455 were upregulated and downregulated, respectively, in the mutant relative to the WT (Figure 7A). In pistils of WT and the osrdr2–3 mutant, 2,957 TEs were expressed. Among them, 438 and 1,072 were upregulated and downregulated, respectively, in osrdr2–3 relative to the WT (Figure 7B). The differentially expressed TEs in stamens and pistils contained a higher ratio of retrotransposons but a lower ratio of miniature inverted repeat TEs (MITEs) than the rice genome (Figure 7C). The differentially expressed TEs did not overlap with more DMRs than all TEs (Supplemental Figure S16A). Among differentially expressed TEs, although the short TEs were the major ones, there were long TEs (Figure 7D), indicating that CHH methylation is unlikely the only factor regulating TE transcription. Consistent with this possibility, 24-nt siRNAs were eliminated on the differentially expressed TEs in both stamens and pistils (Supplemental Figure S16, B and C). The abundance of 21-nt and 22-nt siRNAs on the differentially expressed TEs was not increased (Supplemental Figure S16, D–G), suggesting that post-transcriptional gene silencing unlikely occur for these differentially expressed TEs. These results suggested that CHH methylation, siRNAs, and other epigenetic environments directly or indirectly regulated by OsRDR2 might cooperatively control TE expression.
Figure 7.
TE expression in stamens and pistils of osrdr2. A and B, Box plots showing the transcriptional levels of expressed TEs (exp), upregulated TEs (up), and downregulated TEs (down) in A, stamens, and B, pistils, of the WT and osrdr2–3. (**P < 0.01 by two-tailed z-test). C, Ratio of different types of TEs among the indicated TEs. D, Ratio of TEs with different length among the up- or downregulated TEs in osrdr2 stamen and pistil.
OsRDR2-regulated reproductive development
The osrdr2 mutants were sterile (Figure 3, C–G), probably because of the developmental defects of both male and female reproductive organs (Figure 3H). The anthers of the osrdr2 plants were bent and were smaller and paler than those of control plants (Figure 8A). Scanning electron microscopy (SEM) also showed that the anthers of osrdr2 plants were abnormally small and that some were twisted (Figure 8B). The number of pollen grains produced per anther was significantly lower for osrdr2 than for the WT (Supplemental Figure S17A). Furthermore, iodine-potassium iodide (I2-KI) staining showed that starch filling was significantly less in osrdr2 pollen than in WT pollen (Supplemental Figure S17, B and C). Fluorescein diacetate (FDA) staining showed that pollen viability was significantly lower for osrdr2 than for the WT (Supplemental Figure S17, D and E). We examined transverse sections of WT and osrdr2 anthers at different developmental stages. At Stage 6, when anther morphogenesis is complete, the WT anthers had formed four somatic layers (the epidermis, endothecium, middle layer, and tapetum), and the microsporocytes, which are larger than the surrounding somatic cells, were located at the center of each anther locule (Figure 8C). In osrdr2 mutants at Stage 6, in contrast, some anther lobes had failed to form the organized sporophytic structure, and the center of osrdr2 anthers contained somatic cells rather than microsporocytes (Figure 8C). The anther defects in osrdr2 phenocopied the developmental defects in mutants of ostdl1a (Hong et al., 2012a), mil1 (Hong et al., 2012b), and msp1 (Nonomura et al., 2003). This led us to examine their DNA methylation. The DNA methylation in the CHH context adjacent to those genes was higher in stamens than in seedlings, and the DNA methylation in the CHH context in stamens and panicles was dependent on OsRDR2 (Figure 8D). RT-qPCR analysis showed that the mRNA levels of OsTDL1A, MIL1, and MSP1 were significantly downregulated in osrdr2 stamens relative to WT stamens at Stage 7 (Figure 8E), suggesting that OsRDR2-mediated DNA methylation likely ensures the proper expression of stamen developmental genes.
Figure 8.
Increase of DNA methylation regulates gene expression in stamens. A, Stamen morphology of the WT (Nip) and osrdr2 mutants at Stage 12. Scale bars = 1 mm. B, Scanning electron micrographs of Stage 12 stamens of the WT and osrdr2 mutants. Scale bars = 200 µm. C, Transverse sections of anthers at five stages of development in the indicated genotypes. Msp: microspore, BP: binucleate pollen grain, MP: mature pollen grain, T: tapetum. Scale bar = 20 µm. D, Integrated Genome Browser view of CG, CHG, and CHH methylation levels on OsTDL1A, MIL1, and MSP1 in various organs of the indicated genotypes. E, Relative expression levels of OsTDL1A, MIL1, and MSP1 in stamens of the WT and osrdr2 mutants. Ubq (LOC_Os03g13170) served as the control (**P < 0.01; *P < 0.05; by Student’s t test). Data are means ± sd, n = 3.
Although a small proportion of osrdr2 pollen appeared to be viable (Supplemental Figure S17, B–E), manual crossing revealed that osrdr2 pollen failed to pollinate WT ovaries (Figure 3H), suggesting that even the osrdr2 pollen that stained positive with I2-KI and FDA was abnormal. SEM showed that the surface of round pollen grains was smooth for the WT but wrinkled for the osrdr2 mutants (Supplemental Figure S17F), suggesting that pollen wall development is also defective in osrdr2 mutants. We next examined the mRNA levels of four essential genes for pollen development in Stage 7 stamens of osrdr2 and the WT: EXINE PATTERN DESIGNER 1 (EPAD1), which encodes a lipid transfer protein (Li et al., 2020); Glucan Synthase-Like 5 (GSL5), which regulates callose deposition during microsporogenesis (Shi et al., 2015); Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3), which is required for anther cuticle deposition and pollen exine patterning (Men et al., 2017); and OsGELP34, which encodes a GDSL esterase/lipase protein required for pollen exine formation (Zhang et al., 2020). The mRNA levels of the four genes were significantly downregulated in osrdr2 stamens relative to WT stamens at Stage 7 (Supplemental Figure S17G). Given that we used stamen instead of pollen for RNA isolation, the relative expression levels of those genes would be lower in osrdr2 than in the WT. The DNA methylation in the CHH context near EPAD1, GSL5, OsGPAT3, and OsGELP34 was higher in panicles and stamens than in seedlings, and the DNA methylation was lower in osrdr2 than in the WT (Supplemental Figure S17H), suggesting that DNA methylation on those genes is dependent on OsRDR2. Taken together, the results support the hypothesis that OsRDR2-mediated DNA methylation, by regulating the expression of pollen developmental genes, is required for male fertility.
Meiosis is a crucial step for the formation of functional gametes during sexual reproductive development. In the male sexual lineage of Arabidopsis, loss of RdDM causes mis-splicing of MPS1, which disrupts meiosis (Walker et al., 2018). Mutation in OsRDR6 blocked double-strand break (DSB) formation during meiosis and resulted in complete sterility in rice (Liu et al., 2020). The above two studies motivated us to examine the chromosome behavior in prophase I in microspore mother cells (MMCs) (Supplemental Figure S18A). In osrdr2 mutants, abnormalities were observed at diakinesis (Supplemental Figure S18A). There were univalents in 15.6% of osrdr2–3 MMCs and in 7% of osrdr2–6 MMCs (Supplemental Figure S18B), indicating that meiosis defects are probably partially responsible for the sterility of osrdr2 mutants.
We investigated DNA methylation on rice genes regulating meiosis in seedlings, panicles, and stamens, and isolated eight genes whose DNA methylation level was higher in both stamens and panicles than in seedlings and was dependent on OsRDR2 (Supplemental Figure S18C), indicating the possibility that OsRDR2-mediated DNA methylation regulates their expression. The eight genes include OsSDS which is necessary for formation of DSBs (Wu et al., 2015); PAIR1 which is required for homologous chromosome pairing (Nonomura et al., 2004); P31comet, ZEP1, and CRC1 encoding essential synaptonemal complex components which are essential for meiotic recombination initiation (Miao et al., 2013); OsCOM1 which is required for synaptonemal complex formation, homologous paring, and recombination (Ji et al., 2012); OsMSH4 (Zhang et al., 2014b; Wang et al., 2016) and ZIP4 (Shen et al., 2012) which are required for crossover formation. An RT-qPCR assay showed that their mRNA levels were significantly lower in osrdr2 stamens than in WT stamens at Stage 7 (Supplemental Figure S18D). Given that we used stamen instead of meiotic cells in stamen for RNA isolation, the relative expression levels of those genes would be lower in osrdr2 than in the WT. Even 4′,6-diamidino-2-phenylindole (DAPI) staining could not observe any difference in osrdr2 mutant and WT (Supplemental Figure S18A), it is likely there are some abnormalities for meiotic chromosome behavior from leptotene to pachytene.
The DNA methylation on the eight genes was also higher in pistils than in seedlings, and the DNA methylation in pistils was dependent on OsRDR2 (Supplemental Figure S19A), suggesting that meiosis might also be blocked in osrdr2 ovaries. Abnormal female gametophytes were more frequently observed in osrdr2 mutants than in the WT (Supplemental Figure S19, B and C). In addition, stigma of each gynoecium in osrdr2 lacked one or two hairbrushes (Supplemental Figure S19, D and E). Thus, OsRDR2 regulates both male and female reproductive organs.
Upregulated expression of RdDM machinery genes is responsible for increased CHH DNA methylation
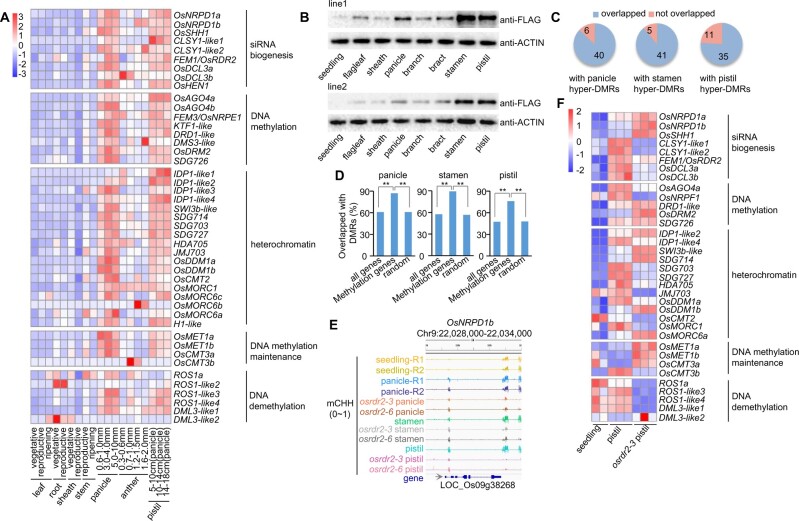
To investigate the mechanism underlying the global increase of DNA methylation in CHH contexts in sexual organs of rice, we examined the expression levels of genes related to DNA methylation (Supplemental Table S6) in seedlings, panicles, pistils, and stamens of the WT. The genes necessary for 24-nt siRNA biosynthesis, including OsRDR2, OsNRPD1, OsSHH1, CLSYs, OsDCL3, and OsHEN1, were upregulated in panicles relative to seedlings, and their mRNA levels were even greater in pistils (Supplemental Figure S20A). Among those genes, however, only CLSY-like 1 expression was increased in stamens (Supplemental Figure S20A). Except the genes related to siRNA biosynthesis mentioned above, many of the genes that function downstream of siRNA in the RdDM pathway, including FEM3/OsNRPE1, OsAGO4, DMS3-like, OsDRM2, and OsMORC6c, were also upregulated in panicles and pistils (Supplemental Figure S20A). To confirm that most RdDM genes were upregulated in the sexual organs of rice, we searched the transcriptomes previously reported for rice seedlings, leaves, shoots, panicles, stamens, and pistils (http://rice.plantbiology.msu.edu/expression.shtml). The search indicated that the expression levels of most RdDM genes were higher in panicles and pistils than in leaves, seedlings, or shoots (Supplemental Figure S20B). Moreover, the expression levels of those genes were much higher in young panicles than in mature panicles (Supplemental Figure S20B), suggesting that the RdDM machinery is probably strengthened early in the development of panicles. Another independent search also demonstrated that most RdDM genes in rice are expressed at higher levels in young panicles and pistils than in leaves, roots, sheaths, or stems (Figure 9A, https://ricexpro.dna.affrc.go.jp/GGEP/index.php).
Figure 9.
Expression levels of RdDM genes are upregulated in reproductive tissues. A, Heatmap showing the relative expression levels of DNA methylation-related genes in various tissues from the Rice XPro database (https://ricexpro.dna.affrc.go.jp/GGEP/index.php). B, Western blot detection of the protein levels of OsRDR2 in various tissues of two independent OsRDR2::OsRDR2-3xFLAG transgenic rice plants. C, DNA methylation-related genes overlapped with panicle-, stamen-, and pistil-CHH hyper-DMRs. D, Percentages of DNA methylation-related genes overlapped with panicle CHH hyper-DMRs (left), stamen CHH hyper-DMRs (middle), and pistil CHH hyper-DMRs (right) (**P < 0.01; by Fisher’s two-sided test). E, Integrated Genome Browser view of CHH methylation levels on OsNRPD1b in various organs of the indicated genotypes. F, Heatmap showing the expression levels of DNA methylation-related genes in WT seedlings, WT pistils, and osrdr2–3 pistils.
To verify the upregulation of OsRDR2 in rice panicles and pistils, we fused 3xFLAG to the C-terminal of native promoter-driven genomic OsRDR2. The homozygous transgenic plants of two independent lines with one locus insertion of OsRDR2::OsRDR2-3xFLAG were used to examine OsRDR2 protein levels. We found that OsRDR2 protein levels were higher in panicles, stamens, and pistils than in seedlings, leaves, or sheaths (Figure 9B), suggesting that OsRDR2 is indeed upregulated in reproductive organs relative to vegetative organs. Overall, these results indicate that the genes encoding RdDM components are upregulated in panicles and pistils, which reinforces the global DNA methylation in sexual organs.
To determine the molecular mechanism underlying the upregulation of RdDM genes in sexual organs of rice, we examined the DNA methylation levels on 46 DNA methylation-related genes in different organs (Supplemental Table S6), and found that the majority of those genes overlapped with three types of hyper-DMRs (40 genes overlapped with panicle CHH hyper-DMRs, 41 genes overlapped with stamen CHH hyper-DMRs, and 35 genes overlapped with pistil CHH hyper-DMRs, Figure 9C). About 90% of the methylation-related genes overlapped with hyper-DMRs, which was significantly higher than the overlap with random genes (Figure 9D). For example, DNA methylation levels in the CHH context on OsRDR2, OsDRM2, and OsNRPD1b were greater in panicles, stamens, and pistils than in seedlings (Figure 9E; Supplemental Figure S20C). The high methylation levels of CHH on those regions were reduced in panicles, stamens, and pistils of osrdr2 mutants (Figure 9E; Supplemental Figure S20C), suggesting that the increase in the methylation of those genes was OsRDR2-dependent. The misregulation of some RdDM genes in osrdr2 pistils and osrdr2 panicles (Figure 9F; Supplemental Figure S20D) indicated a positive feedback between a global increase in DNA methylation and the upregulation of the RdDM machinery in the sexual organs of rice.
Discussion
DNA methylation is an important epigenetic mark that controls the transcription of genes and TEs, and is essential for genome stability. In mammals including humans, cytosine DNA methylation mainly occurs in the CG context, and reprogramming of DNA methylation is important for gamete and embryo development. Mutation in genes controlling DNA methylation and DNA demethylation in mammals often results in death (Smith and Meissner, 2013; Greenberg and Bourc His, 2019). In Arabidopsis, a dicotyledonous plant with a small genome, most DNA-methylation mutants can reproduce normally, and the defects in de novo methylation via the RdDM pathway do not result in any obvious developmental abnormalities (Matzke and Mosher, 2014). These findings with Arabidopsis suggest that de novo DNA methylation may not be crucial for plant development. The results presented here, however, indicate that de novo DNA methylation is crucial for the reproductive development of rice plants.
In the current investigation of rice, a monocotyledonous plant that diverged 150 million years ago and is more representative than Arabidopsis of angiosperms in terms of genome size and TE content, we found that mutation in OsRDR2 substantially reduced CHH methylation and caused total sterility because of developmental defects in both male and female reproductive organs. We demonstrated that the upregulation of RdDM activity including OsRDR2 activity is responsible for the global increases in DNA methylation in sexual organs of rice. These results, therefore, indicate that de novo DNA methylation through enhancement of RdDM is essential for the reproductive development of rice plants. Besides RdDM genes, the expression levels of OsDDM1, OsCMT2, OsCMT3a, OsMET1, and ROS1-like genes were also upregulated in panicles and pistils (Figure 9A), suggesting that other epigenetic mechanisms might regulate reprogramming of DNA methylation in the reproductive organs of rice.
In Arabidopsis, hypomethylation in the vegetative cells releases the expression of many TEs in the pollen grains (Slotkin et al., 2009). The hypomethylation in TEs in the vegetative cells of Arabidopsis pollen might cause the production of easiRNAs (epigenetically activated siRNAs), which are transferred to the sperm cell to repress TE expression (Slotkin et al., 2009; Calarco et al., 2012; Kawashima and Berger, 2014; Martínez et al., 2016; Kim et al., 2019). The paternal easiRNAs of Arabidopsis also function in post-fertilization genome stability and seed viability (Borges et al., 2018; Martinez et al., 2018). The hypomethylation in a locus-specific manner through DNA demethylation in central cells is inherited in the endosperm and results in gene imprinting in rice and Arabidopsis (Park et al., 2016). We hypothesize that the high methylation level in stamens and pistils by enhancement of RdDM might make active or inactive DNA demethylation possible in the central cell and vegetative cell of rice. After DNA demethylation mediated by ROS1 homolog (Kim et al., 2019; Zhou et al., 2021), a specific methylome in various cells of the rice gametophyte is established. The difference in DNA methylation in different cells of the gametophyte is essential for genome stability and seed viability. The role of RdDM during meiosis and mitosis in gamete development of rice warrants additional research.
The mRNA levels of most RdDM genes were higher in rice panicles and pistils than in rice seedlings. We found that DNA methylation increased on the promoters of most RdDM genes in panicles and pistils, which suggests the existence of a positive feedback loop between DNA methylation and RdDM activity in reproductive organs. Considering that DNA methylation in the promoter could enhance gene expression via a protein complex containing DNAJ proteins and SUVH1 and SUVH3, two methylated DNA-binding proteins (Li et al., 2016; Harris et al., 2018; Xiao et al., 2019; Zhao et al., 2019), we hypothesize that RdDM-mediated DNA methylation on the promoters of RdDM machinery genes activates their expression.
With the global increase in methylation in panicles, stamens, and pistils of rice plants, genes encoding RdDM components were upregulated in panicles and pistils but not in stamens. This inconsistency between DNA methylation and expression of RdDM genes in stamens is explained by the finding that the OsRDR2 protein level is increased in stamens. It is possible that, at an early stage of stamen development, genes in the RdDM pathway are upregulated and that their protein level but not their rates of transcription are maintained at high levels, which would ensure high CHH methylation in stamens at later stages of development.
Although OsRDR2 and OsNRPD1 were upregulated in the sexual organs of rice, the abundance of 24-nt siRNAs was not increased on those developmental hyper-DMRs. The lack of positive correlation between 24-nt siRNA abundance and DNA methylation levels might be explained by the finding that hundreds of siren loci produce over 90% of siRNA expression in the ovule and seed coat of Brassica rapa (Grover et al., 2020). We analyzed sRNAs of seedlings, panicles, stamens, and pistils and found hundreds of loci that produce the majority of siRNAs in panicles, stamens, and pistils but not in seedlings, which would explain the inconsistency between the change of siRNA abundance and CHH methylation between reproductive organs and seedlings. In addition, the increased activity of downstream components of RdDM like Pol V, OsAGO4, OsKTF1, OsDRD1, OsDMS3, and OsDRM2 likely contribute to the increase of DNA methylation in reproductive organs.
Materials and methods
Plant materials
OsGA2ox1-overexpression (GAE), OsGA2ox1-silencing (GAS), and fem1 mutants (fem1-1 and fem1-2) are in the background of Taipei 309 (TP309) (O. sativa, japonica). Both GAE and GAS were homozygous for 35S::OsGA2ox1. To genotype the transgene of 35S::OsGA2ox1, we used a forward primer on the 35S promoter and a reverse primer on OsGA2ox1 in order to conduct a PCR assay (Supplemental Table S7). Both fem1-1 and fem1-2 were recessive, and their segregation ratio was 1:3. The osrdr2 mutants (osrdr2–3, osrdr2–4, osrdr2–5, osrdr2–6, and osrdr2–7) were created using clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) technology with the pCBSG03 vector (Zhang et al., 2014a) in the background of Nipponbare (O. sativa, japonica), which was the WT in this study.
Two kinds of panicles (panicle-R1 and osrdr2–3 panicle; R1 indicates WT-Replicate 1) that had grown in a field in Hainan were harvested at anther developmental Stage 12 in April of 2016. Two other kinds of panicles (panicle-R2 and osrdr2–6 panicle; R2 indicates WT-Replicate 2) that had grown in a field in Nanjing were harvested at the same developmental stage in September of 2018. Mature stamens and pistils of the WT and mutants (osrdr2–3 and osrdr2–6) were harvested in Nanjing in August of 2019. The DNA and RNA isolated from panicles were used for high-throughput sequencing. Eighteen-day-old seedlings without roots (seedling-R1, seedling-R2, and osrdr2–6 seedling; R indicates WT-Replicate) grown in Kimura B nutrient solution in a greenhouse (16 h/8 h light/dark with a 28°C/25°C cycle) were collected, and their DNA and RNA were isolated for high-throughput sequencing.
Genetic screen of five elements mountain mutants
GAS seeds (about 50,000) were soaked in water for 24 h at room temperature and then transferred into 1% (v/v) EMS for another 24 h. The EMS-treated seeds were planted in an isolated paddy field. The seeds were separately harvested from each individual plant (more than 23,000 M1 plants). The M2 plants from the same M1 line were planted together. The M2 plants that produced seedlings with GA deficiency symptoms were selected and planted (about 200 M1 lines). When the plants were 3 months old, their OsGA2ox1 mRNA levels and those of GAS plants were assessed. Those dwarf rice plants with OsGA2ox1 mRNA levels that were at least 20 times greater than those of GAS were named fem. The name was inspired by “the journey to the west”, in which Buddha created Five Elements Mountain (“FEM”) to imprison a monkey king who disturbed the hierarchy in heaven (Zheng et al., 2021).
Map-based cloning of FEM1
To clone FEM1, heterozygous fem1 plants were crossed with TN1 plants (cv Taichung Native 1), a Xian/indica variety. Seeds of individual F1 plants were separately collected and planted. The GA-deficient seedlings from the F2 population were selected and served as mapping samples. Using 228 SSR and STS molecular markers, we conducted bulked segregant analysis (BSA) of mutant and WT pools (mutant and WT pools consisted of 30 GA-deficient plants or normal plants, respectively). R1 and R7 were first identified as linked markers in BSA. Fine mapping was then conducted using 140 dwarf plants to narrow the location of fem1-1 between R4 and R5. Finally, a large mapping population consisting of 890 samples was used to determine the 103-kb region that contained fem1-1. A similar procedure was used to map fem1-2. The oligos of molecular markers used for fine mapping are listed in Supplemental Table S7.
Vector construction and plant transformation
To generate the OsGA2ox1 overexpression construct, the 8,515-bp fragment containing the OsGA2ox1 coding region and its promoter and terminator was isolated from BAC OsJNBa0017J22, which was digested by restriction enzyme XbaI (NEB, R0145S). With the T4 DNA ligase (TAKARA, 2011A, Shiga, Japan), the product was ligated to the binary vector pCAMBIA1301-35SN, which was digested by XbaI (NEB, R0145S). The construct with the desired insertion was isolated by colony-PCR (Supplemental Table S7). Agrobacterium EHA105 with the sequenced vector was used for rice transformation.
For the complementary construct, an 8,339-bp DNA fragment composed of the coding region, the 2,098-bp promoter, and the 1,030-bp terminator of OsRDR2 was amplified using high fidelity polymerase (Vazyme, P505-d1, Nanjing, China) and the oligos listed in Supplemental Table S7. The PCR product was cloned into the binary vector pCAMBIA3301, which was digested by EcoRI and HindIII (NEB, R3101S, and R3104S) using the ClonExpress MultiS One Step Cloning Kit (Vazyme, C113-02) according to the manufacturer’s instructions. Agrobacterium EHA105 with the sequenced vector was used for rice transformation.
Two sgRNAs targeting OsRDR2 were designed on http://skl.scau.edu.cn/, and the oligos were synthesized in Genescript (Supplemental Table S7). After gradient annealing, the two oligos were cloned into pCBSG03, which was digested by BsaI (NEB, R0535S) as previously reported (Zhang et al., 2014a). Agrobacterium EHA105 with the sequenced vector was used for rice transformation.
The 7,907-bp genomic DNA fragment of OsRDR2 with the 2,699-bp promoter and without TGA was amplified by PCR using the oligos listed in Supplemental Table S7. The fragment of OsRDR2 was recombined into the pCAMBIA1305-3×FLAG vector (Zhang et al., 2012). The vector was sequenced and transformed into Agrobacterium EHA105 for rice transformation.
Phylogenetic analysis
Protein sequences of RdRP families in rice, maize, and Arabidopsis were downloaded from national center for biotechnology information (NCBI) and changed into the fasta format. The unrooted maximum likelihood tree was constructed by MEGA6 with default parameters (Tamura et al., 2013). Bootstrap analysis was performed using 1,000 replicates.
Pollen staining
Mature anthers at Stage 12 of various genotypes were fixed in 50% FAA (5% formaldehyde, 5% glacial acetic acid, and 63% ethanol) for 24 h. The pollen was then released in a drop of 1% I2-KI on a glass slide. After they were stained for 3 min at room temperature, the pollen grains were examined with a light microscope (Olympus, BX53, Tokyo, Japan).
Fresh mature anthers at Stage 12 of various genotypes were dissected with the aid of a stereoscopic microscope (Olympus, SZ61). After they were released in a drop of 0.01% FDA (prepared with 17% sucrose) on a glass slide, the pollen grains were observed with a microscope (Olympus, BX53).
Semi-thin sections
Anthers of osrdr2 and the WT at various stages (Zhang et al., 2011) were fixed in 50% FAA, and samples were successively dehydrated in 50% ethanol for 30 min, 70% ethanol for 30 min, 70% acetone for 20 min, 80% acetone for 20 min, 90% acetone for 20 min, 95% acetone for 30 min, and finally three times in 100% acetone for 40 min each time. The anthers were then placed in 3:1 acetone: embedding medium (Zhongjingkeyi Technology Co., Ltd., SPI-Pon 812 kit, GP18010) for 2 h, 1:1 acetone: embedding medium for 4 h, and 1:3 acetone: embedding medium for 12 h. The anthers were finally kept at 65°C for 48 h in embedding medium. Microtome sections (2 μm thick) were stained with 0.05% toluidine blue and observed with a light microscope (Olympus, BX53).
SEM
The mature spikelets (anthers at Stage 12) were fixed in 2.5% glutaraldehyde overnight at 4°C and then dehydrated in an ethanol series of 30%, 50%, 70%, 80%, 90%, and 100% (5 min for each step). The anther and pollen were coated with gold particles and observed using a HITACHI SU80I0 scanning electron microscope.
Observation of embryo sacs
Embryo sacs were stained and observed as previously described with slight modification (Zhao et al., 2013). In brief, mature spikelets of the WT and osrdr2 mutants were collected and immediately fixed in 50% FAA for 24 h at room temperature. The samples were then stored in 70% ethanol at 4°C. Before staining, the ovaries were dissected with the aid of a stereomicroscope. The tissue was processed through an ethanol series (50%, 30%, and 15%, 30 min for each concentration) and finally transferred into distilled water for 30 min. The ovaries were successively stained in 2% aluminum potassium sulfate for 20 min, 10 mg/L eosin for 12 h, and 2% aluminum potassium sulfate for 20 min; they were then washed with distilled water for 24 h, followed by three washes with distilled water. The tissue was dehydrated in an ethanol series (30%, 50%, 70%, 80%, 90%, and 100%, 30 min for each step, but three times in 100% ethanol). The ovaries were then placed in 1:1 ethanol:methylsalicylate for >1 h, and were cleared in methylsalicylate three times for 1 h each time. The embryo sacs were examined with a confocal laser scanning microscope (Zeiss, LSM780) at 514-nm wavelength.
Meiotic chromosome preparation
Young panicles (40–60 mm) of the WT and osrdr2 mutants were fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1) for 24 h. Anthers were dissected on glass slides with the aid of a stereoscopic microscope (Olympus, SZ61), and were then squashed in 40% acetic acid between a cover slip and the glass slide. After the preparation was frozen in liquid nitrogen for 5 min, the cover slip was removed. After dehydration at room temperature, the chromosomes were stained within 3 µL of DAPI in the dark for 2 min. The cytological analysis was conducted with a fluorescence microscope (Leica, DMi8).
RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, 15596018, Waltham, MA, USA). About 1 μg of total RNA was used to synthesize cDNA with the HiScript® II Q RT SuperMix kit (Vazyme Biotech Co., Ltd. R223-01, Nanjing, China) according to the manufacturer’s protocol. The OsActin gene (LOC_Os03g50885) and Ubq gene (LOC_Os03g13170) were used as the internal controls in semi-quantitative RT-PCR and real-time PCR, respectively. The gene-specific oligos used for RT-PCR are listed in Supplemental Table S7.
Chop-PCR assay
A 200-ng quantity of genomic DNA was digested by the methylation-sensitive enzyme HaeIII (NEB, R0108S) in a 20-μL reaction mixture at 37°C for 20 h. PCR was performed using 2 μL of the digested DNA as template in a 20-μL reaction mixture with the locus-specific oligos (Supplemental Table S7).
Protein extraction and western blotting
The seeds of OsRDR2::OsRDR2-3xFLAG transgenic rice plants at T0 were harvested and planted. The transgenic rice whose progeny segregated 3:1 for PCR genotyping was allowed to set seeds, and homozygous T2 lines were used for western blotting. Samples of 2-week-old seedlings, flag leaves, sheaths, mature panicles, branches, bracts, stamens, and pistils from homozygous transgenic plants were collected and frozen in liquid nitrogen. The tissues were ground into powder in liquid nitrogen using a pestle and mortar. The powder was suspended in an equal volume of 2× extraction buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), 1 mM ethylenediaminetetraacetic acid (EDTA), and 1 mM dithiothreitol) with 1% protease inhibitor cocktail (04693116001, Roche, Basel, Switzerland). The samples were centrifuged at 4°C for 15 min at 10,000 rpm. The supernatant was mixed with an equal volume of 2× loading buffer and was denaturated at 95°C for 5 min.
Western blotting against FLAG was performed using the FLAG-specific antibody (Sigma, F1804, St Louis, MO, USA) as previously described (Liu et al., 2019). In brief, proteins were separated in a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, IPVH00010, Burlington, MA, USA). The PVDF membrane was washed five times with TBST buffer. After it was blocked with 5% nonfat milk (MDBio, tznf-500g) for 1 h, the membrane was incubated with anti-FLAG antibody (Sigma, F1804, 1:5,000) at 4°C overnight. The membrane was washed five times with TBST buffer. After it was blocked with 5% nonfat milk for 1 h, the membrane was incubated with secondary antibody (CWbio, CW0102S, Cambridge, MA, USA, goat anti-mouse IgG) at 4°C for 2 h. The signal was detected with the eECL Western Blot Kit (Engibody, IF6747) and the Chemiluminescence gel imaging system (Bio-Rad, Chemi DOC XRS+, Hercules, CA, USA).
Small RNA northern blotting
Total RNAs were extracted from leaves of 110-d-old seedlings by using TRIzol (Invitrogen, 15596018). Small RNAs were enriched as previously described (Yang et al., 2016). In brief, an equal volume of PEG 8000 (20% PEG 8000, 1 M NaCl) was added to the total RNAs, and the mixture was centrifuged at 13,000 rpm at 4°C. An equal volume of isopropanol and a 0.1 volume of 3 M NaAC were added to the supernatant, and the mixture was centrifuged at 13,000 rpm at 4°C. The supernatant was discarded, and the pellet was resuspended in diethy pyrocarbonate (DEPC)-treated water to obtain small RNAs. A 70-μg quantity of small RNAs was mixed with an equal volume of Gel Loading Buffer II (Ambion AM8546), and the mixture was denatured at 65°C for 5 min. Denatured sRNAs were subjected to 100-V electrophoresis overnight using a 15% denaturing PAGE gel. After electrophoresis, sRNAs were transferred to a Hybond-N+ membrane (GE Healthcare, RPN303B) with 400 mA for 3 h. The membrane was cross-linked by UV. The dry membrane was hybridized with 32P-labeled probe in hybridization buffer (Sigma, H7033-125ML) at 38°C overnight. The hybridization signals were detected with the Multifunctional Molecular Imager (GE Typhoon, FLA9500). The sequences of the probe are listed in Supplemental Table S7.
WGBS
About 1 μg of genomic DNA was randomly fragmented by ultrasonication (Covaris), and the DNA fragments were separated by agarose gel electrophoresis. The DNA fragments ranging from 200 to 300 bp were cut and purified with the MiniElute PCR Purification Kit (QIAGEN, Hilden, Germany). The fragmented DNA was incubated with End Repair Mix at 20°C, and was purified with the MiniElute PCR Purification Kit. The DNA was incubated with A-Tailing Mix at 37°C, and was purified with the MiniElute PCR Purification Kit. The adapters were ligated to the adenylated DNA and were purified with the MiniElute PCR Purification Kit. The purified DNA was subjected to bisulfite treatment with the Methylation-Gold kit (ZYMO, D5005). The DNA fragments were separated by agarose gel electrophoresis. The fragments ranging from 320 to 420 bp were selected and purified with the QIAquick Gel Extraction kit (QIAGEN). Fifteen cycles of PCR amplification were performed with PCR Primer Cocktail and PCR Master Mix to enrich the DNA fragments. Agarose gel electrophoresis was performed to select DNA fragments ranging in size from 320 to 420 bp. The purified DNA served as the final library. The quality of the library was assessed with the Agilent Technologies 2100 bioanalyzer and the ABI StepOnePlus Real-Time PCR System. The qualified libraries were sequenced on the Hiseq-Xten platform (BGI).
The low-quality reads were removed by NGSQCToolkit version 2.3 (Patel and Jain, 2012). The remaining reads were aligned to release 7 of the rice reference genome (Kawahara et al., 2013) using Bismark version 0.18.1 (Krueger and Andrews, 2011) with 1 mismatch allowed (-N 1). PCR-duplicates were removed by deduplicate_bismark. The cytosine sites were identified by bismark_methylation_extractor on a whole-genome scale. The methylation levels on individual cytosines or regions were calculated as the total number of methylated cytosines divided by the total number of sequenced cytosines of the site or region. The uniquely mapped reads on the chloroplast genome were used to calculate the conversion rate.
The average methylation levels of CG, CHG, and CHH on genes, TEs, and the upstream and downstream regions were analyzed in 100-bp windows. For the average methylation levels of CG, CHG, and CHH on TEs with different lengths, the TE and surrounding region (regions 2-kb upstream and downstream) were divided into 20 windows. The methylation levels were calculated in each window.
DMR identification and analysis
DMRs were identified as previously described method (Stroud et al., 2013b) with modification. Only cytosine covered by at least four reads was considered an effective site. DMRs were searched using a 100-bp sliding window with a 50-bp step size. Windows with at least four effective cytosines were retained. DNA methylation levels were compared pairwise with Fisher’s exact test, and the P-values were adjusted for multiple comparisons using the Benjamini–Hochberg method. Windows with FDR <0.01 were selected. Finally, regions with an absolute methylation level difference of 0.4, 0.2, and 0.1 for CG, CHG, and CHH, respectively, were retained for subsequent analysis. Neighboring windows were merged if the gap was <100 bp as the final DMR. If two DMRs shared one or >1 bp, the two DMRs were defined as overlapped DMRs (bedtools, version 2.25.0).
mRNA sequencing and analysis
About 1 μg of total RNA was used to construct a library with the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530L, NEB). The mRNA was enriched by magnetic beads with oligo dT. The mRNA was broken into short fragments with fragmentation buffer. The fragments were used as templates to synthesize the first strand of cDNA with a hexa-base random primer, 1st Strand Buffer, and 1st Strand Enzyme Mix. The double-stranded DNA was synthesized after adding buffer, dNTPs, RNase H, and DNA polymerase I. The synthesized DNA was purified by AMPure XP beads. The purified DNA was repaired, and A-tailings and ligated adapters were added. The final library was obtained by PCR enrichment. The library was sequenced on the Hiseq-Xten platform (Annoroad Gene Technology Co., Ltd., Beijing, China).
The low-quality reads in the raw data were removed by Trimmomatic version 0.33 (Bolger et al., 2014). The clean reads were then aligned to release 7 of the rice reference genome (Kawahara et al., 2013) using TopHat-2.1.1 (Trapnell et al., 2012) with 1 mismatch allowed (-m 1). After PCR duplicates were removed by samtools rmdup, differential expression analysis (Trapnell et al., 2009, 2012) was conducted with Cuffdiff version 2.2.1. The expression level for each gene was calculated using the fragments per kilobase of exon per million mapped reads (FPKM) method. The DEGs between two samples were selected using the following criteria: the fold-change (FPKM values were averaged for biological replicates) was >2, and the FDR was <0.05. Heatmaps of gene expression levels were generated by R version 3.1.0.
For TE expression analysis, read counts of each TE were calculated by HTSeq version 0.10.0 (Anders et al., 2015) from bam output files of mRNA-seq. The significance (q < 0.05) was calculated by DESeq2 (Love et al., 2014) (version 1.28.1). The expression level of TEs was normalized using the reads per million mapped reads (RPM).
sRNA-seq and analysis
After total RNA was isolated, 1 μg of RNA was subjected to PAGE. The 18- to 30-nt (14–30 ssRNA Ladder Marker, TAKARA) small RNAs were cut, purified and ligated to 3′- and 5′-adapters. The adapted small RNAs were reversely transcribed into cDNA. The cDNA fragments were amplified with 15–18 cycles of PCR. The PCR products between 110 and 130 bp were selected and purified with the QIAquick Gel Extraction Kit (QIAGEN, 28704). The double-stranded DNA was heat-denatured and circularized by the splint oligo. The single-stranded circular DNA served as the final library. The library was validated with the Agilent Technologies 2100 bioanalyzer. Finally, the library was sequenced on the BGISEQ-500 platform (BGI). After the adapters were trimmed by Trim_galore version 0.5.0, the reads with N were removed by script to obtain clean reads. The clean reads were aligned to release 7 of the rice reference genome (Kawahara et al., 2013) using BOWTIE software version 1.1.2 (Langmead, 2010) with 0 mismatch (-v 0). The reads that mapped to structural RNAs (snRNAs, snoRNAs, tRNAs, and rRNAs) were removed. The uniquely mapped 24-nt reads were used for subsequent analysis. The read counts were normalized using the reads per 10 million mapped reads against all mapped reads without structural sRNAs.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: OsRDR2, Os04g0465700; OsGA2ox1, Os05g0158600; OsTDL1A, Os12g0472500; MIL1, Os07g0151100; MSP1, Os01g0917500; EPAD1, Os03g0663900; GSL5, Os06g0182300; OsGPAT3, Os11g0679700; OsGELP34, Os02g0290900; OsSDS, Os03g0225200; PAIR1, Os03g0106300; P31comet, Os05g0251400; ZEP1, Os04g0452500; CRC1, Os04g0479000; OsCOM1, Os06g0613400; OsMSH4, Os07g0486000; ZIP4, Os01g0890900. The accession numbers of DNA methylation related genes were listed in Supplemental Table S6.
Availability of data
Sequencing data have been deposited in the Gene Expression Omnibus (GSE112259, GSE130168, and GSE152155).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The silencing mechanism of GAS.
Supplemental Figure S2. DNA methylation levels of CG, CHG, and CHH in six RdDM loci.
Supplemental Figure S3. Complementation of fem1-1.
Supplemental Figure S4. Protein sequence analysis and allelic test of fem1-1 and fem1-2.
Supplemental Figure S5. Diagrams of knockout OsRDR2.
Supplemental Figure S6. OsRDR2 is involved in maintaining CHH methylation in panicles and seedlings.
Supplemental Figure S7. The abundance of 21-nt and 23-nt siRNAs on CHH hypo-DMRs in osrdr2–3 panicles.
Supplemental Figure S8. OsRDR2 is involved in maintaining CHH methylation in seedlings.
Supplemental Figure S9. OsRDR2 is involved in maintaining CHH methylation in panicles.
Supplemental Figure S10. CHH methylation is higher in panicles than in seedlings.
Supplemental Figure S11. OsRDR2 is involved in maintaining CHH methylation in stamens.
Supplemental Figure S12. OsRDR2 is involved in maintaining CHH methylation in pistils.
Supplemental Figure S13. The hyper-methylation in panicles, stamens, and pistils is largely overlapped.
Supplemental Figure S14. A pathway in addition to RdDM is likely involved in the increase in CHH in reproductive organs.
Supplemental Figure S15. osrdr2 affects gene expression in stamens and pistils.
Supplemental Figure S16. CHH methylation and siRNA abundance in differentially expressed TEs in osrdr2–3.
Supplemental Figure S17. Pollen viability is low in osrdr2 mutants.
Supplemental Figure S18. Meiosis defects in osrdr2 mutants.
Supplemental Figure S19. Developmental defects in osrdr2 pistils.
Supplemental Figure S20. Upregulation of genes related to DNA methylation in reproductive organs.
Supplemental Table S1. Basic information of high-throughput sequencing.
Supplemental Table S2. Panicle CHH hyper-DMRs and osrdr2 CHH hypo-DMRs in panicles and seedlings.
Supplemental Table S3. Stamen CHH hyper-DMRs and osrdr2 CHH hypo-DMRs in stamens.
Supplemental Table S4. Pistil CHH hyper-DMRs and osrdr2 CHH hypo-DMRs in pistils.
Supplemental Table S5. List of DEGs.
Supplemental Table S6. List of DNA methylation-related genes.
Supplemental Table S7. Oligo sequences.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China, the Natural Science Foundation of Jiangsu Province, the National Key Transformation Program, and the Jiangsu Collaborative Innovation Center for Modern Crop Production to DL Yang. We thank Bin Han for providing the BAC clone. The high-throughput sequencing data were analyzed on the high-performance computing platform of the Bioinformatics Center, Nanjing Agricultural University.
Funding
This study was supported by the National Natural Science Foundation of China (31671340), the Natural Science Foundation of Jiangsu Province (BK20170027), the National Key Transformation Program (2016ZX08001002), and the Jiangsu Collaborative Innovation Center for Modern Crop Production to DL Yang.
Conflict of interest statement. The authors declare no conflict of interest.
Y.D.L. conceived of the project and designed the experiments; W.L. conducted most of the experiments and performed the bioinformatic analysis; Z.K. conducted siRNA northern blotting, chop-PCR, and rice plant management; Z.L. conducted the genetic screens; Z.T., Y.Y., and X.D. contributed analyzing agents and tools; Z.H. assisted with the examination of anther and pollen; W.Y. assisted with the bioinformatics analysis; and Y.D.L. and W.L. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Dong-Lei Yang (dlyang@njau.edu.cn).
References
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL (2006) An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442: 295–298 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Feng S, Yu C, Liu W, Kuo HY, Jacobsen EL, Zhai J, Gallego-Bartolome J, Wang L, Egertsdotter U, et al. (2016) DNA methylome of the 20-gigabase Norway spruce genome. Proc Natl Acad Sci USA 113: E8106–E8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15: 1753–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, Pikaard CS (2015) Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Parent J, van Ex F, Wolff P, Martínez G, Köhler C, Martienssen RA (2018) Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat Genet 50: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijo JA, Becker JD, et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL (2000) mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12: 2101–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, Patel DJ (2015) DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16: 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CG, Zhang H, Tang K, Zhu X, Qian W, Hou YJ, Wang B, Lang Z, Zhao Y, Wang X, et al. (2015) Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO J 34: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KJ, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB (2009) RNA polymerase IV functions in paramutation in Zea mays. Science 323: 1201–1205 [DOI] [PubMed] [Google Scholar]
- Gouil Q, Baulcombe DC (2016) DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet 12: e1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC, Bourc His D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20: 590–607 [DOI] [PubMed] [Google Scholar]
- Grover JW, Burgess D, Kendall T, Baten A, Pokhrel S, King GJ, Meyers BC, Freeling M, Mosher RA (2020) Abundant expression of maternal siRNAs is a conserved feature of seed development. Proc Natl Acad Sci USA 117: 15305–15315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CJ, Stonaker JL, Gross SM, Hollick JB (2007) A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol 5: e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CJ, Erhard KJ, Lisch D, Hollick JB (2009) Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, Li X, Chen W, Xue Y, Zhong Z, et al. (2018) A DNA methylation reader complex that enhances gene transcription. Science 362: 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Hong L, Tang D, Shen Y, Hu Q, Wang K, Li M, Lu T, Cheng Z (2012a) MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol 196: 402–413 [DOI] [PubMed] [Google Scholar]
- Hong L, Tang D, Zhu K, Wang K, Li M, Cheng Z (2012b) Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 24: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Tang D, Wang K, Wang M, Che L, Li M, Cheng Z (2012) The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J 72: 18–30 [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (NY) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Berger F (2014) Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet 15: 613–624 [DOI] [PubMed] [Google Scholar]
- Kim MY, Ono A, Scholten S, Kinoshita T, Zilberman D, Okamoto T, Fischer RL (2019) DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc Natl Acad Sci USA 116: 9652–9657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B (2010) Aligning short sequencing reads with Bowtie. Curr Protocol Bioinformatics 11: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AM, Strahl BD, Patel DJ, Jacobsen SE (2013) Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kim YJ, Yang L, Liu Z, Zhang J, Shi H, Huang G, Persson S, Zhang D, Liang W (2020) Grass-specific EPAD1 is essential for pollen exine patterning in rice. Plant Cell 32: 3961–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Li S, Gao L, Zhao Y, Kim YJ, Chen X (2016) SUVH1, a Su(var)3-9 family member, promotes the expression of genes targeted by DNA methylation. Nucleic Acids Res 44: 608–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for Maintenance of CpXpG Methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Liu C, Shen Y, Qin B, Wen H, Cheng J, Mao F, Shi W, Tang D, Du G, Li Y, et al. (2020) Oryza sativa RNA-dependent RNA polymerase 6 contributes to double-strand break formation in meiosis. Plant Cell 32: 3273–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shi Z, Zhang X, Wang M, Zhang L, Zheng K, Liu J, Hu X, Di C, Qian Q, et al. (2019) Inducible overexpression of ideal plant architecture1 improves both yield and disease resistance in rice. Nat Plants 5: 389–400 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, et al. (2006) MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res 16: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O (2013) Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet 45:1029–1039 [DOI] [PubMed] [Google Scholar]
- Martínez G, Panda K, Köhler C, Slotkin RK (2016) Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants 2: 16030. [DOI] [PubMed] [Google Scholar]
- Martinez G, Wolff P, Wang Z, Moreno-Romero J, Santos-González J, Conze LL, DeFraia C, Slotkin RK, Köhler C (2018) Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat Genet 50: 193–198 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJM (2015) RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66: 243–267 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Men X, Shi J, Liang W, Zhang Q, Lian G, Quan S, Zhu L, Luo Z, Chen M, Zhang D (2017) Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J Exp Bot 68: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z (2013) Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh S, Eun CH, Ono A, Asao H, Okano Y, Yamaguchi K, Shimatani Z, Koizumi A, Terada R (2012) Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J 71: 85–98 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. (2008) Distinct size distribution of endogeneous siRNAs in maize: evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci USA 105: 14958–14963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N (2004) The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Kim MY, Vickers M, Park JS, Hyun Y, Okamoto T, Zilberman D, Fischer RL, Feng X, Choi Y, et al. (2016) DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci USA 113: 15138–15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson SE, Gross SM, Hollick JB (2007) Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev Biol 308: 462–473 [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten SO, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Shen Y, Tang D, Wang K, Wang M, Huang J, Luo W, Luo Q, Hong L, Li M, Cheng Z (2012) ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125: 2581–2591 [DOI] [PubMed] [Google Scholar]
- Shi X, Sun X, Zhang Z, Feng D, Zhang Q, Han L, Wu J, Lu T (2015) GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol 56: 497–509 [DOI] [PubMed] [Google Scholar]
- Sidorenko L, Dorweiler JE, Cigan AM, Arteaga-Vazquez M, Vyas M, Kermicle J, Jurcin D, Brzeski J, Cai Y, Chandler VL (2009) A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet 5: e1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdžić M, Becker JD, Feijó JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC (2007) An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220 [DOI] [PubMed] [Google Scholar]
- Stonaker JL, Lim JP, Erhard KJ, Hollick JB (2009) Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet 5: e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Ding B, Simon SA, Feng S, Bellizzi M, Pellegrini M, Wang G, Meyers BC, Jacobsen SE (2013a) Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2: e00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013b) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F, Zhou C, Zhou Q, Zhou S, Yang W, Zhao Y, Li G, Zhou DX (2016) Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol 171: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocol 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J, Gao H, Zhang J, Aldridge B, Vickers M, Higgins JD, Feng X (2018) Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat Genet 50: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang Y, Cheng Z, Zhao Z, Chen J, Sheng P, Yu Y, Ma W, Duan E, Wu F, et al. (2016) The role of OsMSH4 in male and female gamete development in rice meiosis. J Exp Bot 67: 1447–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Gu L, Song X, Cui X, Lu Z, Zhou M, Wang L, Hu F, Zhai J, Meyers BC, et al. (2014) Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc Natl Acad Sci USA 111: 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Pontes O, Pikaard CS, Richards EJ (2007) VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Gene Dev 21: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Ji J, Tang D, Wang H, Shen Y, Shi W, Li Y, Tan X, Cheng Z, Luo Q (2015) OsSDS is essential for DSB formation in rice meiosis. Front Plant Sci 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Zhang J, Li T, Fu X, Satheesh V, Niu Q, Lang Z, Zhu JK, Lei M (2019) A group of SUVH methyl-DNA binding proteins regulate expression of the DNA demethylase ROS1 in Arabidopsis. J Integr Plant Biol 61: 110–119 [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yuan K, Yuan M, Meng X, Chen M, Wu J, Li J, Qi Y (2020) Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol Plant 13: 851–863 [DOI] [PubMed] [Google Scholar]
- Yang DL, Zhang G, Wang L, Li J, Xu D, Di C, Tang K, Yang L, Zeng L, Miki D, et al. (2018) Four putative SWI2/SNF2 chromatin remodelers have dual roles in regulating DNA methylation in Arabidopsis. Cell Discov 4: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Zhang G, Tang K, Li J, Yang L, Huang H, Zhang H, Zhu JK (2016) Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res 26: 66–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Chen Z, Lian B, Rowley MJ, Xia N, Chai J, Li Y, He X, Wierzbicki AT, Qi Y (2016) A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol Cell 61: 222–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, et al. (2015) A one precursor one siRNA model for pol IV-dependent siRNA biogenesis. Cell 163: 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Ning YQ, Zhang SW, Chen Q, Shao CR, Guo YW, Zhou JX, Li L, Chen S, He XJ (2012) IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet 8: e1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lang Z, Zhu JK (2018) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19: 489–506 [DOI] [PubMed] [Google Scholar]
- Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, Ma J, Bai G, Wang P, Zhang SW, Liu ZW, et al. (2013) DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA 110: 8290–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang M, Li Y, Yan W, Chang Z, Ni H, Chen Z, Wu J, Xu C, Deng XW, et al. (2020) GDSL esterase/lipases OsGELP34 and OsGELP110/OsGELP115 are essential for rice pollen development. J Integr Plant Biol 62: 1574–1593 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, et al. (2014a) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12: 797–807 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhu JK (2011) RNA-directed DNA methylation. Curr Opin Plant Biol 14: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tang D, Luo Q, Chen X, Wang H, Li Y, Cheng Z (2014b) Crossover formation during rice meiosis relies on interaction of OsMSH4 and OsMSH5. Genetics 198: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Ku JC, Liu B, Yang D, Yin L, Ferrell TJ, Stoll CE, Guo W, Zhang X, Wang D, et al. (2021) The mop1 mutation affects the recombination landscape in maize. Proc Natl Acad Sci USA 118: e2009475118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao QQ, Lin RN, Li L, Chen S, He XJ (2019) A methylated-DNA-binding complex required for plant development mediates transcriptional activation of promoter methylated genes. J Integr Plant Biol 61: 120–139 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang Y, Liu X, Zhang X, Liu S, Yu X, Ren Y, Zheng X, Zhou K, Jiang L, et al. (2013) A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27: 113–122 [DOI] [PubMed] [Google Scholar]
- Zheng K, Wang L, Zeng L, Xu D, Guo Z, Gao X, Yang DL (2021) The effect of RNA polymerase V on 24-nt siRNA accumulation depends on DNA methylation contexts and histone modifications in rice. Proc Natl Acad Sci USA 118: e2100709118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE (2014) Molecular Mechanism of Action of Plant DRM De Novo DNA Methyltransferases. Cell 157: 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Palanca AMS, Law JA (2018) Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat Genet 50: 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Li X, Liu Q, Zhao Y, Jiang W, Wu A, Zhou DX (2021) DNA demethylases remodel DNA methylation in rice gametes and zygote and are required for reproduction. Mol Plant 14: 1569–1583 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Gene Expression Omnibus (GSE112259, GSE130168, and GSE152155).