REPRISAL allows mapping lignification dynamics in plants based upon a combination of a monolignol chemical reporter strategy, parametric- and AI-based cell wall segmentation, and ratiometrics.
Abstract
This article describes a methodology for detailed mapping of the lignification capacity of plant cell walls that we have called “REPRISAL” for REPorter Ratiometrics Integrating Segmentation for Analyzing Lignification. REPRISAL consists of the combination of three separate approaches. In the first approach, H*, G*, and S* monolignol chemical reporters, corresponding to p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, are used to label the growing lignin polymer in a fluorescent triple labeling strategy based on the sequential use of three main bioorthogonal chemical reactions. In the second step, an automatic parametric and/or artificial intelligence segmentation algorithm is developed that assigns fluorescent image pixels to three distinct cell wall zones corresponding to cell corners, compound middle lamella and secondary cell walls. The last step corresponds to the exploitation of a ratiometric approach enabling statistical analyses of differences in monolignol reporter distribution (ratiometric method [RM] 1) and proportions (RM 2) within the different cell wall zones. We first describe the use of this methodology to map developmentally related changes in the lignification capacity of wild-type Arabidopsis (Arabidopsis thaliana) interfascicular fiber cells. We then apply REPRISAL to analyze the Arabidopsis peroxidase (PRX) mutant prx64 and provide further evidence for the implication of the AtPRX64 protein in floral stem lignification. In addition, we also demonstrate the general applicability of REPRISAL by using it to map lignification capacity in poplar (Populus tremula × Populus alba), flax (Linum usitatissimum), and maize (Zea mays). Finally, we show that the methodology can be used to map the incorporation of a fucose reporter into noncellulosic cell wall polymers.
Introduction
Lignin is a phenolic polymer found in the walls of certain plant cells where it makes up ∼ 25%–30% of the dry weight. Together with cellulose and xylan, it is one of the major constituents of wood and is the second most abundant plant polymer after cellulose on Earth. Both the quantity and the structure of lignin present in plant cell walls have a major effect on the mechanical and chemical properties of numerous plant-derived resources/products including wood, paper, and textiles (Huis et al., 2012). Lignin is also a major factor responsible for the recalcitrance of ligno-cellulose biomass in the industrial production of bio-ethanol for energy. For plants themselves, the presence of lignin in the cell wall contributes positively to mechanical support, facilitates water transport in xylem via its hydrophobicity, and helps to protect them against pathogens and physical injury (Boerjan et al., 2003). The appearance of lignin during evolution has played a major role in ensuring the survival of plants on dry land. The lignin polymer is also an important carbon sink and the formation of this polymer in the wood cells of long-lived tree species, therefore, contributes positively to the struggle against climate change.
Chemically, lignin is formed by the nonenzymatic polymerization of phenoxy radicals mainly derived from three main monomers: p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol that are collectively referred to as “monolignols” (Ralph et al., 2004). Once incorporated into the polymer, these molecules form the hydroxyphenyl (H), guaiacyl (G), and syringyl (S) monomeric units. Lignin can also contain catechyl (C) units derived from caffeyl alcohol in orchids and the cactaceae families (Chen et al., 2012) as well as tricin flavonoids and ferulates in grasses (Grabber et al., 2000; del Río et al., 2012) or hydroxystilbenes in palm fruit (del Río et al., 2017). The amount of lignin and the relative proportion of the different monomers in the cell wall varies according to the botanical group (e.g. gymnosperm lignin contains no/very few S units compared to angiosperm lignin which contains far higher quantities of S units); the organ (leaves are far less lignified than stems/roots), the tissue/cell type—in Arabidopsis (Arabidopsis thaliana), xylem vessels have a lower S/G ratio than fibers, and the cell wall layer (cells with only primary cell walls are generally nonlignified whereas cells with secondary cell walls [SCWs] become lignified) (Campbell and Sederoff, 1996). Monolignols are enzymatically synthesized via the phenylpropanoid pathway and are then exported into the developing cell wall across the plasma membrane. Several hypotheses including the involvement of monolignol-specific transporters, vesicle-related transport, and passive diffusion have all been advanced to explain monolignol transport, but the actual mechanism(s) involved have not as yet been categorically determined and further research is necessary on this point (Perkins et al., 2019). Once in the cell wall compartment, monolignols are oxidized by cell wall-located laccases and/or peroxidases (PRXs) before undergoing polymerization into the growing lignin polymer (Wang et al., 2013).
In order to understand the dynamics of lignification at the plant or organ scale, different analytical techniques have been developed over the last decades (Lupoi et al., 2015). They are generally adapted to the species studied (woody or not) and to the quantities of lignins present in their tissues. For example, the method using acetyl bromide solubilization of lignins (Johnson et al., 1961) is well suited for herbaceous species while the gravimetric Klason method (Effland, 1977) is more suited for woody species. There are also different methods for determining the H, G, and S subunit composition and the S/G ratio of lignins. Historically, thioacidolysis (Lapierre et al., 1986) and nitrobenzene oxidation (Billa and Monties, 1995) methods were used, and are now gradually replaced by pyrolysis coupled with GC/MS (Wagner et al., 2007) or NMR (Mansfield et al., 2012).
All these approaches provide relevant information on the different tissues/organs studied but involve cell homogenization, meaning that it is not possible to get information regarding quantities and compositions at the cellular level. Therefore, histochemical approaches are usually associated with these global analyses such as phloroglucinol staining (Wiesner reaction), which specifically reveals the cinnamaldehyde functions of S and G unit derivatives by producing a red chromogen whose intensity depends on the quantities (Clifford, 1974). The Maüle test uses potassium permanganate and hydrochloric acid to transform G and S residues into catechols, which are transformed into orange-brown quinones for G lignins and purple-red quinones for G-S lignins (Day et al., 2005). Other approaches exploit the autofluorescence of lignins (Donaldson, 2001) or the possibility to visualize them using fluorescent compounds such as auramine (Pesquet et al., 2005) and acriflavine (Donaldson, 2001). All these techniques, therefore, allow the acquisition of relatively fine spatial information about lignin that is already present in the cell wall of the sample analyzed. They do not, however, provide a clear view on the dynamics of lignification and the capacity of the cell wall machinery to incorporate monolignols into lignin since visualization of the active zones of monolignol polymerization in tissues is lacking.
Recent progress in the field of bioimaging has opened the door to novel chemical biology applications (Bukowski et al., 2014; Tobimatsu et al., 2014; Pandey et al., 2015; Simon et al., 2018). Bioorthogonal labeling strategy has emerged as a key technology to support the development of these applications in vitro, ex vivo, and in vivo in plants. Since 2014, several groups have demonstrated the feasibility and the robustness of the established protocols. In this context, we reported the design and characterization of a fully integrated pipeline, demonstrating the study of the dynamics of the plant cell wall formation in four different species: Arabidopsis (A. thaliana), flax (Linum usitatissimum), Nicotiana benthamiana, and poplar (Populus tremula × Populus alba).
The bioorthogonal chemical reporter strategy thus enables the study of de novo lignin formation in living organisms and is complementary to other histochemical/fluorescent approaches. In this two-step strategy, a synthetic derivative (the chemical reporter) of a monolignol is first introduced into the cell wall where it becomes oxidized by cell wall located PRXs and/or laccases and incorporated into the growing lignin polymer. The reporter carries a specific chemical group that enables subsequent interaction with a corresponding probe via a specific bioorthogonal chemical reaction. To enable integration, the group must be small and innocuous so as to minimize its impact on the physicochemical properties of the reporter and allow its recognition by the enzymatic machinery (Rigolot et al., 2021). Once metabolic incorporation has occurred, the reporter group must then react specifically and efficiently with an exogenous molecular probe (e.g. an adequately functionalized fluorophore) while being chemically inert to the surrounding biological environment. Different bioorthogonal reactions have been developed over the past 20 years and are mainly applied to glycan and protein labeling (Wallace and Chin, 2014; Palaniappan and Bertozzi, 2016). In 2014, Ralph and Zhu’s teams simultaneously published the first use of coniferyl alcohol analogues as a lignin chemical reporter in a mono-labeling strategy (Bukowski et al., 2014; Tobimatsu et al., 2014). More recently our group elaborated a double-labeling, and then triple-labeling strategy to visualize active lignification areas in plants. For this, we designed three chemical reporters (G*, H*, and S*) mimicking the three main monolignols (G, H, and S), tagged with an alkyne group, an azide function, and a methylcyclopropene moiety, respectively) and exploited the three main bioorthogonal reactions, namely the copper-catalyzed alkyne-azide cycloaddition (CuAAC), the strain-promoted azide-alkyne cycloaddition (SPAAC) and the inverse electronic demand Diels–Alder (IEDDA) cycloaddition (Lion et al., 2017; Simon et al., 2018). The triple labeling strategy was then used to characterize the dynamics of lignification in flax (L. usitatissimum L.) and we also showed that the method could be transposed to other plant species (Arabidopsis, N. benthamiana, and poplar). This technology provided a powerful approach to investigate lignification at the cell wall/cell wall layer scale in different plant species. However, while a simple visual inspection of different samples is sufficient to detect “differences” in monolignol reporter incorporation profiles, often the sheer complexity of the data obtained makes it more or less impossible to qualify and/or quantify all but the most obvious differences (e.g. lignified versus non/poorly lignified walls) thereby limiting the biological potential of the chemical reporter approach.
We, therefore, decided to develop a segmentation approach that when combined with the chemical reporter strategy can be used for statistical determination of relative reporter incorporation into different cell wall zones based upon a ratiometric method (RM). In this paper, we present this combined methodology that we have called “REPRISAL” for REPorter Ratiometrics Integrating Segmentation for Analyzing Lignification. We describe the development of REPRISAL and demonstrate how it can be used for detailed mapping of lignification in wild-type (WT) Arabidopsis, in the Arabidopsis PRX mutant prx64, as well as in poplar, flax, and maize.
Results and discussion
REPRISAL consists of three separate approaches: 1) bioorthogonal labeling of lignin with the three chemical reporters, 2) parametric or artificial intelligence (AI) segmentation of labeled cell walls, and 3) ratiometric analysis of fluorescence signal intensity (Figure 1). Each approach is described in more detail below.
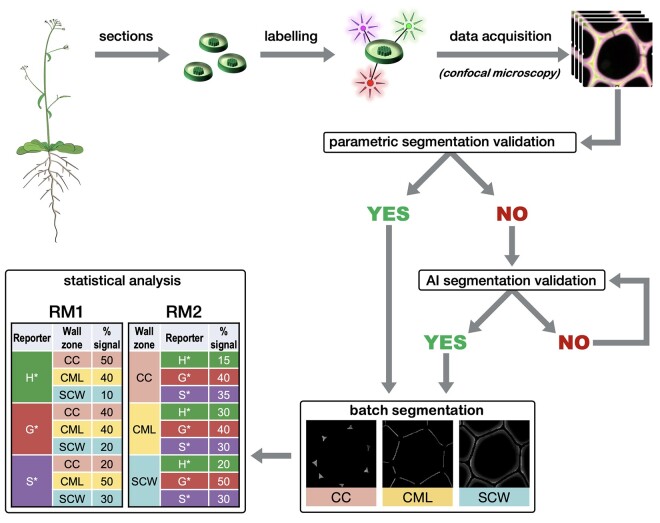
Figure 1.
Global procedure workflow. Plant stem transversal cross-sections are incubated with H*, G*, and S* monolignol reporters that become metabolically incorporated into the growing lignin polymer. Incorporated reporters are identified via a bioorthogonal chemistry strategy specifically linking each type of reporter to a different fluorescent probe allowing imaging by confocal microscopy (see Figure 2). Following image acquisition, a first representative image of the series is selected and the most suitable segmentation method (parametric or AI) is chosen and applied automatically to all acquired images. The data are then extracted for three different cell wall zones (CC, CML, or SCW) and relative reporter distribution and proportion determined by RM1 and RM2. Please see text for more details.
Bioorthogonal labeling and imaging
Three synthetic monolignol surrogates H*, G*, and S* (the chemical reporters) equipped with click-reactive tags (Figure 2) were metabolically incorporated into A. thaliana stem cross-sections ex vivo using our previously reported triple bioorthogonal labeling methodology (Simon et al., 2018). Each incorporated reporter was then specifically linked to a fluorescent probe via a specific bioorthogonal ligation reaction thereby enabling subsequent fluorescence visualization and identification of the tagged substrates. H*-units were labeled with DBCO-Rhodamine Green (SPAAC bioorthogonal reaction), G*-units were labeled with Azide Fluor 545 (CuAAC bioorthogonal reaction), and S*-units were labeled with Tetrazine-Cy5 (IEDDA bioorthogonal reaction). Samples were then visualized by confocal microscopy to produce a set of images based on 3-channel detection (green for H*, red for G*, and magenta for S*) (Figure 2).
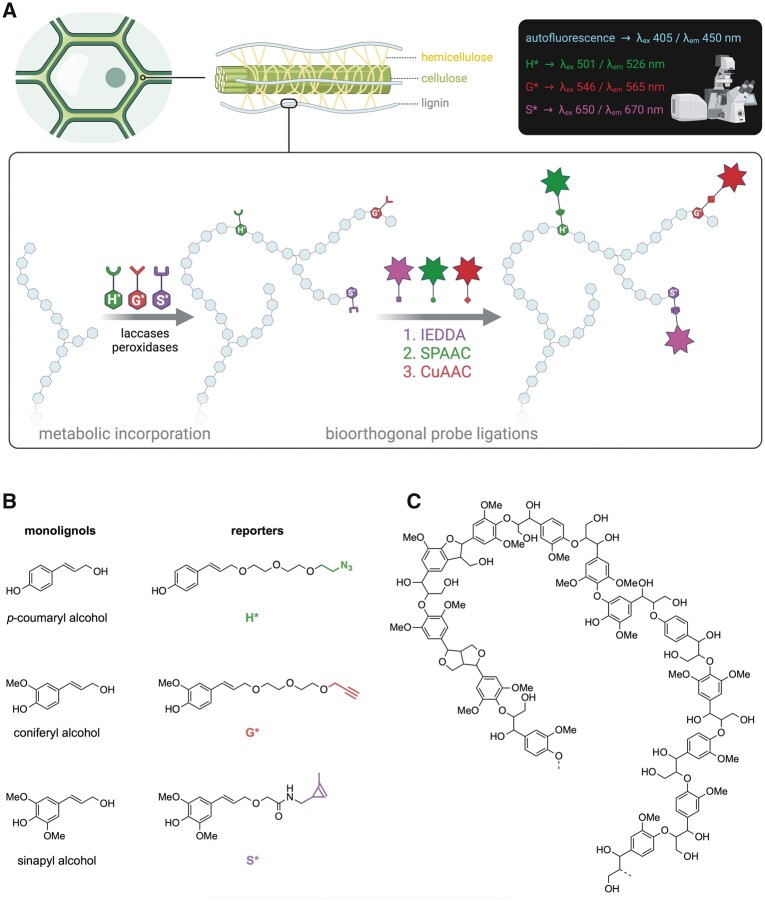
Figure 2.
Lignin bioorthogonal triple labeling and chemical reporter structure. A, Schematic representation of the lignin bioorthogonal triple labeling strategy in plant cell walls. Monolignol chemical reporters H*, G*, and S* are oxidized by cell wall located PRXs and/or laccases and become metabolically incorporated into the growing lignin polymer. Following reporter incorporation, green, red, and magenta fluorescent probes are added and become specifically linked to H*, G*, and S* reporters, respectively, via three sequential bioorthogonal reactions: IEDDA (S*), SPAAC (H*), and CuAAC (G*). Spatial localization of fluorophores is analyzed by confocal laser scanning microscopy (CLSM). B, Chemical structures of native lignin monolignols (left) and corresponding chemical reporters H*, G*, and S* (right). Reporter tags involved in the bioorthogonal reaction with the corresponding fluorophores are shown in color: green—azide group, red—alkyne group, magenta—methyl cyclopropene group. C, Example of typical lignin oligomer structure.
Parametric and AI-based segmentation
To map the localization of the incorporated monolignol reporters we decided to segment the cell wall into three main zones: the cell corner (CC), the compound middle lamella (CML) consisting of the middle lamella and the primary cell wall, and the SCW. Once successfully segmented, pixels can be assigned to the different cell wall sectors and the fluorescence intensity corresponding to each incorporated monolignol reporter determined. Two main strategies can be proposed for the crucial segmentation. Traditional parametric methods are mostly based on the intensity and spatial relationships of pixels, while trainable machine learning methods require the user to “teach” the software which pixel is part of what region. AI-based image analysis is highly complementary to classic parametric segmentation (PS) procedures and has strongly emerged as a valuable asset in the microscopy image analysis field over the last few years. For the REPRISAL methodology, we developed a global algorithm that enables the user to implement both parametric and AI segmentation (Figure 1).
For PS the initial image was acquired according to Nyquist sampling criteria and treated according to the algorithm presented in Figure 3. The transmission image is split into three fluorescence channels (green, red, and magenta) that are transformed into three binary masks using the Otsu threshold method (Otsu, 1979). These masks are then merged so that all the labeled lignin is taken into account in the automatic segmentation. A set of transforms are then applied to this new mask including the Distance map (Legland et al., 2016), Voronoï (Legland et al., 2016), Enhance Local Contrast (Pizer et al., 1987), and Find Maxima transforms (Grishagin, 2015). Binary subtraction operations then generate 3 masks corresponding to the CC, the CML, and the SCW.
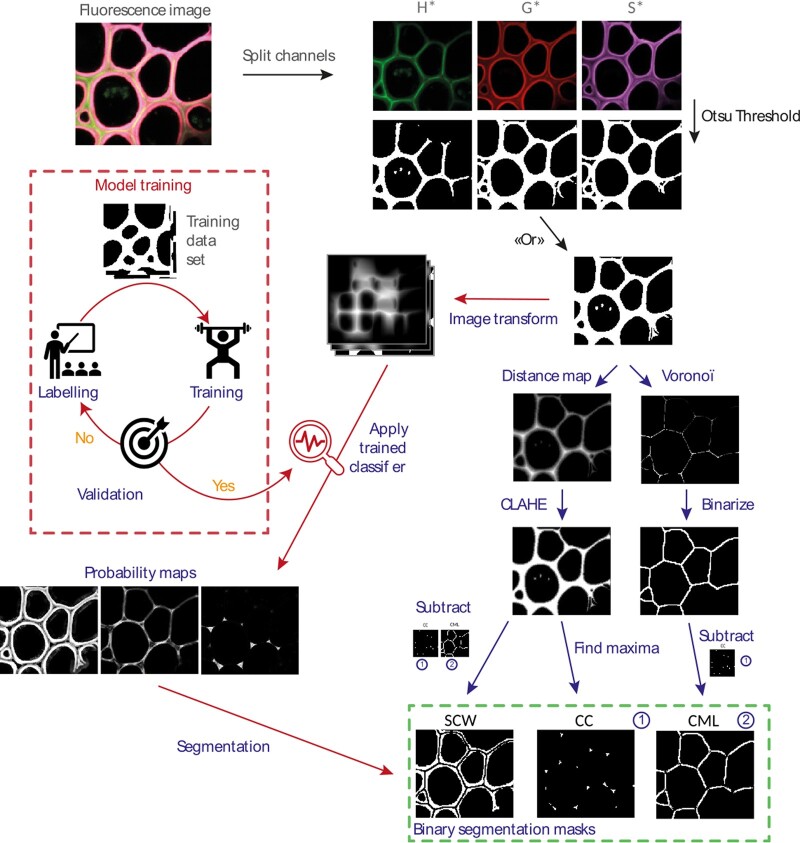
Figure 3.
Scheme of the segmentation algorithm. After transforming the images of the individual channels into binary masks and then combining them, the algorithm allows the user to choose between segmentation based on 1) morphological parameters (blue path) or 2) learning (red path). Once segmented, a mask is created for the SCW, the CML, and the CC. It is then applied to each fluorescent channel corresponding to the three monolignol reporters (H*, G*, and S*). For each fluorescence image, nine fluorescence intensity maps are thereby created and analyzed.
For AI segmentation and in order to obtain an algorithm easily adaptable to different types of lignin fluorescence images, we chose to keep the first steps of binarization and fusion of binary masks used for PS step before implementing the AI segmentation. Images once binarized were manually labeled and then transformed by the Waikato Environment for Knowledge Analysis (WEKA) algorithm (“Gaussian Blur,” “Sobel,” “Hessian,” “Differences of gaussians,” and “membrane projection” with a membrane patch size of 19 and a maximum sigma of 16). Transformed images were then used to train our network (200 initial trees and 4 initialization classes that is, background, CC, CML, and SCW). Once the training was validated on training images, it was applied to a new set of validation images. The classifier we obtained was applied for batch analysis and provides probability maps of each pixel belonging to each category. Probability maps were then converted into binary masks using auto-threshold and despeckle.
To evaluate the efficiency of the developed algorithm, we applied it to confocal microscopy images of Arabidopsis floral stem cross-sections in which lignin was labeled by the bioorthogonal triple marking strategy (Figure 4). Visual inspection of the different areas confirms the efficiency of the segmentation. The form and location of the different objects obtained correspond to expectations: CCs are triangular in shape and located at the interstices where three or more cells are in contact, the CML corresponds to a line formed at the junction between two cells, and the SCW occupies the rest of the total cell wall zone. Overall, the parametric and AI segmentation methods give very similar results although some differences can be detected. The parametric method appears to be more accurate for fine structures whereas the AI method recognizes better the CCs and the middle lamella and follows more closely the curves of the cells. Nevertheless, the percentage distribution of fluorescence intensity values between the three cell wall zones is similar for both methods showing that they can be used interchangeably (Table 1). All subsequent results presented in this paper were obtained using PS.
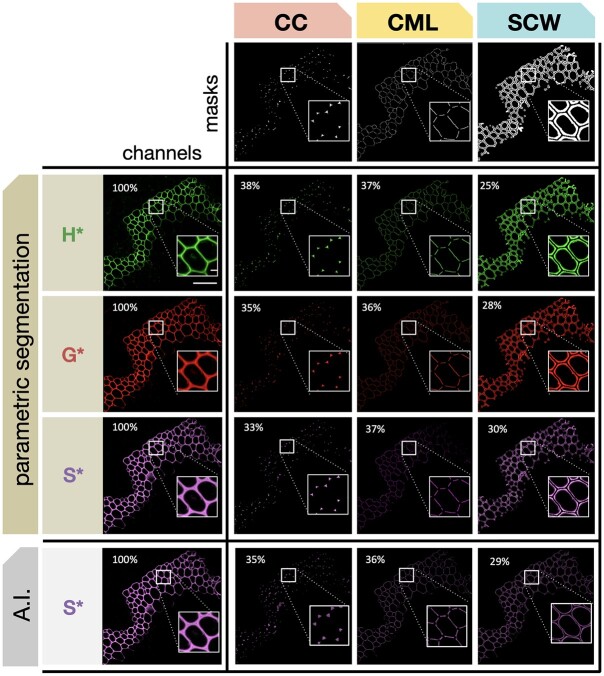
Figure 4.
Automatic segmentation of different cell wall zones. The three masks (CC, CML, or SCW) are applied to cross-sections of Arabidopsis stems in which lignin has been labeled by the triple bioorthogonal strategy. Masks are applied to the three fluorescence channels corresponding to H*, G*, and S* reporters. Percentage values indicate the relative proportion of total reporter signal intensity determined for each cell wall zone by PS (H*, G*, and S*) or AI segmentation where the value for S* is shown for comparison (see Table 1 for corresponding H* and G* values). Scale bar main image = 50 µm, scale bar insert = 5 µm.
Table 1.
Comparison of fluorescence signal distribution in different cell wall zones determined by PS and AI segmentation
| Reporter | CW region | Intensity PSa | Intensity AIa | % Intensity PS | % Intensity AI |
|---|---|---|---|---|---|
| H* | CC | 1,807 | 1,805 | 38 | 40 |
| CML | 1,767 | 1,631 | 37 | 36 | |
| SCW | 1,172 | 1,078 | 25 | 24 | |
| G* | CC | 515 | 543 | 35 | 38 |
| CML | 530 | 503 | 36 | 35 | |
| SCW | 416 | 399 | 28 | 28 | |
| S* | CC | 1,147 | 1,214 | 33 | 35 |
| CML | 1,277 | 1,252 | 37 | 36 | |
| SCW | 1,016 | 983 | 30 | 29 |
H*, G*, and S* are monolignol reporters.
aRaw fluorescence signal intensity.
Ratiometric analysis
During the bioorthogonal reactions, each incorporated monolignol becomes linked to a different fluorophore. Since different fluorophores do not give the same response in terms of the number of photons emitted for the same number of incorporated reporters it is not possible to directly compare the amounts of the different reporters incorporated into the three cell wall zones (Schwartz et al., 2002). To get around this problem and obtain quantitative data that can be statistically treated to provide information about differences in reporter incorporation we developed a ratiometric approach. The rationale for this is based on the fact that the ratios between the fluorescence signal of the different reporters do not change for the same biological sample/condition, thereby allowing the comparison of two or more cell wall zones. While this approach does not allow direct comparisons of the absolute quantity of an incorporated reporter, it does make it possible to determine whether there are changes in the relative incorporation of monolignols reporters incorporated into the cell wall zones of different samples.
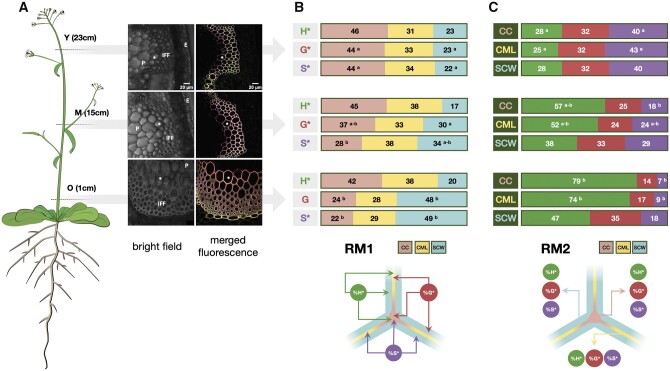
In the ratiometric approach, relative monolignol reporter incorporation is calculated by two methods RM1 & RM2 that provide complementary information (Figure 5). RM1 evaluates the relative distribution of a given reporter in the different segmented cell wall regions (Figure 5, B). Here, the fluorescence intensity of a given reporter (H*, G*, and S*) in a given cell wall region (CC, CML, or SCW) is divided by the total fluorescence intensity of that reporter in all three cell wall zones and expressed as a percentage (e.g. [H*CCintensity]/[∑H*]). RM2 evaluates the relative proportion of each reporter in a given cell wall zone compared to the other reporters (Figure 5, C). Here, the fluorescence intensity of a given reporter (H*, G*, and S*) in a given cell wall region (CC, CML, or SCW) is divided by the total fluorescence intensity of all three reporters in this particular cell wall zone and expressed as a percentage (e.g. [H*CCintensity]/[∑CCintensity]). Our results (Figure 5) illustrate the interest of the combined chemical reporter strategy, segmentation, and ratiometric analysis approach for mapping developmentally related lignification in plants. In this experiment, we compared the capacity of different cell wall regions to incorporate monolignol reporters at different developmental stages by analyzing floral stem cross-sections sampled at three different heights from 7-week-old plants. Visual inspection of merged confocal microscopy images in the three cross-sections (Figure 5, A and Supplemental Figure S1) reveals that the three reporters have incorporated into fiber cell walls at all developmental stages from the “youngest” (23 cm) to the “oldest” (1 cm). Overall, the amount of signal increases during development reflecting reporter incorporation into the SCW. Further interpretation of these images by visual inspection is difficult due to the huge amount of data that they contain and it is, therefore, necessary to apply the segmentation approach.
Figure 5.
Monolignol reporter incorporation profiles in Arabidopsis stem fiber cell walls. A, CLSM images of IFF bundles in Arabidopsis stem cross-sections (Left) cartoon of plant indicating regions used to prepare cross-sections, Y = “young” stem region (23 cm from the base of the floral stem), M = “medium” stem region (15 cm from the base), O = “old” stem region (1 cm from the base), total stem height = 30 cm; (Right) bright-field image of stem region showing fibers analyzed and merged green (H*/DBCO-PEG4-Rhodamine Green), red (G*/Azide-fluor 545), and magenta (S*/tetrazine-Cy5) channels revealing monolignol reporter incorporation in fiber cell wall lignin during metabolic feeding. P, pith, E, epidermis, * indicates the same cell shown in the merged image, scale bar = 20 µm. B, Relative distribution of each reporter incorporated into different fiber cell wall zones in Y, M, and O stem cross-sections analyzed by RM1, figures represent the percentage of total signal for a given reporter incorporated into the different wall zones, different letters indicate significantly different values (ANOVA, P-value < 0.05) between Y, M, and O sections, the absence of a letter indicates that differences are not significant. C, Relative contribution of all reporters to total signal in a given cell wall zone analyzed by RM2, figures represent the percentage contribution of each reporter’s signal to total signal intensity in each cell wall zone, different letters indicate significantly different values (ANOVA, P-value < 0.05) between Y, M, and O sections, the absence of a letter indicates that any differences are not significant.
In young stem sections, the use of RM1 (Figure 5, B) indicated that the relative distribution of the total signal in the different cell wall zones is very similar for all three reporters with the majority of signal being incorporated into the CC, followed by the CML and SCW (CC: 44%–46%; CML: 31%–34%; SCW: 22%–23%). While this distribution pattern did not evolve over time for the H* reporter clear changes could be observed for G* and S* reporters. The relative proportions of total G* and S* signal significantly decreased in the CC during development accompanied by an increase in the percentage of total signal observed in the SCW. No changes were observed in the relative distribution of signal in the CML. The differences in the relative distribution of the G* and S*, but not H* reporters suggest the existence of developmentally related changes in the capacity of the different cell wall zones to incorporate reporters. The increased capacity of the SCW to incorporate G* and S*, but not H* reporters, could indicate differences in the cell wall machinery that specifically favors the incorporation of these two reporters compared to H* reporters. If the increased incorporation capacity was simply due to the fact that the SCW zone is bigger in older sections then one would also expect a similar increase in H* incorporation. The fact that this is not so argues for the existence of a specific mechanism favoring G* and S* incorporation.
Analysis of the same images using RM2 (Figure 5, C) shows that the contribution of H* to the total signal in the CC increases approximately four-fold (28%–79%) in older stem sections compared to younger stem sections. Since the relative distribution of total H* signal measured in the CC remains relatively stable (42%–46%) during development (Figure 5, B), this would suggest that the amounts of G* and/or S* incorporated into the CC decrease during development. This hypothesis is supported by the observations that i) the relative proportions of the total G* and S* signal observed in the CC decrease (Figure 5, B) and ii) that the relative contributions of S* to the total signal also decreases (Figure 5, C). The G* contribution also decreased, but not significantly. The fact that the relative contribution of the S* reporter is more heavily affected (six-fold reduction) than the G* reporter (two-fold reduction) suggests that older CCs are less able to incorporate S* reporters.
Analysis of the CML using RM2 gave rise to a similar result. Once again, the effect was more marked for S* compared with G*. In comparison to the CC and CML regions, the developmental pattern of reporter incorporation in the SCW showed a more complex pattern. RM2 (Figure 5, C) indicated that the contribution of H* to the total signal in this zone increased while the S* contribution decreased as previously observed for the CC and CML regions. In contrast, the relative G* contribution remained unchanged. Although the S* contribution to total signal in the SCW decreased in older stem sections as for the CC and CML zones the extent of the reduction was far less (two-fold) compared to a six-fold reduction (CC) and four-fold reduction (CML). These results are coherent with those obtained by RM1 (Figure 5, B) showing that the relative proportion of the G* and S* total signals increased in the SCW.
Overall, these results indicate that while H* reporters are the most readily incorporated form into all three cell wall regions at all of the developmental stages examined, there is an increasing tendency to incorporate both G*, and especially S* reporters into the SCW of older stem sections. It is possible that these changes are functionally related to the developmentally related occurrence and distribution of lignin-related PRXs/laccases recently reported by the group of Lacey Samuels (Hoffmann et al., 2020). However, it is important to point out that lignification is a complex process, regulated at a number of different levels (e.g. regulation of monolignol biosynthesis genes and enzymes, transport/movement of monolignols across the plasma membrane into the cell wall, oxidative polymerization, etc.). Nevertheless, the observation that differences exist between the incorporation capacity of different cell wall zones when reporters are supplied in equal concentrations hints at the existence of other mechanisms (e.g. redox enzyme-substrate specificity, local supply/transport of H2O2 or O2, differential diffusion of reporters within the polysaccharide matrix, etc.).
In conclusion, these results clearly demonstrate the potential of the REPRISAL methodology that combines chemical reporter + segmentation + ratiometric analysis approaches for mapping developmental lignification in Arabidopsis.
Lignification mapping in the Arabidopsis prx64 mutant
Having demonstrated the interest of REPRISAL in WT plants we decided to use this approach to investigate whether it would be capable of revealing differences in the lignification profile of a lignin mutant compared to WT. For this, we chose the Arabidopsis PRX prx64 mutant (AT5g42180). Several studies show that the AtPRX64 protein is associated with lignification; it is involved in Casparian strip lignification (Lee et al., 2013) and the AtPRX64-mCherry fusion protein becomes localized in the lignified CCs and middle lamella of Arabidopsis fiber cells in a developmentally related pattern (Yi Chou et al., 2018; Hoffmann et al., 2020). The AtPRX64 promoter is also active in both xylary and interfascicular fibers (IFFs; Smith et al., 2017). Despite this apparently clear link between PRX64 and lignification, the stem lignin content has not been investigated in this mutant and we, therefore, decided that it would be a good target to evaluate the efficiency of our methodology. A visual inspection of both UV autofluorescence and merged triple labeling images from WT and mutant plants suggested that lignification was affected in the mutant (Supplemental Figure S2).
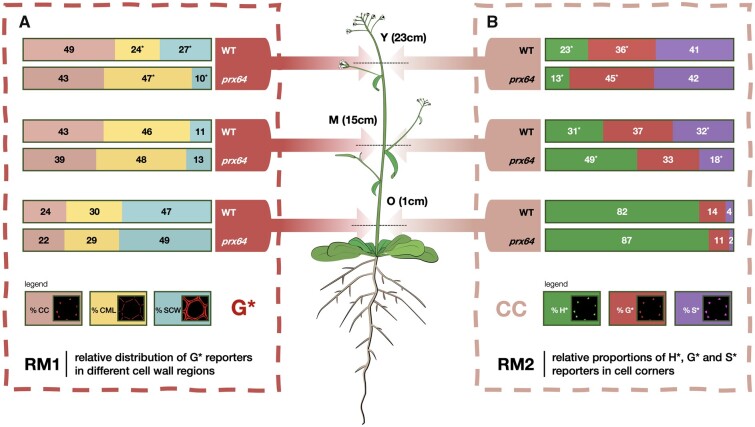
Figure 6 shows the comparison of the application of RM1 for the G* reporter (Figure 6, A) and RM2 for CC (Figure 6, B) in the prx64 mutant compared to WT as an example of the information that can be obtained by our approach. Corresponding data for H* and S*, and CML and SCW are given in Supplemental Figure S3. Analysis by RM1 indicates significant differences in the relative distribution of total signal for all three reporters between mutant and WT samples for the Y (young) stage, but not medium (M) and old (O) stages. In Y samples, significantly higher amounts of all reporters are incorporated in the mutant CML compared with WT. At the same time, significantly lower amounts of H* and G* (but not S*) are incorporated into the SCW. No significant differences are observed for the CC zones. These results would suggest that the capacity of the CML and SCW, but not CC, to incorporate monolignol reporters is modified in the prx64 mutant compared with WT only in Y samples. This is intriguing on several levels; first, studies by the group of Lacey Samuels (Yi Chou et al., 2018; Hoffmann et al., 2020) have shown that the PRX64 protein is localized to the CC region and it could therefore be expected that the mutation would preferentially induce a modification in this zone rather than the others. Secondly, the same group showed that the protein was not detected in Stage 1 (young) fiber CC, but only in the developmentally more advanced Stages 2 and 3 CC. However, our plants were grown in short-day conditions compared to those of Samuels and co-workers, similarly, our Y, M, and O samples were obtained from 30-cm high floral stems of 8-week-old plants (cf. 18 cm high, 5-week-old plants in the other study). As a result, our Y, M, and O samples are not directly comparable to the Stages 1–3 samples used by Samuels and it is possible that differences between the developmental stages analyzed could partly explain the observed discrepancy. Another interesting observation concerns the relative increase of all three reporters incorporated in the mutant CML zone of the mutant compared to WT since it would be expected that the KO of a lignin PRX gene would lead to a reduced reporter incorporation. However, it is also possible that the mutation provokes modifications in the expression profiles of other PRX and/or LAC genes thereby changing the overall incorporation capacity of the different cell wall regions. Future transcriptomics could help to clarify this point.
Figure 6.
Example comparison of monolignol reporter incorporation profiles in WT and prx64 mutant Arabidopsis stem fiber cell walls. A, Relative distribution of G* reporter incorporated into different fiber cell wall zones in Y, M, and O stem cross-sections analyzed by RM1, figures represent the percentage of total G* signal incorporated into the different wall zones, Values marked with * indicate significantly different values (Students t test, P-value < 0.05) between WT and mutant plants for a given stem height (Y, M, and O). B, Relative contribution of all reporters to total signal in the CC analyzed by RM2; figures represent the percentage contribution of each reporter’s signal to the total CC signal intensity, Values marked with * indicate significantly different values (Student’s t test, P-value < 0.05) between WT and mutant plants for a given stem height (Y, M, and O). Corresponding figures for H* and S* probes (RM1) and CML and SCW zones (RM2) are shown in Supplemental Figure S3.
Analysis by RM2 (Figure 6 and Supplemental Figure S3) shows that the relative proportions of the three reporters in each cell wall region also change in the mutant compared with WT. A significantly lower and higher proportion of H* and G* reporters, respectively, are incorporated in all three cell wall regions of the mutant compared to WT in Y samples; the relative proportions of S reporters remain unchanged. In M samples, the trend is inversed with a significantly higher proportion of H* reporter being incorporated in the CC and CML regions of the mutant compared to WT. In the mutant SCW layer, although the relative proportion of H* increases, the difference is not significant. The analysis also shows that the relative proportions of S* reporter incorporated into all three regions increase in the mutant compared with WT; G* reporter proportions are not significantly affected. Finally, in O samples, the only significant change concerns the decrease in S* reporter incorporation in the mutant SCW region compared with WT.
Taken together, our results show that both the RM1 and RM2 analyses, when combined with the bioorthogonal click chemistry and segmentation methodologies, reveal significant complex changes to the spatial lignification profile in the prx64 mutant compared with WT. Such an observation first illustrates the complexity of lignification at the cell wall layer scale. A complete understanding of this process is complicated because of the number of different actors that are involved. In addition to the different redox enzymes such as laccases and PRXs it is necessary to take into account other factors such as the biosynthesis and transport of monolignols to the cell wall, as well as the supply of H2O2 for PRXs as recently demonstrated (Hoffmann et al., 2020). Second, our results also illustrate the enormous potential of the combined labeling + segmentation + ratiometrics to finely dissect changes in the lignification capacity of cell wall mutants, including those where classical approaches have previously failed to detect differences.
Lignification mapping in different plant species
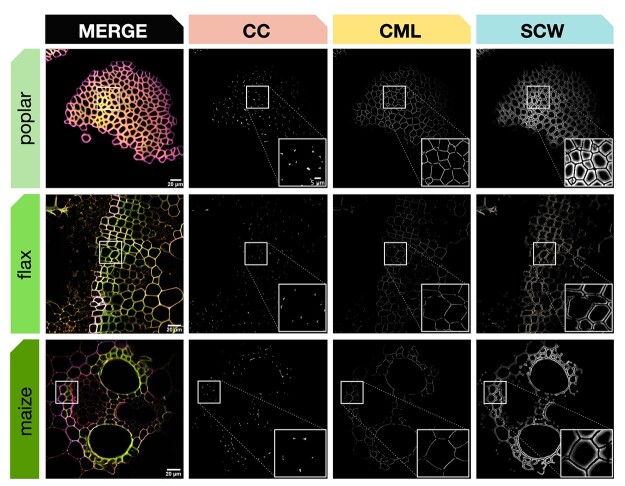
To ensure the robustness of REPRISAL, and to evaluate its interest in mapping lignification we applied it to three other plant biomass species: poplar, flax, and maize (Figure 7 and Table 2). Our results first confirm that the bioorthogonal triple-labeling strategy is applicable to different plant species as previously observed (Simon et al., 2018). Second, they show that the developed macro is capable of successfully segmenting cell walls from different species into the three different zones (CC, CML, or SCW) previously analyzed in Arabidopsis. Closer examination of the percentage distribution of each reporter in the different cell wall zones revealed a number of potentially interesting differences in lignification in the different species.
Figure 7.
Automatic segmentation applied to different plant species. Top line: poplar stem bark fibers, middle line: flax stem xylem, bottom line: maize stem vascular bundle. First column: merged CLSM image of green, red, and magenta fluorescence channels, second to fourth columns: cell wall zone segmentation for CC, CML, and SCW. All samples were labeled by the triple lignin bioorthogonal chemical reporter strategy with H*, G*, and S* monolignol reporters. Square frames indicate the sample region magnified (bottom RHC, Columns 2–4). Scale bar main image = 20 µm, scale bar insert = 5 µm.
Table 2.
Comparison of % total reporter distribution in different cell wall zones for different plant species determined by RM1
| Reporter | CW region | % Int At | % Int Pop | % Int Flax | % Int Maize | Rank At | Rank Pop | Rank flax | Rank maize |
|---|---|---|---|---|---|---|---|---|---|
| H* | CC | 38 | 31 | 31 | 35 | H | L | H | |
| CML | 37 | 28 | 37 | 35 | L | H | H | ||
| SCW | 25 | 40 | 32 | 30 | L | H | L | ||
| G* | CC | 35 | 48 | 35 | 36 | H | H | H | |
| CML | 36 | 23 | 35 | 35 | H | L | H | ||
| SCW | 28 | 29 | 29 | 29 | L | L | L | ||
| S* | CC | 33 | 42 | 29 | 34 | H | L | ||
| CML | 37 | 25 | 39 | 37 | H | L | H | H | |
| SCW | 30 | 32 | 32 | 29 | L | L |
H*, G*, and S* are monolignol reporters. At, A. thaliana; pop, poplar Rank indicates the highest (H) and lowest (L) % values for a given reporter.
In Arabidopsis, poplar and maize, data were obtained on cell walls in functionally similar “support tissues” consisting of IFFs (Arabidopsis), bark sclerenchyma fibers (poplar), and vascular bundle sclerenchyma (maize). Despite their biologically similar roles, the cell walls of poplar fibers incorporated the majority of the H* reporter into the SCW in comparison to Arabidopsis and maize where the reporter was preferentially incorporated into CC lignin. A similar difference was observed for S* reporter that was preferentially incorporated in the CC for poplar compared to the CML for the other two species. In contrast, the distribution of incorporated G* reporter was similar in all three species. While such differences can most probably be related to the developmental stage of the cells/tissues analyzed the results clearly demonstrate the potential of our approach for large-scale and detailed mapping of cell wall lignification at the cell wall layer level in different species. Our results on flax also show that the approach can be successfully applied to lignifying cell walls in other tissues (xylem).
Lignification and noncellulosic polymer mapping
As previously demonstrated (Simon et al., 2018), the triple labeling strategy is also compatible with monosaccharide reporters and we, therefore, decided to test whether REPRISAL could also be successfully implemented to map the incorporation of other cell wall polymers such as pectin or hemicelluloses. For this, we performed a triple labeling experiment on flax stem cross-sections using H* and S* lignin reporters, together with a peracetylated alkyne-tagged fucose reporter that replaced the G* reporter. This monosaccharide is present in the side chains of xyloglucan hemicelluloses and in pectin rhamnogalacturonan I and II (RG-I, RG-II) motifs (Scheller and Ulvskov, 2010; Atmodjo et al., 2013) and has previously been used to label noncellulosic polysaccharides (NCPs) via a bioorthogonal labeling approach (Anderson et al., 2012). Our preliminary results (Supplemental Figure S4) show that it is indeed possible to segment cell walls that have incorporated sugar reporters. As could be expected, monolignol reporters are very largely restricted to differentiating and more mature xylem cell walls but are present in much lower amounts in the phloem/vascular cambium region, whereas the fucose reporter shows the opposite profile. Interestingly, a slightly higher amount of fucose reporter found in the CML and SCW regions could be correlated with higher signal in what appeared to be the pit region between two adjacent cells suggesting that NCP deposition is still active within this specific cell wall structure at this developmental stage. Analyses of relative reporter incorporation using RM1 show that higher amounts of F* are incorporated into the CC and/or CML cell wall zones in tissue regions 1 and 2 in agreement with previous work by our group on NCP localization in flax (Chabi et al., 2017). In region 3 (more mature xylem), F* reporter was almost equally distributed between the CC, CML, and SCW zones. This might be related to the continued NCP deposition in cell wall pits as hypothesized above, although further work would be necessary to confirm this. These results, while preliminary, would suggest that REPRISAL can also be used to map the dynamics of other (nonlignin) cell wall polymers.
Conclusions
In this paper, we have combined three different methods (chemical reporter + segmentation + ratiometric analysis) to generate a robust methodology for analyzing lignification in plants that we have termed REPRISAL. Central to this approach is the development of a segmentation method allowing automatic quantification of the fluorescence signal resulting from lignin bioorthogonal triple labeling in different cell wall zones. Pixel segmentation, that is, their distribution between CCs, CML, and SCW is determined via two methods, one parametric and the other based on AI. As we have shown in Figure 4, these two methods produce very similar results, and while leading to the same biological conclusions, are not identical. On structures of such complexity, the labeling of pixels by two different human experts would also be expected to give slight variations and, in this case, it would be preferable that the analysis of the data set be done by a single expert for the sake of reproducibility. The two experts can also individually analyze all the data before subsequently comparing their interpretations. The same rigor must also be applied when using automated systems such as those proposed and it is therefore important not to mix segmentation methods during analysis even if any systematic bias is most likely minimal.
In the majority of this paper, we have focused on the use of PS to demonstrate its applicability to different conditions and plant models. We chose to validate this method because it is the one which is the least “flexible” in terms of use. While effective PS is usually simple and quick to use when applied to a given type of image, the determined parameters depend upon the size of the pixels, and on the size and shape of the structures. It is therefore often best suited for use at a given magnification, or even to a species or a plant tissue and requires a high expertise in order to be able to apply it to new types of samples. Nevertheless, as we have demonstrated our PS remains adaptable to a large number of situations and is, if possible, the method that should be privileged, because of a greater execution speed (seconds compared to minutes per image analyzed). On the other hand, the WEKA-based (AI) method is in essence adaptable to any sample by training it on new sets of samples representative of the condition to be analyzed. Furthermore, the number of transforms performed by the presented algorithm was deliberately limited to reduce computation time. Adding additional complementary filters such as, “kuwahara,” “Gabor,” or “neighbors” could further improve the ability of the algorithm to distinguish different cell wall areas. Nevertheless, the inclusion of a batch mode in our plugin makes it possible to launch the analysis on a large number of images overnight in order to optimize the analysis pipeline.
Our algorithm combines the advantages of these two methods by first proposing to test a parametric method on the samples. If this is validated, it can then be applied automatically on a large number of images. Otherwise, the user is offered to test a WEKA reference training set which can, in turn, be automated or used to initiate a new complementary training phase. Our overall segmentation methodology, therefore, benefits from the speed of execution of a PS when possible and from the flexibility of WEKA for samples that are too different from our training sets.
The second essential element that contributes to the potential of REPRISAL was the development of a ratiometric approach. Such an approach is necessary because different fluorophores emit different amounts of photons and the measured fluorescence intensities obtained with different fluorophores do not therefore accurately reflect the amount of reporter incorporated. Our approach is based upon two robust and easily interpretable intensity ratios; RM1 evaluates the relative distribution of a given reporter in the different segmented cell wall regions while RM2 evaluates the relative proportion of each reporter in a given cell wall zone. These methods made it possible to extract quantitative information from the complex images generated by the triple monolignol reporter labeling strategy that can then be statistically analyzed to generate useful biological information as discussed above.
The final element necessary to REPRISAL was the implementation of the previously developed triple labeling strategy (Simon et al., 2018). When applied to the Arabidopsis prx64 mutant, REPRISAL was able to detect statistically significant differences in the lignification pattern compared to WT even though a simple visual evaluation was unable to this. In addition to confirming that the AtPRX64 protein does indeed play a role in cell wall lignification in stems, this observation also confirms previous results from our group showing that the chemical reporter approach is able to reveal mutant phenotypes undetectable by classical cell wall analyses (Baldacci-Cresp et al., 2020a).
In this paper, we focused on mapping lignification and we, therefore, used three monolignol chemical reporters. A number of monosaccharide reporters have been successfully used in mono-labeling approaches by other groups and could therefore be implemented for REPRISAL (Anderson et al., 2012; Dumont et al., 2016; Zhu et al., 2016).
Finally, it would also be interesting to adapt REPRISAL, or at least the segmentation component, to the analysis of cell wall images generated by other imaging techniques in order to multiply the information that can be obtained. Possible targets could include, for example, different histochemical reagents such as basic Fuschin, Safranin, Toluidine blue O, or Auramine O that have been shown to be compatible with fluorescence microscopy (Ursache et al., 2018; Baldacci-Cresp et al., 2020b), as well as antibodies capable of recognizing a wide range of cell wall epitopes (Donaldson and Knox, 2012) and other fluorescence-based imaging techniques (DeVree et al., 2021). Another major direction could be to explore the possibilities of combining REPRISAL-type segmentation with the segmentation resulting from multivariate analyses of vibrational spectroscopic data such as Raman (Gierlinger et al., 2012) or Atomic Force Microscopy (Felhofer et al., 2020). Such approaches could be expected to contribute to our understanding, not only of lignification, but also of cell wall biology in general, thereby paving the way for exciting new discoveries in both fundamental and applied areas.
Materials and methods
Plant material
Arabidopsis (A. thaliana, Columbia-0) plants were used for experiments. Seeds were stratified at 4°C in 0.1% Phytagel solution (w/v) for 3 d before being sowed. Plants were grown in growth chambers (GroBank, BB-XXL.3+) under 12-h light cycles (120 µmol photon m−2 s−1) at 23°C during the day and 20°C during the night. The atprx64 insertion mutant (SALK_203548C) was purchased from the Nottingham Arabidopsis Stock Centre.
Flax (L. usitatissimum) and maize (Zea mays) plants were grown in growth chambers (Angelantoni Life Sciences, Massa Martana, Italy) at 22°C with a photoperiod of 16 h/8 h day/night. Poplar (P. tremula × P. alba) plants were grown in a phytotron with a 16‐h light/8‐h dark photoperiod, 24°C/21°C, with 35%–65% hygrometry under white light (HPI Master Plus Philips, metal halide) to maintain a light intensity of 120 µmol m−2 s−1.
Sample preparation and reporter labeling
Sample preparation was adapted from the protocol described in Simon et al. (2018). Sections of 80 µm thickness were made with a vibroslicer (VT-1000S, Leica, Wetzlar, Germany) at different heights (1 cm = O, 15 cm = M, and 23 cm = Y) from the base of the stem of embedded (3.5% w/v agarose) samples of flowering stems of A. thaliana plants (7 weeks). The sections were placed in sterile Murashige and Skoog half-strength (1/2 MS) solution and stored at 4°C prior to incubation with reporters. Maize, flax, and poplar sections were made from the stems of 8-week-old plants.
Bioorthogonal triple labeling of monolignol chemical reporters H*, G*, S* (Figure 2), and F* was performed as previously described (Simon et al., 2018) and outlined in Figure 2. Stem cross-sections were incubated in 300 µL 1/2 MS containing 5 µM of H*, S*, and G*/F* for 20 h in the light at 20°C (Grobank). Control samples were incubated in 300 µL 1/2 MS containing 5 µM corresponding untagged natural monolignols/fucose. After incubation, samples were extensively washed (4 × 1/2 MS) prior to labeling with fluorophores via three sequential bioorthogonal reactions performed in the following order: (1) IEDDA (S*), (2) SPAAC (H*), and (3) CuAAC (G*, F*). Tetrazine Cy5 (Jena Bioscience, Jena, Germany) and Azide Fluor 545 (Sigma–Aldrich, Saint Louis, Missouri, USA) fluorophores were used at 5 µM; DBCO-PEG4-Rhodamine Green fluorophore (Jena Bioscience, Jena, Germany) was used at 2.5 µM. All reactions were carried out in 300 µL 1/2 MS for 1 h in the dark and samples were washed (4 × 1/2 MS) between reactions. Following the final reaction, samples were extensively washed (2 × 1/2 MS, 5 min, 1 × MeOH 70%, 60 min, 1 × 1/2 MS, 5 min, and 3 × 1/2 MS, 10 min) to fully remove any remaining free fluorophore.
Confocal microscopy
Image acquisitions were performed as previously described (Simon et al., 2018). Images from three independent biological replicates for each genotype and tissue were acquired. A Nikon A1R confocal equipped with a 60×/1.4 aperture oil immersion objective (Plan APO VC) and the NIS Element AR version 3.0 software was used (Nikon, Tokyo, Japan). Acquisitions were performed on four channels corresponding to 1) lignin autofluorescence (laser line - ƛex: 405, Collection bandwidth - ƛem: 450/50); 2) H units (ƛex: 488, ƛem: 525/50); 3) G units (ƛex: 561, ƛem: 595/50); 4) S units (ƛex: 561, ƛem: 700/75); and 5) fucose (ƛex: 561, ƛem: 595/50). Laser intensity and detectors gains are adjusted to ensure optimal signal-to-noise-ratio while avoiding photobleaching.
Segmentation
The graphical user interface has been created as a plugin for Fiji using Jython, a Java implementation of Python. Using this language made it possible to use Fiji methods in the Jython script, thereby enabling the creation of the GUI as well as a smooth interaction with Fiji. Fiji macros were used for both AI and PS, allowing the program to run with either type of segmentation. The overall analysis procedure was developed with ImageJ version 1.53 and JAVA version 1.8.
PS
PS was performed using a homemade ImageJ macro. Briefly, the macro first creates a binary mask for 1) SCWs, 2) CCs, and 3) CMLs. The binary mask of each region was applied to each fluorescence channel and fluorescence mean values were extracted for the nine newly created images. A recapitulative montage image was then created to quickly estimate segmentation quality. The ImageJ macro and sample images are available in the Zenodo repository (Supplemental Dataset S1: http://doi.org/10.5281/zenodo.4809980).
AI segmentation
The machine learning approach is based on the “Waikato Environment for Knowledge Analysis” (WEKA) implemented in ImageJ (Witten et al., 2016). We first defined a classification based on four categories: 1) SCW, 2) CCs, 3) CML, and 4) background. Our aim was to provide methods compatible with traditional computers and thus limited the training parameters to gaussian blur, Sobel filter, Hessian, Difference of Gaussians, and membrane projection with a maximum sigma of 16. These training features allowed unambiguous discrimination between the four classes after appropriate training of a random forest classifier with 200 initial trees. Manual labeling was performed on a pixel-by-pixel basis between the four classes. To avoid overlearning, training was performed on confocal images taken from various species, regions and using both WT and mutants. Each image is then segmented based on obtained probability maps (80% probability is used as a threshold) and compared with results achieved through manual segmentation. Once the training is achieved on reference images, the classifier is applied to new sets of images to validate the training step.
For more details, please see the “Results and discussion” section. The ImageJ plugin and sample images are available in the Zenodo repository (Supplemental Dataset S1: http://doi.org/10.5281/zenodo.4809980).
Ratiometric analysis
Two complementary (RMs were used: RM1 evaluates the relative distribution of a given reporter in the different segmented cell wall regions. Fluorescence intensity of a given reporter (H*, G*, S*, and F*) in a given cell wall region (CC, CML, or SCW) is divided by the total fluorescence intensity of that reporter in all three cell wall zones and expressed as a percentage (e.g. [H*CC intensity]/[∑H*]). RM2 evaluates the relative proportion of each reporter in a given cell wall zone compared to the other reporters. Fluorescence intensity of a given reporter (H*, G*, S*, and F*) in a given cell wall region (CC, CML, or SCW) is divided by the total fluorescence intensity of all three reporters in this particular cell wall zone and expressed as a percentage (e.g. [H*CCintensity]/[∑CC intensity]). Statistical differences (P-value < 0.05) between samples were determined by the Student’s t test and analysis of variance (ANOVA). Ten samples (cross-sections) per developmental stage (Y, M, and O) from three individual plants (WT, mutant) were analyzed for data.
Accession numbers
Arabidopsis PRX prx64 gene: AT5g42180.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Dataset S1. Supplemental dataset provided at http://doi.org/10.5281/zenodo.4809980.
Supplemental Figure S1. Bioorthogonal lignin triple (H*, G*, and S*) labeling in WT Arabidopsis floral stems.
Supplemental Figure S2. UV lignin autofluorescence and bioorthogonal lignin triple (H*, G*, and S*) labeling in Arabidopsis WT and prx64 mutant floral stems.
Supplemental Figure S3. Comparison of monolignol reporter incorporation profiles in WT and prx64 mutant Arabidopsis stem fiber cell walls.
Supplemental Figure S4. Automatic segmentation of cell wall zones in flax stem tissues showing relative distribution of lignin and NCP reporters.
Funding
O.M. gratefully acknowledges the joint financial support of the French Hauts-de-France Region and the Austrian Science Fund (Start project Y-728-B16) for a PhD fellowship; this study was partially funded by grants from the EU (CPER/FEDER project ALIBIOTECH).
Conflict of interest statement. None declared.
Supplementary Material
C.B., S.H., and Co.S. conceived the project and supervised, respectively, the chemical, biological, and image data and segmentation experiments. C.L. and Cl.S. designed and synthesized the chemical reporters and elaborated the triple-click labeling method. O.M. performed the experiments and developed an analysis procedure with Co.S. J.S. developed the GUI integrating all algorithms. C.L., F.B.C., G.N., and Co.S. aided design and interpretation of experimental data. O.M., C.L., G.N., C.B., S.H., and Co.S. wrote the article with contributions of all of the authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Christophe Biot (christophe.biot@univ-lille.fr).
References
- Anderson CT, Wallace IS, Somerville CR (2012) Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc Natl Acad Sci USA 109:1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Hao Z, Mohnen D (2013) Evolving views of pectin biosynthesis. Annu Rev Plant Biol 64:747–779 [DOI] [PubMed] [Google Scholar]
- Baldacci-Cresp F, Le Roy J, Huss B, Lion C, Créach A, Spriet C, Duponchel L, Biot C, Baucher M, Hawkins S, et al. (2020a) UDP-GLYCOSYLTRANSFERASE 72E3 plays a role in lignification of secondary cell walls in Arabidopsis. Int J Mol Sci 21:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacci-Cresp F, Spriet C, Twyffels L, Blervacq A-S, Neutelings G, Baucher M, Hawkins S (2020b) A rapid and quantitative safranin-based fluorescent microscopy method to evaluate cell wall lignification. Plant J 102:1074–1089 [DOI] [PubMed] [Google Scholar]
- Billa E, Monties B (1995) Molecular variability of lignin fractions isolated from wheat straw. Res Chem Intermed 21:303–311 [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546 [DOI] [PubMed] [Google Scholar]
- Bukowski N, Pandey JL, Doyle L, Richard TL, Anderson CT, Zhu Y (2014) Development of a clickable designer monolignol for interrogation of lignification in plant cell walls. Bioconjug Chem 25:2189–2196 [DOI] [PubMed] [Google Scholar]
- Campbell MM, Sederoff RR (1996) Variation in lignin content and composition: mechanisms of control and implications for the genetic improvement of plants. Plant Physiol 110:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabi M, Goulas E, Leclercq CC, de Waele I, Rihouey C, Cenci U, Day A, Blervacq A-S, Neutelings G, Duponchel L, et al. (2017) A cell wall proteome and targeted cell wall analyses provide novel information on hemicellulose metabolism in flax. Mol Cell Proteomics 16:1634–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Havkin-Frenkel D, Dixon RA, Ralph J (2012) A polymer of caffeyl alcohol in plant seeds. Proc Natl Acad Sci USA 109:1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford MN (1974) Specificity of acidic phloroglucinol reagents. J Chromatogr A 94:321–324 [Google Scholar]
- Day A, Ruel K, Neutelings G, Crônier D, David H, Hawkins S, Chabbert B (2005) Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta 222:234–245 [DOI] [PubMed] [Google Scholar]
- DeVree BT, Steiner LM, Głazowska S, Ruhnow F, Herburger K, Persson S, Mravec J (2021) Current and future advances in fluorescence-based visualization of plant cell wall components and cell wall biosynthetic machineries. Biotechnol Biofuels 14:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson LA (2001) Lignification and lignin topochemistry - an ultrastructural view. Phytochemistry 57:859–873 [DOI] [PubMed] [Google Scholar]
- Donaldson LA, Knox JP (2012) Localization of cell wall polysaccharides in normal and compression wood of radiata pine: relationships with lignification and microfibril orientation. Plant Physiol 158:642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Lehner A, Vauzeilles B, Malassis J, Marchant A, Smyth K, Linclau B, Baron A, Mas Pons J, Anderson CT, et al. (2016) Plant cell wall imaging by metabolic click-mediated labelling of rhamnogalacturonan II using azido 3-deoxy-d-manno-oct-2-ulosonic acid. Plant J 85:437–447 [DOI] [PubMed] [Google Scholar]
- Effland MJ (1977) Modified procedure to determine acid-insoluble lignin in wood and pulp. Tappi 60:143–144 [Google Scholar]
- Felhofer M, Bock P, Singh A, Prats-Mateu B, Zirbs R, Gierlinger N (2020) Wood deformation leads to rearrangement of molecules at the nanoscale. Nano Lett 20:2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierlinger N, Keplinger T, Harrington M (2012) Imaging of plant cell walls by confocal Raman microscopy. Nat Protocol 7:1694–1708 [DOI] [PubMed] [Google Scholar]
- Grabber JH, Ralph J, Hatfield RD (2000) Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J Agric Food Chem 48:6106–6113 [DOI] [PubMed] [Google Scholar]
- Grishagin I V. (2015) Automatic cell counting with ImageJ. Anal Biochem 473:63–65 [DOI] [PubMed] [Google Scholar]
- Hoffmann N, Benske A, Betz H, Schuetz M, Lacey Samuels A (2020) Laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development. Plant Physiol 184:806–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis R, Morreel K, Fliniaux O, Lucau-Danila A, Fénart S, Grec S, Neutelings G, Chabbert B, Mesnard F, Boerjan W, et al. (2012) Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol 158:1893–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Moore WE, Zank LC (1961) The spectrophotometric determination of lignin in small wood samples. J Tech Assoc Pulp Pap Ind 44:793–798 [Google Scholar]
- Lapierre C, Monties B, Rolando C (1986) Thioacidolyse von pappelholzligninen: nachweis von monomeren syringylverbindungen und charakterisierung von guajacyl-syringyl-ligninfraktionen. Holzforschung 40:113–118 [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153:402–412 [DOI] [PubMed] [Google Scholar]
- Legland D, Arganda-Carreras I, Andrey P (2016) MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32:3532–3534 [DOI] [PubMed] [Google Scholar]
- Lion C, Simon C, Huss B, Blervacq AS, Tirot L, Toybou D, Spriet C, Slomianny C, Guerardel Y, Hawkins S, et al. (2017) BLISS: a bioorthogonal dual-labeling strategy to unravel lignification dynamics in plants. Cell Chem Biol 24:326–338 [DOI] [PubMed] [Google Scholar]
- Lupoi JS, Singh S, Parthasarathi R, Simmons BA, Henry RJ (2015) Recent innovations in analytical methods for the qualitative and quantitative assessment of lignin. Renew Sustain Energy Rev 49:871–906 [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protocol 7:1579–1589 [DOI] [PubMed] [Google Scholar]
- Otsu N (1979) Threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern SMC 9:62–66 [Google Scholar]
- Palaniappan KK, Bertozzi CR (2016) Chemical glycoproteomics. Chem Rev 116:14277–14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey JL, Wang B, Diehl BG, Richard TL, Chen G, Anderson CT (2015) A versatile click-compatible monolignol probe to study lignin deposition in plant cell walls. PLoS ONE 10:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins M, Smith RA, Samuels L (2019) The transport of monomers during lignification in plants: anything goes but how ? Curr Opin Biotechnol 56:69–74 [DOI] [PubMed] [Google Scholar]
- Pesquet E, Ranocha P, Legay S, Digonnet C, Barbier O, Pichon M, Goffner D (2005) Novel markers of xylogenesis in zinnia are differentially regulated by auxin and cytokinin. Plant Physiol 139:1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer SM, Amburn EP, Austin JD, Cromartie R, Geselowitz A, Greer T, ter Haar Romeny B, Zimmerman JB, Zuiderveld K (1987) Adaptive histogram equalization and its variations. Comput vision. Graph image Process 39:355–368 [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl- propanoids. Phytochem Rev 3:29–60 [Google Scholar]
- Rigolot V, Biot C, Lion C (2021) To view your biomolecule, click inside the cell. Angew Chem Int Ed 60:23084–23105 [DOI] [PubMed] [Google Scholar]
- del Río J, Rencoret J, Gutiérrez A, Kim H, Ralph J (2017) Hydroxystilbenes are monomers in palm fruit endocarp lignins. Plant Physiol 174:2072–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río JC, Rencoret J, Prinsen P, Martínez ÁT, Ralph J, Gutiérrez A (2012) Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J Agric Food Chem 60:5922–5935 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61:263–289 [DOI] [PubMed] [Google Scholar]
- Schwartz A, Wang L, Early E, Gaigalas A, Zhang Y-Z, Marti GE, Vogt RF (2002) Quantitating fluorescence intensity from fluorophore: the definition of MESF assignment. J Res Natl Inst Stand Technol 107:83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Lion C, Spriet C, Baldacci-Cresp F, Hawkins S, Biot C (2018) One, two, three: a bioorthogonal triple labelling strategy for studying the dynamics of plant cell wall formation in vivo. Angew Chem Int Ed 57:16665–16671 [DOI] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Karlen SD, Bird D, Tokunaga N, Sato Y, Mansfield SD, Ralph J, Samuels AL (2017) Defining the diverse cell populations contributing to lignification in Arabidopsis stems. Plant Physiol 174:1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y, Van De Wouwer D, Allen E, Kumpf R, Vanholme B, Boerjan W, Ralph J (2014) A click chemistry strategy for visualization of plant cell wall lignification. Chem Commun 50:12262–12265 [DOI] [PubMed] [Google Scholar]
- Ursache R, Andersen TG, Marhavý P, Geldner N (2018) A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J 93:399–412 [DOI] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L (2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA: shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci USA 104:11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S, Chin JW (2014) Strain-promoted sydnone bicyclo-[6.1.0]-nonyne cycloaddition. Chem Sci 5:1742–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chantreau M, Sibout R, Hawkins S (2013) Plant cell wall lignification and monolignol metabolism. Front Plant Sci 4:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten I, Frank E, Hall M, Pal C (2016) Data Mining; Practical Machine Learning Tools and Techniques, 4th edn. Todd Green, Cambridge [Google Scholar]
- Yi Chou E, Schuetz M, Hoffmann N, Watanabe Y, Sibout R, Samuels AL (2018) Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J Exp Bot 69:1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wu J, Chen X (2016) Metabolic labeling and imaging of N-linked glycans in Arabidopsis thaliana. Angew Chemie Int Ed 55:9301–9305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.