Abstract
Cold stress adversely affects plant production, both qualitatively and quantitatively. Banana (Musa acuminata) is sensitive to cold stress and suffers chilling injury (CI) when stored under 11°C, causing abnormal fruit softening. However, the mechanism underlying the abnormal fruit softening due to CI remains obscure. This study uncovered the coordinated transcriptional mechanism of ethylene F-box (EBF1) protein and abscisic acid-insensitive 5 (ABI5)-like protein in regulating chilling-induced softening disorders of Fenjiao banana. Cold stress severely inhibited the transcript and protein levels of EBF1, ABI5-like, and fruit softening-related genes. The ABI5-like protein bound to the promoters of key starch and cell wall degradation-related genes such as β-amylase 8 (BAM8), pectate lyase 8 (PL8), and β-D-xylosidase23-like (XYL23-like) and activated their activities. EBF1 physically interacted with ABI5-like and enhanced the transcriptional activity of the key starch and cell wall degradation-related genes but did not ubiquitinate or degrade ABI5-like protein. This promoted fruit ripening and ameliorated fruit CI in a manner similar to the effect of exogenous abscisic acid treatment. The ectopic and transient overexpression of EBF1 and ABI5-like genes in tomato (Solanum lycopersicum) and Fenjiao banana accelerated fruit ripening and softening by promoting ethylene production, starch and cell wall degradation, and decreasing fruit firmness. EBF1 interacted with EIL4 but did not ubiquitinate or degrade EIL4, which is inconsistent with the typical role of EBF1/2 in Arabidopsis (Arabidopsis thaliana). These results collectively highlight that the interaction of EBF1 and ABI5-like controls starch and cell wall metabolism in banana, which is strongly inhibited by chilling stress, leading to fruit softening and ripening disorder.
The ethylene F-box protein interacts with abscisic acid-insensitive 5-like protein to regulate banana fruit starch and cell wall degradation, mediating cold stress-induced softening disorders.
Introduction
Cold stress is a common abiotic stress that adversely affects plant productivity (Pearce and Fuller, 2001; Guo et al., 2018a; Liu et al., 2018), fruit ripening, and quality (Phothiset and Charoenrein, 2014; Megías et al., 2016). The molecular mechanisms for withstanding cold stress in some model plants have been well studied (Ding et al., 2020). Fruits suffer serious damage both qualitatively and quantitatively under cold stress (Zhao et al., 2013; Albertos et al., 2019). Cold stress causes seed browning, shrinkage (Lee et al., 2020), surface pitting (Lafuente et al., 2017), discoloration, and ripening disorders (Zou et al., 2014; Suo et al., 2018; Song et al., 2019). Therefore, understanding how fruits respond to cold stress can substantially enrich the environmental adaption of plants to cold stress and provide valuable information and genetic resources for improving cold-stress tolerance in fruit crops.
Bananas (Musa acuminata) are among the most important fruit crops in the world. It is the second largest fruit crop (gross production) and among the top 10 highly produced globally (Paul et al., 2016). Banana is a typical climacteric fruit and thus ripens quickly and has a short shelf-life once harvested. This phenomenon substantially shortens its storage period and limits the time allowed for its transportation (Seymour et al., 1993). Being a subtropical fruit, banana is hypersensitive to low temperatures and suffers from chilling injury (CI) when subjected to temperatures below 11°C. CI causes cellular membrane damage, flavor loss, and abnormal fruit ripening (Zhu et al., 2018). Previous studies postulate that CI substantially represses starch degradation in banana (Louro et al., 2013; Song et al., 2019), a phenomenon also observed in potato (Solanum tuberosum L.) (Xie et al., 2018) and melon (Hao et al., 2009). However, the underlying mechanism of abnormal fruit ripening remains unclear.
Fruit ripening is a complex physiological and biochemical process coordinated by an interactive signaling network comprising ethylene, abscisic acid (ABA), other plant hormones, transcription factors (TFs), and external environmental signals (Gu et al., 2019). The hormone levels either activate or inhibit several downstream genes during the post-harvest ripening process (Busatto et al., 2016). Fruit softening is a crucial criterion for determining the ripeness and quality of banana (Xiao et al., 2018). It is mediated by individual phytohormones and through interactions between phytohormones (Perez-Llorca et al., 2019).
Ethylene is an important phytohormone essential for the ripening and softening of climacteric fruits (Giovannoni, 2004; Guo and Ecker, 2004). Two ethylene insensitive 3 (EIN3) binding F-box-1/2 (EBF1/2) proteins play a major role in ethylene transduction (Binder et al., 2007). EBF1/2 functions as an E3 ubiquitin ligase and regulate the stability of EIN3/EIL1 via the 26S-ubiquitin-proteasomal degradation system. EBF1/2 is also a negative player in ethylene signaling in Arabidopsis (Guo and Ecker, 2003; Potuschak et al., 2003; Binder et al., 2007). EBF1/2 silencing stabilized EIN3/EIL1 in Arabidopsis, resulting in enhanced ethylene response. However, plants overexpressing EBF1/2 showed ethylene insensitivity and EIN3 protein destabilization (Guo and Ecker, 2003; Potuschak et al., 2003). Although EBF1 and EBF2 exhibit similar functions, they play overlapping roles in ethylene signaling. EBF1 mainly works during the initial phase of the ethylene response in etiolated seedlings, whereas EBF2 functions during the later stage of the response (Binder et al., 2007). Four EBF genes (SlEBF1, SlEBF2, SlEBF3, and SlEBF4) have been identified in tomato (Solanum lycopersicum). Their expression is induced by exogenous ethylene and increases during the ripening process (Liu et al., 2015). However, silenced mutants show accelerated ripening, whereas overexpressing mutants inhibit fruit ripening and softening (Yang et al., 2010; Deng et al., 2018; Guo et al., 2018b), indicating the complexity of mechanisms linked to SlEBFs in regulating tomato fruit ripening and senescence. The expression of banana MaEBF1 (Wu et al., 2018) and MaEBF2 (Kuang et al., 2013) increases with fruit ripening. MaEBF1 in Fenjiao banana acts as a positive modulator of starch degradation by interacting with MaNAC67-like protein (Song et al., 2019). Overall, these reports indicate that EBFs play an essential role in fruit development and ripening and are specific in different fruit species. The mechanism of EBF1 in mediating fruit softening in banana, therefore, needs to be validated.
ABA is an important phytohormone regulating abiotic and biotic stresses and fruit ripening (Vishwakarma et al., 2017). Abscisic acid-insensitive 5 (ABI5), a plant basic leucine zipper (bZIP) transcription factor, is a vital regulator of the ABA signaling pathway, which controls seed dormancy, germination, plant growth, and flowering time (Yu et al., 2015; Wang et al., 2021). In apple, MdABI5 cooperates with its positive or negative interaction partners to regulate ABA-mediated leaf senescence (An et al., 2020). In banana, MaABI5 participates in ABA-induced cold tolerance by interacting with a RING E3 ubiquitin ligase, MaC3HC4-1 (Chen et al., 2018). Although many ABI5 family members have been identified and characterized in plants, their role in fruit ripening and CI has not been clarified.
Most edible bananas are derived from intraspecific or interspecific hybridization between Musa acuminata (A-genome) and Musa balbisiana (B-genome) species. Combinations of these A- and B-genomes have resulted in various genotypes of cultivated bananas, including diploid (AA, BB, and AB), triploid (AAA, AAB, and ABB), and tetraploid (AAAB, AABB, ABBB) variants. The triploid genotype variants constitute the predominant cultivated varieties planted worldwide, such as BaXi Jiao variety (Musa acuminata L. AAA group cv. Cavendish) (Wang et al., 2019). Fenjiao (Musa ABB Pisang Awak) is a popular banana cultivar that has good flavor, is highly nutritious, and is highly tolerant to abiotic stresses. It has been widely cultivated and consumed in China in recent years (Hu et al., 2015). Studies postulate that the genetic make-up of Fenjiao (ABB) is different from that of Cavendish banana (AAA), specifically in ethylene synthesis and starch degradation characteristics (Wang et al., 2019). Fenjiao banana has an active starch metabolism pathway, which translates to good post-harvest starch metabolism (Wang et al., 2019). Cold stress causes severe CI symptoms on Fenjiao banana, including peel browning and ripening disorders, impeding the complete softening of the pulp, a process closely related to starch degradation (Song et al., 2019). Therefore, Fenjiao banana is an important crop and an excellent material to study the CI-induced ripening disorder.
Herein, we demonstrate that EBF1 interacts with ABI5-like protein to regulate the expression of genes involved in fruit ripening and softening. EBF1 enhances the activation of ABI5-like on the promoters of starch and cell wall degradation-related genes but does not ubiquitinate or degrade ABI5-like protein in banana fruit. Ectopic and transient overexpression (OE) of MaEBF1 and MaABI5-like genes in tomato and Fenjiao banana fruit promotes fruit ripening by activating the expression of starch and cell wall degradation-related genes. Notably, EBF1 interacts with EIL4 but does not degrade EIL4 by ubiquitination, indicating that it may not be a typical EBF1/2 as previously demonstrated in Arabidopsis and tomato. This study established a regulatory module of EBF1/ABI5-like genes in mediating cold stress-induced softening and ripening disorders of Fenjiao banana.
Results
Cold stress causes softening disorder in Fenjiao banana fruit
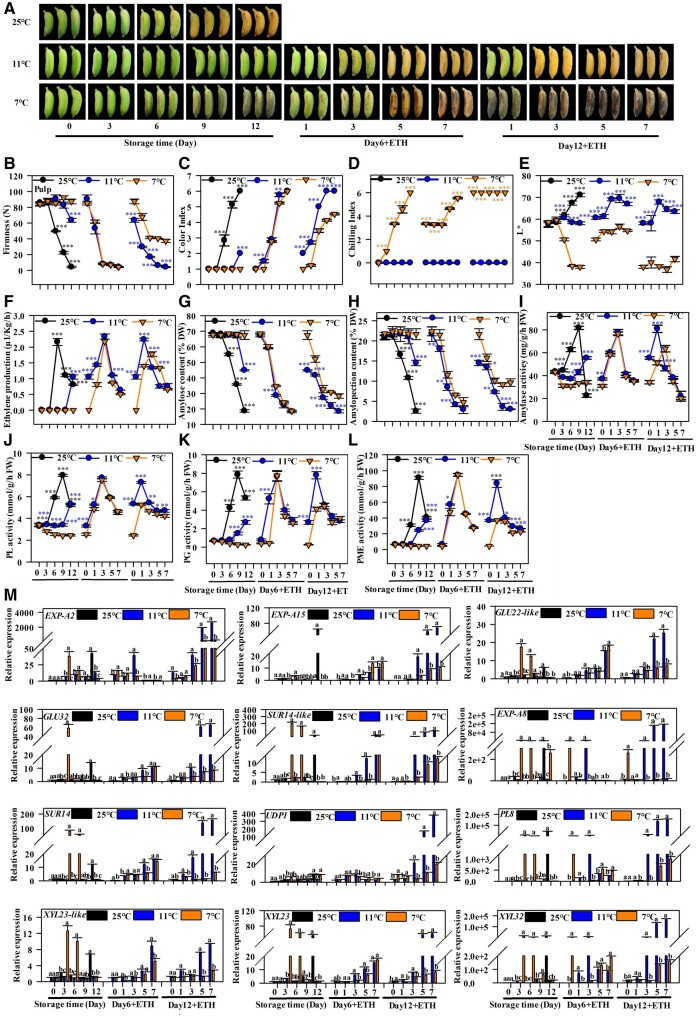
Cold stress (7°C, 12 d) causes the softening disorder of the banana fruit (Song et al., 2019). A suitable Low-Temperature Storage (11°C, LTS) significantly delayed the fruit ripening process by maintaining the fruit firmness and retarding the fruit coloring process (Figure 1, A–C). Fruits under Chilling Temperature Storage (7°C, CTS) developed serious CI after 12 d (Figure 1D). The fruit pulp could not soften completely under CTS after 12 d of storage, even when ripened using ethephon (Figure 1B). Fruits under LTS ripened with normal softening and coloring, without any CI symptoms (Figure 1, A–D). The CI symptoms observed by the L* value are shown in Figure 1E. Moreover, fruits treated with ethephon and exposed to 7°C and 11°C completely softened and turned yellow after 6 d of storage (Figure 1, A–C), indicating that the softening was not affected by mild CI. Fruit after 6 d of CTS showed lower L* value than LTS with mild CI. CTS and LTS repressed ethylene production during storage (Figure 1F). There was no significant difference in ethylene production between LTS and natural ripening of fruits after ethephon treatment. However, significant repressions were observed in CTS fruits (Figure 1F). Fruit softening has an important influence on the taste of banana fruit. Hence, we focused on the softening disorder caused by chilling stress in the present work.
Figure 1.
Effects of cold stress on the physiological indexes of Fenjiao banana fruit. A, Phenotypic variation of Fenjiao bananas under different storage conditions: natural (25°C), LTS (11°C), and CTS (7°C). Fruits were subjected to LTS (11°C) and CTS (7°C) for 6 and 12 d. Fruits were maintained at 25°C upon treatment with ETH. B to F, Changes in physiological indexes during storage and ripening: pulp firmness (B), color index (C), chilling index (D), L* value (E), ethylene production (F). G and H, contents of amylose (G) and amylopectin (H). I–L, activity of amylase (I), PL (J), PG (K), PME (L) in the pulp during storage and ripening. Each value represents the means of biological triplicates (mean ± sd). Differences between various treatments were assessed by ANOVA (analysis of variance) and Duncan’s multiple range test. Asterisks (*, **, and ***) indicate significant differences at P < 0.05, P < 0.01, and P < 0.001 level, respectively. M, The expression of genes related to cell wall degradation after different storage conditions and ripening. The expression levels of each gene at different days are relative to 0 day (freshly harvested fruit) set as 1. Each value represents the means of biological triplicates (mean ± sd). Means with different letters (a–c) are significantly different as determined by Duncan’s multiple range test (P < 0.05). ETH, ethephon.
Cold stress inhibits starch and cell wall degradation in Fenjiao banana fruit
Transmission electron microscopy (TEM) images showed a substantial difference in the number of starch granules in the pulp between fruits under CTS and LTS (Supplemental Figure S1A). Fewer starch granules were observed at the ripening stage in fruits stored at 11°C after ethephon treatment. However, there were numerous starch granules at the full ripening stage of fruits stored at 7°C. The middle layer of the pulp tissues appeared almost completely disintegrated under LTS at the full ripening stage (Supplemental Figure S1B). However, the microfibril structure was still present in the CI fruit (Supplemental Figure S1B).
The amylose and amylopectin content in the natural ripening fruit pulp decreased rapidly (Figure 1, G and H). There was a significantly higher level of amylose and amylopectin in CTS fruits than LTS fruits after 12 d of storage and ethephon treatment; however, no significant difference was observed after 6 d of cold storage, except a slight difference at the third day after ethephon treatment (Figure 1, G and H). Cold storage (7°C and 11°C) significantly repressed the activity of amylase, polygalacturonase (PG), pectate lyase (PL), and pectin pectylhydrolase (PME) during storage (Figure 1, I–L). However, 12 d of CTS severely inhibited the activities of these enzymes compared to LTS. These results indicate that the starch and cell wall degradation process is impaired after 12 d of CTS, explaining the softening disorder caused by the chilling stress.
Cold stress represses the expression of starch and cell wall degradation-related genes
Our previous study showed that 11 starch degradation-related genes are inhibited by cold stress (Song et al., 2019). Herein, 61 cell wall degradation-related genes were selected and characterized based on the RNA-seq assay results (Supplemental Figure S2). The expression of 12 genes involved in cell wall degradation significantly increased during fruit ripening and softening (Figure 1M). These genes include expansin A2 (EXP-A2), expansin A8 (EXP-A8), expansin A15 (EXP-A15), β-glucosidase22-like (GLU22-like), GLU32, PL8, Sugar transporter14 (SUR14), SUR14-like, UDP-galactose transporter1 (UDP1), β-D-xylosidase 23 (XYL23), XYL23-like, and XYL32. There were slight differences in the expression of these genes between fruits under CTS and LTS after 6 d, with a similar transcript level. However, the expression of these genes was severely repressed in fruits under CTS compared with those under LTS and natural ripening conditions after 12 d of storage and ethephon treatment, where the expression of several genes varied up to thousands of times between CTS and LTS (Figure 1M;Supplemental Figure S2). These results indicate that these genes play essential roles in cold stress-induced fruit softening and ripening disorders.
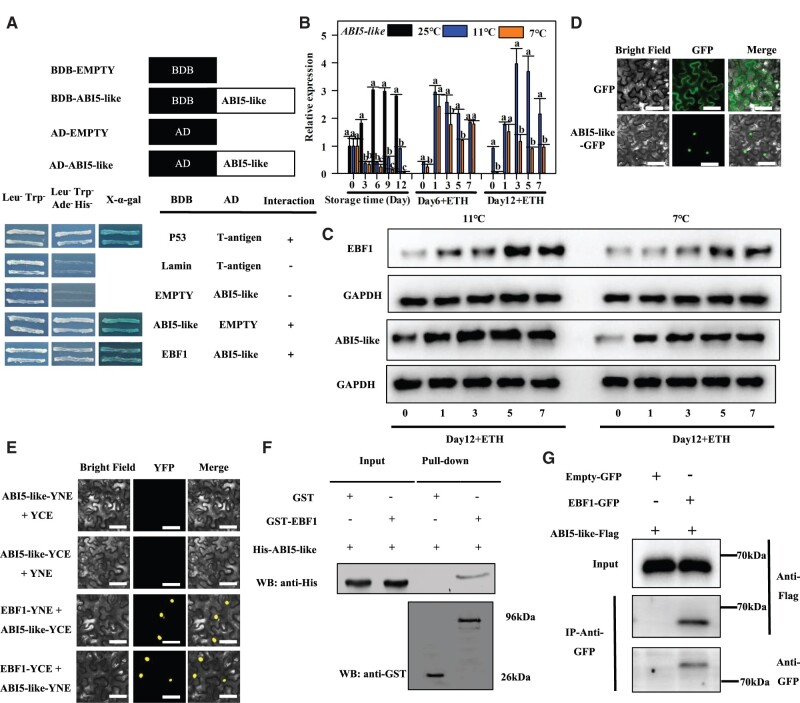
EBF1 interacts with ABI5-like both in vitro and in vivo
Transcriptomic data have shown that EBF1 is significantly differentially expressed after cold storage and 1-methylcyclopropene (1-MCP) treatments, suggesting that it may play important roles in Fenjiao banana fruit ripening (Song et al., 2019, 2020b). Our previous work showed that the expression of ABI5-like and EBF1 was closely related to Fenjiao fruit ripening under normal temperature condition, which increased with fruit ripening, and significantly repressed in ripening-delayed samples by 1-MCP (Song et al., 2020b). They are important candidate genes in regulating fruit ripening. So far, five EBF genes (MaEBF1-5) have been isolated from banana fruit (Jourda et al., 2014). Phylogenetic analysis of EBF proteins from different plant species showed that MaEBF1 maintains more protein sequence identity with other EBFs from banana, but not with other plant species. For example, MaEBF1 shares 50.23% and 47.87% amino acid identity with Arabidopsis thaliana AtEBF1 and AtEBF2, and 51.37% and 53.28% identity with SlEBF1 and SlEBF2 from tomato, respectively (Supplemental Figure S3). Among the five EBFs gene families in banana, only EBF1 has been identified via differential expression analysis. Therefore, it was used as a bait protein to screen the interacting proteins in the cDNA library (Song et al., 2019). Herein, ABI5-like protein was identified, and its interaction with EBF1 was validated using the yeast two-hybrid (Y2H) assay (Figure 2A). A total of 22 differentially expressed genes were identified in the ABA signaling pathway from the RNA-seq data, including ABI5-like (Supplemental Figure S4). Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) analysis showed that ABI5-like gene expression was remarkably increased under the natural ripening conditions. However, cold storage (7°C and 11°C) significantly inhibited the expression of ABI5-like gene, especially in CTS fruits. There were no significant differences in the ripening process between CTS and LTS fruits after 6 d of storage. However, ABI5-like gene expression was remarkably repressed in CTS fruits than in LTS fruits during ripening after 12 d of storage (Figure 2B). Western blot analysis revealed that both EBF1 and ABI5-like protein levels increased during fruit ripening under LTS but were severely repressed under CTS (Figure 2C).
Figure 2.
The expression profiles of ABI5-like and EBF1, and the interaction between EBF1 and ABI5-like proteins. A, The Y2H assay confirms the interaction between EBF1 and ABI5-like proteins. Yeast cells grew on sd/-Leu-Trp-Ade-His- with 125 μM Aureobasidin A. They turned blue in the presence of 4 mg mL−1 X-α-Gal, which served as a positive control. B, The transcript levels of ABI5-like in the pulp during storage and ripening. The expression levels of each gene at different days are relative to 0 day (freshly harvested fruit) set as 1. Each value represents the means of biological triplicates (mean ± sd). Means with different letters (a–c) are significantly different as determined by Duncan’s multiple range test (P < 0.05). C, The protein levels of EBF1 and ABI5-like during ripening after 12 d of storage. Fruits were maintained at 25°C upon treatment with ethephon. D, The subcellular localization of ABI5-like in N. benthamiana leaves. The fusion protein (ABI5-like) and the GFP (positive control) were transiently expressed into N. benthamiana leaves via Agrobacterium infiltration. Localization of fluorescent proteins was observed 36–72 h after infiltration using an upright fluorescence microscope (Axio Imager D2, Zeiss, Germany). Images were obtained using Axio Vision SE64 software in a dark field for green fluorescence, while the cell outlines of the same images were photographed in a bright-field microscopy. The overlays of fluorescent and bright field images were also obtained. The left column shows the bright-field image; the middle column shows green fluorescent signal; and the right column shows the merged image. Bars = 50 μM. The experiment was repeated at least three times. Bars = 50 μM. E, BiFC assay confirming the interactions between EBF1 and ABI5-like proteins in N. benthamiana leaf epidermal cells. EBF1 and ABI5-like genes were cloned with N- and C-terminus of YFP, respectively. The fusion protein (ABI5-like) and N- and C-terminus of YFP alone (negative control) were transiently expressed in N. benthamiana leaves via Agrobacterium infiltration. After 36–72 h, fluorescent signals were observed and captured using a fluorescence microscope (Axio Imager D2, Zeiss, Germany). The images were captured under dark field for fluorescence signal and bright-field for the cell outlines. Bars = 50 μM. F, GST Pull-down assay validation of the interaction between EBF1 and ABI5-like proteins. ABI5-like-His protein was incubated with either EBF1-GST or GST, and the bound proteins were detected by immunoblot assays using the anti-His and anti-GST antibodies, respectively. G, CoIP assay validation of the interaction between EBF1 and ABI5-like. Nicotiana benthamiana leaves co-expressing EBF1-GFP and ABI5-like-Flag, or empty-GFP and ABI5-like-Flag were IP with the anti-GFP antibody, and the immunoblot was performed with the anti-GFP and anti-Flag antibodies, respectively.
The ABI5-like gene consists of a 1044 bp open reading frame encoding a polypeptide of 347 amino acids containing a b-ZIP domain (Supplemental Figure S5A). Phylogenetic analysis further revealed that the ABI5-like gene is closely related to the ABI5 from Arabidopsis (Supplemental Figure S5B). ABI5-like gene is located in the nucleus similar to EBF1 (Figure 2D). Bimolecular fluorescence complementation (BiFC) (Figure 2E), glutathione S-transferase (GST) pull-down (Figure 2F), and co-immunoprecipitation (CoIP) assays (Figure 2G) revealed that EBF1 interacts physically with ABI5-like protein. Y2H experiments were conducted to check further if ABI5-like protein could interact with other EBFs in banana. The results showed no interaction between ABI5-like protein and other banana EBFs, including EBF2, EBF3, EBF4, and EBF5 (Supplemental Figure S6).
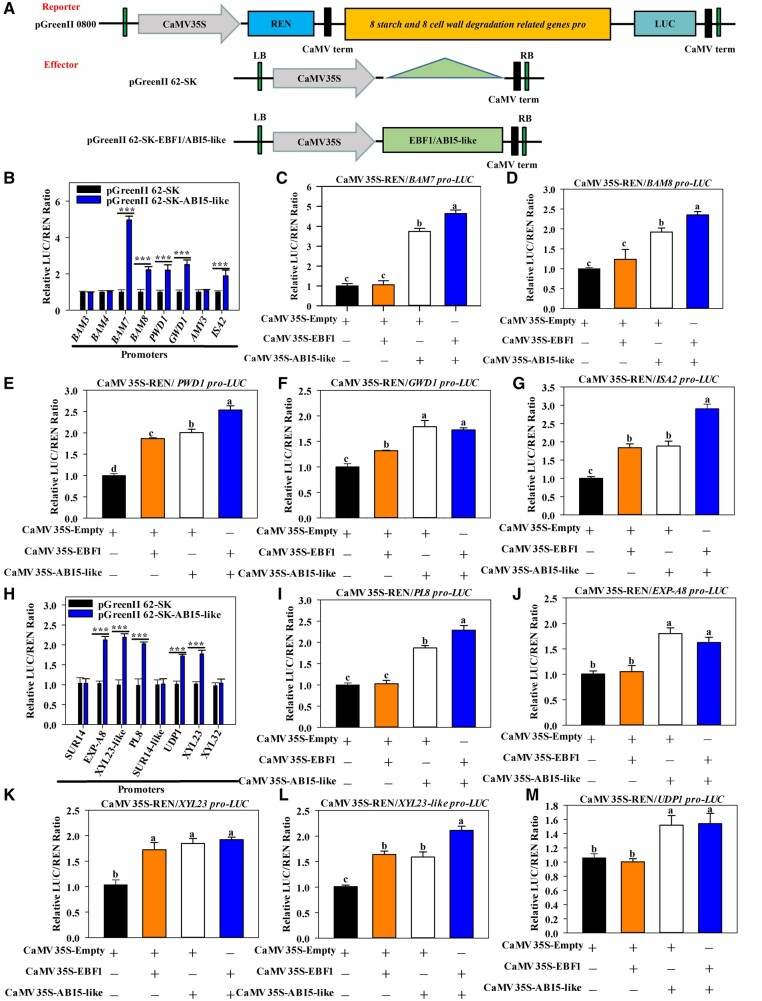
ABI5-like cooperates with EBF1 to activate starch and cell wall degradation-related genes
The promoter sequences of the 12 cell wall and 11 starch degradation-related genes were obtained and analyzed. Several ABRE/G-box motifs recognized by ABI5 (Sibéril et al., 2001) were identified in the promoter sequences of eight starch and eight cell wall degradation-related genes (Supplemental Figure S7). The starch degradation-related genes included gene encoding β-amylase (BAM3, BAM4, BAM7, BAM8), α-amylases (AMY3), starch debranching enzymes (ISA2), α-glucan water dikinases (GWD1) and phosphoglucan water dikinase (PWD1), whereas the cell wall degradation-related genes were EXP-A8, PL8, SUR14, SUR14-like, UDP1, XYL23, XYL23-like, and XYL32. The promoters of these genes were ligated to the pGreenII 0800-LUC reporter vectors, whereas the full-length CDS of EBF1 and ABI5-like were ligated into the pGreenII 62-SK effector vector to determine their activities (Figure 3A). As expected, the promoter activities of five starch (BAM7, BAM8, PWD1, GWD1, and ISA2) and five cell wall (EXP-A8, XYL23-like, PL8, UDP1, and XYL23) degradation-related genes were significantly induced by ABI5-like protein. The genes had a significantly higher LUC (firefly luciferase)/REN (renilla luciferase) ratio than the control (Figure 3, B and H). Intriguingly, transient OE of MaEBF1 alone could also induce the transcription of PWD1, GWD1, ISA2, XYL23, and XYL23-like, except for BAM7, BAM8, PL8, EXP-A8, and UDP1, implying that EBF1 might regulate gene expression via an uncharacterized machinery, possibly through interaction with unknown partners like other TFs. When both EBF1 and ABI5-like were co-transformed, the LUC/REN ratio of BAM7, BAM8, PWD1, MaISA2, MaPL8, and MaXYL23-like were more significantly induced compared to transformation with ABI5-like or EBF1 alone (Figure 3, C–E, G, I, and L), except for GWD1, EXP-A8, XYL23, and UDP1 (Figure 3, F, J, K, and M).
Figure 3.
EBF1 and ABI5-like proteins activate the expression of genes involved in starch and cell wall degradation. A, Schematic representation of the reporter and effector constructs used in the dual-luciferase reporter assay. B and H, The interactions between ABI5-like protein and the promoters of genes related to the degradation of starch (B) and cell wall (H) obtained from transient assays in N. benthamiana leaves in vivo. Asterisks indicate significant differences as determined by Duncan’s multiple range test at ***P < 0.001 level. C–G, Transcriptional activity of EBF1, ABI5-like and ABI5-like + EBF1 on the promoter activities of five starch degradation genes: BAM7 (C), BAM8 (D), PWD1 (E), GWD1 (F), and ISA2 (G). I–M, Transcriptional activity of EBF1, ABI5-like and ABI5-like + EBF1 on the promoter activities of five cell wall degradation genes: PL8 (I), EXP-A8 (J), XYL23 (K), XYL23-like (L), and UDP1 (M). The ratio of LUC to REN of the empty vector. The promoter vector was used as a calibrator (set as 1). Each value was calculated from the means of six biological replicates. The values represent the means ± se. Different letters indicate significant differences as determined by Duncan’s multiple range test between groups (P < 0.05).
The yeast one-hybrid (Y1H) assay was performed to investigate whether ABI5-like protein binds directly to the promoters of the starch and cell wall degradation genes. ABI5-like bound to the promoter of these genes in yeast (Figure 4A). Furthermore, electrophoretic mobility shift assay (EMSA) experiments were conducted using purified recombinant GST-ABI5-like proteins (Supplemental Figure S8C), which showed that the ABI5-like protein directly binds to the promoter ABRE/G-box motifs of the five starch and five cell wall degradation-associated genes (Figure 4B). The chromatin immunoprecipitation (ChIP)-qPCR analysis further revealed that the promoter regions of these genes were specifically enriched by an anti-ABI5-like rather than a nonspecific antibody (pre-immune rabbit IgG) (Figure 4C). These results indicate that ABI5-like protein binds directly to the promoter sequences of the fruit softening genes (Figures 3 and 4). The interaction between EBF1 and ABI5-like proteins further enhances the activation of the promoter activities.
Figure 4.
ABI5-like protein binds to the promoters of five starch and five cell wall degradation genes in vitro and in vivo. A, Yeast growth assays for determining the binding ability of ABI5-like protein to promoters of genes related to starch and cell wall degradation. ABI5-like effector or empty (AD, negative control) plasmids were transformed into Y1H reporter strains. Their interaction was assessed based on the growth conditions of transformed yeast on sd medium supplemented with ABA but lacking Leu-. B, EMSA analysis showed the binding of ABI5-like to the ABRE/G-box motif in the promoters of the five starch and five cell wall degradation genes in vitro. The probe sequences corresponding to each target gene promoter are denoted with red letters representing the ABRE/G-box and the mutant ABRE/G-box. The purified GST or fusion GST-ABI5-like protein was mixed with probes, and the protein–DNA complexes separated on native polyacrylamide gels. Triangles indicate increasing amounts of unlabeled or mutated probes for competition. “−” represents absence, “+” represents presence. C, ChIP-qPCR assay confirmed the direct binding of ABI5-like proteins to the promoters of five starch and five cell wall degradation genes. The promoter structure of the five starch and five cell wall degradation genes is shown in the left and above. The red lines indicate the cis-elements of ABI5-like gene. The underlined fragment was used for ChIP-qPCR. Values represent the percentage of DNA fragments co-IP with anti-ABI5-like antibodies or the nonspecific antibody (anti-IgG) relative to the input DNA. Each value was calculated from the means of three biological replicates. The values represent the means ± se. Asterisks indicate significant differences as determined using ANOVA followed by Duncan’s multiple range test at ***P < 0.001 level.
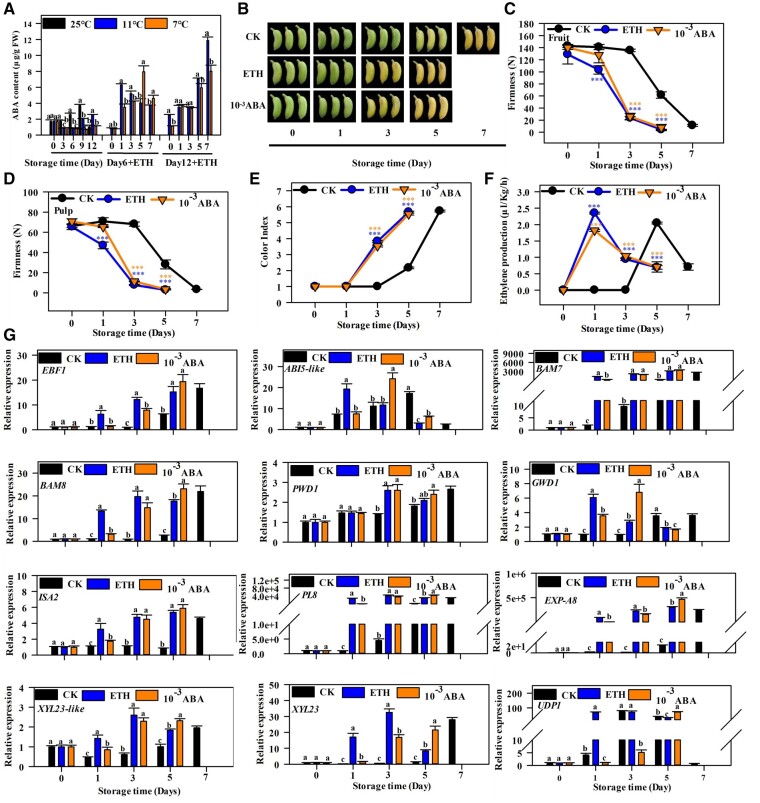
Exogenous ABA application promotes ripening and softening of Fenjiao banana fruits
The endogenous ABA content increased with fruit ripening and declined with fruit senescence in the naturally ripened fruits (Figure 5A). Cold storage (7°C and 11°C) significantly reduced endogenous ABA content, especially CTS. The endogenous ABA content increased rapidly to a constant level during the ripening of both CTS and LTS fruits after 6 d. However, there were significantly lower levels of endogenous ABA in CTS than in LTS after 12 d. These results indicate that the ABA content is closely related to fruit ripening and CTS-induced ripening disorder.
Figure 5.
ABA treatment promotes the ripening of Fenjiao banana fruits. A, Cold stress inhibits endogenous ABA content. Each value represents the means of three biological replicates (mean ± sd). Different letters indicate significant differences as determined by Duncan’s multiple range test between groups (P < 0.05). B, The natural ripening process (25°C) and ripening process of fruit after 1 ml·L−1 ETH and 10−3 M ABA (10−3 ABA) treatments. C–F, Changes in the firmness of the whole fruit (C) and pulp (D), color index (E), ethylene production (F) during fruit ripening. Each value represents the means of three biological replicates (mean ± sd). Asterisksindicate significant differences as determined using ANOVA followed by Duncan’s multiple range test at ***P < 0.001 level. G, The transcript levels of EBF1, ABI5-like, and the corresponding genes related to starch and cell wall degradation during ripening. The expression levels of each gene at different days are relative to 0 day (freshly harvested fruit) set as 1. Each value represents the means of three biological replicates (mean ± sd). Different letters indicate significant differences between groups as determined using ANOVA following Duncan’s multiple range test (P < 0.05). ETH, ethephon.
Further analysis showed that fruit ripening was accelerated by exogenous ABA (10−3 M) and ethylene treatment (Figure 5B). ABA and ethylene had similar effects on fruit ripening. They accelerated the decrease of fruit and pulp firmness, color index, and ethylene production (Figure 5, C–F). ABA and ethylene treatments significantly activated the expression of EBF1, ABI5-like, and genes related to starch and cell wall degradation compared to natural ripening (Figure 5G), as well as the marker genes related to the ethylene and ABA biosynthesis and signaling pathways (Supplemental Figure S9). These results demonstrate that exogenous ABA promotes the ripening and softening of Fenjiao banana by inducing the expression of EBF1, ABI5-like, and fruit softening-related genes.
Exogenous ABA relieves CTS-induced CI symptoms in Fenjiao banana fruits
Exogenous ABA application on CTS (5°C) fruits was conducted to validate the effect of ABA on the CI in Fenjiao banana fruits. Exogenous ABA treatment significantly relieved the CI symptoms of Fenjiao banana (Supplemental Figure S10, A and B), resulting in a low chilling index, fruit and pulp firmness, as well as higher L* and color index (Supplemental Figure S10, B–F). Moreover, exogenous ABA induced the expression of EBF1, ABI5-like, and the starch and cell wall degradation-related genes compared to the control condition (CTS; 5°C) fruits (Supplemental Figure S10G). These results confirm that exogenous ABA relieves CI symptoms and promotes the ripening and softening of Fenjiao banana by inducing EBF1, ABI5-like, and genes related to starch and cell wall degradation.
OE of MaEBF1 and MaABI5-like in tomato promotes fruit ripening and softening
MaEBF1 and MaABI5-like OE lines were generated in the background of Micro-Tom tomato to test further the function of MaEBF1 and MaABI5-like in fruit ripening. More than 10 independent transgenic lines obtained in each case exhibited increased ripening of the tomato fruits.
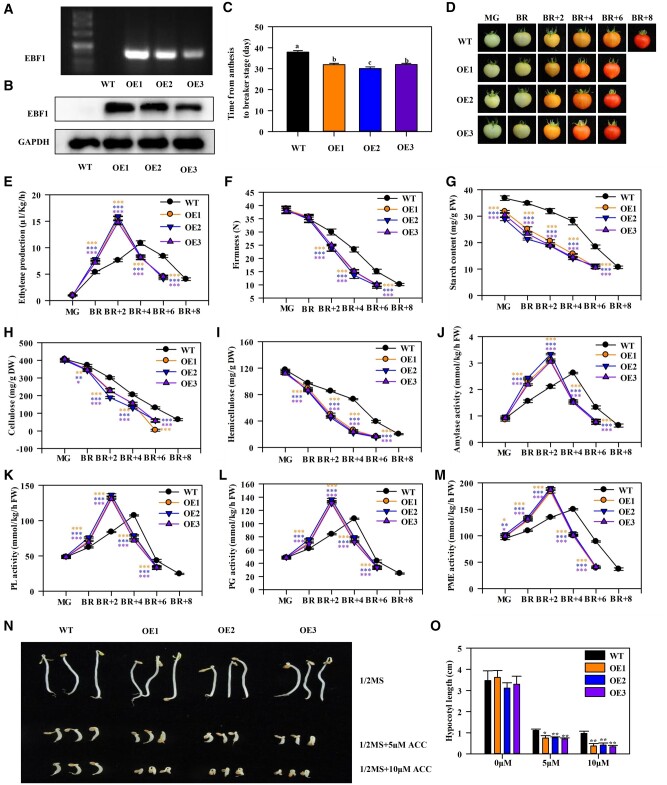
Three representative lines (OE1, OE2, and OE3) with high MaEBF1 transcript and protein levels were selected for further investigation (Figure 6, A and B; Supplemental Figure S11A). All the MaEBF1-OE lines exhibited faster tomato fruit ripening (Figure 6, C and D). The onset of fruit ripening in the MaEBF1-OE lines was significantly advanced by 6 d compared to the wild-type (WT) fruits (Figure 6C). The breaker (BR) stage occurred at around 32 d post-anthesis in the MaEBF1-OE lines compared to 38 d post-anthesis in WT fruit. Their ripening process from BR stage to full ripening was accelerated by about 2 d compared with the WT lines (Figure 6D). MaEBF1-OE fruits showed a more rapid ethylene increase and a higher ethylene level than the WT fruits during ripening (Figure 6E). The MaEBF1-OE lines showed a more rapid decrease in firmness than the WT lines (Figure 6F). Moreover, there were significantly lower starch, cellulose, and hemicellulose contents in the OE1/2/3 fruits compared to the WT fruits during ripening (Figure 6, G–I). The amylase activity and enzymes associated with fruit softening, including PL, PG, and PME, were significantly higher in MaEBF1-OE fruits than in the WT fruits (Figure 6, J–M). These results show that overexpressing MaEBF1 in tomato promotes fruit ripening and softening. Additionally, OE1/2/3 seedlings grown in the dark displayed an enhanced ethylene triple response when treated with 5 and 10 μM 1-aminocyclopropane-1-carboxylic acid (ACC) than the WT seedlings, with significantly shorter hypocotyl elongation than the WT (Figure 6, N and O), suggesting the enhanced ethylene sensitivity in the transgenic lines.
Figure 6.
OE of EBF1 in tomato promotes fruit ripening, induces starch and cell wall degradation, and enhances the ethylene hypersensitivity phenotype in seedlings. A–C, The expression profiles of EBF1 in fruits of WT and EBF1-OE lines as determined by RT-PCR (A), and western-blot (B). Pericarp tissues of EBF1-OE lines and WT at 30 d post anthesis (DPA) were used for detection. For western-blot analysis, total proteins were extracted and probed with an anti-EBF1 polyclonal antibody. GAPDH was used as the internal reference. C, Time from anthesis to BR stage of WT and EBF1-OE lines. Each value represents the means of 30 biological replicates (mean ± sd). Means with different letters (a–c) are significantly different as determined by ANOVA and Duncan’s multiple range tests (P < 0.05). D, The fruit ripening process of WT and EBF1-OE lines at MG, BR, BR + 2 d, BR + 4 d, BR + 6 d, and BR + 8 d stages. E–M, Ethylene production (E), changes in fruit firmness (F), the content of starch (G), cellulose (H), and hemicellulose (I), and the activities of amylase (J), PL (K), PG (L), and PME (M) in WT and EBF1-OE lines during ripening. N, The triple response of WT and EBF1-OE lines with and without ACC treatments. O, The hypocotyl length of seedlings in response to ACC. Seedling length was measured using image J software (https://imagej.nih.gov/ij/). Each value represents the means of three biological replicates (mean ± sd). The ANOVA and Duncan’s multiple range tests were conducted to compare the differences between the means of various conditions. Asterisks indicate significant differences at *P < 0.05, **P < 0.01, and ***P < 0.001 level, respectively.
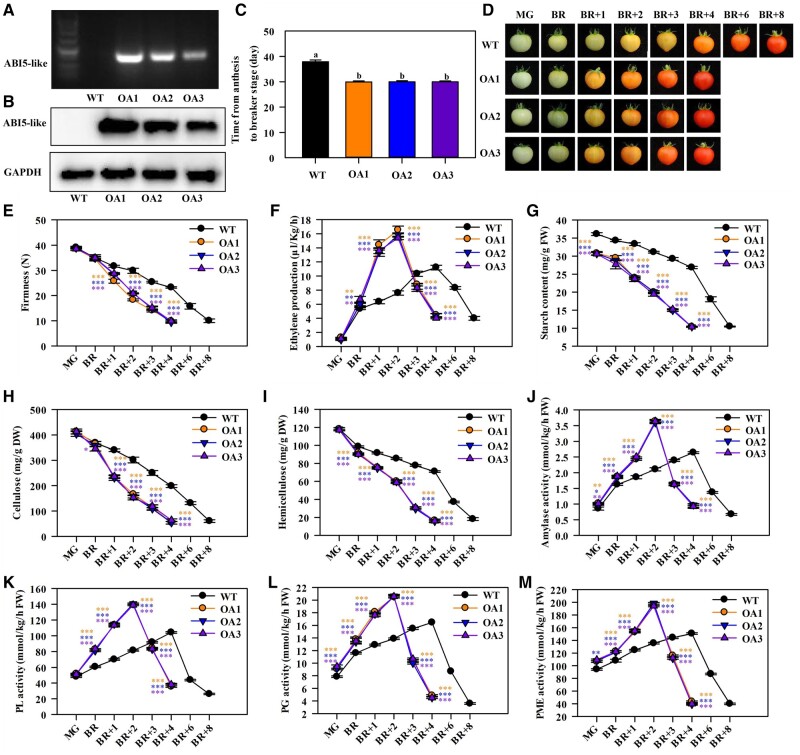
Similar results were obtained from the MaABI5-like-overexpressing lines. Three representative lines (OA1, OA2, and OA3) with high MaABI5-like transcript and protein levels were selected for further investigation (Figure 7, A and B; Supplemental Figure S11B). All the lines displayed a rapid maturation phenotype by about 8 d compared to the WT fruits (Figure 7C). The ripening process of the MaABI5-like-OE lines from the BR stage to full ripening was also accelerated by about 4 d compared to the WT line (Figure 7D). The MaABI5-like-OE lines showed a more rapid decrease in firmness, a higher level of ethylene production, and significantly lower levels of starch, cellulose, and hemicellulose content compared with the WT fruit during ripening (Figure 7, E–I). The amylase, PL, PG, and PME activity were significantly higher and increased more rapidly in the MaABI5-like-OE fruits than in the WT fruits (Figure 7, J–M). This phenomenon explained the increased fruit ripening and softening phenotype.
Figure 7.
OE of ABI5-like in tomato promotes fruit ripening and induces starch and cell wall degradation. A and B, The expression of ABI5-like in WT fruit and ABI5-like-OE lines as determined by RT-PCR (A), and western-blot (B). Pericarp tissues of ABI5-like-overexpressing lines and WT at 30 DPA were used for detection. For western-blot analysis, total proteins were extracted and probed with an anti-ABI5-like polyclonal antibody. GAPDH was used as the internal reference. C, Time from anthesis to BR stage of WT and ABI5-like-OE lines. Each value represents the means of 30 biological replicates (mean ± sd). Means with different letters (a–c) are significantly different as determined by ANOVA and Duncan’s multiple range tests (P < 0.05). D, The fruit ripening process of WT and ABI5-like-OE lines at MG, BR, BR + 1 d, BR + 2 d, BR + 3 d, BR + 4 d, BR + 6 d, and BR + 8 d stages. E–M, Changes in fruit firmness (E), ethylene production (F), the content of starch (G), cellulose (H), and hemicellulose (I), and the activities of amylase (J), PL (K), PG (L), and PME (M) in WT and ABI5-like-OE lines during ripening. Each value represents the means of three biological replicates (mean ± sd). Asterisks indicate significant differences as determined by ANOVA and Duncan’s multiple range tests at *P < 0.05, **P < 0.01, and ***P < 0.001 level, respectively.
OE of MaEBF1 and MaABI5-like alters the transcriptional profiles of genes related to fruit ripening and softening
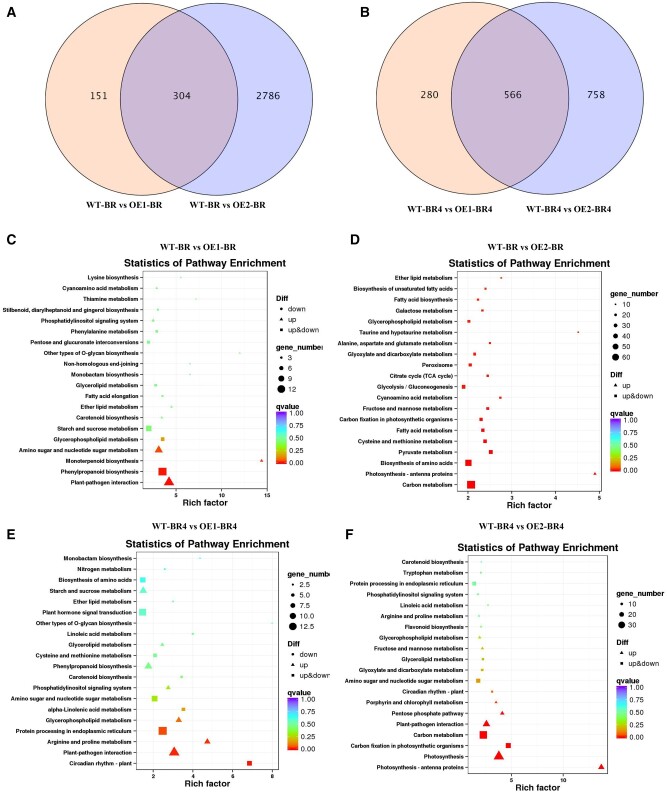
WT, OE1, and OE2 samples at BR and BR + 4 d were subjected to RNA-seq analysis to further investigate the effects of MaEBF1 OE on fruit ripening. A total of 3241 and 1604 differentially expressed genes (DEGs) were detected in the OE1/2 and the WT samples at BR and BR + 4 stage, respectively (Figure 8, A and B). The DEGs detected in the WT and OE1/2 were enriched in phenylpropanoid biosynthesis and metabolism of starch, sucrose, carbon, fructose, and mannose at the BR stage (Figure 8, C and D). In the same lines of OE1/2, the DEGs were enriched in amino and nucleotide sugar metabolism, phenylpropanoid biosynthesis, plant hormone signal transduction, starch, and sucrose metabolism, photosynthesis, and carbon metabolism at the BR + 4 stage (Figure 8, E and F).
Figure 8.
Comparison of the DEG of the WT and EBF1- OE lines. A and B, Venn diagram analysis of the WT and EBF1-OE lines at BR (A) and BR + 4 d (BR4) (B) stages. The library from the WT samples was used as a calibrator to normalize the DEGs of the other four libraries. C and E, Top 20 KEGG enrichment pathways comparing the DEGs of WT and OE1 lines at BR (C) and BR + 4 (E) stage. D and F, Top 20 KEGG enrichment pathways comparing the DEGs of WT and OE2 lines at BR (D) and BR + 4 (F) stage.
Similar results were obtained from the RNA-seq analysis of MaABI5-like-OE lines. Numerous DEGs were detected in the OA1/2 and the WT at the BR and BR + 2 stage (Supplemental Figure S12, A and B). The DEGs were mainly enriched in phenylpropanoid biosynthesis, photosynthesis, and metabolism of starch, sucrose, carbon, fructose, and mannose (Supplemental Figure S12, C–F).
Ethylene, auxin, and ABA are essential hormones that regulate fruit ripening. Various genes involved in the ethylene, auxin, and ABA pathways were significantly upregulated in both MaEBF1 and MaABI5-like transgenic lines compared to the WT lines (Supplemental Figures S13, A–C, and S14, A–C). Genes involved in chlorophyll, starch, and cell wall degradation pathways were also significantly upregulated in the transgenic lines than the WT lines (Supplemental Figures S13, D–F, and S14, D–F). Notably, SlEBFs were significantly downregulated in MaEBF1 transgenic lines (Supplemental Figures S13A and S15). However, SlABI5s (SlABI5-like2 and SlABI5-like7) were upregulated in the MaABI5-like transgenic lines (Supplemental Figure S14C).
Transient OE of EBF1 or ABI5-like promotes ripening and softening of Fenjiao banana fruit
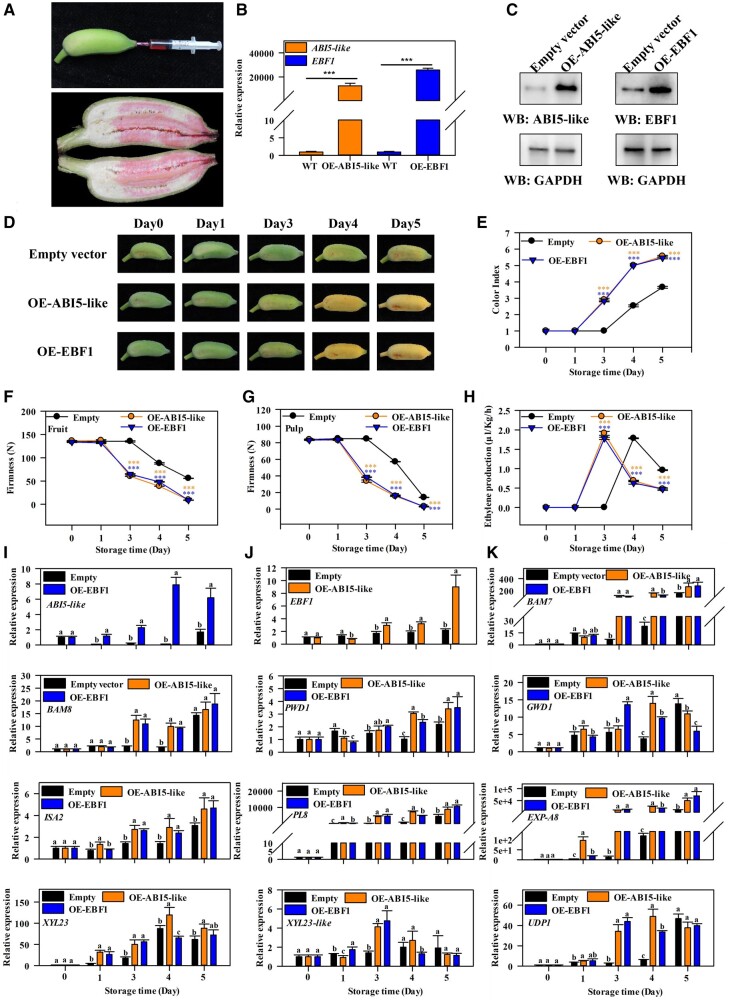
The roles of EBF1 and ABI5-like proteins in regulating fruit ripening and softening were investigated by their transient OE in Fenjiao banana fruit (Figure 9A). The banana fruits showed high expression levels of EBF1 and ABI5-like genes and proteins (Figure 9, B and C). OE of EBF1 and ABI5-like accelerated fruit ripening, characterized by more rapid yellowing of the peels than the nontransformed fruits (Figure 9D). Moreover, OE of EBF1 and ABI5-like increased the fruit color index, decreased the fruit and pulp firmness, and increased ethylene production compared to the control fruits (Figure 9, E–H). The expression of EBF1 and ABI5-like was induced in ABI5-like-OE and EBF1-OE lines, respectively (Figure 9, I and J). Because ABI5-like protein targets the genes related to starch and cell wall degradation, the expression of these genes was tested in the transient OE fruits. According to the results, genes related to starch and cell wall degradation, including BAM7, BAM8, PWD1, GWD1, ISA2, EXP-A8, XYL23-like, PL8, UDP1, and XYL23, were expressed at significantly higher levels in EBF1- and ABI5-like-OE fruits compared to the control fruits (Figure 9K). These results show that EBF1- and ABI5-like-OE promotes the ripening and softening of Fenjiao banana fruits by inducing the expression of softening-related genes. This further confirms the function of ABI5-like and EBF1 in regulating the ripening and softening of banana fruit.
Figure 9.
Effects of transient OE of EBF1 and ABI5-like in Fenjiao banana fruit on ripening, and starch, and cell wall degradation. A, Schematic diagram showing the injection of A. tumefaciens solution into Fenjiao banana pulp through the distal end of the fruit. B and C, The OE of EBF1 and ABI5-like in Fenjiao banana fruit of the empty vector as determined by RT-qPCR (B) and western-blot (C). The expression levels of each gene are relative to WT, which is set as 1. Each value represents the means of biological triplicates (mean ± sd). Each value represents the means of biological triplicates (mean ± sd). Asterisks indicate significant differences as determined by ANOVA and Duncan’s multiple range tests at ***P < 0.001 level. D, Fenjiao banana fruit ripening process of the empty vector, EBF1-, and ABI5-like-OE lines. E–H, Changes in the color index (E), firmness of whole fruit (F) and pulp (G), and ethylene production (H) during fruit ripening. Differences between various treatments were assessed by ANOVA and Duncan’s multiple range tests (***P < 0.001). I–K, The transcript levels of ABI5-like (I), EBF1 (J), and corresponding genes related to starch and cell wall degradation (K) during ripening. The expression levels of each gene at different days are relative to that of 0 day (freshly harvested fruit) set as 1. Each value represents the mean of three biological replicates (mean ± sd). Different letters indicate significant differences between groups as determined by ANOVA and Duncan’s multiple range tests (P < 0.05).
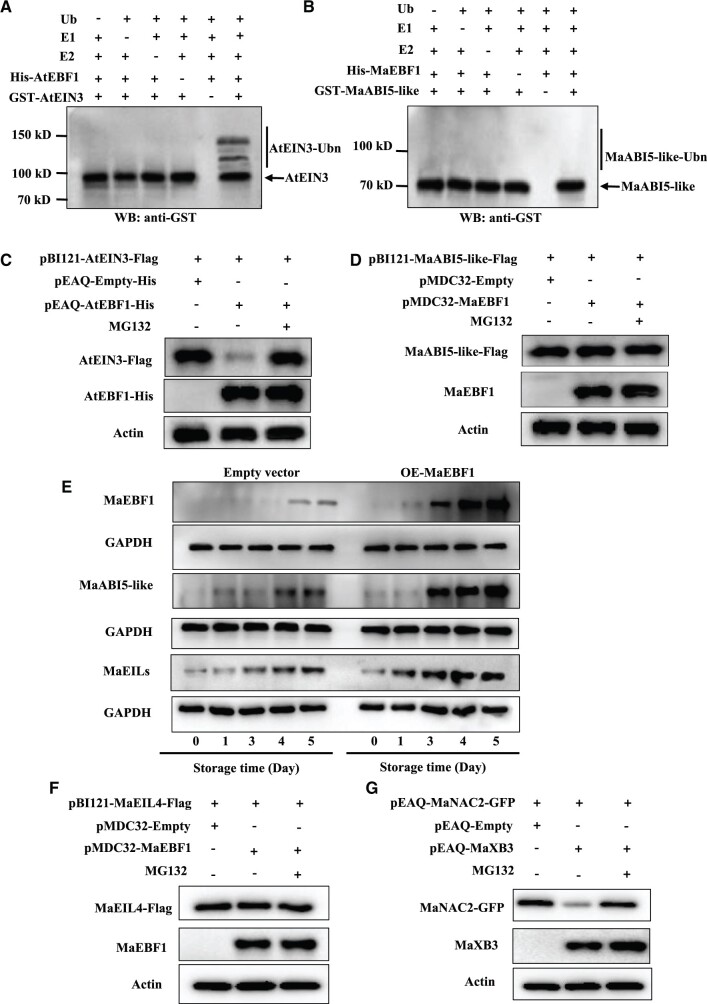
EBF1 does not ubiquitinate or degrade ABI5-like protein via the ubiquitin/proteasome pathway
EBF1 has been shown to function as an E3 ubiquitin ligase in Arabidopsis. It is an F-box protein that targets EIN3/EIL1 for degradation via the ubiquitination pathway in Arabidopsis; thus it plays a negative role in ethylene signaling (Guo and Ecker, 2003; Potuschak et al., 2003; Binder et al., 2007). To test the role of EBF1 in banana fruit ripening, we investigated whether EBF1 could ubiquitinate ABI5-like protein. The ubiquitin–proteasome system of Arabidopsis EBF1 proteins (AtEBF1–AtEIN3) was introduced into the assay as a positive control. Co-incubation of recombinant GST-AtEIN3 (Supplemental Figure S8A) and His-AtEBF1 proteins (Supplemental Figure S8B) in the presence of Ub, ATP, E1, and E2 yielded poly-ubiquitinated chains products with higher molecular-mass bands. However, the ubiquitinated band was not observed in the absence of Ub, E1, or E2 (Figure 10A). Notably, the ubiquitinated band was not produced during the co-incubation of recombinant GST-MaABI5-like (Supplemental Figure S8C) and His-MaEBF1 proteins both in the presence and in the absence of Ub, E1, or E2 (Supplemental Figure S8F; Figure 10B). These results indicate that MaEBF1 cannot ubiquitinate MaABI5-like protein in vitro. In vivo ubiquitination assays also showed that AtEBF1 could ubiquitinate and degrade AtEIN3 (Figure 10C;Supplemental Figure S16B). However, MaEBF1 could not ubiquitinate MaABI5-like (Supplemental Figure S16A), and the stability of the MaABI5-like protein was not affected by MaEBF1 in the plant cells (Figure 10D). The level of ubiquitination in the OE-MaEBF1 banana, fruit and empty-vector control was also assessed using an anti-ubiquitin antibody. The results showed that the ubiquitin signal increased during the later fruit ripening stage, but no difference was observed between the empty vector control and the OE-MaEBF1 fruit (Supplemental Figure S17). This result indicated that overexpressed MaEBF1 did not enhance the ubiquitination level of fruit. In addition, the total protein of OE-MaEBF1 banana fruit was extracted and immunoprecipitated (IP) using the anti-ABI5-like antibody, then detected by anti-Ub, anti-EBF1, and anti-ABI5-like antibody, respectively. As shown in Supplemental Figure S18, after IP using the anti-ABI5-like antibody, the EBF1 and ABI5-like proteins were detected by immunoblotting using the anti-EBF1 and anti-ABI5-like antibodies, respectively. However, no ubiquitin signal was detected when immunoblotting using the anti-Ub antibody (Supplemental Figure S18). These results indicated that EBF1 interacted with ABI5-like, but not ubiquitinated ABI5-like in banana fruit. The above findings were further confirmed by assessing the levels of MaABI5-like protein in banana fruits transiently overexpressing MaEBF1. Transient OE of MaEBF1 accelerated fruit ripening compared to the empty control (Figure 9D). MaEBF1 protein levels increased with fruit ripening, and the transient OE fruits showed substantially more MaEBF1 protein than the control (Figure 10E). Notably, the levels of MaABI5-like protein also increased with fruit ripening, and substantially more MaABI5-like protein was detected in MaEBF1 OE samples. These findings indicate that the levels of MaABI5-like and MaEBF1 proteins were positively associated in the MaEBF1 transient OE lines (Figure 10E). These results show that MaEBF1 does not ubiquitinate or degrade MaABI5-like protein through the ubiquitin/proteasome pathway.
Figure 10.
MaEBF1 does not ubiquitinate and degrade MaABI5-like and MaEIL proteins. A, AtEBF1 ubiquitinates AtEIN3 in vitro. Recombinant His-AtEBF1 protein was co-incubated with GST-AtEIN3 protein in the presence or absence of ubiquitin (Ub), triphosadenine (ATP), human E1, and human E2 UbcH5b at 30°C for 2 h. B, MaEBF1 does not ubiquitinate MaABI5-like in vitro. Recombinant His-MaEBF1 protein was co-incubated with GST-MaABI5-like protein in the presence or absence of Ub, ATP, human E1, and human E2 UbcH5b at 30°C for 2 h. The reaction mixture was analyzed by immunoblot with an anti-GST antibody. Ubiquitination in a heterogeneous collection of higher molecular mass proteins detected using anti-GST antibody. The molecular weight (kilodarcy) is indicated on the left side of the gel. C and D, MaEBF1 does not mediate the proteasomal degradation of the MaABI5-like protein in plant cells. Technique control was done using AtEBF1–AtEIN3 as mentioned above. As indicated, AtEIN3 fused with Flag was expressed alone or co-expressed with AtEBF1-His in N. benthamiana leaves in the presence or absence of MG132 (a potent covalent inhibitor of the aldehyde proteasome pathway, forms a hemiacetal with the hydroxyl of the active site threonines and inhibits proteasome function) (C). MaABI5-like fused with Flag was expressed alone or co-expressed with MaEBF1 in N. benthamiana leaves in the presence or absence of MG132 (D). The resulting protein extracts were analyzed using an anti-Flag, anti-His or MaEBF1 antibody. Actin was used as the loading control. The molecular weight (kilodarcy) is indicated on the right side of the gel. E, The protein level of MaEBF1, MaABI5-like, and MaEILs in MaEBF1 transient OE line and empty vector. F–G, MaEBF1 does not mediate the proteasomal degradation of MaEIL4 protein in plant cells. As indicated, MaEIL4 fused with Flag was expressed alone or co-expressed with MaEBF1 in N. benthamiana leaves in the presence or absence of MG132 (F). Technique control using MaNAC2-MaXB3 as reported by Shan et al. (2020). As indicated, MaNAC2 fused with GFP was expressed alone or co-expressed with MaXB3 in N. benthamiana leaves in the presence or absence of MG132 (G). The resulting protein extracts were analyzed using an anti-Flag or anti-GFP antibody. Actin was used as the loading control. The molecular weight (kilodarcy) is indicated on the right side of the gel.
EIL4 interacts with EBF1 but is not ubiquitinated and degraded by it
We were curious to understand why EBF1 does not ubiquitinate and degrade ABI5-like protein via the ubiquitin/proteasome pathway but functions as a typical EBF in the ethylene signal pathway that degrades EIN3/EILs. To answer this question, we first assessed EIL protein levels during the ripening process of banana fruits transiently overexpressing MaEBF1 using the AtEIN3 antibody. MaEILs protein was detected using the AtEIN3 antibody (Supplemental Figure S19A). MaEILs and MaEBF1 protein levels increased with fruit ripening. Substantially higher levels of MaEILs protein were detected in MaEBF1 OE samples compared with the control fruits (Figure 10E), indicating that EBF1 did not degrade ethylene-insensitive 3-like (EILs) protein. Furthermore, the ubiquitination assays performed in vivo showed that MaEBF1 could not ubiquitinate or degrade MaEIL4. MaEBF1 interacts with MaEIL4 in yeast but not with MaEIL1/2/3/5 (Supplemental Figure S20A). The interaction was further validated by BiFC assay (Supplemental Figure S20B). MaEIL4 expression was similar to the softening-related genes and MaABI5-like, which remarkably increased under the natural ripening condition but were severely repressed by cold storage (7°C and 11°C) (Supplemental Figure S20C). However, the in vivo ubiquitination assays showed that MaEBF1 could not ubiquitinate MaEIL4 (Supplemental Figure S20D), and the stability of the MaEIL4 protein was not affected by MaEBF1 in the plant cells (Figure 10F). The technique control included in the assays showed that MaXB3 could ubiquitinate and degrade MaNAC2 (Figure 10G, Supplemental Figure S20E). The transient OE of MaEBF1 in banana fruit induced the expression of MaEIL4 and several marker genes in the ethylene signaling pathways (Supplemental Figure S21). These results indicate that MaEBF1 is not a true ortholog of EBF1/2 of Arabidopsis and tomato but harbors a different function in regulating fruit ripening in banana.
Discussion
Fruit firmness is an important phenotypic indicator of fruit ripening. Starch and cell wall degradation play a critical role in banana fruit softening (Fan et al., 2018; Xiao et al., 2018; Song et al., 2019). Herein, CTS caused a softening disorder that impaired the fruit ripening quality (Figure 1). The softening of banana pulp is attributed to starch and cell wall degradation during ripening (Shiga et al., 2011). The amylose and amylopectin contents decrease while amylase activity increases during fruit softening. However, cold stress significantly repressed these activities (Figure 1, G–I). The starch degradation process in banana involves multiple enzymes. Currently, 38 genes encoding starch degradation-related enzymes have been identified and characterized (Xiao et al., 2018). Our previous study identified 38 genes encoding starch degradation-related enzymes through RNA-seq analysis, including 11 starch degradation genes severely repressed by CTS and closely associated with Fenjiao banana ripening disorders (Song et al., 2019). Cell wall modification is a primary factor for maintaining fruit firmness. It involves nonenzymatic and enzymatic degradation of cell wall components (Li et al., 2010). Herein, the TEM assay revealed that the cell wall of CTS fruits was not completely degraded (Supplemental Figure S1B). PG, PL, and PME activities increased during fruit softening. However, their activities were severely repressed by cold stress (Figure 1, J–L). Previously, 23 cell wall-modifying genes closely related to banana softening have been identified (Asif et al., 2014). Herein, 61 cell wall degradation-associated genes were identified by RNA-seq analysis. Among them, 12 were closely related to ripening disorder (Figure 1M;Supplemental Figure S2). Cold stress-induced fruit softening disorders are attributed to the repression of ethylene production. In this study, cold stress severely inhibited ethylene production (Figure 1F). Previously, ethylene production was reported to increase the expression of 27 starch and 11 cell wall degradation genes (Feng et al., 2016; Xiao et al., 2018). For example, the transcript levels of AdBAM3.1/3L/9 and AdAMY1 remarkably increased with the ripening of kiwifruits (Hu et al., 2016). In papaya, CpPG1, CpPG2, CpEXP1, CpEPX2, and CpPME1/2/3 were upregulated during fruit ripening (Ding et al., 2019). These reports indicate that ethylene controls fruit softening by regulating the genes related to starch and cell wall degradation.
ABI5 is a plant leucine zipper (bZIP) transcription factor that functions in the ABA signaling pathway (Yu et al., 2015). In A. thaliana, AtABI5 regulates seed germination, TAG (triacylglycerol) biosynthesis, adaptation to abiotic stress, chlorophyll degradation, leaf senescence, and growth of lateral roots (Anna et al., 2016). ABI5 also interacts with the components of gibberellic acid (GA) (Xi et al., 2010), cytokinins (Wang et al., 2011), jasmonic acid (Chen et al., 2012), auxins (Yuan et al., 2014), and brassinosteroids pathways to regulate various biological processes (Cai et al., 2014). ABI5 is regulated by various transcript factors at the transcriptional level, including MYB7, WRKY18, WRKY40, WRKY60, ABI3, and ABI4 (Anna et al., 2016; Wang et al., 2021). At the posttranslational level, ABI5 activity is regulated via phosphorylation/dephosphorylating and ubiquitination process (Anna et al., 2016). In plum fruit, ABI5-like protein binds to the ACS1 promoter to enhance ethylene synthesis, accelerating fruit ripening (Sadka et al., 2019). Herein, ABI5-like protein interacted with EBF1 and activated the expression of starch and cell wall degradation-related genes by directly binding to their promoters (Figures 2–4). Exogenous ABA treatment hastened the ripening and softening process and relieved the CI symptoms and ripening disorders caused by the cold stress. Also, the treatment induced the expression of EBF1, ABI5-like, and genes related to starch and cell wall degradation, thus hastening the fruit ripening processes (Figure 5; Supplemental Figure S10). Ethylene synthesis and starch and cell degradation genes were significantly upregulated in ABI5-like-OE lines compared to the WT lines (Supplemental Figure S14). Notably, five starch and five cell wall degradation genes were significantly upregulated in EBF1- and ABI5-like-OE fruits compared to the nontransformed lines (Figure 9K). These results indicate possible crosstalk between the ethylene and ABA signaling pathways in regulating fruit ripening and softening in Fenjiao banana. Both ethylene and ABA play important roles in fruit ripening and softening and regulating cold stress-induced ripening disorder.
EBF1 interacts with NAC67-like protein to regulate fruit ripening and softening. Cold stress causes ripening disorder in Fenjiao banana fruits (Song et al., 2019). Notably, NACs (NAM, ATAF1/2 and CUC2) TF play important roles in the ripening of Cavendish banana fruits. MaNAC1/2 expression is closely related to the fruit ripening process. The protein interacts with MaEIL5, an important component of the ethylene signaling pathway (Shan et al., 2012). MaEBF2 has also been shown to interact with MaEIL5 in Cavendish banana fruit (Kuang et al., 2013). Evidence suggests that EBFs/EIL5/NACs interaction network may play an important role in banana fruit ripening. MaNAC1/2 regulates the expression of MaXB3 and interacts with MaXB3 protein involved in the multilayered regulatory cascade of MaXB3/MaNACs/MaERF11/MaACS1/MaACO1 that controls ethylene biosynthesis during banana fruit ripening (Shan et al., 2020). Herein, a group of SlNAC gene family members was differentially expressed in the MaEBF1-OE and WT lines (Supplemental Figure S22A). The SlNACs genes, especially those close to MaNAC67-like in the evolutionary tree, were significantly upregulated in the MaEBF1-OE lines (Supplemental Figure S22B). These results indicate that MaEBF1 OE in tomato fruits potentially alters SlNAC expression to regulate fruit softening.
Substantial evidence indicates that EBFs play a vital role in ethylene signal transduction as negative regulators in the ethylene signaling pathway (Guo and Ecker, 2003; Potuschak et al., 2003). In tomato, SlEBF1 and SlEBF2 genes were significantly upregulated during the BR stage and maintained a high expression level during the ripening process. Their expression was induced by exogenous ethylene. However, their corresponding silenced mutants showed accelerated ripening (Yang et al., 2010). Similarly, the expression of SlEBF2-like and SlEBF3 increased with fruit ripening. However, OE of SlEBF2-like and SlEBF3 reduced ethylene production and inhibited fruit ripening and softening in tomato (Deng et al., 2018; Guo et al., 2018b). In apple, the expression of MdEBF1/2 increases sharply during fruit ripening. The interaction of MdEBF1 with MdEILs represses the trans-activation of MdPG1 (Tacken et al., 2012). In papaya, the expression of CpEBF1 increases with fruit ripening, thus activating the activity of CW-degradation gene promoters by interacting with CpMADS1/3 (Ding et al., 2019). CpEBF1 ubiquitinated and degraded CpEIL1 via the 26S proteasome pathway, which mediates the transcription of fruit ripening-associated genes and regulates papaya fruit ripening (Zhang et al., 2020). Five EBF genes were isolated from the banana genome, and all could respond to ethylene treatment, especially MaEBF3/4 (Jourda et al., 2014). The expression of MaEBF1 (Wu et al., 2018) and MaEBF2 (Kuang et al., 2013) in bananas increases with fruit ripening and is positively regulated by ethylene. Ethylene induces the promoter activity of MaEBF2, further supporting its involvement in fruit ripening (Kuang et al., 2013). In our previous study, differential expression analysis of RNA-seq data of Fenjiao banana fruit only screened out MaEBF1, which may be involved in fruit ripening (Song et al., 2019, 2020b). Herein, MaEBF1 was shown to interact with MaABI5-like and regulate the starch and cell wall degradation (Figures 2–4). However, no interaction was observed between MaABI5-like and MaEBF2, MaEBF3, MaEBF4, and MaEBF5 proteins (Supplemental Figure S6). OE of MaEBF1 in tomato promoted ripening by enhancing ethylene production and the expression of genes involved in ethylene biosynthesis and signaling pathways, as well as cell wall and starch degradation (Figure 6; Supplemental Figure S13). The MaEBF1 OE lines also showed enhanced triple response when treated with ACC, indicating a higher sensitivity to ethylene (Figure 6, N and O). However, these results differed from those obtained with the SlEBF3 OE lines, which showed less sensitivity to ethylene than the WT lines (Deng et al., 2018). SlEBF3 OE lines showed delayed fruit ripening phenotype, reduced ethylene production and lower PL gene expression level (Deng et al., 2018). SlEBF2-like OE lines also showed significantly delayed fruit development and ripening process in tomato (Guo et al., 2018b). These results suggest that MaEBF1 may play a different role from SlEBFs in tomato. Intriguingly, as a non-TF, EBF1 could also induce the transcription of some cell wall and starch degradation-related genes. It was hypothesized that EBF1 might act as a transcriptional co-factor to regulate gene expression. It may interact with partners other than ABI5-like and affect their transcriptional activation to regulate fruit ripening. Similar results also showed that transient OE of non-TFs, such as OsMFT2, MaMPK2, and CpEBF1 alone could induce the transcription of target genes in rice, banana, and papaya, respectively (Ding et al., 2019; Wu et al., 2019; Song et al., 2020a). These results also indicate that EBF1 plays important roles in fruit ripening.
This study also found that EBF1 cannot ubiquitylate and degrade EIL4 in vivo (Figure 10F, Supplemental Figure S20D). EBF1 interacts with EIL4, but not with other EILs (EIL1/2/3/5) (Supplemental Figure S20, A and B). Phylogenetic analysis of EIL proteins indicates that EIL4 is more similar to other EILs in banana (EIL3/1/5) than those in other species, and is more closely related to SlEIL3 in tomato and AtEIL1 in Arabidopsis (Supplemental Figure S23). Similar results were reported by Jourda et al. (2014). EIL4 expression was closely related to fruit ripening (Supplemental Figure S20) and was induced by the transient OE of EBF1 in banana fruit (Supplemental Figure S21). The phenotype difference between banana and tomato was further validated using the RNA-seq data and RT-qPCR results. SlEBF1, SlEBF1-like, and SlEBF2 were repressed in the MaEBF1 OE lines (Supplemental Figures S12, A and S14), suggesting that the lines partially served as SlEBF1, SlEBF1-like, and SlEBF2 knockdown mutants, and showed an accelerated ripening phenotype as previously reported (Yang et al., 2010). These results indicate that MaEBF1 is not a functionally conserved EBF ortholog in Arabidopsis and tomato, but harbors a different function in regulating fruit ripening in banana. This function of MaEBF1 could be due to the expansion of EIL genes in banana, which is also paralleled by the expansion of EBF genes that control EIL protein levels in banana (Jourda et al., 2014). This gene expansion phenomenon potentially led to the functional redundancy or subfunctionalization of MaEBF and MaEIL genes to ensure finer control of ethylene signaling (Jourda et al., 2014).
Our results also show that EBF1 cannot ubiquitinate ABI5-like protein (Figure 10, B and D). In the banana fruits transiently overexpressing EBF1, ABI5-like and EBF1 protein levels substantially increased with fruit ripening and were positively associated, indicating that ABI5-like is not degraded by EBF1 ubiquitination (Figure 10E).
Based on our results, we proposed a model deciphering the roles of EBF1 and ABI5-like proteins in regulating the ripening and softening of Fenjiao banana fruits (Supplemental Figure S24). Both ethylene and ABA hormones play an important role in regulating fruit ripening by inducing the expression of EBF1 and ABI5-like. ABI5-like binds to the promoters of starch and cell wall degradation genes, thereby activating their expression. Additionally, EBF1 interacts with ABI5-like protein, thus activating the starch and cell wall degradation genes to promote fruit ripening and softening. However, cold stress (7°C, CTS) reduces ethylene production and ABA content, thereby severely repressing EBF1 and ABA5-like expression. This phenomenon inhibits starch and cell wall metabolism genes, resulting in fruit softening failure. Although the mechanism of interaction between EBF1 and ABI5-like in regulating fruit ripening needs further investigation, this study provides insights into climacteric fruit ripening and softening, and offers potential techniques of inducing tolerance to chilling stress in banana.
Materials and methods
Plant materials, treatments, and growth conditions
Mature-green Fenjiao banana (Musa ABB Pisang Awak cv. “Guangfen NO.1”) fruits were harvested at the 85%–90% plump stage from a local commercial farm in Guangzhou city, Guangdong province, China, in August 2018. The fruits were then pretreated as described in a previous study (Song et al., 2019).
Three independent experiments were conducted after pretreatment. They included cold storage, controlled ripening by ethylene and ABA, and exogenous ABA application on fruit under cold storage dubbed experiments 1, 2, and 3, respectively. For experiment one, three storage temperatures were set, including a control (natural ripening at 25°C), 7°C, and 11°C. Fruits stored at 7°C and 11°C were treated with 1 μl·L−1 of ethephon for 1 min after 6 and 12 d of storage, and then stored at 25°C and 90% relative humidity to favor ripening. Supplemental details of the experiment are as reported previously (Song et al., 2019). Experiments two and three are described in Supplemental Method S1.
Nicotiana benthamiana plants were planted in soil and cultivated in a growth chamber at 22°C under long-day conditions (16-h light/8-h dark) and maintained for 4–6 weeks. They were then used for Agrobacterium tumefaciens-mediated transient expression assays. Tomato (Solanum lycopersicum cv. Micro-Tom) was used for transformation. Tomato seedlings were grown in a phytotron with a 16-h light (25°C)/8-h dark cycle.
The physiological parameters examined are described in Supplemental Method S2.
Y2H assay
The Y2H assay was conducted using the Matchmaker Gold Yeast Two-Hybrid System (Clontech, Cat. No. 630489). The full coding sequence (CDS) of EBF1 was ligated into the pGBKT7 vector and used as the bait protein to screen the candidate interacting proteins in the cDNA library. The ABI5-like and EIL4 proteins were screened out. The full-length CDS of ABI5-like and EIL1-5 was separately inserted into pGBKT7 and pGADT7 vectors. The yeast strains were then co-transformed with EBF1 + ABI5-like and EBF1 + EIL1-5 to confirm their interaction.
BiFC assay
EBF1 was cloned into a pBIFC vector as described by Song et al. (2019). Meanwhile, the full CDS of ABI5-like and EIL4 without the stop codon was ligated into the pBIFC vectors using Gateway technology. The BiFC experiment was then performed as described by Ding et al. (2019).
GST pull-down assay
The EBF1-GST protein was purified as described by Song et al. (2019). The full-length CDS of ABI5-like without the stop codon was cloned into a PET-28a vector. The fused vector was then transformed into BM Rosetta (DE3) cells, and the expression of the ABI5-like-His protein-induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG, catalog no. 9030, TAKARA, Japan) at 20°C for 6 h. The cells were subsequently harvested and the ABI5-like-His protein purified using the Ni-NTA His Purification Kit (Clontech, Cat. No. 635658). The GST pull-down experiment was performed as described by Song et al. (2019).
CoIP assay
CoIP experiments were conducted as described by Sun et al. (2019). The full CDS of EBF1 and ABI5-like genes without the stop codons were inserted into the pEG302 and pMDC43 vectors. The constructs were then introduced into A. tumefaciens strain GV3101 and infiltrated into the abaxial side of the 4- to 6-week-old N. benthamiana leaves using a 1 mL needleless syringe as described by Sainsbury et al. (2009).
The N. benthamiana leaves were then ground in liquid nitrogen after infiltration for 48 h. Total proteins of 35S: EBF1-GFP and 35S: ABI5-like-Flag of N. benthamiana leaves were extracted using IP buffer (10% [v/v] Glycerol, 25 mM Tris–HCl pH 7.5, 1-mM EDTA, 150-mM NaCl, 0.1% [v/v] Triton-100, 2 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 × protease inhibitors). The two protein sources were mixed and incubated for 2–3 h at 4°C after centrifugation at 13,000 rpm for 20 min. The mixture was then incubated with the anti-GFP rabbit polyclonal antibody 10004D (Thermo Fisher, CA, USA) for another 2 h. The beads were then washed four times with IP buffer and subsequently boiled in 60 μL 2 × SDS buffer. The protein mixture was then separated on a 10% (v/v) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), and the bands were probed using anti-Flag antibodies at a 1:1000 dilution. The banding signals were detected using the CanoScan LiDE 100 scanner (Canon, Japan).
Dual-luciferase transient expression assays
The promoter sequences of 11 starch and 12 cell wall degradation genes were cloned into the pGreenII 0800-LUC double-reporter vector. The full CDS of EBF1 and ABI5-like were then inserted into the pGreenII 62-SK effector vector, followed by co-infiltration of the reporter and effector plasmids into N. benthamiana leaves (Song et al., 2019). The Dual-Luciferase Assay Kit (Promega, Madison, WI, USA) was used to analyze the luciferase activity. The results were expressed as the LUC (firefly luciferase)/REN(renilla luciferase) ratio as an average of six replicates.
Polyclonal antibody preparation and immunoblot analysis
The coding sequences of EBF1 and ABI5-like genes were cloned into pET-28a to generate His-EBF1 and His-ABI5-like constructs, which were subsequently transformed into Escherichia coli strain BL21. The recombinant His-EBF1 and His-ABI5-like proteins were expressed with 0.5-mM IPTG at 20°C for 6 h, and affinity-purified using His60 Ni Superflow Resin (Clontech, Cat. No. 635658). His-fusion proteins were separated using SDS–PAGE. Bands of interest were excised and used as antigens for antibody production. The antibodies were produced in rabbits by HuaAn Biotechnology Company (Hangzhou, China).
Immunoblot analysis was performed using the polyclonal antibodies of EBF1 and ABI5-like at 1:500 dilution with Tris-buffered saline with Tween-20 (TBST) buffer. The secondary antibodies (anti-rabbit horseradish peroxidase-conjugated, 1 mg/mL; Cat. No. CW0103; CW Biotech) were diluted 1:1,000 with TBST buffer. AtEIN3 antibody was used as described by Guo and Ecker (2003). The anti-EBF1 and anti-ABI5-like antibody specificity was confirmed by immunoblot analysis using in vitro-translated recombinant protein and total protein extracts from banana fruit pulp (Supplemental Figure S19). The anti-ubiquitin antibody (Sigma) was used for examining the ubiquitination level of banana fruit transiently overexpressing MaEBF1.
Protein expression analysis and electrophoretic mobility shift assay
The coding sequence of the ABI5-like and EIL4 genes was ligated into the pGEX-4T-1 vector. The plasmids were then transformed into E. coli strain BM Rosetta (DE3) cells and subsequently treated with 0.5-mM IPTG at 28°C for 6 h before cell harvesting. The ABI5-like-GST and EIL4-GST proteins were then purified using the GST-Tagged Protein Purification Kit (Clontech, Cat. No. 635619) following the manufacturer’s protocol.
The probes, including the ABI5-like binding site (ABRE/G-box) originating from the promoters of starch and cell wall degradation genes, were labeled with biotin at the 5′-end using the Pierce™ Biotin 3′-End DNA Labeling Kit (Thermo Scientific, Cat: No. 89818). The unlabeled probes with the same sequence DNA fragment were used as competitors, and the ABRE/G-box base changed to A as a mutant. EMSA was performed using the Light Shift Chemiluminescent EMSA Kit (Thermo Scientific, Cat: No. 20148), as described by Xiao et al. (2018).
ChIP-qPCR analysis
The ChIP-qPCR assay was conducted as described by Fan et al. (2018). The banana fruit pulp was submerged in 1% (v/v) formaldehyde to crosslink genomic DNA and protein. The chromatin was sheared to an average length of 500 bp by sonication. Immunoprecipitation of ABI5-like crosslinked DNA was then performed using the affinity-purified polyclonal antibody of ABI5-like for 12 h at 4°C with rotation. A reaction with pre-immune serum IgG or without the antibody was used as the negative control. The DNA–protein–antibody complex was captured on Protein A/agarose beads after incubation for 1 h at 4°C. The beads were pelleted and washed sequentially for 10 min at 4°C with a low-salt wash buffer, high-salt wash buffer, lithium chloride wash buffer, and TE buffer. The IP material was then eluted through gentle rotations for 15 min at 65°C. Crosslinking of IP DNA was reversed by overnight incubation in 0.2 M NaCl at 65°C. The IP DNA was then purified and eluted after treatment with proteinase K. The precipitated DNA fragment was quantified using RT-qPCR as described in Supplemental Method S3.
In vitro ubiquitination assay
The coding sequences of AtEBF1 and AtEIN3 genes were cloned into pET-28a or pET-GST to generate His-AtEBF1 and GST-AtEIN3 constructs. The constructs were then transformed into E. coli strain BL21 using the electroporation method. The recombinant His-AtEBF1 and GST-AtEIN3 proteins were expressed with 0.5 mM IPTG at 16°C for 16 h, and affinity-purified using His60 Ni Superflow Resin (Clontech, Cat. No. 635658) and GST-Tagged Protein Purification Kit (Clontech, Cat. No. 635619). The ubiquitination assay was performed as described by Shan et al. (2020). Briefly, reactions (30 mL) containing 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 0.05 mM ZnCl2, 1 mM ATP (Sigma-Aldrich), 0.2 mM dithiothreitol, 10 mM phosphocreatine, 0.1 unit of creatine kinase (Sigma-Aldrich), 50 ng of human E1 (Boston Biochem), 250 ng of human E2 (Boston Biochem), 2 mg of ubiquitin (Boston Biochem), 500 ng of His-AtEBF1 or His-MaEBF1, and 500 ng of GST-AtEIN3 or -MaABI5-like were incubated at 30°C for 2 h. The reactions were stopped by adding the sample buffer and analyzed by SDS–PAGE followed by immunoblotting using anti-GST antibody (Sigma-Aldrich).
Tomato transformation
The full CDS of MaEBF1 and MaABI5-like was cloned into the PMDC32 vector to generate the OE plants. The constructed plasmid was then introduced into the A. tumefaciens strain GV3101 by electroporation. Tomato (Solanum lycopersicum L. cv. Micro-Tom) plants were transformed following standard methods (Fillatti et al., 1987). The positive transformants were selected on a hygromycin-containing medium and confirmed using the transcript and protein levels of MaEBF1 and MaABI5-like. Three independent transformed lines (T2) were used for the experiments. The ethylene content, firmness, and color index of the WT tomato fruits with overexpression of MaEBF1 were determined at mature green (MG), BR, BR + 2 d, BR + 4 d, and BR + 6 d. Similarly, the ethylene content, firmness, and color index of the WT tomato fruits with overexpression of MaABI5-like were determined at MG, BR, BR + 1 d, BR + 2 d, BR + 3 d, and BR + 4 d.
Transient OE analysis in Fenjiao banana fruit
The full CDS of EBF1 and ABI5-like were cloned into the pMDC32 vector. The constructed plasmids were then introduced into the A. tumefaciens strain GV3101 by electroporation. The A. tumefaciens suspension containing pMDC32-EBF1 and pMDC32-ABI5-like were separately introduced into Fenjiao fruits during the 85%–90% plump stage by injection through the distal end of the fruits (Shan et al., 2020). Transformed fruits were stored at 22°C and 90% relative humidity for 5 d. The fruit firmness, color index, ethylene production, gene expression, and protein accumulation were determined on days 0, 1, 3, 4, and 5 as previously described (Song et al., 2019).
All primers used herein are listed in Supplemental Table S1. Other experimental methods are listed in Supplemental Methods S3–S10.
Data analysis
All experiments were conducted in triplicate in a completely randomized design. Data are presented as means ± standard deviation (sd) for three or six independent biological replicates. Statistical differences between samples were determined using ANOVA followed by the Duncan's multiple range test with software SPSS 16.0. For all tests, data were considered statistically significant when P < 0.05, P < 0.01, or P < 0.001.
Accession numbers
The raw sequence data for RNA-seq were deposited in the NCBI Sequence Read Archive (SRA) under accession nos PRJNA699982 and PRJNA699750. Sequence data from this article can be found in the NCBI Database (https://www.ncbi.nlm.nih.gov/). EBF1 (XM_009405188.2), ABI5-like (XM_009413679.2), BAM3 (XP_009384530.1), BAM4 (XP_009399963.1), BAM7 (XP_009397011.1), BAM8 (XP_009403535.1), PWD1 (XP_009416602.1), GWD1 (XM_009394010.2), AMY3 (XP_009412382.1), ISA2 (XP_009404709.1), PL8 (XM_009387592.2), EXP-A2 (XM_009401634.2), EXP-A8 (XM_009386403.2), UDP (XM_009395179.2), XYL32 (XM_009387774.2), GLU22-like (XM_009419457.2), GLU32 (XM_009384393.2), SUR14-like (XM_009405502.2), SUR14 (XM_009419658.2), XYL23 (XM_009403289.2), XYL23-like (XM_009419212.2), EXP-A15 (XM_009405806.2).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Microstructural assessment of fruit starch and cell wall by transmission electron microscopy (TEM).
Supplemental Figure S2. Expression of 61 cell wall degradation-associated genes in RNA-seq analysis under three different storage and ripening conditions.
Supplemental Figure S3. Phylogenetic analysis of EBF proteins.
Supplemental Figure S4. Differentially expressed genes in abscisic acid signalling pathway analyzed by RNA-seq data.
Supplemental Figure S5. Phylogenetic analyses of ABI5-like sequences.
Supplemental Figure S6. The Y2H assay to test the interaction between EBF2-5 and ABI5-like proteins.
Supplemental Figure S7. The location of ABRE/G-box motifs in the promoters of eight starch and eight cell wall degradation genes.
Supplemental Figure S8. SDS–PAGE gel stained with Coomassie blue demonstrating affinity purification of the recombinant proteins.
Supplemental Figure S9. The transcript levels of some mark genes in ethylene and ABA biosynthesis and signal transduction pathways after exogenous ethephon or ABA treatment during fruit ripening process.
Supplemental Figure S10. Exogenous ABA relieves chilling injury symptoms in Fenjiao banana fruits caused by cold stress (5°C).
Supplemental Figure S11. The expression profiles of MaEBF1 and MaABI5-like in fruits of WT and overexpression lines as determined by RT-qPCR.
Supplemental Figure S12. Comparison of the differentially expressed genes (DEG) of WT and MaABI5-like-overexpression lines.
Supplemental Figure S13. The expression profiles of the differentially expressed genes in WT and MaEBF1-overexpression lines.
Supplemental Figure S14. The expression profiles of the differentially expressed genes in WT and MaABI5-like-overexpression lines.
Supplemental Figure S15. Gene expression analysis of SlEBF1 in WT and MaEBF1-overexpression lines conducted by RT-qPCR.
Supplemental Figure S16. EBF1 does not ubiquitinate ABI5-like in vivo.
Supplemental Figure S17. Immunoblotting analysis showing the level of ubiquitination in MaEBF1-overexpressing and control banana fruit pulp.
Supplemental Figure S18. EBF1 does not ubiquitinate ABI5-like in banana fruit.
Supplemental Figure S19 . Immunoblot analysis of the specificity of polyclonal antibodies.
Supplemental Figure S20. EBF1 interacting with EIL4 but not mediating EIL4 protein degradation via ubiquitin-proteasome system.
Supplemental Figure S21. The transcript levels of several mark genes in ethylene biosynthesis and signal transduction pathways in transient overexpression of EBF1 banana during fruit ripening.
Supplemental Figure S22. A comparison of the differentially expressed SlNAC genes in MaEBF1-overexpression lines and WT lines.
Supplemental Figure S23 . Phylogenetic analysis of EIL proteins.
Supplemental Figure S24. A proposed working model of EBF1 and ABI5-like regulation of starch and cell wall degradation, and the fruit ripening disorder caused by cold stress.
Supplemental Method S1. Ripening treatment by ethylene and ABA.
Supplemental Method S2. Measurement of physiological parameters.
Supplemental Method S3. Gene expression analysis.
Supplemental Method S4. RNA-seq analysis.
Supplemental Method S5. Subcellular localization analysis.
Supplemental Method S6. Yeast One-Hybrid (Y1H) assay.
Supplemental Method S7. In vivo ubiquitination assay.
Supplemental Method S8. In vivo banana ubiquitination assay.
Supplemental Method S9. In vivo proteasomal degradation assay.
Supplemental Method S10. Ethylene triple-response assay.
Supplemental Table S1. Primers used in present study.
Supplementary Material
Acknowledgements
We would like to thank Prof Hongwei Guo for kindly providing the AtEIN3 antibody and Prof Jianye Chen for the generous gift of constructs of pEAQ-MaNAC2-GFP, pEAQ-MaXB3-His, pEAQ-MaXB3 vectors, and MaXB3 antibody. We thank Christian Marzars for proof reading of the manuscript.
Funding
This research was supported by China Agriculture Research System of MOF and MARA (grant no. CARS-31), Guangdong Banana and Pineapple Industry Technology System Innovation Team (grant no. 2020KJ109), Pearl River Talent Program for Young Talent (grant no. 2017GC010321), the International Science and Technology Cooperation Major Project Cultivation Special Fund of SCAU (2019SCAUGH05), and National Key Research and Development Program (grant no. 2016YFD0400103).
Conflict of interest statement. The authors declare no conflicts of interest.
X.Z., W.C., and Xu.L. conceived and designed the experiment. Z.S., Xi.L., Y.Y., J.Q., X.D. and Q.Z. performed the experiments. X.Z. and Z.S. carried out the analysis. Z.S. and X.Z. wrote the manuscript. Xu.L., W.C. and X.P. revised the manuscript. All the authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Xiaoyang Zhu (xiaoyang_zhu@scau.edu.cn).
References
- Albertos P, Wagner K, Poppenberger B (2019) Cold stress signalling in female reproductive tissues. Plant Cell Environ 42:846–853 [DOI] [PubMed] [Google Scholar]
- An J, Zhang X, Liu Y, Zhang J, Wang X, You C, Hao Y (2020) MdABI5 works with its interaction partners to regulate abscisic acid-mediated leaf senescence in apple. Plant J 13:1566–1581 [DOI] [PubMed] [Google Scholar]
- Anna S, Agata DG, Iwona S (2016) The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front Plant Sci 7:1884–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif MH, Lakhwani D, Pathak S, Gupta P, Bag SK, Nath P, Trivedi PK (2014) Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC Plant Biol 14:316–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleeker AB, Vierstra RD (2007) The Arabidopsis EIN3 Binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signalling. Plant Cell 19:509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto N, Tadiello A, Trainotti L, Costa F (. 2016) Climacteric ripening of apple fruit is regulated by transcriptional circuits stimulated by cross-talks between ethylene and auxin. Plant Signaling Behav 12:e1268312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y, Pan S, Dong R, Tang G, Barajas-Lopez J, et al. (2014) GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci USA 111:9651–9656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Li F, Wu Q, Jiang Y, Yuan D (2018) Banana MaABI5 is involved in ABA-induced cold tolerance through interaction with a RING E3 ubiquitin ligase, MaC3HC4-1. Sci Hortic 237:239–246 [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24:2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Pirrello J, Chen Y, Li N, Zhu S, Chirinos X, Bouzayen M, Liu Y, Liu M (2018) A novel tomato F-box protein, SlEBF3, is involved in tuning ethylene signaling during plant development and climacteric fruit ripening. Plant J 95:648–658 [DOI] [PubMed] [Google Scholar]
- Ding X, Zhu X, Ye L, Xiao S, Wu Z, Chen W, Li X (2019) The interaction of CpEBF1 with CpMADSs is involved in cell wall degradation during papaya fruit ripening. Hortic Res 6:2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S (2020) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol 222:1690–1704. [DOI] [PubMed] [Google Scholar]
- Fan Z, Ba L, Shan W, Xiao Y, Lu W, Kuang J, Chen J (2018) A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J 96:1191–1205 [DOI] [PubMed] [Google Scholar]
- Feng B, Han Y, Xiao Y, Kuang J, Fan Z, Chen J, Lu W (2016) The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J Exp Bot 67:2263–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat Biotechnol 5:726–730 [Google Scholar]
- Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell 16:170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Jia S, Huang X, Wang L, Fu W, Huo G, Gan L, Ding J, Li Y (2019) Transcriptome and hormone analyses provide insights into hormonal regulation in strawberry ripening. Planta 250:145–162 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115:667–677 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7:40–49 [DOI] [PubMed] [Google Scholar]
- Guo X, Liu D, Chong K (2018a) Cold signaling in plants: insights into mechanisms and regulation. J Integr Plant Biol 609:745–756 [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang Y, Tu Y, Wang Y, Cheng W, Yang Y (2018b) Overexpression of an EIN3-binding F-box protein2-like gene caused elongated fruit shape and delayed fruit development and ripening in tomato. Plant Sci 272:131–141 [DOI] [PubMed] [Google Scholar]
- Hao J, Li T, Meng S, Zhao B, Sun L (2009) Effects of night low temperature on sugar accumulation and sugar-metabolizing enzyme activities in melon fruit. Sci Agric Sin 10:3592–3599 [Google Scholar]
- Hu X, Kuang S, Zhang A, Zhang W, Chen M, Yin X, Chen K (2016) Characterization of starch degradation related genes in post-harvest kiwifruit. Int J Mol Sci 17:2112–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Zuo J, Hou X, Yan Y, Wei Y, Liu J, Li M, Xu B, Jin Z (2015) The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci 6:742–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourda C, Cardi C, Mbéguié-A-Mbéguié D, Bocs S, Garsmeur O, D'Hont A, Yahiaoui N (2014) Expansion of banana (Musa acuminata) gene families involved in ethylene biosynthesis and signalling after lineage-specific whole-genome duplications. New Phytol 202:986–1000 [DOI] [PubMed] [Google Scholar]
- Kuang J, Chen L, Shan W, Yang S, Lu W, Chen J (2013) Molecular characterization of two banana ethylene signaling component MaEBFs during fruit ripening. Postharvest Biol Technol 85:94–101 [Google Scholar]
- Lafuente M, Establés-Ortíz B, González-Candelas L (2017) Insights into the molecular events that regulate heat-induced chilling tolerance in citrus fruits. Front Plant Sci 8:1113–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yi, Choi J, Lee E (2020) Analyses of targeted/untargeted metabolites and reactive oxygen species of pepper fruits provide insights into seed browning induced by chilling. Food Chem 332:127406. [DOI] [PubMed] [Google Scholar]
- Li X, Xu C, Korban S, Chen K (2010) Regulatory mechanisms of textural changes in ripening fruits. Crit Rev Plant Sci 29:222–243 [Google Scholar]
- Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M (2015) Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol 169:2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li L, Li B, Zhan G, Wang Y (2018) Evaluation of low-temperature phosphine fumigation for control of oriental fruit fly in loquat fruit. J Econ Entomol 111:1165–1170 [DOI] [PubMed] [Google Scholar]
- Louro RP, Nascimento JR, Purgatto E, Tavares MI, Lajolo FM, Cordenunsi BR (2013) The cold storage of green bananas affects the starch degradation during ripening at higher temperature. Carbohyd Polym 96:137–147 [DOI] [PubMed] [Google Scholar]
- Megías Z, Martínez C, Manzano S, Manzanoa S, Garcíaa A, Rebolloso-Fuentesb M, Valenzuelaa JL, Garridoc D, Jamilenaa M (2016) Ethylene biosynthesis and signaling elements involved in chilling injury and other postharvest quality traits in the non-climacteric fruit of zucchini (Cucurbita pepo). Postharvest Biol Technol 113:48–57 [Google Scholar]
- Paul J, Khanna H, Kleidon J, Hoang P, Geijskes J, Daniells J, Zaplin E, Rosenberg Y, Rosenberg A, Mlalazi B, et al. (2016) Golden bananas in the field: elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol J 15:520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RS, Fuller MP (2001) Freezing of barley studied by infrared video thermography. Plant Physiol 125:227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Llorca M, Munoz P, Muller M, Munne-Bosch S (2019) Biosynthesis, metabolism and function of auxin, salicylic acid and melatonin in climacteric and non-climacteric fruits. Front Plant Sci 10:136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phothiset S, Charoenrein S (2014) Effects of freezing and thawing on texture, microstructure and cell wall composition changes in papaya tissues. J Sci Food Agric 94:189–196 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115:679–689 [DOI] [PubMed] [Google Scholar]
- Sadka A, Qin Q, Feng J, Farcuh M, Shlizerman L, Zhang Y, Toubiana D, Blumwald E (2019) Ethylene response of plum ACC synthase 1 (ACS1) promoter is mediated through the binding site of abscisic acid insensitive 5 (ABI5). Plants 8:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7:682–693 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Taylor JE, Tucker GA (1993) Biochemistry of fruit ripening. Ind J Agric Biochem 18:51–60 [Google Scholar]
- Shan W, Kuang J, Chen L, Xie H, Peng H, Xiao Y, Li X, Chen W, He Q, Chen J, et al. (2012) Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J Exp Bot 63: 5171–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Kuang J, Wei W, Fan Z, Deng W, Li Z, Bouzayen M, Pirrello J, Lu W, Chen J (2020) MaXB3 modulates MaNAC2, MaACS1, and MaACO1 stability to repress ethylene biosynthesis during banana fruit ripening. Plant Physiol 184:1153–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga TM, Soares CA, Nascimento JR, Purgatto E, Lajolo FM, Cordenunsi BR (2011) Ripening-associated changes in the amounts of starch and non-starch polysaccharides and their contributions to fruit softening in three banana cultivars. J Agric Food Chem 91:1511–1516 [DOI] [PubMed] [Google Scholar]
- Sibéril Y, Doireau P, Gantet P (2001) Plant bZIP G-box binding factors modular structure and activation mechanisms. Eur J Biochem 268:5655–5666 [DOI] [PubMed] [Google Scholar]
- Song S, Wang G, Wu H, Fan X, Liang L, Zhao H, Li S, Hu Y, Liu H, Ayaad M, et al. (2020a) OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J 103:532–546 [DOI] [PubMed] [Google Scholar]
- Song Z, Qin J, Yao Y, Lai X, Zheng W, Chen W, Zhu X, Li X (2020b) A transcriptomic analysis unravels key factors in the regulation of stay-green disorder in peel of banana fruit (Fenjiao) caused by treatment with 1-MCP. Postharvest Biol Technol 168:111290 [Google Scholar]
- Song Z, Qin J, Zheng Q, Ding X, Chen W, Lu W, Li X, Zhu X (2019) The involvement of the banana F-Box protein MaEBF1 in regulating chilling-inhibited starch degradation through interaction with a MaNAC67-like protein. Biomolecules 9:552–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Wang S, Xu G, Kang X, Zhang M, Ni M (2019) SHB1 and CCA1 interaction desensitizes light responses and enhances thermomorphogenesis. Nat Commun 10:3110–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J, Li H, Ban Q, Han Y, Meng K, Ming J, Zhang Z, Rao J (2018) Characteristics of chilling injury-induced lignification in kiwifruit with different sensitivities to low temperatures. Postharvest Biol Technol 135:8–18 [Google Scholar]
- Tacken EJ, Ireland HS, Wang YY, Putterill J, Schaffer RJ (2012) Apple EIN3 binding F-box 1 inhibits the activity of three apple EIN3-like transcription factors. AoB Plants 2012:pls034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma K, Upadhyay N, Kumar N, Yadav G, Singh J, Mishra RK, Kumar V, Verma R, Upadhyay RG, Pandey M (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front Plant Sci 8:161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Huang S, Zhang A, Guo P, Liu Y, Xu C, Cong W, Liu B, Xu Z (2021) JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol 230:567–584 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Zhao S, Wu Y (2011) Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J 68:249–261 [DOI] [PubMed] [Google Scholar]
- Wang Z, Miao H, Liu J, Xu B, Yao X, Xu C, Zhao S, Fang X, Jia C, Wang J, et al. (2019) Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat Plants 5:810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Shan W, Liang S, Zhu L, Guo Y, Chen J, Lu W, Li Q, Su X, Kuang J (2019) MaMPK2 enhances MabZIP93-mediated transcriptional activation of cell wall modifying genes during banana fruit ripening. Plant Mol Biol 101:113–127 [DOI] [PubMed] [Google Scholar]
- Wu Q, Li T, Chen X, Wen L, Yun Z, Jiang Y (2018) Sodium dichloroisocyanurate delays ripening and senescence of banana fruit during storage. Chem Cent J 12:131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22:1733–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Kuang J, Qi X, Ye Y, Wu Z, Chen J, Lu W (2018) A comprehensive investigation of starch degradation process and identification of a transcriptional activator MabHLH6 during banana fruit ripening. Plant Biotechnol J 16:151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Onik JC, Hu X, Duan Y, Lin Q (2018) Effects of (S)-Carvone and gibberellin on sugar accumulation in potatoes during low temperature storage. Molecules 23:3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wu Y, Pirrello J, Regad F, Bouzayen M, Deng W, Li Z (2010) Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J Exp Bot 61:697–708 [DOI] [PubMed] [Google Scholar]
- Yu F, Wu Y, Xie Q (2015) Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci 20:569–575 [DOI] [PubMed] [Google Scholar]
- Yuan T, Xu H, Zhang K, Guo T, Lu Y (2014) Glucose inhibits root meristem growth via ABA INSENSITIVE 5, which represses PIN1 accumulation and auxin activity in Arabidopsis. Plant Cell Environ 37:1338–1350 [DOI] [PubMed] [Google Scholar]
- Zhao M, Wang J, Shan W, Fan J, Kuang J, Wu K, Li X, Chen W, He F, Chen J, et al. (2013) Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ 36:30–51 [DOI] [PubMed] [Google Scholar]
- Zhang T, Li W, Xie R, Xu L, Zhou Y, Li H, Yuan C, Zheng X, Xiao L, Liu K (2020) CpARF2 and CpEIL1 interact to mediate auxin–ethylene interaction and regulate fruit ripening in papaya. Plant J 13:1318–1337 [DOI] [PubMed] [Google Scholar]
- Zhu X, Li Q, Li J, Luo J, Chen W, Li X (2018) Comparative study of volatile compounds in the fruit of two banana cultivars at different ripening stages. Molecules 23:2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Zhang L, Rao S, Zhu X, Ye L, Chen W, Li X (2014) The relationship between the expression of ethylene-related genes and papaya fruit ripening disorder caused by chilling injury. PLoS One 9:e116002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.