Abstract
Shade-intolerant plants rapidly elongate their stems, branches, and leaf stalks to compete with neighboring vegetation, maximizing sunlight capture for photosynthesis. This rapid growth adaptation, known as the shade-avoidance response (SAR), comes at a cost: reduced biomass, crop yield, and root growth. Significant progress has been made on the mechanistic understanding of hypocotyl elongation during SAR; however, the molecular interpretation of root growth repression is not well understood. Here, we explore the mechanisms by which SAR induced by low red:far-red light restricts primary and lateral root (LR) growth. By analyzing the whole-genome transcriptome, we identified a core set of shade-induced genes in roots of Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum) seedlings grown in the shade. Abiotic and biotic stressors also induce many of these shade-induced genes and are predominantly regulated by WRKY transcription factors. Correspondingly, a majority of WRKY genes were among the shade-induced genes. Functional analysis using transgenics of these shade-induced WRKYs revealed that their role is essentially to restrict primary root and LR growth in the shade; captivatingly, they did not affect hypocotyl elongation. Similarly, we also found that ethylene hormone signaling is necessary for limiting root growth in the shade. We propose that during SAR, shade-induced WRKY26, 45, and 75, and ethylene reprogram gene expression in the root to restrict its growth and development.
Low red:far-red shade induces genes in roots regulated by WRKY transcription factors and ethylene, restricting primary and lateral root growth.
Introduction
Plants are exposed to various environmental challenges throughout their life cycles, such as suboptimal access to sunlight, low water, nutrient availability, extreme temperatures, presence of competitors, herbivores, and pathogens (Casal, 2012). Plants exhibit incredible plasticity to withstand these adverse conditions and respond by locally modifying growth rhythms, metabolism, and reproduction to best adapt to their environment (Chory, 2010; Kohnen et al., 2016). An excellent example of adaptive phenotypic plasticity is the shade-avoidance response (SAR). In shade-intolerant plants, SAR is triggered when they are in close proximity to other plant competitors or under a canopy by activating a series of morphological changes to maximize sunlight capture and ensure reproductive fitness (Smith, 1982). The characteristic phenotypes of SAR include rapid stem and petiole elongation, leaf hyponasty, accelerated reproduction, apical dominance, and reduced root growth and development (Salisbury et al., 2007; Casal, 2012). The molecular mechanisms controlling gene expression changes leading to the phenotypic alterations in the shoot organs during SAR are well understood in the model plant Arabidopsis (Arabidopsis thaliana; Casal, 2012; Li et al., 2012; Galvão and Fankhauser, 2015; Pedmale et al., 2016). But the impact of shade on the growth of underground root systems and the molecular account leading to this phenomenon are poorly understood.
Under a dense canopy, plants sense vegetational shading by detecting either a reduction in the ratio of red to far-red (R:FR) light, blue light, or photosynthetically active radiation (PAR; Keller et al., 2011; Keuskamp et al., 2011; Hornitschek et al., 2012). Any changes in the R: FR and blue light in the environment are largely perceived by the R/FR light-sensing phytochrome B (PHYB) and UV-A/blue light-sensing cryptochrome (CRY) 1 and 2 photoreceptors, respectively. In seedlings, CRY- and PHY-mediated shade perception induces the expression of growth-promoting genes in the hypocotyl, such as those involved in hormone biosynthesis and cell-wall remodeling proteins and enzymes, which are both required for the rapid stem elongation (Kohnen et al., 2016; Pedmale et al., 2016; de Wit et al., 2016; Paik et al., 2017).
A handful of studies have linked root growth and development with the SAR, and those have mainly focused on the lateral root (LR) emergence and development (Salisbury et al., 2007; Chen et al., 2016; van Gelderen et al., 2018, 2021). Salisbury et al. (2007) showed that mainly PHYB induces LR formation via auxin signaling and suggested that the inhibition of LR number under low R:FR might be caused by decreased auxin transport or responsiveness in the roots. Findings from Chen et al. (2016) demonstrated that LR development is induced by shoot illumination regardless of the light conditions in which the roots are cultivated, suggesting that a long-distance signal produced in the shoots causes LR formation. It was suggested that ELONGATED HYPOCOTYL 5 (HY5) transcription factor (TF), which is stabilized in the shoots under shade (Pacín et al., 2016), is transported to the roots, where it induces its own expression and regulates LR formation. Based on this observation, van Gelderen et al. (2018) demonstrated that HY5 locally represses LR development in the shade by controlling auxin-dependent pathways at the LR primordia. In a recent study, it was reported that the expression of hypocotyl-localized HY5 was insufficient to complement the LR growth defects seen in hy5 mutant Arabidopsis plants (Burko et al., 2020).
SAR imparts an important adaptive function to a plant under suboptimal conditions by allowing plants to compete for light. However, such adaptation comes at a cost. For instance, plants prioritize rapid stem and petiole elongation over immunity defense response to herbivores in the shoot, and thus shaded plants are more susceptible to microbial diseases and herbivory (Ballaré, 2014). This prioritization of growth responses over defense is likely to make use of the limited resources efficiently. The presence of pathogens or herbivores activate pattern-recognition receptors present on the cell surface to activate pattern-triggered immunity (PTI), which leads to the induction of salicylic acid (SA) and jasmonic acid (JA)-mediated pathways as a defense response (Ballaré et al., 2012). It has been demonstrated that defense responses including JA signaling are lowered in phyB mutant and WT plants exposed to low R:FR shade (Leone et al., 2014; Ortigosa et al., 2020).
Plant disease resistance or biotic stress and abiotic stress responses are primarily mediated by WRKY TFs (Pandey and Somssich, 2009). They constitute the largest family of plant-specific transcriptional regulators, acting as either repressors or activators (Bakshi and Oelmüller, 2014). Accumulating evidence shows that a large number of WRKY genes take center stage to regulate various aspects of plant innate immunity by responding to herbivores, PTI elicitors, regulation of defense-related SA and JA hormones, synthesis of defense-related compounds, and phytoalexins (Chi et al., 2013). Apart from their role in stress responses, WRKYs also have diverse biological functions in many plant processes not limited to nutrient homeostasis, seed and trichome development, embryogenesis, seed dormancy, senescence, etc. (Eulgem et al., 2000; Skibbe et al., 2008; Mao et al., 2011; Birkenbihl et al., 2018; Karkute et al., 2018; Viana et al., 2018; Chen et al., 2019). WRKY proteins are largely identified by the presence of a conserved WRKY DNA-binding domain defined by the WRKYGQK amino acid sequence. Apart from the WRKY domain, these TFs contain an atypical zinc-finger domain in their carboxyl-terminus (Rushton et al., 2010; Chen et al., 2019). WRKY TFs primarily bind to the W-box cis-elements in the promoter of their target genes (Ciolkowski et al., 2008; Rushton et al., 2010). Thus, WRKYs are essential regulators in responding to internal and external developmental signals as well as stresses.
To understand the molecular account of how low R:FR shade leads to the inhibition of primary root growth, we analyzed the whole genome transcriptome of the roots of Arabidopsis and tomato (Solanum lycopersicum) seedlings grown in the shade. We identified a core set of shade-induced genes in the roots of shaded plants, and most of them were also induced by abiotic and biotic stressors. The majority of the shade-induced genes contain W-box promoter elements and are considered the targets of WRKYs. Many WRKY family members were also significantly upregulated in the roots of shaded plants. To decipher the contribution of individual WRKYs in controlling root growth during the SAR, we overexpressed in Arabidopsis a large number of shade-induced WRKYs. We identified that WRKY26 and WRKY45 overexpression led to a constitutive-shaded, short primary root phenotype even in the absence of shade. In contrast, the overexpression of WRKY75 lead to a decrease in the LR number in the shade but did not affect the primary root growth. Interestingly, the overexpression of these WRKYs affected only the roots, and it did not lead to any hypocotyl elongation defects seen during the SAR. Similarly, like WRKYs, our study implicates ethylene hormone to be necessary to limit root growth but was insignificant for hypocotyl growth in the shade. In summary, we found that low R:FR shade induces a large number of WRKYs, particularly to restrict root growth and development. We hypothesize that the reduced growth of root organs helps the plant divert its critical resources to the elongating organs in the shoot to ensure competitiveness under limiting photosynthetic radiation.
Results
Shade-induced genes in the roots resemble biotic and abiotic stress-induced transcriptome
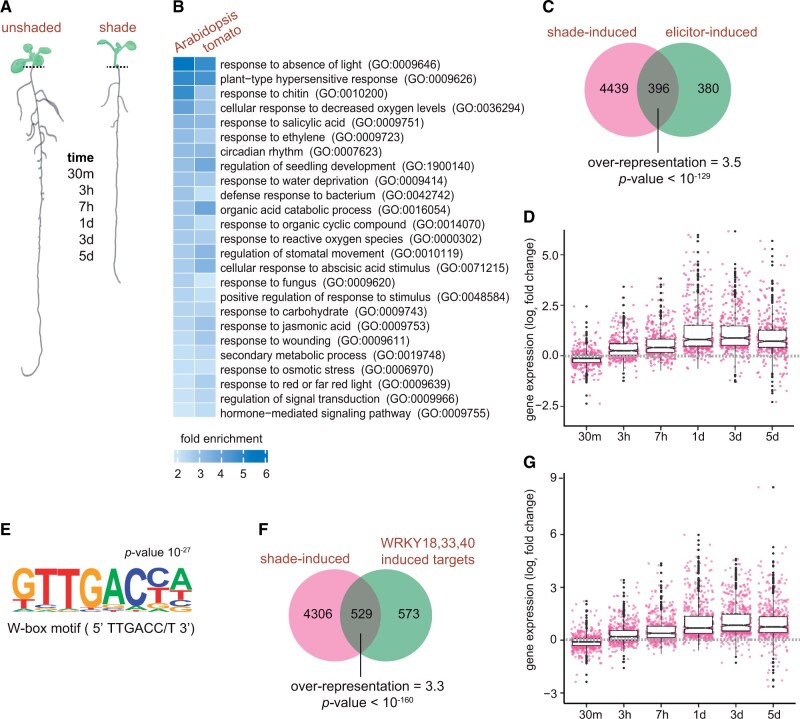
To determine how the low R:FR of vegetational shade affects root growth and development, we had performed a whole genome transcriptomic analysis using RNA-seq as a time course on the roots excised from Arabidopsis seedlings grown in white light (WL, unshaded; high R:FR) and white light supplemented with FR (shade; low R:FR) conditions (Figure 1A;Supplemental Figure S1A). The 5-d-old WL grown Arabidopsis seedlings were transferred to shade or mock-treated, then their roots were harvested after 30 min, 3 h, 7 h, 1 d, 3 d, and 5 d of treatment duration. Similarly, we performed a comparable experiment in 7-d tomato seedlings, and the root tissue was harvested from them after 3 h, 6 h, 12 h, and 24 h. Total RNA was isolated from these root tissues and the whole genome transcriptome analysis (RNA-seq) was performed using short-read sequencing. Gene expression matrices and statistically significant (false discovery rate; FDR <0.05) differentially expressed genes (DEG) were determined by comparing the shade and unshaded samples to its own developmental time point (Supplemental Table S1). Pearson’s correlation coefficient calculated from the fragments per kilobase of exon per million reads (FPKM) values for each biological replicate indicated a very high correlation (R > 0.9) between them (Supplemental Figure S1B). We identified a total of 4,835 DEGs that were induced along the time-course in Arabidopsis, and 2,523 in tomato in all the time points combined (Supplemental Figure S1C and Supplemental Table S1). Henceforth, we will refer these upregulated DEG as shade-induced genes. Next, we subjected the shade-induced genes up to 24-h time point for gene ontology (GO) analysis to assign them a biological function. Our GO analysis on the shade-induced genes in the roots was largely enriched and overrepresented with GO terms related to stress responses, defense against pathogens, and innate immune responses in both Arabidopsis and tomato (Figure 1C;Supplemental Table S2).
Figure 1.
Shade-induced genes in the roots resemble biotic and abiotic stress-induced transcriptome. A, Phenotypic representation of a 9-d-old Arabidopsis seedling grown under constant white light (unshaded) and shade (low R:FR); and the time-points used for RNA-seq analysis. Dashed lines indicate the region where the root tissue was excised from the shoot for RNA-seq analysis. B, GO terms of biological processes, commonly enriched among the genes upregulated in Arabidopsis and tomato roots during the first 24 h (30 m, 3 h, 7 h, 1 d; and 3 h, 6 h, 12 h, 24 h, respectively) under shade. C, Arabidopsis genes induced by the shade in the roots along the time-course (up to 5 d) and by various PTI-elicitors from a prior study (Bjornson et al., 2021). Overrepresentation and P-value were calculated based on hypergeometric distribution and Fisher’s exact test. D, Expression profile of common Arabidopsis shade-induced and elicitor-induced genes along the time-course. Values represent log2 fold change in the shade relative to unshaded control. E, de novo-enriched cis-motif element found in the promoters of the 396 genes induced by both shade and elicitors as determined in a previous study (Bjornson et al., 2021). F, Arabidopsis genes induced by the shade in the roots along the time-course (up to 5 d) and by WRKY18, WRKY33, and WRKY40 in response to immune response flg22 elicitor from a prior study (Birkenbihl et al., 2017). Overrepresentation and P-value were calculated based on hypergeometric distribution and Fisher’s exact test. G, Expression profile of common Arabidopsis shade-induced and WRKY-induced genes along the time-course. Values represent log2 fold change in the shade relative to the unshaded control. D, G, Boxplot components correspond to: box limits, first and third quartile; central line, median; whiskers, upper and lower 1.5× interquartile range; pink dots, individual genes; black dots, outliers.
As our GO analysis revealed that the shade-induced genes were also induced during biotic and abiotic stress, and plant’s defense against pathogens, therefore, we compared our dataset with publicly available published RNA-seq datasets, especially related to immunity and defense responses. In one of the comparisons, we chose a study in Arabidopsis that identified 776 common genes that are induced when treated with seven separate elicitors (3-OH-FA, flg22, elf18, nlp20, CO8, OGs, and Pep1) of PTI (Bjornson et al., 2021). More than half (51%) or 396 of the 776 elicitor-induced genes overlapped with our shade-induced genes (Figure 1C), representing an enrichment of 3.5-fold over the number of genes that would be expected by random chance (P < 10−129). These 396 genes, commonly induced by shade and elicitors of PTI displayed an increased temporal expression in roots of Arabidopsis seedlings exposed to shade (Figure 1D), suggesting that prolonged exposure to shade activates defense-like responses in the roots in the absence of pathogens.
Promoters of the shade-induced genes contain W-box elements
To obtain further insights on the nature of the genes that are universally responding to shade stimuli in the roots, we sought to identify the conserved cis-elements in the promoters of shade-induced genes. We performed de novo cis-motif analysis on the promoter sequences (500-bp upstream and 50-bp downstream) of the transcription start site of the shade-induced genes in Arabidopsis (Supplemental Figure S1C and Supplemental Table S1), as well as those overlapping with the PTI elicitor-induced genes (Figure 1C). We identified W-box motif [TTGACC/T] as one of the top enriched cis-element among the promoters of the shade-induced genes (P < 10−17; Supplemental Figure S2A), as well among the shade and PTI elicitor-induced genes (P < 10−27; Figure 1E). Approximately 33% of the promoters of the shade-induced genes in Arabidopsis roots contained W-box motifs. Interestingly, we did not identify the W-box motif in the promoters of the downregulated genes (Supplemental Figure S2B). Similarly, W-box is among the top enriched motifs in the promoters of tomato shade-induced genes (Supplemental Figure S2C). Therefore, considering that WRKYs are central to both biotic and abiotic stresses, we hypothesized that they are likely responsible for the induction of stress and defense-related gene expression program that we observed in the roots of shaded plants (Figure 1B). Consistently, 48% of the genes known to be directly regulated by WRKY18, WRKY33, and WRKY40 TFs, were also induced by shade (Figure 1, F and G;Birkenbihl et al., 2017). Among them, well-characterized defense marker genes, CYP71A12, MYB51, and PIP1 (Lakshmanan et al., 2012; Hou et al., 2014; Birkenbihl et al., 2017), were found to be substantially induced in the roots of Arabidopsis under shade (Supplemental Figure S2D). Combined, these results suggest that the shade induces genes in the roots that are also upregulated when a plant encounters abiotic and biotic stress, and a large proportion of these genes contain W-box promoter elements, which are binding sites for WRKY TFs.
Large number of WRKY TFs are induced in response to shade
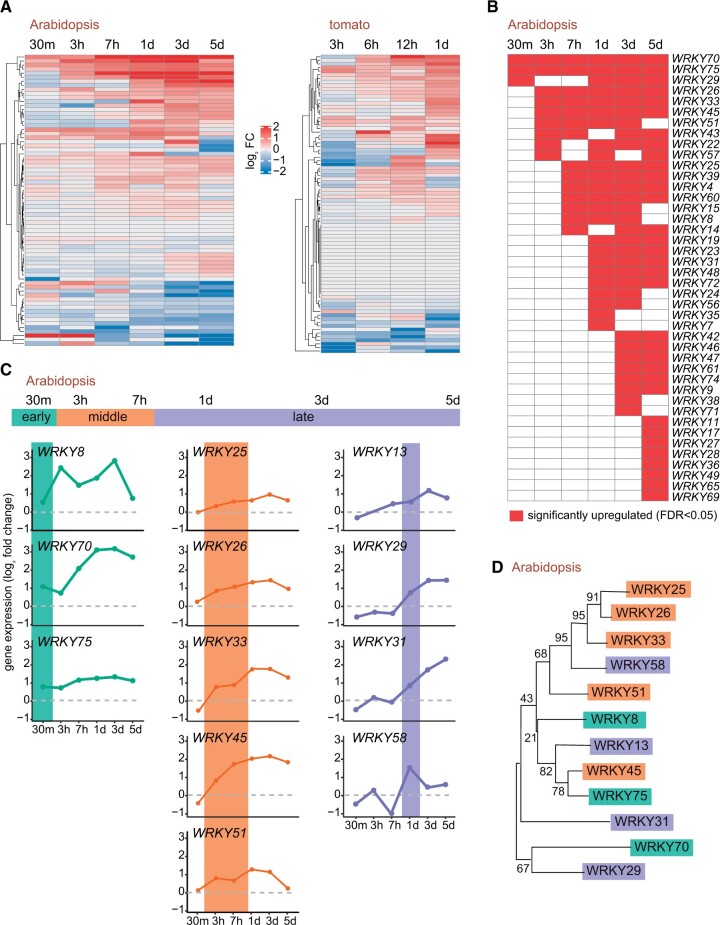
The RNA-seq and GO analysis on the shade-induced genes, and the discovery of W-box promoter elements in them, indicated the involvement of WRKY TFs in mediating root responses to shade. To test this hypothesis, first we surveyed the expression of all the WRKY genes in Arabidopsis and tomato in our transcriptomic data. We found a large number of WRKYs were induced along the time-course in response to shade. Thirty-three out of 74 WRKYs in Arabidopsis were significantly expressed (FDR <0.05) in one or more time points, similarly, 21 out of 83 WRKYs were upregulated in tomato (Figure 2, A and B; Supplemental Figure S3A).
Figure 2.
A Large number of WRKY transcription factors are induced in response to shade. A, Global transcriptional profile of WRKY gene family members in Arabidopsis and tomato roots in the shade. Values represent log2 fold expression of WRKY genes in the shade compared to its unshaded developmental control. B, WRKY genes that are significantly upregulated in the Arabidopsis roots in shade relative to its unshaded control (FDR ≤0.05). C, Transcriptional profile of the selected WRKYs consistently induced by shade, i.e. those upregulated above the threshold of 0.5 log2 fold relative to the unshaded control. Expression groups are color coded and characterized by the timing of their significant induction in the shade during the time course. D, Dendrogram of selected Arabidopsis WRKY proteins, classified as early, middle, and late as in (B). The phylogenetic tree was inferred by Maximum Likelihood and JTT model. Branch lengths are scaled according to number of substitutions per site. Numbers represent the percentage of trees in which the associated proteins clustered together considering 500 bootstrap replicates.
Next, to perform in-depth functional analysis of the shade-induced WRKYs in Arabidopsis, we sought to narrow down the candidates as large number of them were induced (Figure 2B). In order to do this, we selected WRKYs with a minimum threshold of 0.5 log2 fold-change induction relative to the unshaded control along the time-course. Using this parameter, we identified 12 WRKYs that were consistently upregulated in the shade (Figure 2C). We further classified these 12 genes in to three groups, namely, “early”, “middle”, and “late”, based on the time they were upregulated post shade treatment. WRKY8, WRKY70, and WRKY75 were upregulated within the first 30 min of the shade treatment, where we classified them as immediate early-induced genes. Next, WRKY25, WRKY26, WRKY33, WRKY45, and WRKY51 were classified as intermediate middle, as their expression was seen between 3 and 7 h of shading. Lastly, WRKY13, WRKY29, WRKY31, and WRKY58 were classified as late-induced genes as they were expressed only after 24 h of shading (Figure 2C). We also compared the Arabidopsis WRKY expression profile to its putative orthologs in tomato (Supplemental Figure S3, B and C). There were some similarities between tomato and Arabidopsis WRKY temporal expression patterns; however, several WRKYs showed statistically significant induction only at 24 h time point in the shade. It is possible that the temporal expression of WRKY genes could be delayed in tomato when compared to Arabidopsis and, thus, a longer time-course experiment in tomato could be more suitable to address ortholog-specific similarities. Nevertheless, it is clear that shade activates similar pathways in these two distantly related species.
Focusing on Arabidopsis, we investigated whether the early, middle, and late shade-induced WRKY genes were closely related phylogenetically. For this, we constructed a maximum likelihood phylogenetic tree based on the polypeptide sequences of the 12 WRKYs that were classified above and induced by shade. We observed that the pattern of WRKY expression did not reflect the phylogenetic relationship between them. For instance, WRKY75 and WRKY45 that are closely related to each other (Figure 2D), were induced in the middle and at earlier time points (Figure 2C). An exception to this was the branch comprising WRKY25, WRKY26, and WRKY33, which are phylogenetically close and were induced in the middle of the time course (Figure 2C). This was consistent with previous studies, which indicated that these WRKYs (25, 26, and 33) act redundantly in Arabidopsis’ response to high temperature, gibberellin (GA), and abscisic acid (Li et al., 2011; Zhang et al., 2015). It is not surprising, however, that closely related genes, such as WRKY45 and WRKY75, have different expression profiles. It is very common that paralogous genes initially diverge by changes in the promoter region, which leads to sub- or neo-functionalization of duplicated genes (Rosado et al., 2016; Teufel et al., 2016). Collectively, our results suggest that a large number of WRKY TFs are specifically induced in both Arabidopsis and tomato roots in response to the shade stimuli.
Shade-induced WRKY proteins accumulate in the roots and largely absent in the shoot
As known with the large gene families, various members of the WRKY genes are paralogous, and are documented for functional redundancy due to gene duplications, which complicates genetic analysis to determine the role of individual WRKY TFs (Eulgem et al., 2000; Zhang et al., 2015). In this scenario, we decided that the best strategy for studying the role of the shade-induced WRKYs (Figure 2C) in root growth during the SAR is by overexpressing them. It is documented that the overexpression of WT genes can also cause mutant phenotypes; be used to assess the impact of genetic alterations and gene activity in generating phenotypes and is comparable to traditional loss-of-function methods (Palatnik et al., 2003; Chua et al., 2006; Prelich, 2012; KaiserLi et al., 2015). Therefore, we generated transgenic Arabidopsis lines overexpressing the selected shade-induced 12 WRKYs as an mCitrine fluorescent protein fusion (WRKYox) under the control of the constitutive Arabidopsis UBIQUITIN 10 (UBQ10) promoter. We identified multiple independent transgenic lines with a single insertion of the transgene and we selected a minimum of three lines for further analysis, except for WRKY8ox and WRKY33ox, as we could not recover stable transgenic lines for them.
First, we performed immunoblot analysis to ensure that the transgenic lines for rest of the 10 shade-induced WRKYs were expressing full-length mCitrine fusion proteins, and not partial fusions or free mCitrine alone. Using total protein lysates obtained from the whole 5-d-old transgenic seedling grown in unshaded or exposed to the shade for 3–24 h, we performed immunoblot analysis using an anti-GFP antibody. Immunoblot analysis could detect the presence of full-length mCitrine fusion with WRKY 25, 26, 31, 45, 51, and 75 among the independent transgenic lines (Supplemental Figure S4). Differential protein accumulation was not observed in the shade and unshaded growth conditions in these transgenes. However, the specific protein for WRKY 13, 29, 58, and 70 and some indicated independent lines could not be detected in the immunoblot. This could be due to one or more reasons; transgenic protein expression was below the detection limit of the antibody used in the immunoblot assay, low expression of the transgene, dilution of the specific signal due to the use of the whole seedling lysates, or instability of the protein.
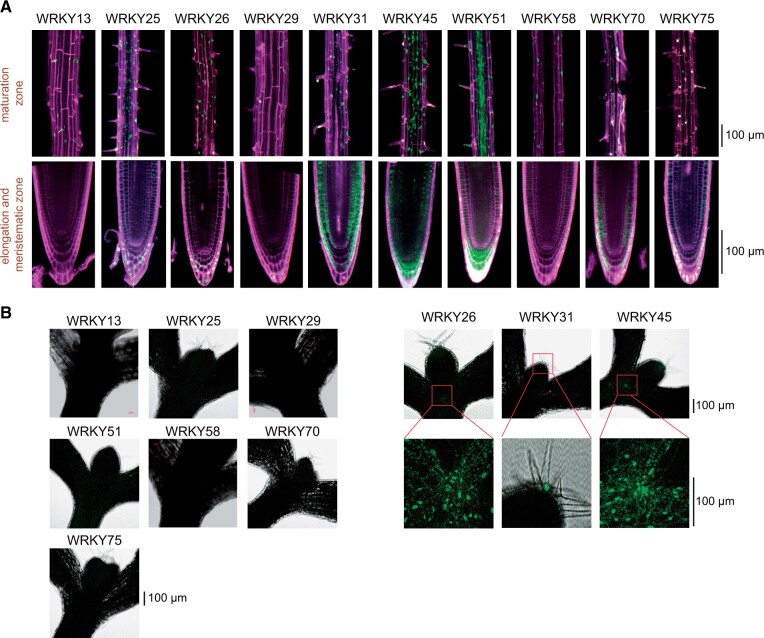
Nevertheless, we performed confocal microscopy to visualize WRKY-mCitrine fusion proteins in three independent transgenic lines for each of the 10 WRKYox that were exposed to a minimum of 24 h of shade. For all of the 10 WRKYs, we observed them to be present in the nucleus of the root epidermal cells within the maturation zone in shade (Figure 3A), consistent with their role as a nuclear-localized TF (Eulgem et al., 2000). As we had used a constitutive UBQ10 promoter to express these WRKYs, however, their protein expression and distribution varied considerably within the cell types of the roots (Figure 3A, upper part). For example, in the elongation and meristematic zones of the root (Figure 3A, lower part), WRKY25, WRKY45, and WRKY51 were detected in both the epidermis and cortex. In the meristematic zone, WRKY26 was detected only in the columella cells and WRKY58 was observed in the LR cap. In the shoots (Figure 3B), we detected fluorescence signal for WRKY26 and WRKY45, whereas WRKY31 signal was observed only in the trichomes. Remarkably, we did not detect any signals for rest of the WRKY proteins in the shoot. This varied expression profile might be due to regulation of these WRKYs post-transcriptionally or post-translationally. However, it is also likely that technical limitations of confocal microscopy or poor expression levels impede us from detecting WRKY-mCitrine fusions in all cell types.
Figure 3.
Shade-induced WRKY proteins accumulate in the roots. A and B, Confocal microscopy images of the indicated 5-d-old shaded Arabidopsis WRKYox (fused with mCitrine protein) transgenic seedlings. Prior to the shade treatment, the seedlings were grown in unshaded light for 4 d. A, Arabidopsis roots and B, Arabidopsis shoots largely encompassing the hypocotyl, petiole, and the first true leaves. Magenta color indicates the propidium iodide counterstaining of the cell wall and the green signal indicates mCitrine fluorescence.
Shade-induced WRKYs affect primary root and LR growth in shaded and unshaded conditions
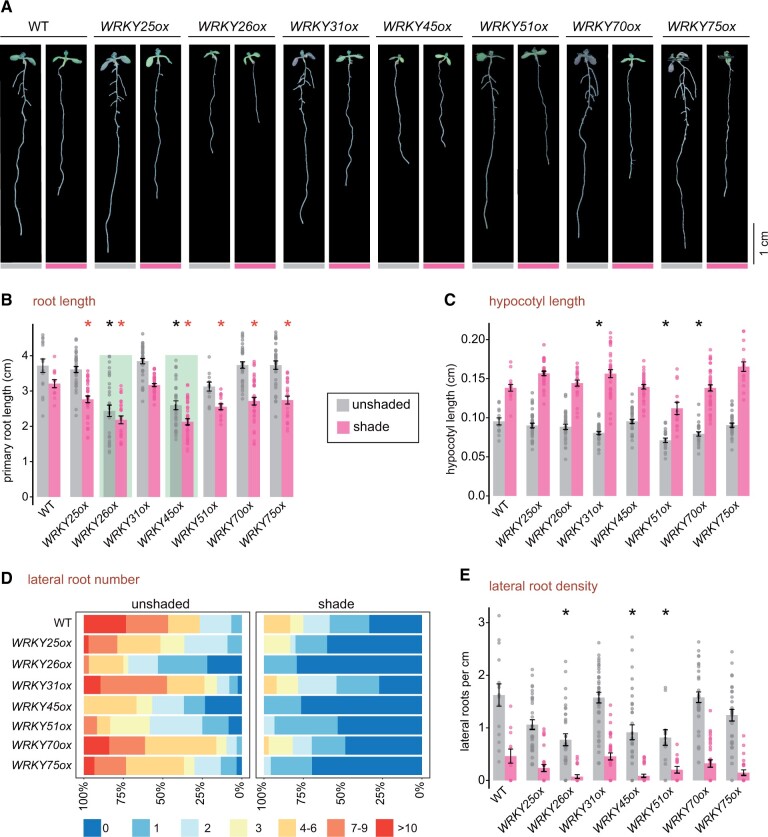
Since many WRKYs are upregulated in the roots of Arabidopsis and tomato seedlings in the shade, we sought to assess their functional contribution in regulating root growth. We chose to determine the effect of their overexpression in the low R:FR-mediated SAR, specifically on hypocotyl elongation and root growth inhibition. For each of the shade-induced selected WRKY overexpressors, we used three independent Arabidopsis transgenic lines (described in Figure 3) for phenotyping, except for WRKY51, for which we recovered only two separate lines (Supplemental Figure S5). We analyzed four phenotypic traits under unshaded and shade light conditions: length of the primary root, LR number, LR density, and hypocotyl length. We report the average phenotypic values in Figure 4, by combining the measurements from all the independent transgenic lines employed for each of the WRKYs.
Figure 4.
Shade-induced WRKYs primarily affect root growth in the shade and have no effect on the hypocotyl growth during the SAR. A, Phenotype of representative 9-d-old WT and WRKYox seedlings under unshaded light and shade (low R:FR). B, Primary root length of the indicated genotypes in cm; C, Hypocotyl length of the indicated genotypes in cm; D, Frequency of seedlings with 0–10, or more LRs; E, LR density as the number of LR per 1 cm of the primary root under unshaded or shade conditions in 9-d-old seedlings;. B–E represent means ± se of combined independent transgenic lines (UBQ10::WRKYox) for each candidate gene; dots represent individual data points; 26–72 individual seedlings were used per genotype. Asterisks represent significant difference (P < 0.05) with the WT for each light condition (black for unshaded and red for shade) in two-tailed t test with Benjamini–Hochberg correction for multiple testing. Green shading highlights root length reduction in WRKY26ox and WRKY45ox.
The 5-d-old seedlings grown in unshaded condition were transferred to the shade or mock-treated for 4 d and then their primary root length, LR number, and LR density were measured. Most of the WRKY overexpressing seedlings produced primary roots whose length was comparable to the WT (Figure 4, A and B). However, WRKY26 and WRKY45 overexpressors displayed a constitutive primary root growth and LR branching inhibition even in the absence of the shade stimulus (Figure 4, A and B). The primary root length of WRKY26 and WRKY45 did undergo a modest decrease in size in the shade compared to other WRKY overexpressors and the WT (Figure 4B; Supplemental Figure S5A). However, the root and hypocotyl growth of mutants that harbor T-DNA insertion in the protein coding region of WRKY26 and WRKY45 were indistinguishable from WT in the shade and unshaded light (Supplemental Figure S6, A–C). Consistently, transgene expression levels in the WRKY26ox and WRKY45ox did not vary with the light conditions as determined by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) analysis using transgene specific oligos that hybridize to the mCitrine tag. We observed an overall increase in WRKY26 and WRKY45 expression levels in the root as opposed to the whole seedling in response to shade by RT-qPCR analysis using oligos that recognize both the transgene and the WT gene (Supplemental Figure S6D).
Surprisingly, all the WRKY overexpressing transgenic seedlings did not have any measurable defects in their hypocotyl length and were comparable to the WT (Figure 4C; Supplemental Figure S5B) in the shade and non-shading control conditions. WRKY26ox and WRKY45ox, which displayed hypersensitive shorter roots in non-shading conditions, did not have any obvious hypocotyl growth defects in the shade. However, the expression of WRKY26 and WRKY45 protein was detected in the hypocotyl (Figure 3B;Supplemental Figure S6E). So, the lack of hypocotyl defects in WRKY26ox and WRKY45ox likely reflects their specialized role in the avoidance response, which is restricted in limiting root growth.
In all the 10 WRKY overexpressing seedlings, we observed reduced LR number in the shaded seedling compared to their unshaded counterparts, similar to the WT (Figure 4D; Supplemental Figure S5C). In WRKY26ox and WRKY45ox seedlings, significantly reduced to nearly absent LRs were noted (Figure 4, A and D), in the shade and nonshading conditions, similar to its constitutive primary root growth inhibition in these conditions (Figure 4, A and B). During the duration of our assay (9 d) in the shade, the LRs were not detected for WRKY26ox and WRKY45ox seedlings; in contrast, up to six LR could be seen in each WT seedlings (Figure 4DSupplemental Figure S5C). Although WRKY51ox and WRKY75ox did not influence the inhibition of the primary root growth, we observed a marked decrease in their number of LR in the shade (Figure 4D;Supplemental Figure S5C).
Of note, we could only detect a few statistically significant differences in the number of LRs in our transgenic lines. This is likely due to variation in LR produced by individual seedlings and accounted for by WRKY’s expression levels in the cells. Significant differences were not observed in the LR density, measured as a number of LR per 1 cm of the primary root (Figure 4E;Supplemental Figure S5D). This result was not particularly unexpected, considering that most WRKYox seedlings did not have much of an impact on the primary root growth and LR number, except for WRKY26, WRKY45, and WRKY75. WRKY51ox seedlings also displayed a slight shortening of the primary root and the hypocotyl, but these transgenic lines also showed variable and delayed seed germination, and other pleiotropic defects. Therefore, other confounding factors could be influencing the resulting phenotype of WRKY51ox in the shade (Figure 4, A–C).
The overexpression of WRKY13, WRKY29, and WRKY58 showed a slight increase in their root length, LR production, and LR density compared to the WT in the shade and nonshading conditions (Supplemental Figure S7, A–C). However, these observations were reinforced only in the WRKY13ox independent lines, but not on WRKY29ox and WRKY58ox individual seedlings (Supplemental Figure S7, D–F). Considering that these three WRKYs are shade-induced in the “late” stages of our time-course analysis (Figure 2C), it is plausible that they could have a secondary role or oppose the shade-mediated repression of root growth.
Together, our results here indicate the significance of WRKYs, especially the upregulation of WRKY26 and WRKY45 in the roots of shaded plants to limit their growth and not that of the hypocotyl. The overexpression of WRKY26 and WRKY45 resulted in reduced primary root growth and LR number under the control unshaded condition, as to resemble the phenotype of WT roots in the shade. The overexpression of WRKY75 had no substantial role in repressing the primary root growth, but had an effect by repressing LR emergence in the shade and not in unshaded growth.
Ethylene is required for root growth inhibition in the shade
“Response to ethylene” was one of the top enriched GO terms among the shade-induced genes in both Arabidopsis and tomato (Figure 1B). WRKY TFs integrate ethylene hormone responses along with environmental and developmental signals (Koyama, 2014). Furthermore, ethylene and components of the ethylene signaling pathway are required for efficient resistance towards certain plant pathogens. For example, ethylene-insensitive Arabidopsis mutant ein2 was more susceptible than WT plants to infection by Botrytis cinerea, a fungal pathogen (Thomma et al., 1999). Involvement of ethylene signaling and multiple WRKYs in response to senescence and high temperature have been documented (Li et al., 2011; Koyama et al., 2013; Koyama, 2014). Ethylene also regulates root growth by mostly restricting cell elongation (Růžička et al., 2007). Previous studies have determined the importance of ethylene in petiole elongation but not for hypocotyl elongation during SAR (Pierik et al., 2009; Das et al., 2016), although ethylene biosynthetic pathway was particularly upregulated in the hypocotyl in response to shade (Kohnen et al., 2016). Therefore, in light of this knowledge and observation, we tested the effect of ethylene and the components of ethylene signaling on root growth during shade avoidance.
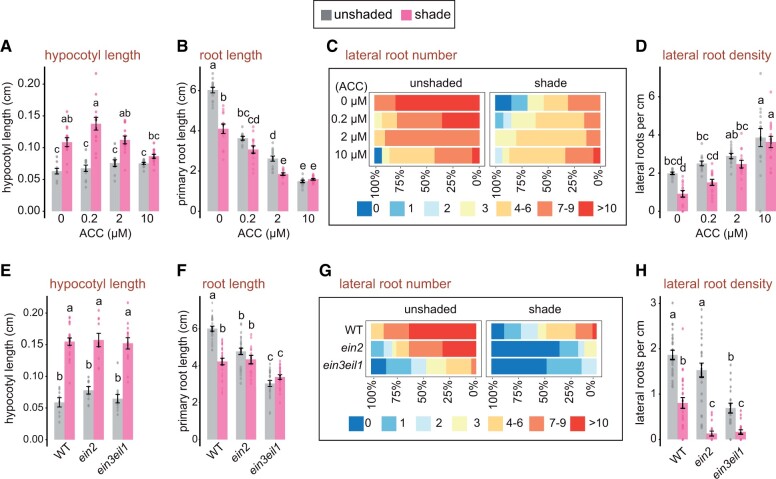
We grew WT seedlings in unshaded and shaded conditions on a growth media supplemented with different concentrations of 1-aminocyclopropane-1-carboxylic acid (ACC), a routinely used biosynthetic precursor of ethylene (Růžička et al., 2007). In the shade, a lower dose of 0.2 µM ACC had a stimulatory effect on the hypocotyl elongation. In contrast, higher doses of 2 and 10 µM had a moderate impact on the hypocotyl elongation (Figure 5A), confirming previous results (Das et al., 2016). But ACC treatment profoundly affected the primary root growth in the seedlings grown in both shade and unshaded conditions (Figure 5B). At 0.2 µM ACC, the primary root growth of unshaded seedlings was indistinguishable from that of shaded seedlings, having a shorter root length which was comparable to untreated roots in the shade. Increasing concentrations of ACC led to further attenuation of the root growth in both unshaded and shade conditions with similar root length, especially at 10 µM of ACC. ACC treatment, notably at 10 µM, led to a modest reduction in LR number in unshaded seedlings (Figure 5C). However, ACC had the opposite effect on shaded seedlings, as we observed increased LR density at 10 µM ACC (Figure 5D). At 2 and 10 µM ACC, the LR density in shaded and unshaded seedlings was similar (Figure 5D). Therefore, these results indicate that ethylene is required for root growth inhibition in the shade.
Figure 5.
Ethylene hormone signaling is required for the repression of root growth and development in the shade. A–D, Phenotype of unshaded and shaded WT seedlings treated with the indicated concentrations of ACC. E–H, Seedlings of the WT and ethylene signaling mutants grown in shaded and unshaded light. The phenotypic measurement represents 4-d-old seedlings grown in unshaded light and then transferred to the shade or mock-treated for 4 d. A and E, Hypocotyl length in cm; B and F, Primary root length in cm; C and G, Frequency of seedlings with 0–10, or more lateral roots; D and H, Lateral root density is the number of lateral roots per cm of the primary root. Values represent means ± se of 8–15 seedlings; dots represent individual data points. Different letters denote significant differences in post hoc Tukey’s test (analysis of variance P < 0.05).
Next, to further explore the importance of ethylene in regulating root growth in the shade, we analyzed mutants defective in ethylene signaling, namely, ein2 and ein3eil1 double mutant, respectively. ETHYLENE INSENSITIVE 2 (EIN2) is a crucial signaling transducer, and ETHYLENE INSENSITIVE 3 (EIN3) and EIL1 (EIN3-LIKE 1) are critical downstream TFs in the ethylene response (Dolgikh et al., 2019). Hypocotyl growth defect was not observed in ein2 and ein3eil1 seedlings in the shade (Figure 5E), agreeing with a previous study (Das et al., 2016). Interestingly, the primary roots of both ein2 and ein3eil1 mutants did not respond to shade-induced growth inhibition, regardless of the shade or unshaded growth conditions (Figure 5F). The root phenotypes of ein2 and ein3eil1 further resembled WRKY26ox and WRKY45ox seedlings (Figure 4, B–E). Accordingly, ein2 and ein3eil1 mutants presented much fewer LRs and LR density compared to the WT, both in the shade and nonshading conditions (Figure 5, G and H). Together, the data presented in Figure 5 indicate that ethylene and its associated signaling are required to restrict root growth in the shade. Also, the data support the requirement of ethylene signaling along with WRKYs, analogous with the plant defense responses.
Roots of WRKY26ox and WRKY45ox are sensitive to ACC
To further dissect the nexus between WRKYs and ethylene, we tested whether ethylene can trigger the expression of WRKY45 and WRKY26, whose overexpression resulted in root-specific phenotypes in the shade and unshaded light (Figure 4B). WT Arabidopsis seedlings were treated with 2 µM ACC in the shade or unshaded light and RNA was extracted from the whole seedlings or from dissected roots. RT-qPCR was performed to measure the expression levels of WRKY26 and WRKY45 along with the ethylene responsive ERF1B marker gene. ERF1B displayed increased expression in the root after ACC treatment when compared to the mock control and unshaded condition (Figure 6A). Likewise, we observed an increased expression of WRKY45 in the unshaded and shaded WT seedlings treated with ACC (Figure 6A). However, the increased expression of WRKY26 was not statistically supported in our RT-qPCR assay on the RNA from the roots. In whole seedlings, ACC triggered the expression of ERF1B in WT seedlings, but not in WRKY26ox and WRKY45ox. But we did not observe increased expression of WRKY26 and WRKY45 in whole seedlings (Figure 6B). Next, we measured the primary root length of WRKY26ox and WRKY45ox supplemented with different amounts of ACC. The roots of WRKY26ox and WRKY45ox showed increased sensitivity toward ACC in both shaded and unshaded light when compared to the WT (Figure 6C). ACC treatment also affected LR number and density in WRKY26ox and WRKY45ox (Supplemental Figure S8, A and B). These results indicate that ACC, precursor of ethylene has the ability to induce WRKY45 in the root in the shade as well as in unshaded light. In addition, ACC further reduced the root length of WRKY26ox and WRKY45ox transgenic lines indicating that ethylene and WRKYs likely work together or in parallel to inhibit root growth.
Figure 6.
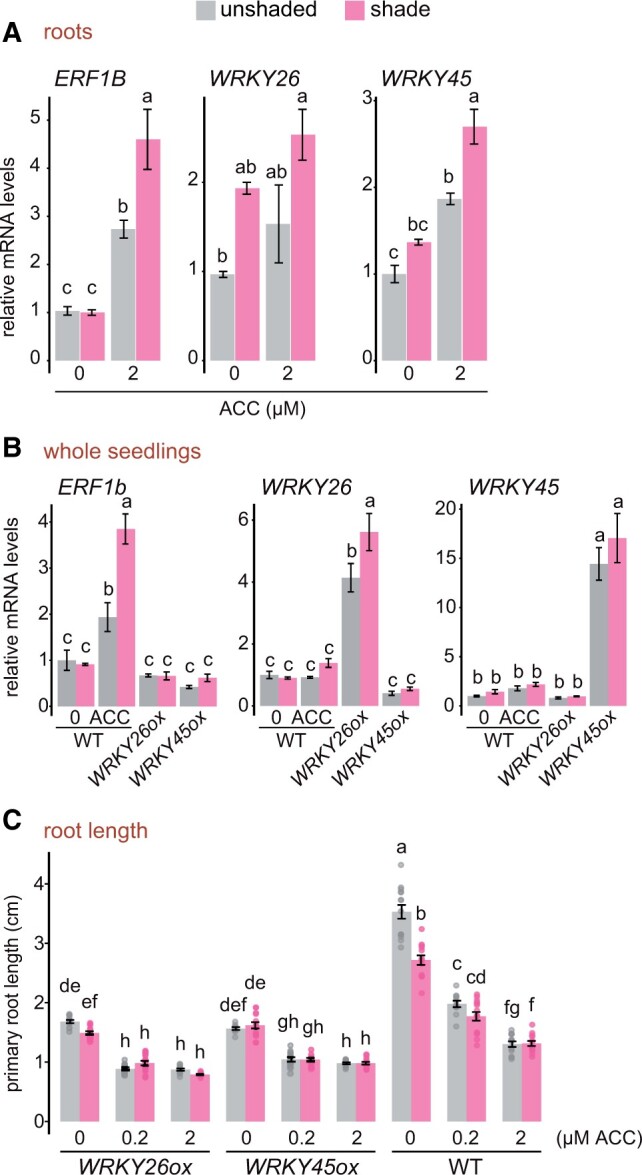
Ethylene further represses root growth in WRKY26ox and WRKY45ox lines. A and B, RT-qPCR analysis in 4-d-old seedlings grown in unshaded light and then treated with shade and/or ACC (2 µM) as indicated; C, Primary root length in centimeter of unshaded and shaded seedlings treated with the indicated concentrations of ACC. The phenotypic measurement represents 4-d-old seedlings grown in unshaded light and then transferred to the shade or mock-treated for 3 d. Values represent means ± se of three biological replicates (composed of 10–20 seedlings each) for A and B, and 9–15 seedlings for C. Dots represent individual data points. Different letters denote significant differences in post hoc Tukey’s test (analysis of variance P <0.05).
Discussion
Mechanisms underlying stem and petiole elongation under shade have been widely studied for several decades (Hornitschek et al., 2009; Pierik et al., 2009; Li et al., 2012; Pedmale et al., 2016). However, our understanding of how shade perceived by the above-ground shoots leads to the reduced growth of the belowground primary root and LR has been limited. This study presents evidence that many WRKY genes are transcriptionally upregulated in the roots of shaded Arabidopsis and tomato plants. We further demonstrate that several WRKYs (26, 45, 75) and ethylene function in restricting root and LR growth but did not affect hypocotyl elongation in the shade.
We discovered genes induced by biotic and abiotic stressors overlapped with a large proportion of the shade-induced genes in the roots of Arabidopsis and tomato seedlings grown in the shade (Figure 1B). PTI-induced genes also coincided with a significant portion of the shade-induced genes in the roots, which are known to be regulated by WRKY TFs (Figure 1, C–G). Importantly we found W-box promoter elements in a large number of shade-induced genes in the roots (Figure 1E; Supplemental Figure S2A), suggesting the involvement of WRKYs in the reprogramming of the gene expression to restrict root growth, typically observed during SAR. Furthermore, a large number of WRKY genes were significantly (FDR <0.05) upregulated in the roots of both shaded Arabidopsis and tomato, progressively increasing through the time course in the shade (Figure 2; Supplemental Figure S2). To identify the contribution of shade-induced WRKYs in regulating root growth in the shade, we performed functional analysis on a select 10 WRKY members by overexpressing them. We chose overexpression as an alternate yet powerful tool to generate mutant phenotypes (Chua et al., 2006; Prelich, 2012) and also to overcome known functional and genetic redundancy among the WRKY gene family members and potential gene-compensation (Rushton et al., 2010; Zhang et al., 2015). The overexpression of WRKY26 and WRKY45 led to a retarded root growth and LR emergence, irrespective of shade or unshaded light. WRKY26ox and WRKY45ox seedlings had a constitutive shade avoiding shorter primary root and reduced LR in unshaded light, mimicking a WT seedling in the shade. Importantly, in WRKY75ox seedlings, there was no effect on the primary root length, but a marked reduction in LR number was seen, similar to WRKY26 and WRKY45 overexpressors. But, none of the 10 shade-induced WRKYs that we characterized affected hypocotyl elongation, indicating that the roles of these WRKYs are primarily limited to regulate root growth during the SAR.
Our results here demonstrate that phenotypic activation of WRKY TFs is feasible as a general approach to identify their functional roles in plant growth and development (Figure 4). Previous studies have shown enhanced tolerance toward pathogens, salt, and drought by overexpressing WRKYs that were under investigation in Oryza sativa (rice), Glycine max (soybean), and Arabidopsis (Rushton et al., 2010). However, apart from this study, only a few prior reports have associated WRKYs with light signaling and adaptation. For instance, Arabidopsis WRKY18 and WRKY40 have been shown to co-localize with PHYB and PHYTOCHROME-INTERACTING FACTORS (PIFs) in the nuclear speckles or photobodies, but their role in red/far-red light signaling is unknown (Geilen and Böhmer, 2015). WRKY40 is required for adaptation towards high light stress in Arabidopsis (Aken et al., 2013), and WRKY22 is involved in dark-induced senescence (Zhou et al., 2011).
We characterized root-specific and shade-induced WKRYs by overexpressing them in Arabidopsis. We reasoned that overexpression of a WT protein can also cause mutant phenotypes, which is an alternative yet powerful tool to traditional loss-of-function analysis to infer gene function, especially when loss-of-function opportunities are not available (Palatnik et al., 2003; Prelich, 2012; KaiserLi et al., 2015). However, the overexpression of several shade-induced WRKYs produced no observable mutant primary root and LR growth phenotypes. Out of the 10 WRKYox lines that we characterized, we observed primary root-specific phenotypes only in two of them, for WRKY26 and WRKY45 respectively. Importantly, hypocotyl growth was not affected in WRKY26ox and WRKY45ox in the shade indicating their specificity in regulating root growth in spite of overexpression. Among the independent transgenic plants for a given WRKY, we observed variation in their protein expression. For example, among the three independent lines we characterized for WRKY26ox, line #2 had the strongest root phenotype (Supplemental Figure S5A) and protein levels in whole seedlings (Supplemental Figure S4). The variation in protein expression in these transgenic lines could be due to the positional effect of the transgene in the genome. WRKY51ox seedlings had detectable protein levels by immunoblot analysis, but they exhibited poor seed germination and we were unable to phenotype them adequately due eliminate confounding interpretation. Our microscopic analysis revealed that WRKY26 and WRKY45 were expressed in most of the cell types in the shoot. However, even with the use of constitutive expression, transgenic protein in several WRKYox lines was not detected or they were below the detection limit in our immunoblot assay and confocal microscopy. This lack of protein detection could be attributed to post-translational degradation or post-transcriptional regulation in some of the WRKYox lines. Therefore, we cannot completely rule out the lack of phenotypes in many WRKY overexpressing lines (e.g. in WRKY13ox, WRKY29ox, WRKY58ox, and WRKY70ox) could be attributed to lack of optimal protein expression or accumulation. The lack of phenotypes in some WRKYs (e.g. WRKY31 and WRKY75), could be due to the absence of required activating factor, which had to be overexpressed along with the WRKYs. Several WRKYs are also known to be regulated by Ca2+ and bind to 14-3-3 proteins (Rushton et al., 2010). Another aspect could be the feedback loops, which could interfere with WRKYs as several reports point that WRKYs are capable of binding to their own promoters or of other WRKY genes in response to stress (Skibbe et al., 2008; Rushton et al., 2010; Li et al., 2011, 2015; Birkenbihl et al., 2018). Expression and post-translational modification could further limit the function of the shade-induced WRKYs when overexpressed. Epitope tags are routinely fused with TFs, and they rarely interfere with the function of the TFs as determined by various in vitro and in vivo experiments such as chromatin immunoprecipitation-sequencing, etc. However, we cannot rule out completely whether the fusion of the WRKYs with mCitrine could be interfering with their activity. However, in future studies, tissue or organ-specific expression of WRKYs, analysis of multi-genic or higher-order mutants of WRKYs will help decipher the exact contribution of shade-induced WRKYs in the SAR.
Previously, WRKY26 was identified as a positive regulator of thermotolerance in Arabidopsis plants, working synergistically with WRKY25, WRKY33, and ethylene signaling (Li et al., 2011). In this literature, overexpression of WRKY26 led to the reduction of fresh weight in adult Arabidopsis plants, mirroring our results, where its overexpression led to shorter roots in the shade and unshaded light (Figure 4). Arabidopsis WRKY45 is a positive regulator of GA-mediated age-induced leaf senescence (Chen et al., 2017), and has been implicated in the activation of PHOSPHATE TRANSPORTER1;1 (PHT1;1) in the roots of plants undergoing phosphate deficiency (Wang et al., 2014). Wang et al. (2014) further showed that WRKY45ox lines had shorter primary roots in the presence of arsenate. This result is similar to our study, where WRKY45ox led to constitutive primary root length shortening in the shade and unshaded light. Furthermore, we report that WRKY75 overexpression had no effect on the primary root length but caused a reduction in LR frequency in seedlings grown only in the shade but not in unshaded conditions (Figure 4E; Supplemental Figure S5D). Silencing of WRKY75 by RNAi resulted in more LR than WT Arabidopsis but no differences in the primary root length (Devaiah et al., 2007). Also, WRKY75, like WRKY45, can promote leaf senescence, participating in a positive feedback loop with hydrogen peroxide and SA to accelerate leaf senescence (Guo et al., 2017). WRKY75 is also a positive regulator of GA-mediated control of flowering time (Zhang et al., 2018). Surprisingly, although WRKY45 and WRKY75 have overlapping roles in phosphate acquisition, leaf senescence, and GA-mediated signaling, yet, only WRKY45ox seedlings displayed inhibition of root length, indicating that many WRKYs share many overlying as well as divergent functional roles.
Ethylene is necessary for petiole elongation during the SAR but had a limited effect on hypocotyl elongation (Pierik et al., 2009; Das et al., 2016). Our results (Figure 5) show that ethylene and its associated signaling are required to restrict root growth in the shade, suggesting an organ-specific role of ethylene signaling in the shade. Ethylene signaling is critical for a plants’ defense responses and molecular links between this hormone and WRKY activity have been proposed (Bakshi and Oelmüller, 2014). Ethylene induces the expression of several WRKYs, and WRKY25, 26, and 33, induce the expression of EIN2, forming a positive feedback loop (Li et al., 2011, 2013). Here, ACC, precursor of ethylene was capable to induce the expression of WRKY45 in absence of the shade in the root. Expression of EIN3 is induced by ethylene via EIN2, and also by the defense-hormone JA, and by light via PIF4 and PIF5 basic helix-loop-helix TFs (Li et al., 2013; Sakuraba et al., 2014). Our data showed that ethylene-treated WT seedlings and ein2 and ein3eil1 had similar root phenotypes to WRKY26ox and WRKY45ox lines in the shade (Figures 4 and 5; Supplemental Figure S5). In addition, WRKY26ox and WRKY45ox were more sensitive to ACC compared to the WT. WRKYs can function up, and downstream of the many phytohormone pathways (Antoni et al., 2011). Therefore, it is conceivable that ethylene and the shade-induced WRKYs act in concert to antagonize root growth and development, and signaling dependent upon auxin, brassinosteroid, and cytokinin hormones in the shade.
WRKYs are plant-specific TFs and are central components of the plant’s resistance to pathogens and responses to abiotic and biotic stresses (Bakshi and Oelmüller, 2014). Arabidopsis and tomato genomes have 74 and 83 WRKY genes, respectively, and a great diversity of WRKYs allows plants to cope with various adverse conditions (Bakshi and Oelmüller, 2014). WRKYs have been well studied in their involvement with biotic stress compared to abiotic responses (Bakshi and Oelmüller, 2014). SAR is also stressful for the plant, as it leads to photosynthetic impairment and reduced carbon acquisition, reducing the overall fitness of the plant (Smith, 1982; Smith, 2000). In addition, low R:FR SAR downgrades plant defense against pathogens and herbivorous insects in the shoots (Moreno et al., 2009; Ballaré et al., 2012; Courbier and Pierik, 2019). Likewise, other studies have shown that many WRKY genes are rapidly induced in response to wounding, drought, salinity, osmotic, cold, carbon starvation, and heat stress (Pandey and Somssich, 2009; Bakshi and Oelmüller, 2014; Rinerson et al., 2015; Viana et al., 2018). Aptly, our finding on the speedy upregulation of WRKY gene expression in the roots of shaded plants is conceivable. During SAR, stress-like responses likely help the plant to relocate critical resources from the root to the growing shoot organs in order to maintain competitiveness. Future studies will be required to determine the nature of the signal downstream of the photoreceptors that leads to the induction of many WRKY genes in the shade. Also, which are the specific gene targets of the shade-induced WRKYs required to repress root growth and development in the shade?
Conclusions
Low R:FR shade represses root growth and development by triggering gene expression changes in tomato and Arabidopsis, mainly resembling the transcriptional changes caused by biotic and abiotic stressors. These shade-induced genes in the roots are known to be regulated by WRKY TFs, and many WRKYs were significantly upregulated in the shade. Here, we report the involvement of crucial WRKY TFs, namely WRKY26, WRKY45, and WRKY75, along with ethylene in the SAR to inhibit primary root and LR growth. These factors had no involvement in regulating hypocotyl elongation.
Materials and methods
Plant material, growth conditions, and light treatments
Arabidopsis (A. thaliana) genotypes used in this work are in Columbia WT (Col-0) ecotype background. Seeds of ein2-5 and ein3-1 eil1-1 double mutant were obtained from Dr Hong Qiao (University of Texas, Austin). T-DNA insertional mutant for wrky26 (SAIL_121_C01C1) and wrky45 (CS333963/GK-684G12) were obtained from Arabidopsis Biological Resource Center (ABRC; Ohio). For phenotyping of Arabidopsis seedlings, seeds were surface sterilized with 70% (v/v) ethanol and 0.1% triton X-100 (v/v) and rinsed several times with sterile water. Seeds were plated on 0.5× Linsmaier and Skoog (LS) medium (HiMedia Laboratories) pH 5.8 containing 0.8% phyto agar (w/v). Shaded and unshaded light conditions were used as described previously (Tao et al., 2008). Briefly, Petri plates containing growth medium with the seeds were then stratified at 4°C for 4–5 d before being placing them vertically in the growth chamber (Percival) with constant white growth light from a LED source (unshaded; PAR ∼120 µmol m−2 s−1) at 22°C. After 4 d, seedlings were either kept under unshaded control light or transferred to constant stimulated low R:FR shade where growth light was supplemented with FR (R:FR ratio of 0.35) for 1 d (microscopy and immunoblot analysis) or 3–5 d (phenotypic measurements). Light measurements were obtained using a full quantum sensor (Apogee Instruments). For phenotypic measurements, Petri plates with 7- to 9-d-old seedlings were scanned using a flatbed scanner (Epson V600), primary root length and later root (LR) numbers (counted as emerged LR) were obtained with SmartRoot (Lobet et al., 2011) and hypocotyl length was measured using NIH ImageJ software.
Cloning and generation of transgenic lines in Arabidopsis
Arabidopsis root tissue cDNA library was used as a template to amplify and clone the coding sequence of WRKY13, WRKY25, WRKY26, WRKY29, WRKY31, WRKY45, WRKY51, WRKY70, and WRKY75; while Arabidopsis genomic DNA was used as a template to amplify WRKY58. PCR was performed using KOD polymerase (Toyobo) with the primers listed in Supplemental Table S3 and introduced in to the Gateway donor vectors, either pDONR207 or pDONR221 (Thermo Fisher Scientific) using BP clonase II enzyme (Thermo Fisher Scientific). Multisite Gateway reaction using LR clonase II mix (Thermo Fisher Scientific) was performed to combine the donor constructs with either pB7m34GW or pK7m34GW binary vector (Karimi et al., 2007) along with the UBQ10 promoter in pDONR P4-P1R donor vector and Citrine in pDONR P3-P1R vector (destination vectors used for each WRKY gene are listed in Supplemental Table 2). Destination constructs were introduced in Agrobacterium tumefaciens to transform Arabidopsis using the floral dip method (Clough and Bent, 1998). T1 transgenic Arabidopsis plants were selected on 0.5× LS medium supplemented with either kanamycin or basta according to the vector used for transformation (Supplemental Table S3). Segregation analysis was performed on T2 plants grown on the selective agar media and lines carrying a single copy of the transgene were propagated further and the T3 seeds were used for experiments and other analysis.
Immunoblot analysis of transgenic lines
Arabidopsis seedlings were grown under constant white light for 4 d and either kept in unshaded conditions or shade for additional 24 h. Twenty excised roots or whole seedlings were harvested from 5-d-old plants and immediately frozen in liquid nitrogen. Samples were ground to a fine powder, resuspended in 2× lithium dodecyl sulfate (LDS) sample buffer (53-mM Tris–HCl, 70.5-mM Tris base, 1% (v/v) LDS, 5% (v/v), glycerol, 0.255-mM ethylenediaminetetraacetic acid (EDTA), 0.11-mM Serva Blue G250, 0.0875-mM phenol red, pH 8.5) with 50-mM TCEP and heated to 90°C for 10 min, cooled to room temperature and then centrifuged for 5 min to obtain the total protein lysate. For electrophoretic separation of proteins, equal amount of total protein was loaded on 10% (v/v) Bis–Tris polyacrylamide gel and electrophoretically separated using 3-(N-morpholino)propanesulfonic acid-sodium dodecyl sulfate (MOPS-SDS) buffer (2.5-mM MOPS, 2.5-mM Tris base, 0.005% [w/v] SDS, 0.05-mM EDTA). Separated proteins were then transferred electrophoretically to a nitrocellulose membrane (GE Lifesciences) using transfer buffer (10% [v/v] methanol, 1.25-mM bicine, 1.25-mM Bis–Tris base, 0.05-mM EDTA). After transfer, the membrane was stained with ponceau red, imaged, and blocked with 5% (w/v) nonfat dry milk prepared in Tris-buffered saline with 0.05% (v/v) Tween-20 (TBST) for 30 min. Next, membranes were incubated for 1 h in 1% (w/v) milk prepared in TBST with anti-GFP antibody (Roche). The blot was washed three times with TBST and incubated for 30 min in 1% (w/v) milk prepared in TBST with anti-mouse HRP conjugate (Bio-Rad). Chemiluminescent detection was performed using SuperSignal West Dura Extended Duration (Thermo) HRP substrate to detect the WRKY-mCitrine fusion protein.
Confocal microscopy
Stable transgenic Arabidopsis seedlings expressing UBQ10::WRKY-Citrine in their T3 generation were grown in unshaded light for 4–5 d and transferred to shade for 24 h. Seedlings were stained with 5 µg·mL−1 propidium iodide (PI) and then mCitrine and PI fluorescence were detected in a high-resolution laser scanning confocal microscope (LSM900 with Airyscan2, Zeiss) using 488 and 561 nm lasers along with BP 620/60 emission filter.
Phylogenetic analysis
Protein sequences of the 12 candidate Arabidopsis WRKYs were retrieved from The Arabidopsis Information Resource (TAIR) database (Garcia-Hernandez et al., 2002). Sequences were aligned with Clustal-W algorithm, and the phylogenetic tree was inferred by using the Maximum Likelihood method and JTT matrix-based model in Mega X software (Kumar et al., 2018).
Short-read mRNA-sequencing (RNA-seq) and analysis
The raw short-read sequencing data and expression files are available in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database with accession number GSE175963. In Arabidopsis, as described earlier, 5-d-old seedlings grown in continuous unshaded light were mock-treated or exposed to simulated shade (R:FR ∼0.6) at the indicated time points and the roots were harvested using a razor blade and flash frozen in liquid nitrogen. For tomato (S. lycopersicum, M82 cultivar), seedlings were grown in the dark for 3 d followed by 4 d in constant unshaded light at 25°C before mock-treatment or shade treatment (R:FR ∼0.3) and then roots were collected at the indicated time points and flash frozen in liquid nitrogen. Total RNA from Arabidopsis roots was extracted using RNeasy Micro kit (Qiagen). Total RNA from tomato roots was extracted using ReliaPrep RNA kit (Promega). We prepared short-read sequencing libraries from two biological replicates for each time point and treatment with either Ultra II DNA Library Prep Kit for Illumina (New England Biolabs) or TruSeq RNA-seq library kit (Illumina) according to manufacturer’s instruction. All sequencing libraries for each species were pooled together and sequenced using 1 × 76 bp in NextSeq 500. Single short reads were mapped against Arabidopsis (TAIR 10) or tomato (ITAG 4.0) reference genomes using STAR v2.7.2 (Dobin et al., 2013). Assembly and quantification of transcripts was done with Cufflinks v2.1.1 (Trapnell et al., 2012). Pearson’s correlation (Supplemental Figure S1B) was used with the FPKM values to evaluate reproducibility of biological replicates, which displayed very high correlation (R < 0.98 in most cases, with lowest R < 0.94) which exceeds the ENCODE standard of 0.90 (Dunham et al., 2012). Relative gene expression was calculated as log2 fold-change in shade relative to unshaded control for each time point using Cuffdiff (part of Cufflinks v2.1.1). Genes were considered differentially expressed if FDR-adjusted P-value <0.05.
De novo identification of cis-motifs
De novo cis-motifs in the promoters of differentially expressed genes were identified with HOMER (Heinz et al., 2010) by analyzing the 500-bp upstream and 50-bp downstream of the transcriptional start site.
Computational analysis and graphics generation
GO term enrichment was performed on Panther Classification System (Mi et al., 2020) using Fisher’s exact test and Bonferroni correction to obtain Biological Processes enriched relative to Arabidopsis reference genome. Since tomato GO annotation is comparatively poorer, we generated a list of Arabidopsis and tomato orthologs (Supplemental Table S2). We then used the Arabidopsis accessions corresponding to tomato genes upregulated by shade for the GO analysis. We then compared the list of significantly (Bonferroni correction; P < 0.05) enriched GO terms between the species and manually filtered based on hierarchy to remove term redundancy. All GO terms retrieved are reported in Supplemental Table S2. R environment (R Foundation) and its packages (ggplot2, pheatmap, ComplexHeatmap, dendsort, rstatix, ggpubr, dplyr, reshape2, tidyverse, RColorBrewer, circlize, ggfortify, gridExtra) were used for statistical analysis and to visualize results.
ACC treatment
For ACC treatments, WT, WRKY26ox, or WRKY45ox seedlings were grown for 4 d as previously described in unshaded conditions, then transferred to media containing 0, 0.2, 2, or 10 µM of ACC (Sigma). Then, they were either kept in unshaded or shaded conditions for 3–4 d, as indicated. Seedlings on the Petri plates were then scanned for phenotypic analyses.
RT-qPCR analysis
Whole seedlings or roots of Arabidopsis WT, WRKY26ox or WRKY45ox were used for RT-qPCR analysis. RNA was extracted using Trizol (Thermo) and DNaseI (New England Biolabs) was used to eliminate genomic DNA contamination. cDNA was synthesized using iScript cDNA Synthesis Kit (BioRad) according to manufacturer instructions. qPCR was carried out in the QuantStudio 6 Pro PCR system (Thermo) using Power SYBR Green Master Mix (Thermo) in a 10 µL reaction volume. Expression values were normalized with ACTIN7 and PP2A reference genes using the 2–ΔΔCt method. All primers and accession numbers can be found in Supplemental Table S3.
Statistical analysis
Most statistical analyses were performed in RStudio. Two-way analysis of variance and pairwise post hoc Tukey’s analysis were performed in InfoStat statistical software (InfoStat). Phenotypic data were analyzed by comparing the means between treatments or genotypes according to the test specified at the figures.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers WRKY13 (AT4G39410), WRKY25 (AT2G30250), WRKY26 (AT5G07100), WRKY29 (AT4G23550), WRKY31 (AT4G22070), WRKY45 (AT3G01970), WRKY51 (AT5G64810), WRKY58 (AT3G01080), WRKY70 (AT3G56400), WRKY75 (AT5G13080), EIN2 (AT5G03280), EIN3(AT3G20770), EIL1(AT2G27050), Actin7 (AT5G09810), PP2A (AT1G13320), ERF1B (AT3G23240), SlWRKY04 (Solyc05g012770), SlWRKY05 (Solyc03g104810), SlWRKY06 (Solyc02g080890), SlWRKY13 (Solyc04g051540), SlWRKY16 (Solyc02g032950), SlWRKY29 (Solyc08g081610), SlWRKY31 (Solyc06g066370), SlWRKY33 (Solyc09g014990), SlWRKY38 (Solyc02g094270), SlWRKY51 (Solyc04g051690), SlWRKY71 (Solyc02g071130), SlWRKY75 (Solyc05g015850), SlWRKY80 (Solyc03g095770), SlWRKY81 (Solyc09g015770).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcriptional changes induced by shade in the roots.
Supplemental Figure S2. Shade induces transcriptional changes via similar TFs in Arabidopsis and tomato.
Supplemental Figure S3. Shade induces the expression of a large group of WRKYs in tomato.
Supplemental Figure S4. WRKYox–Citrine protein levels are unaffected by shade.
Supplemental Figure S5. Phenotype of independent Arabidopsis WRKYox transgenic lines in the shade.
Supplemental Figure S6. Expression dynamics of WRKY26ox and WRKY45ox.
Supplemental Figure S7. Overexpression of WRKY13, 29, and 58 does not affect root growth.
Supplemental Figure S8. Ethylene further represses root growth in WRKY26ox and WRKY45ox lines.
Supplemental Table S1. Differential expression in Arabidopsis and tomato roots in response to shade.
Supplemental Table S2. GO terms enriched among the shade-induced genes in roots of Arabidopsis and tomato.
Supplemental Table S3. Oligonucleotide sequences and plasmids used in this work.
Supplementary Material
Acknowledgments
We thank Jillian Arber and Jaynee Hart for the technical assistance, cloning, and plant care. We thank Hong Qiao for ein2 and ein3eil1 seeds.
Funding
This work was funded by National Science Foundation (NSF) Division of Integrative Organismal Systems grant IOS-1755355 to U.V.P. National Institutes of Health (NIH) grants R35GM125003, GM12500303S1, and GM12500304S1 to U.V.P. São Paulo Research Foundation (FAPESP) grant no. 2016/01128-9 and the Brazilian National Council of Scientific and Technological Development (CNPq) funded M.R. O.S. was supported by David and Fanny Luke Fellowship.
Conflict of interest statement. None declared.
U.V.P. conceived the study. D.R., M.R., and U.V.P. designed experiments. D.R. performed most of the experiments and analyzed the data; A.A. and O.S. performed genotyping, generation of transgenic plants, and immunoblots, D.R. and U.V.P. wrote the paper and collected contributions of all authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Ullas V. Pedmale (pedmale@cshl.edu).
References
- Aken OV, Zhang B, Law S, Narsai R, Whelan J (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162: 254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R, Rodriguez L, Gonzalez-Guzman M, Pizzio GA, Rodriguez PL (2011) News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr Opin Plant Biol 14: 547–553 [DOI] [PubMed] [Google Scholar]
- Bakshi M, Oelmüller R (2014) WRKY transcription factors. Plant Signal Behav 9: e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL (2014) Light regulation of plant defense. Annu Rev Plant Biol 65: 335–363 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Mazza CA, Austin AT, Pierik R (2012) Canopy light and plant health. Plant Physiol 160: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Roccaro M, Somssich IE (2017) Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29: 20–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Ross A, Kramer K, Finkemeier I, Somssich IE (2018) Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP‐triggered immunity. Plant J 96: 487–502 [DOI] [PubMed] [Google Scholar]
- Bjornson M, Pimprikar P, Nürnberger T, Zipfel C (2021) The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat Plants 7: 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y, Gaillochet C, Seluzicki A, Chory J, Busch W (2020) Local HY5 activity mediates hypocotyl growth and shoot-to-root communication. Plant Commun 1: 100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ (2012) Shade avoidance. Arabidopsis Book 2012: e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D (2017) Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant 10: 1174–1189 [DOI] [PubMed] [Google Scholar]
- Chen X, Li C, Wang H, Guo Z (2019) WRKY transcription factors: evolution, binding, and action. Phytopathol Res 1: 13 [Google Scholar]
- Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X (2016) Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol 26: 640–646 [DOI] [PubMed] [Google Scholar]
- Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu J-Q, Chen Z (2013) Protein–protein interactions in the regulation of WRKY transcription factors. Mol Plant 6: 287–300 [DOI] [PubMed] [Google Scholar]
- Chory J (2010) Light signal transduction: an infinite spectrum of possibilities. Plant J 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR (2006) Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci USA 103: 12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol 68: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Courbier S, Pierik R (2019) Canopy light quality modulates stress responses in plants. Iscience 22: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Onge KRSt, Voesenek LACJ, Pierik R, Sasidharan R (2016) Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol 172: 718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Keuskamp DH, Bongers FJ, Hornitschek P, Gommers CMM, Reinen E, Martínez-Cerón C, Fankhauser C, Pierik R (2016) Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr Biol 26: 3320–3326 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgikh VA, Pukhovaya EM, Zemlyanskaya EV (2019) Shaping ethylene response: the role of EIN3/EIL1 transcription factors. Front Plant Sci 10: 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C (2015) Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol 34: 46–53 [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Berardini T, Chen G, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller LA, et al. (2002) TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomic 2: 239–253 [DOI] [PubMed] [Google Scholar]
- Geilen K, Böhmer M (2015) Dynamic subnuclear relocalisation of WRKY40 in response to Abscisic acid in Arabidopsis thaliana. Sci Rep-UK 5: 13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H (2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López‐Vidriero I, Franco‐Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non‐DNA binding bHLH heterodimers. Embo J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Pietro AD, Zhang W (2014) The secreted peptide PIP1 amplifies immunity through receptor-like Kinase 7. Plos Pathog 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Páldi K, O’Donnell L, Batalov O, Pedmale UV, Nusinow DA, Kay SA, Chory J (2015) Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev Cell 35: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkute SG, Gujjar RS, Rai A, Akhtar M, Singh M, Singh B (2018) Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 13: 8–17 [Google Scholar]
- Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, Ballaré CL (2011) Cryptochrome 1 and phytochrome B control shade‐avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J 67: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Vos I, Peeters AJM, Voesenek LACJ, Pierik R (2011) Blue‐light‐mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67: 208–217 [DOI] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Sénéchal F, Müller-Moulé P, Maloof J, Xenarios I, Fankhauser C (2016) Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T (2014) The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front Plant Sci 5: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Nii H, Mitsuda N, Ohta M, Kitajima S, Ohme-Takagi M, Sato F (2013) A regulatory cascade involving Class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol 162: 991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V, Kitto SL, Caplan JL, Hsueh Y-H, Kearns DB, Wu Y-S, Bais HP (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160: 1642–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Keller MM, Cerrudo I, Ballaré CL (2014) To grow or defend? Low red: far‐red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol 204: 355–367 [DOI] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung H-S, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Gene Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhang J, Li J, Zhou G, Wang Q, Bian W, Erb M, Lou Y (2015) Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. e Life 4: e04805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fu Q, Chen L, Huang W, Yu D (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233: 1237–1252 [DOI] [PubMed] [Google Scholar]
- Li Z, Peng J, Wen X, Guo H (2013) ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G, Pagès L, Draye X (2011) A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol 157: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell Online 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Ebert D, Muruganujan A, Mills C, Albou L-P, Mushayamaha T, Thomas PD (2020) PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res 49: gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballaré CL (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortigosa A, Fonseca S, Franco‐Zorrilla JM, Fernández‐Calvo P, Zander M, Lewsey MG, García‐Casado G, Fernández‐Barbero G, Ecker JR, Solano R (2020) The JA‐pathway MYC transcription factors regulate photomorphogenic responses by targeting HY5 gene expression. Plant J 102: 138–152 [DOI] [PubMed] [Google Scholar]
- Pacín M, Semmoloni M, Legris M, Finlayson SA, Casal JJ (2016) Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol 211: 967–979 [DOI] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim J-I, Huq E (2017) Expanding roles of PIFs in signal integration from multiple processes. Mol Plant 10: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Djakovic-Petrovic T, Keuskamp DH, de Wit M, Voesenek LACJ (2009) Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiol 149: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G (2012) Gene overexpression: uses, mechanisms, and interpretation. Genetics 190: 841–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinerson CI, Rabara RC, Tripathi P, Shen QJ, Rushton PJ (2015) The evolution of WRKY transcription factors. BMC Plant Biol 15: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado D, Gramegna G, Cruz A, Lira BS, Freschi L, de Setta N, Rossi M (2016) Phytochrome interacting factors (PIFs) in Solanum lycopersicum: diversity, evolutionary history and expression profiling during different developmental processes. Plos One 11: e0165929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Růžička K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang M-Y, Kim J, Paek N-C, Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5: 4636. [DOI] [PubMed] [Google Scholar]
- Salisbury FJ, Hall A, Grierson CS, Halliday KJ (2007) Phytochrome coordinates Arabidopsis shoot and root development. Plant J 50: 429–438 [DOI] [PubMed] [Google Scholar]
- Skibbe M, Qu N, Galis I, Baldwin IT (2008) Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H (1982) Light quality, photoperception, and plant strategy. Ann Rev Plant Physiol 33: 481–518 [Google Scholar]
- Smith H (2000) Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer J-L, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel AI, Liu L, Liberles DA (2016) Models for gene duplication when dosage balance works as a transition state to subsequent neo- or sub-functionalization. BMC Evol Biol 16: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Tierens KFM-J, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2Gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, van Kang C, Li P, Pierik R (2021) Regulation of lateral root development by shoot-sensed far-red light via HY5 is nitrate-dependent and involves the NRT2.1 nitrate transporter. Front Plant Sci 12: 660870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Paalman R, Keuskamp DH, Hayes S, Pierik R (2018) Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30: tpc.00771.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana VE, Busanello C, da Maia LC, Pegoraro C, de Oliveira AC (2018) Activation of rice WRKY transcription factors: an army of stress fighting soldiers? Curr Opin Plant Biol 45: 268–275 [DOI] [PubMed] [Google Scholar]
- Wang H, Xu Q, Kong Y-H, Chen Y, Duan J-Y, Wu W-H, Chen Y-F (2014) Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol 164: 2020–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen L, Yu D (2018) Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol 176: 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gu L, Ringler P, Smith S, Rushton PJ, Shen QJ (2015) Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci 236: 214–222 [DOI] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D (2011) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.