Abstract
Arabidopsis (Arabidopsis thaliana) primary and lateral roots (LRs) are well suited for 3D and 4D microscopy, and their development provides an ideal system for studying morphogenesis and cell proliferation dynamics. With fast-advancing microscopy techniques used for live-imaging, whole tissue data are increasingly available, yet present the great challenge of analyzing complex interactions within cell populations. We developed a plugin “Live Plant Cell Tracking” (LiPlaCeT) coupled to the publicly available ImageJ image analysis program and generated a pipeline that allows, with the aid of LiPlaCeT, 4D cell tracking and lineage analysis of populations of dividing and growing cells. The LiPlaCeT plugin contains ad hoc ergonomic curating tools, making it very simple to use for manual cell tracking, especially when the signal-to-noise ratio of images is low or variable in time or 3D space and when automated methods may fail. Performing time-lapse experiments and using cell-tracking data extracted with the assistance of LiPlaCeT, we accomplished deep analyses of cell proliferation and clonal relations in the whole developing LR primordia and constructed genealogical trees. We also used cell-tracking data for endodermis cells of the root apical meristem (RAM) and performed automated analyses of cell population dynamics using ParaView software (also publicly available). Using the RAM as an example, we also showed how LiPlaCeT can be used to generate information at the whole-tissue level regarding cell length, cell position, cell growth rate, cell displacement rate, and proliferation activity. The pipeline will be useful in live-imaging studies of roots and other plant organs to understand complex interactions within proliferating and growing cell populations. The plugin includes a step-by-step user manual and a dataset example that are available at https://www.ibt.unam.mx/documentos/diversos/LiPlaCeT.zip.
A pipeline and the plugin “Live Plant Cell Tracking” (LiPlaCeT) coupled to the publicly available ImageJ image analysis program allow 4D cell tracking and lineage analysis.
Introduction
To generate functional organs and tissues, cell proliferation in multicellular organisms needs to be tightly regulated during growth and morphogenesis. The dynamics of cell proliferation within Arabidopsis (Arabidopsis thaliana) primary root apical meristem (RAM), and lateral root (LR) primordia (LRPs), constitute two models in which the analysis of cell population over time can shed light into the complex regulatory networks imposed both at cellular and tissue level during the organogenesis. Extensive knowledge has been gathered concerning the gene regulatory and hormonal networks involved in root development (Azpeitia et al., 2010; Garay-Arroyo et al., 2012; Petricka et al., 2012; De Lucas and Brady, 2013; Lavenus et al., 2015; Du and Scheres, 2017; García-Gomez et al., 2017; Di Mambro et al., 2018; Trinh et al., 2018; Torres-Martínez et al., 2019). However, analysis of whole cell populations of dividing cells through time is still challenging, but necessary to understand the emergent properties of these local networks interacting in complex 3D organs.
In Arabidopsis, LRs are developed from LRPs that start their formation deep inside the root tissues (Malamy and Benfey, 1997; Dubrovsky et al., 2000; Beeckman et al., 2001; Dubrovsky et al., 2001; Casimiro et al., 2003). Soon after the cells are displaced from the RAM, some pericycle cells become primed to be specified as the first founder cells (De Smet et al., 2007; Moreno-Risueno et al., 2010; Van Norman et al., 2013; Toyokura et al., 2019). Before or during the founder cell specification, a parent root zone called the “oscillation zone” is established where auxin response oscillates at a transcriptional level and where many genes are expressed in-phase and in anti-phase with auxin response (Moreno-Risueno et al., 2010; Wachsman et al., 2020; Perianez-Rodriguez et al., 2021). In long-term time-lapse experiments, it has been established that auxin acts as a morphogenetic trigger of founder cell identity acquisition (Dubrovsky et al., 2008; Benková et al., 2009), and when the first founder cell is specified, it starts to divide contributing to LRP formation. During the LRP formation, the founder cell descendants follow a stereotypical pattern of cell division (Malamy and Benfey, 1997; Napsucialy-Mendivil and Dubrovsky, 2018; Torres-Martínez et al., 2019). We recently showed that LR initiation is a gradual, multistep, and nonstereotypical process accompanied with the recruitment of new founder cells (Torres-Martínez et al., 2020). The analysis of LRP morphogenesis permits the establishment of certain morphogenetic rules that suggest self-organizing properties in the developing LRP (von Wangenheim et al., 2016; Fujiwara et al., 2021; Schütz et al., 2021). Importantly, the analysis of LRP morphogenesis and complex gene regulatory networks involved in its control would not be possible without understanding cellular bases of LR formation. All these studies require a 4D analysis of the LRP morphogenesis in time-lapse and are time consuming.
Arabidopsis primary RAM contains a stem cell niche (SCN), composed of an organizer, or quiescent center (QC), surrounded by initial (stem) cells, which in turn act as progenitors of cells that will enter the cell proliferation domain (Ivanov and Dubrovsky, 2013; García-Gómez et al., 2020), also called the transit-amplifying domain (TAD). Cells within the proliferation domain actively proliferate a limited number of cell cycles before they are displaced to the RAM transition domain and subsequently to the elongation and differentiation zones (Verbelen et al., 2006; Ivanov and Dubrovsky, 2013); in the differentiation zone, they attain the final differentiation state (Galinha et al. 2007; Scheres, 2007; Shishkova et al., 2008; Doonan and Sablowski, 2010; Perilli et al., 2013; Rodriguez et al., 2015; Rahni and Birnbaum, 2019). The balance between cell proliferation and transition to differentiation within these cell populations is highly responsive to developmental cues, and they are capable of integrating endogenous signals like hormonal and metabolic status of the plant in order to modulate growth and root system architecture (Blilou et al., 2005; Osmont et al., 2007; Ingram and Malamy, 2010; Garay-Arroyo et al., 2012; Vanstraelen and Benková, 2012; Reyes-Hernández et al., 2014, 2019; Morris et al., 2017; Salvi et al., 2020).
Light-sheet and confocal microscopy time-lapse studies of whole RAM (Campilho et al., 2006; Maizel et al., 2011; Sena et al., 2011; Busch et al., 2012; Keinath et al., 2015; von Wangenheim et al., 2017; Baesso et al., 2018; Rahni and Birnbaum, 2019) show promising results because their spatial and temporal resolution makes it possible to record every cell division event, as well as the shape and position of every cell within a developing tissue. In this work, we describe two setups, one for the analysis of LRP morphogenesis and another one for the RAM studies. The first one consists of a confocal microscopy setup appropriate for the study of developing LRP with a high magnification objective and a short working distance that permits the visualization of individual nuclei and cell plasma membrane within an Arabidopsis root. The second setup for RAM studies was designed based on a Macro Zoom microscope (see “Materials and methods”) that allows observation and data collection on roots growing vertically over the agar surface and under physiological conditions similar to those used for Arabidopsis grown in Petri dishes. To facilitate the 4D analysis, we developed the Live Plant Cell Tracking (LiPlaCeT) ImageJ open-source plugin that permits uncovering different facets of the dynamic behavior of cell populations over long time-lapse experiments, one in the developing LRP and the other in the RAM.
Once the data have been collected for cell dynamics of a whole population in time-lapse experiments, the information obtained consists of a large amount of microscopy images. 3D reconstructions of root tissue are laborious and time consuming, and a number of tools, such as the intrinsic root coordinate system (Schmidt et al., 2014, Lavrekha et al., 2017) or MorphoGraphX (De Reuille et al., 2015; Strauss et al., 2021) have been developed. A 3D analysis in time (4D) is even more complex and transition from 3D to 4D analysis is not straightforward and may contain different types of errors (Schiegg et al., 2013). An exhaustive revision (Ulman et al., 2017) of several tracking algorithms reported a very good overall performance for images with high signal-to-noise ratio (SNR); however, for images with low SNR, a substantial amount of manual work would be needed to correct when an algorithm is used (Ulman et al., 2017). There are several automatic or semi-automatic tools developed for tracking objects of interest in 3D+t (Jaqaman et al., 2008; Winter et al., 2016; Berg et al., 2019; Trullo et al., 2020); however, with these approaches it is difficult to correctly identify what subset of data requires a postprocessing or manual correction; see Emami et al. (2021) for a detailed review of computerized cell tracking methods. TrackMate (Jaqaman et al., 2008; Tinevez et al., 2017; Tosi and Campbell, 2019) is an automated tool developed for particle tracking where first blob-like structures are detected using a Laplacian of Gaussian or Difference of Gaussian filters and then a tool allows the elimination of erroneous spots based on quality, intensity, filter response, size, etc. Finally, the detected blob-like structures are connected across time using a linear assignment problem. Although this is a very flexible approach, successful in several applications, it has difficulties in its application for objects having simultaneously low and high SNR regions of interest (see “Discussion”). Another approach used for cell tracking is Massive Multiview Tracker “MaMuT” (Wolff et al., 2018) designed as a Fiji plugin that combines the TrackMate and BigData capabilities. This platform is a powerful tool allowing the cell tracking in multi-view and multi-terabyte datasets; it also enables automated annotations (Wolff et al., 2018, see also Supplemental Table S1). Despite all these features, for internal plant tissues, due to variable SNR, automated annotations are impossible (see also “Discussion”). von Wangenheim et al. (2016) developed a tool for manual tracking using an in-house created Mathematica program which allows 3D in time analysis. The manual tracking permits detection of division events, cell cycle number, cell cycle duration, and other parameters. However, this approach requires the knowledge of Wolfram Language. The summary of these tracking methods and their features are presented in Supplemental Table S1.
In this article, we present a complete workflow designed to annotate manually and automatically computed descriptors for cell lineage, cell proliferation events, cell size, and cell dynamics such as growth rate, displacement rate, and clonal relations. In a step-by-step user manual, we described how to visualize clonal information and cell dynamics in 2D, 3D, and 3D+ time representations (4D).
The methodological approaches described here allowed us to analyze cell proliferation and growth at the whole population level, particularly, cell cycle dynamics in the developing LRP and in TAD of the RAM. Furthermore, cell tracking of the LRP cells allowed us to better understand the impact of cell proliferation for the LRP morphogenesis and to reconstruct genealogical trees of cells starting from an early developmental stage.
Results
To be able to track cells in the acquired 3D images of live root captured as Z-stacks of 2D images in time-lapse experiments (3D in time, 4D), we designed “Live Plant Cell Tracking” (LiPlaCeT) plugin to be used with the open-source image processing package Fiji (ImageJ; Schindelin et al., 2015; Rueden et al., 2017).
The LiPlaCeT plugin was successfully used in a study of LRP morphogenesis (Torres-Martínez et al., 2020). First, we addressed the challenge of identifying a single cell and its progeny through time within a multicellular environment. The plugin was designed to work with one or two channel images, and this allowed us the use of Arabidopsis lines with fluorescent protein labeled nuclei or plasma membranes. We used a double transgenic F1 line of a cross between a line expressing histone H2B fused to RED FLUORESCEN PROTEIN1 driven by the constitutive promoter 35S of the cauliflower mosaic virus (p35S::H2B-RFP; Federici et al., 2012) and a Wave 131 line expressing a plasma membrane protein NPSN12 (At1g48240) fused to YELLOW FLUORESCENT PROTEIN under the constitutive promoter UBIQUITIN 10 pUBQ10::NPSN12-YFP (Geldner et al., 2009); in this cross all the nuclei and plasma membranes of cells are visualized with the aid of laser scanning confocal microscopy showing red and green emissions, respectively (Figure 1).
Figure 1.
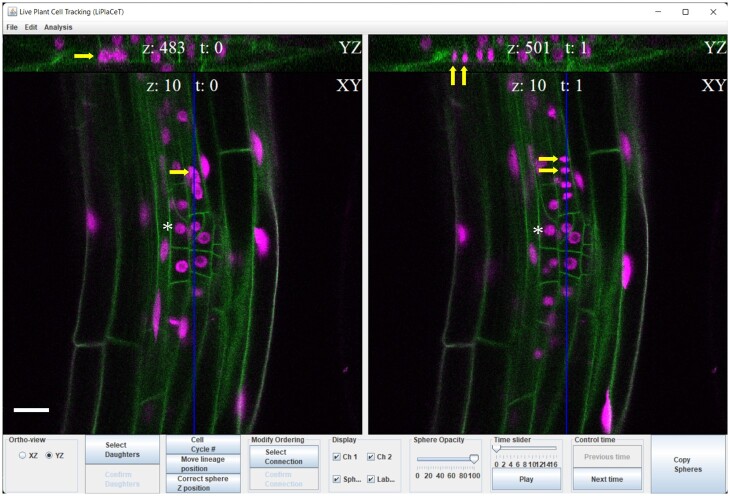
LiPlaCeT plugin interface for user-friendly cell tracking of 3D images in a time-lapse experiment. LiPlaCeT plugin displays two consecutive frames (t = n and t = n + 1). Within each time frame, a longitudinal section of the root is displayed (XY plane), and the user can choose to display an orthogonal YZ (as shown at the top of the image) or XZ in order to facilitate the identification of a single cell within a 3D image (see also Figure 2). An LRP of a p35S::H2B-RFP pUBQ10::NPSN12-YFP F1 seedling is shown. Asterisks indicate the nucleus of a single cell that was not divided between these two time frames. Yellow arrows on the left (t = n), show a cell before it divided and it can be identified in both XY and YZ orthogonal panels; yellow arrows on the right panel (t = n + 1), show the resulting daughter cells. The blue line in XY shows position of YX section. Scale bar = 20 μm.
The LiPlaCeT interface and its capabilities were designed to facilitate the tracking of clonally related cell populations whose 3D distribution emerges during development. For this purpose, multiple orthogonal views are displayed for two subsequent time points, ensuring that each cell can be identified within a growing organ by analyzing defined longitudinal, XY, transversal, YZ (Figures 1 and 2C), or tangential, XZ (Figure 2A) sections generated by the LiPlaCeT algorithm. Therefore, it is straightforward to identify manually the position of an individual cell, which can be marked with a color-coded circle (Figure 2) or can be given an identification number, or both. Once multiple cells have been identified and marked within a time frame, all the markings can be transferred as a group to a subsequent time frame. The user can then adjust the 3D position of each label to identify the new location of each marked cell in the next time frame. Analysis in 3D requires a constant change between time frames and from one Z-section to another. As a result, simultaneous visualization of consecutive time points is a very valuable feature of LiPlaCeT. The LiPlaCeT plugin is designed in a way that allows the synchronization of Z slices in both windows between consecutive time points, which facilitates the verification of the cell annotations.
Figure 2.
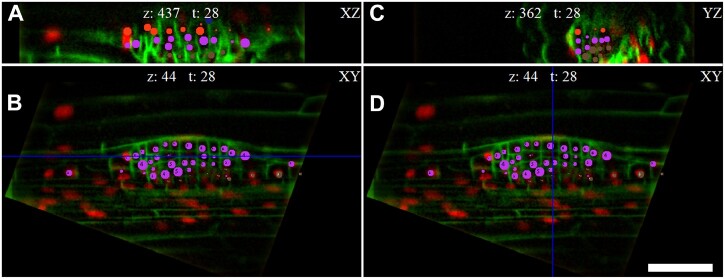
User-friendly cell identification across time frames within multiple orthogonal views reconstructed by the LiPlaCeT algorithm. A tangential (perpendicular to the radial axis, XZ) view is displayed in (A), a longitudinal section of the root is displayed (XY plane) in (B) and (D) and a transversal section, YZ, is shown in (C). Different points of view of the LiPlaCeT windows help to follow complex LRP cell lineages. The LRP of p35S::H2B-RFP pUBQ10::NPSN12-YFP F1 seedling is shown. Cell progeny formed from pericycle founder cells in the same cell file is marked with the same color-coded circles. The blue line in (B) shows position of ZX section; the blue line in (D) shows position of YZ section. Scale bar = 50 μm.
When a mitotic figure is identified, cell division can easily be documented (Figure 1). However, when mitosis occurs between captured time frames, recently divided sister cells can be identified using the relative position of their neighbors. Thus, at the beginning of the analysis, it is important to establish the relative position of all the cells (their nuclei, if possible) in the subset to be analyzed. As neighboring plant cells grow, they do not slide one along another and, after a division, they maintain the relative position of their cell walls in relation to the cell walls of their neighbor cells; a phenomenon called symplastic growth (Sinnott, 1960; Erickson, 1986). Since mitotic events are not synchronized within RAM or LRP populations, if time frames are spaced close enough so that only a minority of mitotic events occur between them, the undivided cells can be recognized between subsequent captures (Figure 1, asterisks), and the extra cells can be labeled as daughter cells associated with their most likely mother cell (Figure 1, yellow arrows). As a result, cells clonally related to a single progenitor share common numeric and color-coded annotations. In this way, the same cells can be linked between different time frames and when they divide, a branched link between mother cell and daughters is constructed. This capability of LiPlaCeT plugin permits a detailed tracking of the whole cell population within a developing organ (Figure 2), and cell genealogy trees (Figures 3 and 4) can be subsequently constructed using ParaView software (see “Materials and methods”). Annotations of each cell are stored and associated to the metadata including time, position, and lineage information, which make it possible to reconstruct the dynamics of cell division and growth patterns for the whole cell population. If some error occurs in the cell annotation, the LiPlaCeT plugin is designed to create a new or eliminate an existent color-coded annotation with one click of the mouse.
Figure 3.
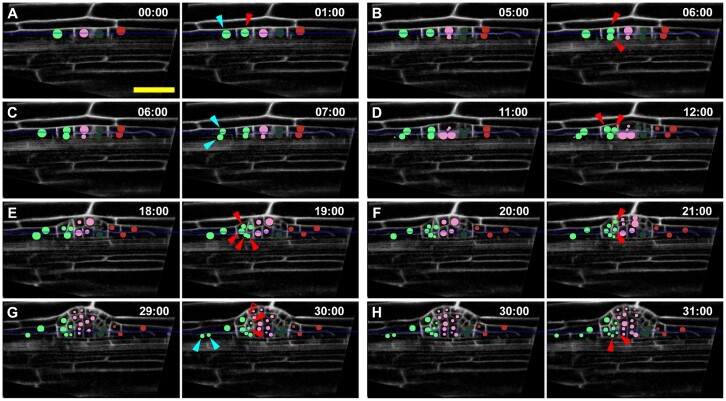
An example of accurate cell lineage tracking of a developing LRP in a time-lapse experiment using LiPlaCeT plugin. A–H, A “green cell” lineage tracking (green circles) of a developing LRP in p35S::H2B-RFP pUBQ10::NPSN12-YFP F1 seedlings; YFP signal is pseudo colored in gray. When the “green cell” in (A) divides, it produces a central domain (red arrowhead) and a flanking domain (blue arrowhead), the daughters. Each subsequent panels show time points when new daughters of central and flanking daughters are formed, they are marked by arrowheads of corresponding color. All panels show the same Z section. Unfilled red arrowhead in (G) points a location of invisible descendant cells of the upper cell formed in (F); these cells are invisible because they are displaced to a different Z-slice. Numbers indicate time in hours; 00 h is the time of the beginning of the time-lapse analysis. Only some time points of a 48-h experiment are shown, images were acquired each hour. See also Supplemental Movie S2. Scale bar = 50 μm.
Figure 4.
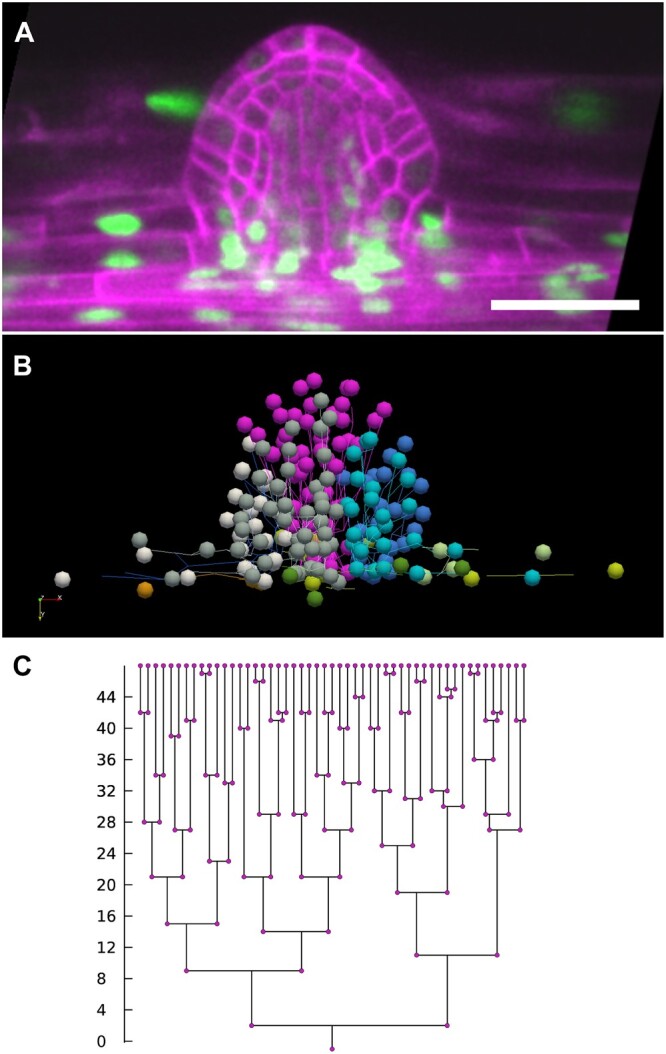
Analysis of cell lineages in the developing LRP performed with the LiPlaCeT plugin. A, Longitudinal section of the last time point (48 h) of a developing LRP in p35S::H2B-RFP pUBQ10::NPSN12-YFP F1 seedling. Pseudo colors are shown; red nuclei and green plasma membrane colors have been inverted. B, 3D visualization of the same LRP shown in (A), made with ParaView software, of every cell lineage tracked with LiPlaCeT. Different colors indicate different cell lineages produced in a 48-h time-lapse experiment; image stacks were acquired each hour. See also Supplemental Movie S1. C, Cell genealogy tree of a pink cell lineage shown in (B). Scale bar = 50 μm.
In summary, the designed LiPlaCeT Fiji (ImageJ) plugin has several valuable features that permit cell tracking in 4D such as: (1) multiple orthogonal views; (2) simultaneous visualization of consecutive time points in two windows; (3) synchronized change of slices in both windows between consecutive time points; (4) one click color-coded spot creation and its deletion; (5) easy link of the same cells between consecutive time points and construction of a branched link between mother cell and daughters; (6) copying current state to the next time point; (7) capability to display one or two channels or only to display annotated spots; and (8) capability to save and load tracking analysis metadata. LiPlaCeT plugin is freely available (see “Materials and methods” for requirements and installation details in Supplemental Material S1).
Next, we illustrate how we tracked cell populations through time within Arabidopsis LRP and RAM cells using the LiPlaCeT plugin, and subsequently used ParaView to semi automatically perform cell lineage analysis, determine and calculate quantitative parameters regarding cell proliferation, and estimate individual cell elongation and displacement in the context of growing 3D tissues. A step-by-step user manual for cell tracking and cell population analysis using LiPlaCeT plugin and ParaView software is available (Supplemental Material S1).
Application of LiPlaCeT plugin to understand LRP morphogenesis in live roots
Here we illustrate the application of LiPlaCeT Fiji (ImageJ) plugin analyzing LRP formation. For this purpose, we performed 48 h of time-lapse experiments using laser scanning confocal microscopy (Torres-Martínez et al., 2020), and here we show how the analysis was performed. In the experiment depicted in Figure 3 (see also Supplemental Movies S1 and S2), cell tracking starts from a one-cell-layer Stage I LRP (Malamy and Benfey, 1997). If we follow the LPR cell marked in green (0 h), we can see that it divides 1 h later and the daughter cells can be seen in the central domain (Figure 3, 1 h, red arrowhead) and in the longitudinally flanking domain, respectively (Figure 3, 1 h, blue arrowhead). Five hours after the beginning of the experiment, the centrally located cell, born in the first hour has not been divided yet, but we could find two daughter cells at 6 h (Figure 3B). Therefore, the duration of the cell cycle was between 4 and 5 h, or ∼4.5 h. Cycle times evaluated in this way in the central domain of the LRP shown in Figure 3 were 4.5, 5.5, 6.5, 12.5, 11.5, 8.5, and 8.5 h. This example illustrates that the cell tracking permitted us to conclude that the centrally located cell passed through up to four cell cycles over a period of 30 h with an average cycle time of 8.2 h. On the other hand, the daughter cell produced by the first anticlinal division at 1 h (Figure 3A) that is located at the LRP flanking domain, divided subsequently at 7 h (Figure 3C, blue arrowheads) and the daughter at the bottom divided at 30 h (Figure 3G, blue arrowheads). Therefore, the daughter cell located in the flanking domain passed only through two division rounds of the cell cycle, one division at ∼5.5 h and the second at 12.5 h. Analysis of other developing LRPs showed the same pattern when cell lineages produced in flanking versus central domain are compared. From these analyses, we can conclude that the flanking daughter cell shows a much lower proliferation potential (P = 0.008, Mann–Whitney rank sum test). By on average, 31.2 ± 5.0 h from the beginning of the experiment (early Stage I LRP), the centrally located cells passed through on average 3.0 cell cycles, while during the same time the flanking cells passed through 1.5 ± 0.8 cell cycles (n = 6, mean ± sd). These examples clearly show how cell tracking can help to establish different morphogenetic potential of LRP cells located at central and flanking domains. Moreover, with the use of LiPlaCeT, it is possible to record the number of cell cycles undergone within a clone to produce a specific cell in any given time frame and this feature can be available automatically if the position of each cell and its progeny are curated.
Using this approach, a progeny of each cell in a developing LRP can be reconstructed (Figure 4). In this example, tracking analysis started from Stage I LRP. Once cell tracking has been made with the aid of LiPlaCeT, the nucleus position along time and space of a developing LRP, can be visualized with ParaView software as 3D objects in time (each lineage tracked in time, Figure 4B). Populations of identified cells and their properties curated by the user can be displayed as vector maps (Figure 4C) and metadata associated with parameters for each cell such as time frame, coordinates, lineage, and cell division times can be exported as a *.cvs file (Supplemental Material S1). With this analysis we were able to construct cell genealogical trees (Figure 4C) for each LRP cell from Stage I onwards and extract information on the number of cell cycles and cell cycle duration from each cell lineage produced during developing LRP. Cell lineage analysis also permits the calculation of the principal growth directions (Supplemental Movie S3). This analysis allows the dynamic study of changes from 2D to 3D growth during LRP formation.
Overall, this analysis showed that, similar to previous findings (von Wangenheim et al., 2016), LRP formation takes ∼48 h during which cells pass through approximately seven cell cycles and the application of LiPlaCeT substantially facilitates this analysis. Interestingly, the genealogical tree in this example showed that the first three cell cycles are relatively short while the later cell cycles are longer (Figure 4C). We also could appreciate that daughter cells frequently have different cell cycle times (Figure 4C). Differences in the number of cell cycles in the central and flanking domains (Figure 3) clearly show that proliferation potential within the developing LRP strongly depends on cell position.
LiPlaCeT plugin for analysis of cell proliferation within the primary RAM
In addition to cell lineage and cell cycle analyses on developing LRP study described above, our LiPlaCeT algorithm permits the extraction of many other descriptors in a developing 3D tissue in time. To illustrate them, we aimed to visualize individual and whole-tissue cell dynamics of cell division, cell elongation, and cell displacement with respect to one cell type of the primary RAM. To do this, we devised a method to track every meristematic endodermis cell using the pSCR::H2B-YFP (Xu et al., 2006) marker line, in which endodermis nuclei are marked with yellow fluorescent proteine (YFP). Nuclear positions were used in this experiment to estimate several dynamic parameters for endodermis cells within the RAM of a growing root through time.
Compared to LRP time-lapse experiments, RAM analysis imposes additional challenges because (1) the primary root growth involves helical root tip movement known as circumnutation (Taylor et al., 2021) and therefore it continuously changes its trajectory and (2) the cell cycle dynamics and cell displacements within the RAM and the elongation zone are continuous and highly responsive to changes in growth conditions. To ensure that cell population dynamics in our study were similar to those of roots growing under standard in vitro culture conditions, we designed an experimental setup that allowed visualization of Arabidopsis seedlings growing vertically under in vitro physiological standard conditions. Our experimental design was based on an AZ100 Multi Zoom epifluorescence microscope (Nikon Instruments Inc. Melville, NY, USA) adapted to a horizontal position (Figure 5A, see “Materials and methods”). This microscope combines macro lenses with on-axis image formation pathway and internal optical zoom, which are adequate to generate planar images of complete RAM endodermis cell layer, in which fluorescently labeled nuclei can be identified (Figure 5B). Unlike experimental setups adapted for high-resolution microscope lenses, in which objective working distances usually are very short, from a few micrometer region to a fraction of mm, we opted for a system based on macro lenses with working distances ranging from 15 to 45 mm. This setup allowed us to position a closed growth chamber with specimens within the focal plane without the need to design a chamber that would allow to maintain root growth limited to a region of a fraction of mm apart from the coverslip, which is often the case in experimental designs based on objectives with relatively short working distances (e.g. Maizel et al., 2011; Sena et al., 2011; Rahni and Birnbaum, 2019). Our setup included automated control of the sample position with resolution of 1 μm in X, Y, and Z-axis.
Figure 5.
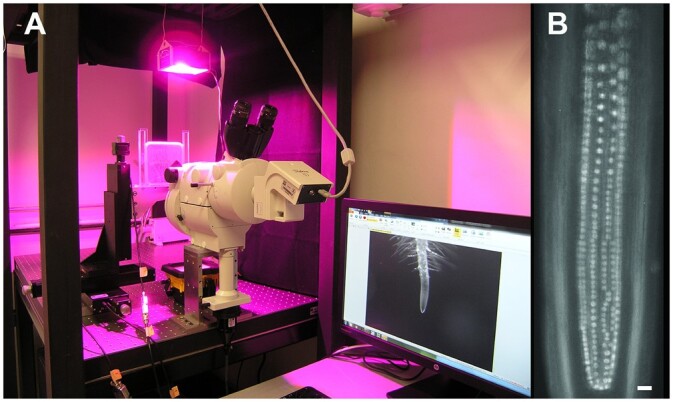
Experimental setup designed for time-lapse analysis of endodermis nuclei within the TAD of the RAM. A, Nikon AZ100 Multi Zoom with epifluorescence supported in horizontal position. Plants were maintained in homemade chambers prepared within a Petri dish (see “Materials and methods”), and roots grew on a surface of agar growth medium. Environmental light and temperature conditions were controlled to ensure growth conditions similar to A. thaliana growth room environments. Chambers were positioned in front of the microscope lenses and supported by automated translation stage base plates to control X, Y, and Z positions of the specimen. This setup allowed roots to grow parallel to the gravity axis. B, The Multi Zoom system permitted alternating between magnifications to capture either images of the root apex as in (A), or images of individual nuclei, as in (B). Scale bar = 20 μm. These images were taken in the Laboratorio de Microscopía y Microdisección Láser (LabMicroLas) at the Instituto de Ecología, UNAM, Mexico.
Here we show the results of a proof-of-concept study in which the whole meristematic endodermis population was monitored during 71 h. Seedlings from the pSCR::H2B-YFP line were analyzed from 2 to 5 d postgermination (dpg). In a semi-automated setup (see “Materials and methods”), we recorded every endodermis nucleus within the transit-amplifying cell population every 4–9 h (day–time captures were taken approximately every 4 h, while the period between the last capture of each day and the following morning, expanded to 8–9 h). During this time-lapse, seedling roots grew at an average rate of 0.25 mmh−1, which was comparable to published root growth rates of seedlings with the same genetic background grown in Petri dishes, similar age, and maintained under similar conditions (Dello Ioio et al., 2007; López-Bucio et al., 2014; Rodriguez et al., 2015). Consistently with previous experiments with young seedlings (Beemster and Baskin, 1998; Dello Ioio et al., 2007; López-Bucio et al., 2014; Rodriguez et al., 2015), qualitative inspection of RAM showed that its length increased throughout the experiment.
Using LiPlaCeT interface, we were able to track individual endodermis cells and their proliferation events through time within the RAM TAD. As LiPlaCeT algorithm records 3D positions of each nucleus through time, as well as time and location of every proliferation event, it generates a wealth of data that can be analyzed using ParaView to study a wide range of cell dynamics parameters in the context of a complex meristematic population.
To understand the complex balance of proliferation, cell growth, and cell displacement necessary to maintain an active SCN while ensuring root growth in response to physiological and environmental stimuli, cell length profiles within cell files and the distribution of proliferation activity are widely studied parameters (Kumpf et al., 2014; Pacheco-Escobedo et al., 2016; Rahni and Birnbaum, 2019). Our analytic pipeline is designed to automate assessment of nuclear position to visualize approximations for cell length and position and to represent the resulting values as 3D and 3D + t vector maps. Furthermore, we were able to use these data to assess dynamic cell descriptors such as the rate of nucleus displacement, which can be interpreted as a proxy for cell growth and displacement. Although intracellular variability of the nuclear position limits the precision of these assessments, particularly regarding individual cells, this inherent measuring error will have opposite effects in consecutive cells (e.g. over-estimation in one cell results in under-estimation in the following one) making it possible to visualize the dynamic changes within the population. The methodology used for evaluating these parameters is described in Figure 6 and Supplemental Figure S1, and 2D representations of them are shown in Figure 7. Step-by-step instructions to perform these analyses are included in Supplemental Material S1.
Figure 6.
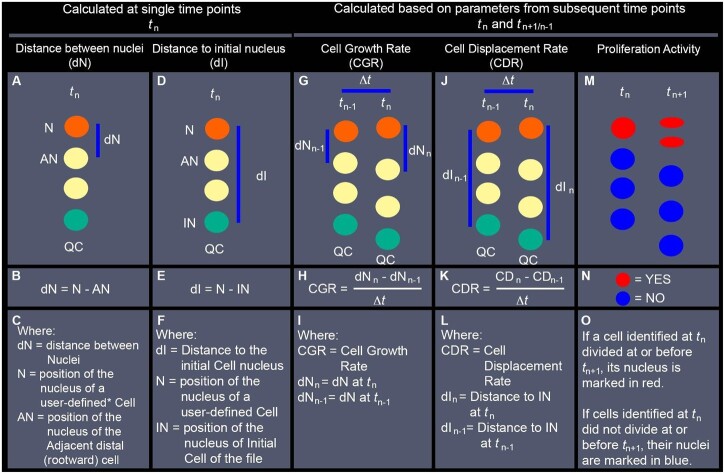
Description of nuclear position-based parameters evaluated with LiPlaCeT plugin followed by analyses with ParaView. Simplified diagrams represent nuclei of cells within a single cell file. A–C, Estimation of the distance between adjacent nuclei within the same file (dN) at the displayed time point (tn). D–F, Distance to the nucleus of the initial cell of the file (dI), estimated as the distance between a user-defined Nucleus (N)* and the nucleus of the initial cell of the file (IN), at the displayed time point (tn). G–I, CGR, approximation calculated as the rate at which estimated dN changed during a period between the displayed time (tn) and a previous time point (tn–1) (The Methodology followed for evaluation of CGR in scenarios when cell division occurs between tn–1 and tn, is described in Supplemental Figure S1). J–L, CDR, approximation calculated as the rate at which each nucleus was displaced away from the IN during a period between the displayed time (tn) and a previous time point (tn−1). M–O, Distribution of cell proliferation events that took place between the displayed time point (tn) and the following timepoint (tn+1). QC position is indicated in all diagrams as a positional reference, but its position is not used in the calculations of the described parameters. Instead, the position of the nucleus of the initial cell (IN, green circles) is used as a reference for the distal limit of each cell file. * a user-defined cell (mentioned in (C)), is an individual cell identified manually using the LiPlaCeT plugin. Every nucleus manually marked acquires a unique ID containing its position and lineage information at different time points. This information is in turn analyzed and displayed for each user-defined nucleus using ParaView.
Figure 7.
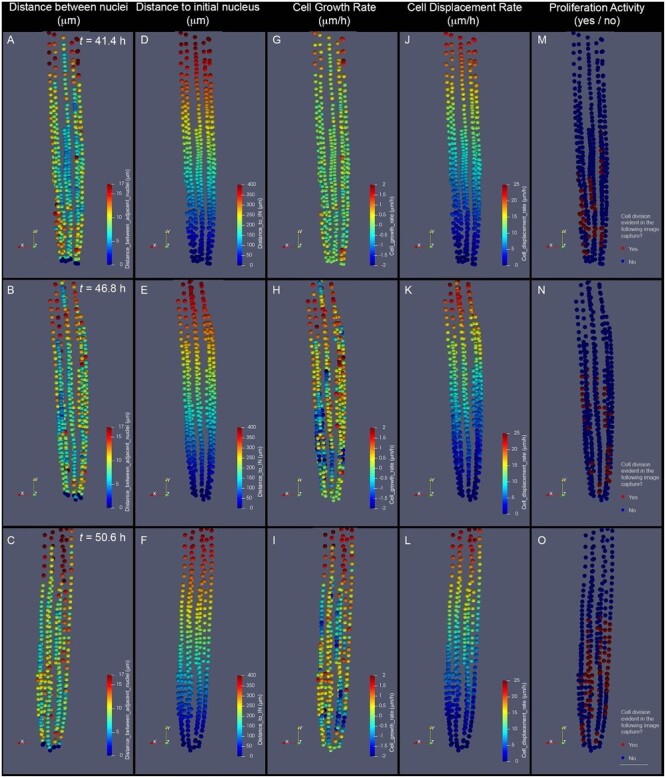
Parameters of cell dynamics extracted from cell tracking and evaluated as 3D descriptions. A–C, dN. D–F, Distance to the IN (nucleus of an initial cell). G–I, CGR. J–L, CDR. M–O, Cell proliferation distribution. The depicted images represent the RAM of the same growing root, imaged at three subsequent time points after beginning a time-lapse experiment: 43.4 h (A, D, G, and J), 46.8 h (B, E, H, and K), and 50.6 h (C, F, I, and L). Our analysis pipeline integrates this information and generates color-coded representations of each evaluated nucleus. Quantitative parameters are represented as intensity scales to facilitate whole-tissue visualization. dN, measured in micrometer (A–C), is estimated by measuring the distance between each nucleus and the adjacent distal (rootward) nucleus within a cell file. Distance to IN measured in micrometers (D–F) is estimated by measuring the distance between each nucleus and the nucleus of the initial cell of each cell file. CGR, measured in micrometers per hour (G–I), is estimated by comparing dN values in the displayed time (tn) with those of the previous time point (tn−1). This value represents an approximation of the rate at which cells grew during the last evaluated time frame. CDR, measured in micrometers per hour (J–L), is estimated by comparing values of dI in the displayed time (tn) with those of the previous time point (tn−1). This value represents an approximation of the rate at which each cell was displaced from the most distal cell of the file during the last evaluated time frame. Proliferation activity, evaluated as a yes (red label) or no (blue label) value (M–O), represents whether or not each depicted nucleus belongs to a cell that will divide before or at the following time point evaluated (tn+1). To calculate CGR and CDR of cells depicted in (G) and (J), tn−1 corresponds to 33.8 h (data not shown). To determine Proliferation activity of cells depicted in (O), tn+1 corresponds to 54.2 h (not shown). The images shown are 2D representations of 3D whole-tissue reconstructions. For clarity purposes, the color density and size of the dots that represent nuclei were adjusted in these images to obscure nuclei behind them. Scale bar = 50 μm.
To illustrate the complex patterns that emerge within the endodermis cell files during RAM growth, we selected three consecutive time frames (∼4 h apart from each other) from a proof-of-concept time-lapse experiment (Figure 7). The 3D distribution of two cell descriptors evaluated at a single time point are shown here: distance between nuclei (dN) profile, representing an approximation of cell length profiles, and Distance to the nucleus of the initial cell profile (Figure 7, A–F).
In addition to 3D-evaluated distances between nuclei and a distance to the nucleus of the initial cell, we can use recorded time information to compute the rates at which these two parameters change (Figure 6, G–L), thereby generating information regarding how cell growth rate (CGR) and cell displacement rate (CDR) patterns change as individual cells transit through different meristematic regions (Figure 7, G–L). The methodology was also used to estimate CGR when the displayed images included recently divided cells, and it is explained in Supplemental Figure S1. Using information on tracking of cell proliferation events generated with the LiPlaCeT plugin, it is possible to generate 3D maps of distribution of Proliferation activity for each time frame (Figures 6, M–O and 7, M–O). Therefore, it is possible to analyze correlation between the patterns and dynamics of different parameters for cell populations over time for the same region, or interaction between different meristematic regions. Our proof-of-concept experiment demonstrates that this workflow is suitable to generate data and visual representations necessary to study complex patterns that emerge within a meristematic population in 3D as it develops through time. The source code is freely available (see “Materials and methods”).
Discussion
In this work, we presented the LiPlaCeT ImageJ plugin developed to allow an easier 4D analysis of massive data collected from time-lapse experiments on live plant tissues with the purpose of understanding cell proliferation dynamics, cell growth, cell displacement, and other cellular parameters within a developing organ. Our plugin uses multi-view (XY, XZ, and YZ planes) and the visualization of two time points (time t and t+1) to navigate across the hyper-stack. The multiview approach allows us to accurately identify the 3D position of cells, and it is also computationally efficient compared to approaches using 3D volume rendering. Furthermore, volume rendering may hide important data for images with a high number of cells and low contrast like those found deep inside the tissues as in a developing LRP. The two timepoints visualization approach allows the user to easily identify an object of interest, for example, mitotic events, without changing the time point view. Instead, approaches using a single time point visualization require the user to change the current view for the next time to identify changes (e.g. mitotic events) which need iterating several times to identify all the mitotic events; besides, if needed, our plugin can also be used in this mode. Other approaches require saving the images in a specific file format (h5 file format; Wolff et al., 2018) to be opened by its corresponding viewer (BigDataViewer). LiPlaCeT only requires the hyperstack to be loaded in ImageJ. The Graphical User Interface from our plugin allows an easier interaction with the user by implementing the most common tools that can be handled with the mouse. Creating and removing spheres (cell or nuclear identifier) can be achieved with a single click and changing the sphere position only requires dragging the sphere to the new position. Increasing or decreasing sphere size requires moving the mouse wheel up or down. Besides, mitotic events can be marked with mouse clicks. The navigation across the slices of a stack can be performed also by moving the mouse wheel up or down. Some other important features, such as movement to the next or the previous time point, zoom in and out, copy spheres positions from a current time point to the next one, among others, can be performed with the keyboard. In addition, to visually inspect different qualitative and quantitative parameters, some of which are exemplified in Figures 4 and 6, ParaView *.vtk files can be generated. These parameters include, but they are not limited, to cell cycle number, position of mitotic events, color-coded cell lineages, a tree representation of cell lineage in time scale, CDR, cell distance to a point of reference (e.g. RAM initial cell), cell length, CGR, cell division direction, average cell growth direction, and cell trajectory; all these metadata can be saved as a *.csv file. Finally, LiPlaCeT has been implemented as an ImageJ plugin and its distribution is open-source, and freely available. Users can modify the code to implement their own parameters and measurements or add additional features. LiPlaCeT is well described in a user manual addressing in detail step-by-step operations for each type of analysis (Supplemental Material S1).
It is important to consider specific features of LiPlaCeT in comparison with other programs for cell tracking of live plant cells (see Supplemental Table S1). One such program is TrackMate (Jaqaman et al., 2008; Tinevez et al., 2017). TrackMate allows automatic tracking. To do this, a local maximum of a signal indicates the localization of a spot; a fixed threshold value is used to discard spots with low intensity and the estimated spot diameter is required in advance to search for spots of similar size. However, one serious limitation is that automatically detecting spots can be a difficult or unachievable task because (1) a threshold can be well suited for a given time point but it cannot be good enough for a different time point as the signal strength is changing with time and (2) for tissues deep inside of a plant organ, SNR is always variable, being lower in the most internal tissues. Heterogeneity of signal strength in time-lapse studies is present both in experiments performed with light-sheet (Figure 1 in von Wangenheim et al., 2016, Figure 2 in von Wangenheim et al., 2020), and in laser scanning confocal microscopy (e.g. see Figure 4 in this work; find also examples in Figures 4–6 in Goh et al., 2016; Supplemental Figure S4 in Shimotohno et al., 2018; additional file 12 in Rahni and Birnbaum, 2019) and in both, wild-type and mutant backgrounds. Because of heterogeneity of signal strength in live plant material, or also when only a fraction of the object can be analyzed, manual curation is always a requirement (e.g. Goh et al. (2016) analyzed with TrackMate the progeny of only the outer layer of St II LRP). Even with other subjects, TrackMate does not always permit automatic tracking. For example, Tinevez et al. (2017, p. 84) comment on TrackMate: “There are many use cases where a fully automatic strategy is not possible due to factors such as variable signal to noise (SNR) in the images.” We tested a spot detection algorithm of TrackMate’s for a representative stack of Z-sections of an LRP, and the spot detection was good for the initial time points where the spot intensity was high, however, the SNR of fluorescently marked nuclei decreased with time, and the algorithm failed to detect the spots (see Supplemental Figure S2). This comparison clearly shows that in time-lapse experiments, the automatic function of TrackMate is only partially suitable for tissues deep inside the root, and that LiPlaCeT is appropriate, even under these conditions, to follow a cell lineage and collect other parameters.
Another approach for plant cell tracking is MorphoGraphX (De Reuille et al., 2015; Strauss et al., 2021). This software permits quantitative analysis in 3D in time (4D) and works very well for high contrast images, for example, when cell wall contrast is sufficient to perform the segmentation of a region of interest and when an external tissue, such as abaxial leaf epidermis (e.g. Kierzkowski et al., 2019) is studied. In this case, a user must select an adequate threshold parameter for correcting under-segmentation and over-segmentation errors. However, as mentioned above, for tissues deep inside a plant organ it is impossible, at least with currently available molecular markers, to achieve equal SNR in all cells making segmentation problematic. We tested this approach, and the results are acceptable for most cells, but not for all, due to heterogeneity of SNR of the regions of interest (Supplemental Figure S3). The comparison of LiPlaCeT with other approaches is summarized in Supplemental Table S1. While the previously mentioned automated approaches can be switched to manual mode to correct erroneously generated data, LiPaCeT was designed for an ad hoc ergonomic and easy manual cell tracking, containing specific curating tools, especially useful when SNR of images is low or variable in time or 3D space and when automated methods may fail; moreover, our program is more appropriate for internal tissues of an organ and permits obtaining reliable data on material with heterogeneous fluorescence signals of an object.
The application of LiPlaCeT was very practical for addressing LRP morphogenesis and the role of auxin signaling and transport in founder cell recruitment (Torres-Martínez et al. 2020). Here with this approach, we analyzed cell cycle time in the LRP central and longitudinally flanking domains and found differences in cell proliferation potential in these domains. Previously, it was shown that cells derived from tangentially peripheral founder cell files have a longer cycle time compared to those derived from the centrally located cell, but no formal analysis of differences in the proliferation of central and flanking domain cells was addressed (von Wangenheim et al., 2016). Here we have shown that Stage I cells over a period of 31 h, passed twice the lower number of cycles in the longitudinally flanking domain compared to the central one. Consistently with this finding, it has recently been shown that the gradient between periclinal cell divisions in the LRP central versus flanking domain is essential for correct LRP morphogenesis (Fujiwara et al., 2021). In addition to quantitative analysis, identification of cell clones, representing descendants of certain LRP cells, helps to understand how LRP shaping takes place (Figure 4). Overall, LiPlaCeT facilitates the analysis of the cell cycle duration and morphogenesis in developing plant organs. This certainly will be useful to decipher genetic control of LR morphogenesis.
During this work, we designed an experimental setup, shown in Figure 5, appropriate for visualizing cell dynamics of the TAD cells in the RAM. Median cell cycle duration of RAM cells evaluated in a time-lapse experiment was found to be 12.6 h (Rahni and Birnbaum, 2019). Therefore, to visualize subsequent cell division events and to trace cell lineage and cell population changes require several days long-term time-lapse experiments. To visualize whole populations of TAD cells over time under standard for Arabidopsis in vitro culture conditions, our setup was performed with roots growing vertically toward the gravity axis on top of a layer of agar containing standard growth medium (see “Materials and methods”). Our setup was adapted by inverting to a horizontal position a Nikon AZ100, as was previously successfully used for the evaluation of the role of AUX1-mediated auxin transport and cytokinin signaling during gravitropic response (Pernisova et al., 2016). The lenses available in this microscope (see “Materials and methods”) allowed visualization of the whole meristem within a single Z-stack; however, it required human intervention to locate and center the region of interest in each capture, and this limited the temporal resolution of the proof-of-concept experiment presented here.
Till now, the experimental analysis of cell proliferation dynamics in long-term experiments was only possible for a distal RAM portion (Rahni and Birnbaum, 2019) or for the whole RAM of relatively small size (von Wangenheim et al., 2017). Our design allowed us to maintain standard growth conditions, avoid oxygen deprivation while roots grew over an agar layer, and to capture images of endodermis nuclei from the whole RAM region. Presented proof-of-concept experiment made on the whole RAM illustrates both applicability of our experimental design and the convenience and potential of the LiPlaCeT plugin that we developed. Since roots continue to grow during the experiment, human intervention to locate and center the region of interest (RAM region) in each capture was required. For this reason, no specific programming for fixed time captures was used. This limitation can be overcome with application of approaches similar to the TipTracker program that automatically recognizes and follows root tips during growth (von Wangenheim et al., 2017). Our material was collected before this program was available. Here we present an example of data collection and its analysis with LiPlaCeT only for a single cell type, endodermis. If multiple fluorescent markers are used, or all RAM cells are marked, this approach can also be successfully applied. Our goal was to perform a proof-of-principle experiment that appears to be successful.
Low temporal resolution in our proof-of-concept experiment limited our capacity to identify unambiguously every cell through time, but visual landmarks of endodermal nuclei, such as longitudinal divisions and their progeny and visual evidence of mitotic events, provided us with a set of data necessary to implement the LiPlaCeT analytic pipelines. Cell tracking using the LiPlaCeT plugin proved instrumental to facilitate identification and annotation of a subset of cells through time. Analysis of relative nuclei position of each nucleus within cell files over time allowed us to generate visual approximations for both cell length patterns at each time point, and cell growth patterns over time (Figures 6 and 7). Cell length profile within a single file at a single time point provides valuable information regarding the regions where cells are actively proliferating, or transiting out of mitotic cycles to endoreduplication, rapid elongation, and eventual terminal differentiation (French et al., 2012; Ivanov and Dubrovsky, 2013; Pacheco-Escobedo et al., 2016). However, individual cell lengths can be strongly influenced by dynamic changes in neighboring tissues such as cell proliferation and/or growth, by root bending, and other factors. The initial cells of each cell file are part of the root SCN. As cells are displaced away from the SCN, they transit through chemical gradients and physical environments, which regulate their cell cycle and overall gene expression patterns (Wendrich et al., 2017). Theoretical models indicate that anisotropic growth across root regions is key for both, self-organization of root meristems and growth plasticity (Nakielski and Lipowczan, 2012, 2013). The ability to visualize how cell length profiles change as they are displaced away from the initial cell (due to the proliferation and growth of more distal cells) can be useful to understand how the dynamics of different meristematic regions interact through time. Therefore, it is useful to be able to associate changes in cell lengths of individual cells, with distance from each cell to initial cells, both for individual cells and the whole cell population level (Figure 7).
In the future, LiPlaCeT could be improved. For example, to save time, it could be helpful to add the capability of importing tracking metadata from other software. We have selected ParaView software to visualize the tracking data. It would be helpful to use the 3D visualization capabilities of ImageJ to visually inspect the different cell lineage properties. Recently, deep learning approaches have gained a lot of attention due to their high efficiency in resolving several tasks such as object detection, segmentation, and tracking (Lugagne et al., 2020; Isensee et al., 2021; von Chamier et al., 2021; Wen et al., 2021). The main difficulty of a deep learning approach is the large amount of curated data required to get a good performance. Our plugin is a good starting point to generate manually curated data to be used for training a convolutional neuronal network, CNN (He et al., 2017). This CNN model will allow the user to save time in creating a cell lineage by only editing the errors of the CNN. Nevertheless, the developed plugin described here is a powerful tool to address various aspects of behavior and morphogenesis of populations of live cells in 4D and can be applied to different plant organs.
Materials and methods
Plant material and growth conditions for LRP visualization
The transgenic Arabidopsis (A. thaliana) lines pSCR::SCR-H2B-YFP (Xu et al., 2006) p35S::H2B-RFP (Federici et al., 2012), and pUBQ10::NPSN12-YFP (Geldner et al., 2009), all in Col-0 background, were used to visualize endodermis nuclei, all the nuclei, and plasma membrane, respectively. Seedlings were surface-sterilized, stratified for 2 or 3 d, and germinated in vitro on growth medium in Petri dishes maintained in vertical position. The medium contained 0.2× Murashige and Skoog (MS) salts prepared from Linsmaier and Skoog medium (L477; Phytotechnology Laboratories, Lenexa, KS, USA), pH 5.7, and was supplemented with vitamins (0.1 mg L−1 pyridoxine, 0.1 mg l−1 nicotinic acid), 1% (w/v) sucrose, and 0.8% agar (w/v, Bacto Agar; BD Difco, Sparks, MD, USA). Plants were grown at 21°C, under a 16-h photoperiod with a light intensity of 105 μm m−2 s−1.
Growth conditions for RAM visualization
Two dpg seedlings were transplanted to 12 × 12 cm square Petri dishes in which 2 × 4 cm rectangular orifices were cut out of the base and covered with coverslips attached with general use silicon F109 (SISTA, Mexico). This resulted in wells within the base of the Petri dish, which were filled with 2 mL of 0.2× MS salts, 1% sucrose and 0.8% agar. Humidity within these chambers was maintained by moist pieces of sterile cotton placed inside the dish by its borders before sealing it with micropore. From this time on, seedlings were incubated inside the microscopy room and environmental conditions were controlled as follows. Temperature was maintained at 22°C ± 2°C. Plants were grown at a 16-h photoperiod under a LED green/red Procyon lamp of 12 V DC 4.2 Amp (Home Grown Lights, USA). We adjusted the distance between the light source and the sample to ensure that seedlings were exposed in the microscopy room to the similar light intensity as in growth chambers. The growth of plants and image acquisition was performed in the Laboratorio de Microscopía y Microdisección Láser (LabMicroLas) at the Instituto de Ecología (UNAM).
Microscopy for RAM visualization
The experimental setup assembled for time-lapse experiments on vertically growing root apex was based on a Nikon Advanced Zoom Macro Microscope system AZ100 (NIKON CORPORATION, Tokyo, Japan); https://d33b8x22mym97j.cloudfront.net/phase4/literature/Brochures/2ce-mrvh-4.pdf?mtime=20180725121028&focal=none) with on-axis image capturing capability, equipped with epifluorescence and DIC illumination. For the visualization of the endodermis, YFP labeled-nuclei, we used a PlanFluor 5× macro lens, working distance (WD) 15 mm, numerical aperture (NA) 0.5 and an additional optical magnification of 4× provided by the microscope zoom. The epifluorescence illumination source was an X-CITE XLED1, Lumen Dynamics (Excelitas Technologies, http://www.excelitas.com/Pages/Product/X-Cite-XLED1.aspx), filtered with an EGFP/FITC/Cy2/Alexa Fluor 488 filter cube (480/30×, 535/40 m, 505DC, Chroma).
To free the space in front of the objectives, we did not include in our setup the microscope base designed by-default for episcopic or diascopic illumination. Instead, the microscope stand was attached in a horizontal position to a home-made aluminum base. The microscope stage was replaced with a purpose-designed support for large square Petri dishes, whose position was controlled in the X, Y, and Z-axis by three NRT100, 100-mm motorized translation stages and stepper motors with 1-μm step in X, Y, and Z-axis, respectively (Thorlabs, https://www.thorlabs.com/thorproduct.cfm?partnumber=NRT100). The acquisition of Z-stacks with 1-μm step resolution were automated by controlling the translation stages, but time-lapse captures required human intervention to identify the RAM before each capture.
Microscopy for time-lapse analysis of LRP
Given that LR growth is not affected by horizontal growth of the primary root, LRP initiation events were observed on an inverted confocal laser scanning microscopy setup previously described in Reyes-Hernández et al. (2019). Briefly, a confocal system was built around Zeiss Axiovert 200M microscope (Oberkochen, Germany) that consisted of a high-speed galvo-resonant scanner for visible wavelengths (SCAN-VIS), a 488-nm laser source, a filter cube with 525/45 nm, and 630/92 nm bandpass filters for green fluorescent protein and red emission fluorescence, respectively; a dual-channel photomultiplier tube (PMT) module, a linear motor travel XY Stage and a Z-axis piezo stage with controllers, all from Thorlabs, Inc. (Newton, NJ, USA). Laser intensity used was 10%, and gain was 100%. A Zeiss C-APO ×63, 1.2NA W objective (Oberkochen, Germany) was used. As cell cycle time in early LRP ranges from on average 4.1–7.3 h from the second to the fourth cycle of the founder cell (Torres-Martínez et al., 2020), most experiments on the LRP development in this work were programmed to take optical sections each 1 h during 48 h. For each time point, a total of 51 μm of a developing LRP thickness was scanned, 101 sections, 0.5 μm each. Time-lapse analysis starts by marking each cell of interest at the beginning of experiment with a colored circle and then is continued following a lineage (see Supplemental Material S1, LiPlaCeT User Manual).
Five dag seedlings were placed in a 1-well Thermo Scientific Nunc Lab-Tek II Chamber Slide System with a cover glass #1.5 at the bottom (ThermoFisher Scientific, Waltham, MA, USA). A rectangular piece (∼3-mm thick) of 0.2× MS agar medium was used to cover the primary root. Seedlings were maintained in the horizontal position for 1 h before the beginning of experiments. The Chamber Slide System was placed on the motorized microscope stage and the shoot was illuminated through an FP600ERT, 600-µm core multimode fiber from a light source of fiber-coupled LED MWWHF2, 4,000 K, 16.3 mW (Thorlabs, Inc., Newton, NJ, USA). To maintain a 16 h photoperiod, the LED source was connected through a timer. The fiber illumination of the shoot did not interfere with laser scanning. Temperature was maintained at 21°C.
LiPlaCeT development
LiPlaCeT was developed over several years and adapted to biologist needs. Plant biology-oriented co-authors of this work suggested features that will allow them to speed up the manual cell lineage and tracking analyses, and those features were implemented in LiPlaCeT. Several rounds of iterations were required to arrive to the last version of the LiPlaCeT. It was developed as an ImageJ Plugin because this open-source software platform is highly used by the scientific community (Schindelin et al., 2015; Rueden et al., 2017). During its development, UNIX platform (Ubuntu 18.04 LTS and Ubuntu 20.04 LTS), Matlab software, and Java programming language was used. The source code is freely available at https://github.com/paul-hernandez-herrera/LiPlaCeT and the ImageJ plugin including a dataset example and the User Manual can be downloaded from https://www.ibt.unam.mx/documentos/diversos/LiPlaCeT.zip.
Statistical analysis
The statistical analysis was performed using SigmaPlot version 12 (Systat Software, San Jose, CA, USA).
Supplemental data
The following supplemental materials are available in the online version of this article.
Supplemental Figure S1. Methodology followed to estimate CGR in different scenarios of cell proliferation between the displayed time (tn) and the reference time (tn−1).
Supplemental Figure S2. Testing the performance of the spot detection algorithm of TrackMate.
Supplemental Figure S3. Testing the performance of MorphoGraphX.
Supplemental Table S1. Feature comparison between LiPlaCeT and TrackMate, MorphoGraphX, and MaMuT.
Supplemental Movie S1. Example of cell tracking of a developing LRP in p35S::H2B-RFP pUBQ10::NPSN12-YFP F1 seedlings starting from Stage I.
Supplemental Movie S2. An example of cell lineage tracking of a developing LRP in a time-lapse experiment with the use of LiPlaCeT plugin.
Supplemental Movie S3. An example of LiPlaCeT application for the analysis of principal growth directions of a developing LRP.
Supplemental Material S1. LiPlaCeT user manual.
Supplementary Material
Acknowledgments
We thank B. Scheres, L. Laplaze, and N. Geldner for seed donation and S. Napsucialy-Mendivil and J. M. Hurtado-Ramírez for technical help. We also thank Diana Belén Sánchez and Diana Romo for their logistical support and facilities from LabMicroLas at the UNAM, where the time-lapse experiments of the RAM were performed.
Funding
We acknowledge support from Dirección General de Asuntos del Personal Académico (DGAPA)-Universidad Nacional Autónoma de México (UNAM) scholarship CJIC/CTIC/4556/2020 to P.H.H., DGAPA-Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT)-UNAM (grant IN204221 to J.G.D., grants IN206220, IN203220, IN200920, and IN211721 to A.G.A., B.G.P., M.P.S., and E.R.Á.B., and grant IV100420 to G.C. for partial funding), and Mexican Consejo Nacional de Ciencia y Tecnología (CONACyT grant A1-S-9236 to J.G.D., grant 225680 to E.R.Á.B., grant 102987 to M.P.S., and grant 102959 to A.G.A.). E.R.Á.B. and Y.U.C. acknowledge CONACYT (grant No. 225680) for partial funding. H.H.T.-M. was supported by a Ph.D. fellowship from CONACyT and had a CONACyT postdoctoral support (A1-S-9236).
Conflict of interest statement. The authors declare that they have no conflict of interest.
E.R.Á.B., G.C., and J.G.D. planned and conceived the research. E.R.Á.B., B.G.P., M.P.S., A.G.A., Y.U.C., G.C., and J.G.D. designed cell tracking experiments and approaches that served to substantiate the pipeline presented here. Y.U.C., A.V.A., and J.E.C.V. designed and performed time-lapse experiments analyzing primary root meristem using the Nikon AZ100 horizontally adapted microscope in the LabMicroLas at the UNAM and H.H.T.M. and J.G.D. designed and performed time-lapse experiments analyzing lateral root formation; P.H.H. designed the original version of the Fiji plugin software and together with H.H.T.M. and Y.U.C. improved it. P.H.H., H.H.T.M., Y.U.C.; J.G.D., E.R.Á.B., B.G.P., M.P.S., A.G.A., and G.C. discussed and analyzed data as input for plugin design. P.H.H., H.H.T.M., Y.U.C., and J.G.D. wrote the plugin user manual. Y.U.C., P.H.H., H.H.T.M., J.G.D., and G.C. wrote the article draft. All authors revised and contributed to the final version of the article. J.G.D. and G.C. supervised and completed the writing. E.R.Á.B., G.C., and J.G.D. agree to serve as authors responsible for contact and ensure communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Gabriel Corkidi (gabriel.corkidi@ibt.unam.mx).
References
- Azpeitia E, Benítez M, Vega I, Villarreal C, Alvarez-Buylla ER (2010) Single-cell and coupled GRN models of cell patterning in the Arabidopsis thaliana root stem cell niche. BMC Syst Biol 4: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baesso P, Randall RS, Sena G (2018) Light sheet fluorescence microscopy optimized for long-term imaging of Arabidopsis root development. Methods Mol Biol 1761:145–163 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inzé D (2001) The peri-cell-cycle in Arabidopsis. J Exp Bot 52:403–411 [DOI] [PubMed] [Google Scholar]
- Beemster GT, Baskin TI (1998) Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116:1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Ivanchenko MG, Friml J, Shishkova S, Dubrovsky JG (2009) A morphogenetic trigger: is there an emerging concept in plant developmental biology? Trend Plant Sci 14:189–193 [DOI] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Kreshuk A, et al. (2019) Ilastik: interactive machine learning for (bio) image analysis. Nat Methods 16:1226–1232 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44 [DOI] [PubMed] [Google Scholar]
- Busch W, Moore BT, Martsberger B, Mace DL, Twigg RW, Jung J, PruteanuMalinici I, Kennedy SJ, Fricke GK, Clark RL, et al. (2012) A microfluidic device and computational platform for high-throughput live imaging of gene expression. Nat Methods 9:1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campilho A, Garcia B, Toorn HV, Wijk HV, Campilho A, Scheres B (2006) Time-lapse analysis of stem-cell divisions in the Arabidopsis thaliana root meristem. Plant J 48:619–627 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trend Plant Sci 8:165–171 [DOI] [PubMed] [Google Scholar]
- De Lucas M, Brady SM (2013) Gene regulatory networks in the Arabidopsis root. Curr Opin Plant Biol 16:50–55 [DOI] [PubMed] [Google Scholar]
- De Reuille PB, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A (2015) MorphoGraphX: a platform for quantifying morphogenesis in 4D. eLife 4:e05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134:681–690 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares Scaglia F, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17:678–682 [DOI] [PubMed] [Google Scholar]
- Di Mambro R, Sabatini S, Dello Ioio R (2018) Patterning the axes: a lesson from the root. Plants (Basel) 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan JH, Sablowski R (2010) Walls around tumours—why plants do not develop cancer. Nat Rev Cancer 10:794–802 [DOI] [PubMed] [Google Scholar]
- Du Y, Scheres B (2017) Lateral root formation and the multiple roles of auxin. J Exp Bot 69:155–167 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colón-Carmona A, Rost TL (2000) Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol 124:1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colón-Carmona A, Doerner PW (2001) Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana Planta 214:30–36 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105:8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami N, Sedaei Z, Ferdousi R (2021) Computerized cell tracking: current methods, tools and challenges. Vis Inform 5: 1–16 [Google Scholar]
- Erickson RO (1986) Symplastic growth and symplasmic transport. Plant Physiol 82:1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici F, Dupuy L, Laplaze L, Heisler M, Haseloff J (2012) Integrated genetic and computation methods for in planta cytometry. Nat Methods 9:483–485 [DOI] [PubMed] [Google Scholar]
- French AP, Wilson MH, Kenobi K, Dietrich D, Voß U, Ubeda-Tomás S, Pridmore TP, Wells DM (2012) Identifying biological landmarks using a novel cell measuring image analysis tool: cell-o-Tape. Plant Methods 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Goh T, Tsugawa S, Nakajima K, Fukaki H, Fujimoto K (2021) Tissue growth constrains root organ outlines into an isometrically scalable shape. Development 148:dev196253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057 [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, De La Paz Sánchez M, García-Ponce B, Azpeitia E, Álvarez-Buylla ER (2012). Hormone symphony during root growth and development. Dev Dynamics 241:1867–1885 [DOI] [PubMed] [Google Scholar]
- García-Gómez ML, , Ornelas-Ayala D, Garay-Arroyo A, García-Ponce B, Sánchez MP, Álvarez-Buylla ER (2020) A system-level mechanistic explanation for asymmetric stem cell fates: Arabidopsis thaliana root niche as a study system. Sci Rep 10: 3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez ML, Azpeitia E, Álvarez-Buylla ER (2017) A dynamic genetic-hormonal regulatory network model explains multiple cellular behaviors of the root apical meristem of Arabidopsis thaliana. PLoS Comput Biol 13:e1005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59:169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Toyokura K, Wells DM, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett MJ (2016) Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development 143:3363–3371 [DOI] [PubMed] [Google Scholar]
- He K, Gkioxari G, Dollár P, Girshick R (2017) Mask R-CNN. IEEE International Conference on Computer Vision (ICCV), Venice, Italy, pp 2980–2988
- Ingram PA, Malamy JE (2010) Root system architecture. InJean-Claude K, Michel D, eds, Advances in Botanical Research, Vol 55. Academic Press, Cambridge, MA, pp 75–117 [Google Scholar]
- Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211 [DOI] [PubMed] [Google Scholar]
- Ivanov VB, Dubrovsky JG (2013) Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends Plant Sci 18:237–243 [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G (2008) Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods 5:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Waadt R, Brugman R, Schroeder JI, Grossmann G, Schumacher K, Krebs M (2015) Live cell imaging with R-GECO1 sheds light on flg22- and chitinInduced transient [Ca(2+)]cyt patterns in Arabidopsis. Mol Plant 8:1188–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzkowski D, Runions A, Vuolo F, Strauss S, Lymbouridou R, Routier-Kierzkowska AL, Wilson-Sánchez D, Jenke H, Galinha C, Mosca G, et al. (2019) A growth-based framework for leaf shape development and diversity. Cell 177:1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R, Thorstensen T, Rahman MA, Heyman J, Nenseth HZ, Lammens T, Herrmann U, Swarup R, Veiseth SV, Emberland G, et al. (2014) The ASH1-RELATED3 SET-domain protein controls cell division competence of the meristem and the quiescent center of the Arabidopsis primary root. Plant Physiol 166:632–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Guyomarc’h S, Hill K, Lucas M, Voß U, Kenobi K, Wilson MH, Farcot E, Hagen G (2015) Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell 27:1368–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrekha VV, Pasternak T, Ivanov VB, Palme K, Mironova VV (2017) 3D analysis of mitosis distribution highlights the longitudinal zonation and diarch symmetry in proliferation activity of the Arabidopsis thaliana root meristem. Plant J 92:834–845 [DOI] [PubMed] [Google Scholar]
- López-Bucio JS, Dubrovsky JG, Raya-González J, Ugartechea-Chirino Y, LópezBucio J, de Luna-Valdez LA, Ramos-Vega M, León P, Guevara-García AA (2014) Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J Exp Bot 65:169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugagne JB, Lin H, Dunlop MJ (2020) DeLTA: automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput Biol 16:e1007673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, von Wangenheim D, Federici F, Haseloff J, Stelzer EH (2011) High resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J 68:377–385 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana Development 124:33–44 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329:1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, Mairhofer S, Burr-Hersey J, Goh T, Von Wangenheim D, Atkinson B, Sturrock CJ, Lynch JP (2017) Shaping 3D root system architecture. Curr Biol 27:R919–R930 [DOI] [PubMed] [Google Scholar]
- Nakielski J, Lipowczan M (2012) A method to determine the displacement velocity field in the apical region of the Arabidopsis root. Planta 236:1547–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielski J, Lipowczan M (2013) Spatial and directional variation of growth rates in Arabidopsis root apex: a modelling study. PLoS One 8:e84337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napsucialy-Mendivil S, Dubrovsky JG (2018) Genetic and phenotypic analysis of lateral root development in Arabidopsis thaliana. InBarbez E, Ristova D, eds, Root Development: Methods and Protocols, Methods in Molecular Biology, Vol 1761. Springer, Berlin, Germany, pp 47–75 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS (2007) Hidden branches: developments in root system architecture. Ann Rev Plant Biol 58:93–113 [DOI] [PubMed] [Google Scholar]
- Pacheco-Escobedo MA, Ivanov VB, Ransom-Rodríguez I, Arriaga-Mejía G, Ávila H, Baklanov IA, Pimentel A, Corkidi G, Doerner P, Dubrovsky JG, et al. (2016) Longitudinal zonation pattern in Arabidopsis root tip defined by a multiple structural change algorithm. Ann Bot 118:763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perianez-Rodriguez J, Rodriguez M, Marconi M, Bustillo-Avendaño E, Wachsman G, Sanchez-Corrionero A, De Gernier H, Cabrera J, Perez-Garcia P, Gude I (2021) An auxin-regulable oscillatory circuit drives the root clock in Arabidopsis. Sci Adv 7:eabd4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Perez-Perez JM, Di Mambro R, Peris CL, Díaz-Triviño S, Del Bianco M, Pierdonati E, Moubayidin L, Cruz-Ramírez A, Costantino P, et al. (2013) RETINOBLASTOMA-RELATED protein stimulates cell differentiation in the Arabidopsis root meristem by interacting with cytokinin signaling. Plant Cell 25:4469–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisova M, Prat T, Grones P, Harustiakova D, Matonohova M, Spichal L, Nodzynski T, Friml J, Hejatko J. (2016) Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol 212: 497–509 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Ann Rev Plant Biol 63:563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahni R, Birnbaum KD (2019) Week-long imaging of cell divisions in the Arabidopsis root meristem. Plant Methods 15:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Hernández BJ, Srivastava AC, Ugartechea-Chirino Y, Shishkova S, Ramos-Parra PA, Lira-Ruan V, Díaz de la Garza RI, Dong G, Moon JC, Blancaflor EB, et al. (2014) The root indeterminacy-to-determinacy developmental switch is operated through a folate-dependent pathway in Arabidopsis thaliana. New Phytol 202:1223–1236 [DOI] [PubMed] [Google Scholar]
- Reyes-Hernández BJ, Shishkova S, Amir R, Quintana-Armas AX, NapsucialyMendivil S, Cervantes-Gamez RG, Torres-Martínez HH, Montiel J, Wood CD, Dubrovsky JG (2019) Root stem cell niche maintenance and apical meristem activity critically depend on THREONINE SYNTHASE1. J Exp Bot 70:3835–3849 [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, Cools T, De Veylder L, Benfey PN, Palatnik JF (2015) MicroRNA miR396 regulates the switch between stem cells and transit amplifying cells in Arabidopsis roots. Plant Cell 27:3354–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT SJ, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017) ImageJ2: imageJ for the next generation of scientific image data. BMC Bioinformatics 18: 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi E, Rutten JP, Di Mambro R, Polverari L, Licursi V, Negri R, Ioio RD, Sabatini S, Ten Tusscher K (2020) A Self-organized PLT/Auxin/ARR-B network controls the dynamics of root zonation development in Arabidopsis thaliana. Dev Cell 53:431–443 [DOI] [PubMed] [Google Scholar]
- Scheres B (2007) Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8:345–354 [DOI] [PubMed] [Google Scholar]
- Schiegg M, Hanslovsky P, Kausler BX, Hufnagel L, Hamprecht FA (2013) Conservation tracking. IEEE International Conference on Computer Vision, Sydney, NSW, Australia, pp 2928–2935
- Schindelin J, Rueden CT, Hiner MC, Eliceiri KW (2015) The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Pasternak T, Liu K, Blein T, Aubry-Hivet D, Dovzhenko A, Duerr J, Teale W, Ditengou FA, Burkhardt H, et al. (2014) The iRoCS Toolbox – 3D analysis of the plant root apical meristem at cellular resolution. Plant J 77:806–814 [DOI] [PubMed] [Google Scholar]
- Schütz LM, Louveaux M, Barro AV, Bouziri S, Cerrone L, Wolny A, Kreshuk A, Hamprecht FA, Maizel A (2021) Integration of cell growth and asymmetric division during lateral root initiation in Arabidopsis thaliana. Plant Cell Physiol 62: 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Frentz Z, Birnbaum KD, Leibler S (2011) Quantitation of cellular dynamics in growing Arabidopsis roots with light sheet microscopy. PLoS One 6:e21303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A, Heidstra R, Blilou I, Scheres B (2018) Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev 32:1085–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott EW (1960) Plant morphogenesis. Plant Morphogenesis. McGraw-Hill Book Co., New York, Toronto and London [Google Scholar]
- Shishkova S, Rost TL, Dubrovsky JG (2008) Determinate root growth and meristem maintenance in angiosperms. Ann Bot 101:319–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss S, Runions A, Lane B, Eschweiler D, Bajpa N, Trozzi N, Routier-Kierzkowska AN, Yoshida S, Rodrigues-da-Silveira S, Vijayan A, et al. (2021) MorphoGraphX 2.0: providing context for biological image analysis with positional information. BioRxiv 10.1101/2021.08.12.456042 [DOI] [PMC free article] [PubMed]
- Taylor I, Lehner K, McCaskey E, Nirmal N, Ozkan-Aydin Y, Murray-Cooper M, Jain R, Hawkes EW, Ronald PC, Benfey PN, et al. (2021) Mechanism and function of root circumnutation. Proc Natl Acad Sci 118:e2018940118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J-Y, Perry N, Schindelin J, HoopesGM, , ReynoldsGD, , Laplantine E, BednarekSY, , ShorteSL, , Eliceiri KW (2017) TrackMate: An open and extensible platform for single-particle tracking. Methods 115: 80–90 [DOI] [PubMed] [Google Scholar]
- Torres-Martínez HH, Rodríguez-Alonso G, Shishkova S, Dubrovsky JG (2019) Lateral root primordium morphogenesis in Angiosperms. Fron Plant Sci 10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Martínez HH, Hernández-Herrera P, Corkidi G, Dubrovsky JG (2020) From one cell to many: morphogenetic field of lateral root founder cells in Arabidopsis thaliana is built by gradual recruitment. Proc Natl Acad Sci 117:20943–20949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi S, Campbell K (2019) 3D tracking of migrating cells from live microscopy time-lapses. Methods Mol Biol 2040:385–395 [DOI] [PubMed] [Google Scholar]
- Toyokura K, Goh T, Shinohara H, Shinoda A, Kondo Y, Okamoto Y, Uehara T, Fujimoto K, Okushima Y, Ikeyama Y (2019) Lateral inhibition by a peptide hormone-receptor cascade during Arabidopsis lateral root founder cell formation. Dev Cell 48:64–75 [DOI] [PubMed] [Google Scholar]
- Trinh CD, Laplaze L, Guyomarc'h S (2018) Lateral root formation: building a meristem de novo. Ann Plant Rev 1:847–890 [Google Scholar]
- Trullo A, Dufourt J, Lagha M (2020). MitoTrack, a user-friendly semi-automatic software for lineage tracking in living embryos. Bioinformatics 36:1300–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulman V, Maška M, Magnusson KE, Ronneberger O, Haubold C, Harder N, Matula P, Matula P, Svoboda D, Radojevic M, et al. (2017) An objective comparison of cell-tracking algorithms. Nat Methods 14:1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN (2013) To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140:4301–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E (2012) Hormonal interactions in the regulation of plant development. Ann Rev Cell Dev Biol 28:463–487 [DOI] [PubMed] [Google Scholar]
- Verbelen JP, De Snodder T, Le J, Vissenberg K, Baluška F (2006) The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities. Plant Signal Behav 1:296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Chamier L, Laine RF, Jukkala J, Spahn C, Krentzel D, Nehme E, Lerche M, Hernández-Pérez S, Mattila PK, Henriques R, et al. (2021). Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat Commun 12:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EH, Maizel A (2016) Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr Biol 26:439–449 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Hauschild R, Fendrych M, Barone V, Benková E, Friml J (2017) Live tracking of moving samples in confocal microscopy for vertically grown roots. eLife 6:e26792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D, Banda J, Schmitz A, Boland J, Bishopp A, Maizel A, Stelzer EH, Bennett M (2020) Early developmental plasticity of lateral roots in response to asymmetric water availability. Nat Plants 6:1–5 [DOI] [PubMed] [Google Scholar]
- Wachsman G, Zhang J, Moreno-Risueno MA, Anderson CT, Benfey PN (2020) Cell wall remodeling and vesicle trafficking mediate the root clock in Arabidopsis. Science 370:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C, Miura T, Voleti V, Yamaguchi K, Tsutsumi M, Yamamoto K, Otomo K, Fujie Y, Teramoto T, Kimura KD, et al. (2021). 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. eLife 10:e59187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich JR, Möller BK, Li S, Saiga S, Sozzani R, Benfey PN, De Rybel B, Weijers D (2017) Framework for gradual progression of cell ontogeny in the Arabidopsis root meristem. Proc Natl Acad Sci USA 114:E8922–E8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M, Mankowski W, Wait E, Temple S, Cohen AR (2016) LEVER: software tools for segmentation, tracking and lineaging of proliferating cells. Bioinformatics 32:3530–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C, Tinevez JY, Pietzsch T, Stamataki E, Harich B, Guignard L, Preibish S, Shorte S, Keller PJ, Tomancak P, et al. (2018) Multi-view light-sheet imaging and tracking with the MaMuT software reveals the cell lineage of a direct developing arthropod limb. eLife 7:e34410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B (2006) A molecular framework for plant regeneration Science 311:385–388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.