Confocal live-imaging provides a quantitative atlas for analyzing cellular growth dynamics underlying stamen organogenesis in Arabidopsis thaliana.
Abstract
Development of multicellular organisms is a complex process involving precise coordination of growth among individual cells. Understanding organogenesis requires measurements of cellular behaviors over space and time. In plants, such a quantitative approach has been successfully used to dissect organ development in both leaves and external floral organs, such as sepals. However, the observation of floral reproductive organs is hampered as they develop inside tightly closed floral buds, and are therefore difficult to access for imaging. We developed a confocal time-lapse imaging method, applied here to Arabidopsis (Arabidopsis thaliana), which allows full quantitative characterization of the development of stamens, the male reproductive organs. Our lineage tracing reveals the early specification of the filament and the anther. Formation of the anther lobes is associated with a temporal increase of growth at the lobe surface that correlates with intensive growth of the developing locule. Filament development is very dynamic and passes through three distinct phases: (1) initial intense, anisotropic growth, and high cell proliferation; (2) restriction of growth and proliferation to the filament proximal region; and (3) resumption of intense and anisotropic growth, displaced to the distal portion of the filament, without cell proliferation. This quantitative atlas of cellular growth dynamics provides a solid framework for future studies into stamen development.
Introduction
One major question in biology is how multicellular organisms acquire their diverse forms. In plants, organ shapes emerge from complex interactions among cellular behaviors controlling growth, patterning, and differentiation that occur within mechanically connected tissues (Sablowski, 2015; Coen et al., 2017; Echevin et al., 2019). During the development, there are multiple possibilities to modulate these processes in terms of both space and time to generate diverse shapes (Kierzkowski et al., 2019; Whitewoods et al., 2020). Furthermore, distinct developmental patterns can produce comparable geometries (Green et al., 1970; Coen et al., 2004; Rebocho et al., 2017; Solly et al., 2017). Therefore, it is impossible to understand organ development solely by observing their final shape. Inferring growth from developmental snapshots of fixed samples or from clonal analysis has provided quantitative information about various cellular parameters underlying organogenesis (Silk et al., 1989; Sauret-Güeto et al., 2013; Bassel et al., 2014; Eldridge et al., 2016; Dong et al., 2020; Whitewoods et al., 2020; Vijayan et al., 2021). However, deep understanding of organ development requires direct measurements of spatiotemporal behaviors of individual cells to precisely link them with changes in tissue geometry (Bassel and Smith, 2016; Hong et al., 2016; Fox et al., 2018; Sapala et al., 2018; Kierzkowski et al., 2019; Ripoll et al., 2019; Zhang et al., 2020, 2021).
Flowers are composed of homologous, yet very diverse, organs that are believed to be modified versions of a leaf-like structure (Pelaz et al., 2001). So far, quantitative growth data at cellular resolution have been acquired only for sepals, the most external floral organs (Tauriello et al., 2015; Hervieux et al., 2016; Tsugawa et al., 2017; Zhu et al., 2020). The general trends of growth for internal floral organs have been inferred from clonal analysis (Rolland-Lagan et al., 2003; Sauret-Güeto et al., 2013; Eldridge et al., 2016), which does not reveal the cellular dynamics of developmental processes. Some quantitative live-imaging studies have been performed on the gynoecium; however, they focused on relatively late developmental stages, mostly postfertilization (Ripoll et al., 2019; Dong et al., 2020). Live-imaging of internal floral organs is technically challenging, since they are hidden by sepals inside the floral bud.
Stamens are male reproductive floral organs that emerge in the second innermost whorl of the flower and play a crucial role in plant sexual reproduction during pollination. They are composed of long filaments topped with anthers. The mature anther in Arabidopsis thaliana presents a complex four-lobed shape, which in the transverse section resembles a butterfly. The two pairs of lobes are separated by a central connective tissue, and each lobe contains four concentric tissue layers surrounding one locule, where pollen grains are formed (Goldberg et al., 1993; Sanders et al., 1999; Walbot and Egger, 2016). The anther anatomy is set up by very specific patterns of cell division in its internal tissues (Goldberg et al., 1993; Sanders et al., 1999; Zhao, 2009). Although considered less complex than the anthers, filaments are also of great importance for plant reproduction since their elongation is responsible for the anthers reaching the stigma for self-pollination or for making pollen accessible for pollinators. Despite stamen importance for reproduction, its developmental dynamics are largely unknown. Stamen morphogenesis has been extensively studied in fixed tissues, providing only a glimpse of the developmental processes taking place inside the floral bud (Smyth et al., 1990; Sanders et al., 1999; Scott et al., 2004; Cecchetti et al., 2008).
In this study, we monitored the development of stamens in A. thaliana. To this end, we developed a confocal live-imaging method that allows long-term observation and quantification of stamen growth parameters at cellular resolution during the entire development of stamen. Using complete lineage tracing, we precisely identified the origin of the stamen primordia as well as the timing of filament/anther specification. Our analysis indicates that the outgrowth of the anther lobes correlates with an intensive growth of the developing locules. We also found that filament elongation is very dynamic and passes through three distinct phases with different cell growth and cell proliferation patterns.
Results
A live-imaging method to follow stamen development
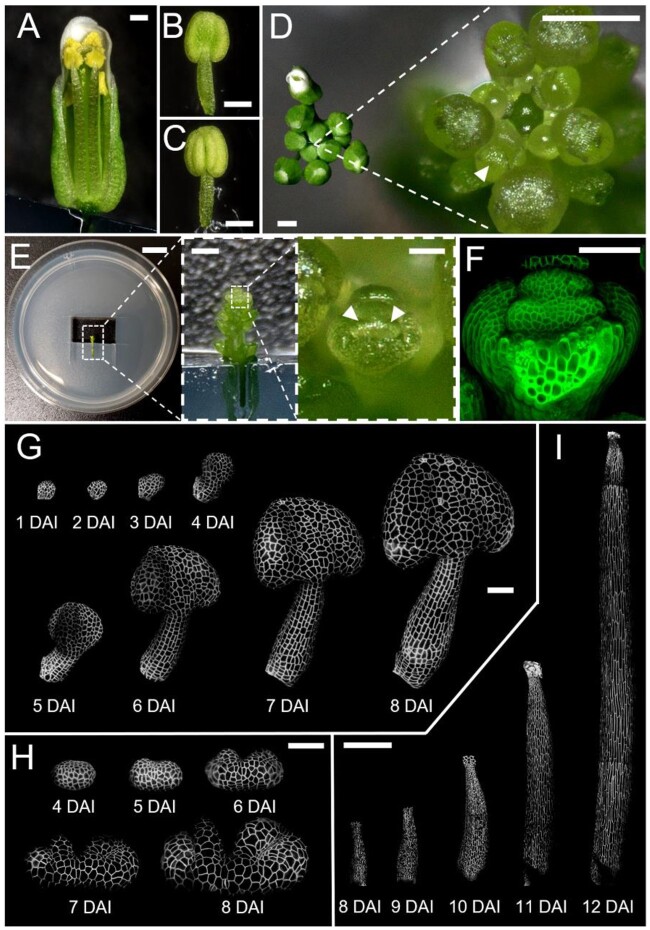
To precisely characterize stamen organogenesis, we developed a confocal live-imaging procedure that allows quantitative observation of complete stamen development from primordium initiation until the acquisition of its final morphology (Figure 1). We first carefully dissected sepal primordium at floral stage 5 (Smyth et al., 1990; Figure 1D) to uncover the primordia of long (abaxial) stamens. We then mounted dissected flowers on an in vitro medium and imaged the plasma membrane tagged with a fluorescent marker using a confocal microscope with an interval of 24 h (Figure 1, E and F; see “Materials and methods” for more details).
Figure 1.
A live-imaging method enables long-term observation of stamen development in A. thaliana. A, Wild-type flower after anthesis when stamens reach the stigma; sepal and petals are removed to access the internal organs. B and C, Abaxial (B) and adaxial (C) views of the mature stamen before anthesis. D, Top view of inflorescence before (left) and after (right) dissection of older flowers. Arrowhead indicates the flower bud (at stage 5) to be dissected for imaging. E, Dissected inflorescence (after removal of abaxial sepal from the flower bud) mounted on one-half MS medium. Arrowheads indicate long stamen primordia. F, Confocal image of dissected flower bud shown in (E) with plasma membrane marker in green. G and H, Time-lapse series of the developing stamen primordia, imaged at 24 h intervals. Imaging from the abaxial side (G) and from the top (H). I, Time-lapse series of filament elongation, imaged at 24 h intervals. White signal in (G–I) represents plasma membrane marker in the epidermis projected on the digitally extracted organ surface. DAI indicates days after primordium initiation. Scale bars, 250 µm in (A–D and I); 1 cm, 500 µm, and 50 µm from left to right in (E); 50 µm in (F–H).
Stamens developed normally for up to 11 days of imaging (Figure 1, G–I), undergoing the same sequence of developmental stages as previously described, including the formation of lobes and notches in the anther (Figure 1H) and filament elongation (Figure 1I;Smyth et al., 1990; Sanders et al., 1999). The timing of developmental events we observed was comparable to the duration of floral stages in intact plants (Smyth et al., 1990; Sanders et al., 1999). This suggests that our dissection, in-vitro culture, and imaging methods had no substantial impact on the overall development of stamens.
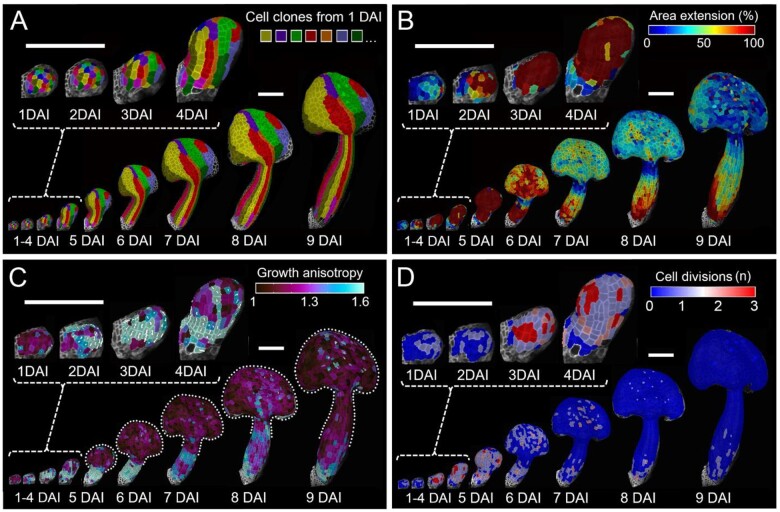
The quality of our 3D time-lapse data was sufficient to quantify growth parameters at cellular resolution for the entire development of the stamen using MorphoGraphX (Barbier de Reuille et al., 2015). We computed the complete lineage data to reveal the contribution of cells in the early primordium to the development of each stamen region (Figure 2A;Supplemental Figure S1). Using lineage relationships, we quantified cellular parameters contributing to organ shape, including cellular growth rates (cell area increase), cell growth anisotropy (ratio of expansion along the maximal and minimal directions of growth), and number of cell divisions (Figure 2, B–D). We also computed cell sizes (expressed as cell surface area), cell shapes (i.e. cell lobeyness), and overall organ curvature to assess the progression of organ and cell differentiation (see “Materials and methods” for details).
Figure 2.
Developmental patterns of A. thaliana stamen. A, Cell-lineage tracing in the young stamen. Colors indicate clones developing from single cells at 1 DAI displayed on the digitally extracted organ surface. B–D, Heat maps of area extension (B), growth anisotropy (C), and cell proliferation (D). B–D, Heat maps generated between two consecutive time points are displayed on the digitally extracted organ surface at the earlier time point. Dotted lines in (C) mark stamen outlines for clarity. White lines in (C) indicate cell-growth orientation where anisotropy >40%. Scale bars, 100 µm.
Initiation and stamen region specification
Stamen primordia emerge at the third whorl of the floral meristem and become apparent when sepals have not yet totally enclosed the floral bud (around stage 5; Smyth et al., 1990; Sanders et al., 1999). To determine how many cells at the flower meristem give rise to a stamen primordium, we followed the growth of incipient stamen primordia from the stage when no apparent bulge is present on the surface of the flower meristem until a clear protrusion is visible 24 h later (Figure 3A). Using clonal analysis, we determined that stamen primordia emerge from approximately 28 ± 2 epidermal cells (Figure 3B).
Figure 3.
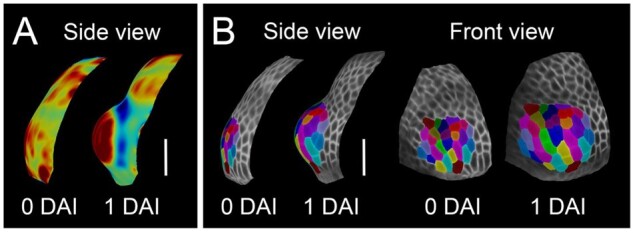
Stamen initiates from 28 ± 2 epidermal cells. A, Heat maps of Gaussian curvature from red (positive) to blue (negative) of the initiating primordium (displayed on the digitally extracted organ surface). B, Cell-lineage tracing in the initiating primordium. Colors indicate clones developing from single cells when stamen primordium initiates displayed on the digitally extracted organ surface. Scale bars, 20 µm.
We next quantified growth patterns leading to stamen differentiation from this small primordium (Figure 2). During the first 3 days after initiation (DAI), cellular growth rates intensified and cells grew anisotropically (i.e. faster in one direction) along the proximal–distal axis of the organ, leading to the progressive elongation of the early primordium (Figure 2, B and C). Intensive growth at this stage corresponded with progressive increase in frequency of cell divisions (Figure 2D). At 4 DAI, regional differences in growth anisotropy started to emerge. Although the proximal portion of the primordium continued to grow faster along the main axis of the organ, cells in the distal part switched toward more isotropic growth, leading to its broadening (Figure 2C). This indicates that the filament and anther regions start to differentiate around 4 DAI.
To precisely determine the stage when the anther and filament are specified, we followed cellular clones originating from individual cells at early developmental stages until clearly defined anther and filament regions were present at 10 DAI (Supplemental Figure S1). Cellular clones originating from the early primordium at 4 DAI were entirely located on either the filament or anther, confirming that complete specification of those two regions occurred between 4 and 5 DAI (Supplemental Figure S1C).
Based on cellular clones developing from 4 DAI, we assigned cells to either filament or anther regions (Figure 4A) and quantified their cellular dynamics separately (Figure 4, D–F). Growth and cell division rates in both the anther and filament were initially very high (4–6 DAI; Figure 2, B and D; Figure 4, D and F). Note that while the directionality of growth (i.e. anisotropy) clearly differed between the future anther and filament starting from 4 DAI, their growth rates remained comaprable between 4 and 5 DAI (Figure 2, B and C; Figure 4, D and E), suggesting that cellular growth directions could be regulated independently from areal expansion. As the stamen progressed through its development (6–10 DAI), growth and division rates gradually decreased in both regions (Figure 2, B and D; Figure 4, D and F). The developing filament displayed gradients of cell dynamics, with higher rates of growth and division at the base and nearly no growth observed at the very top (Figure 2, B and D; Figure 4, D and F). From 4 to 7 DAI, cells in the filament elongated around three times faster along the main axis of the organ in comparison to cells located in the anther (Figures 2C and 4E). Although growth anisotropy progressively decreases at later stages, the cumulative growth of the filament between 4 and 10 DAI is highly anisotropic, while the anther expands more uniformly in all directions (Figure 4B). This is further confirmed by the presence of jigsaw-puzzle cells in the anther epidermis at 10 DAI, which are known to develop in tissues growing more isotropically (i.e. equally in all directions; Sapala et al., 2018; Figure 4C). Overall, our data suggest an early differentiation of the anther and filament, which then follow distinct developmental pathways.
Figure 4.
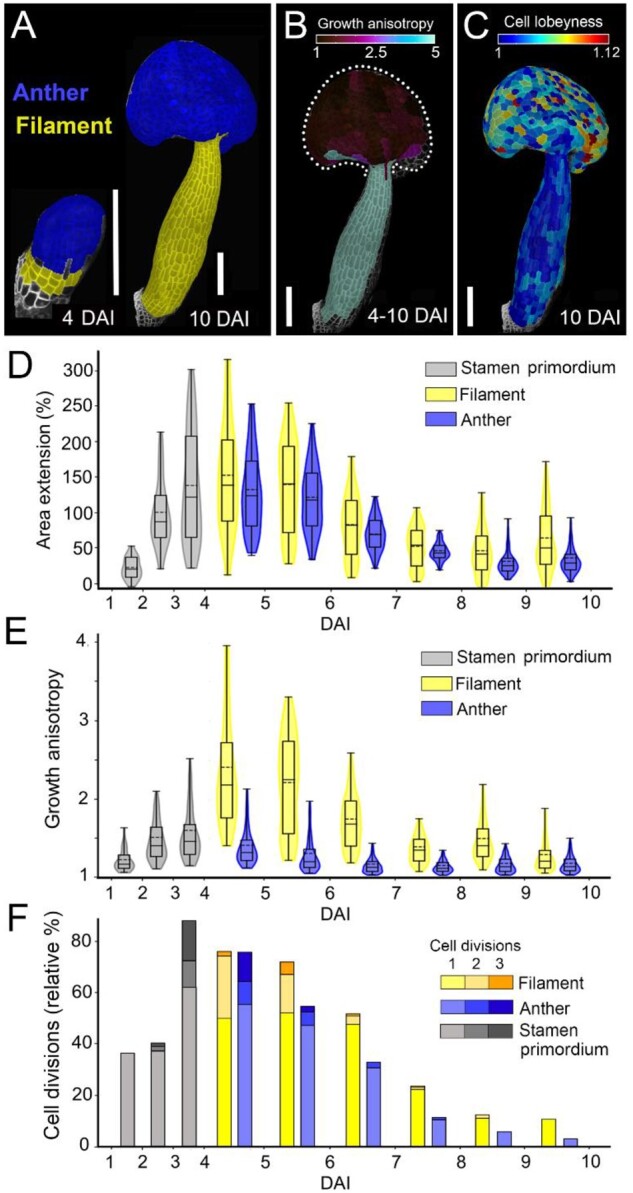
Anther and filament are specified at 4 days after stamen initiation and display different growth characteristics. A, Lineage tracing of anther (blue) and filament (yellow) between 4 and 10 DAI displayed on the digitally extracted organ surface. B, Heat map of growth anisotropy from 4 to 10 days after stamen initiation displayed on the digitally extracted organ surface. Dotted line marks stamen outline for clarity. C, Heat map of cell lobeyness at 10 DAI. D–F, Quantification of area extension (D), growth anisotropy (E), and cell divisions (F). Violin plots contain 95% and boxplots 90% of the values, respectively. The boxes indicate the range between the first and the third quartile and the whiskers include 90% of the values. Lines represent median and dashed lines represent the mean (three independent time-lapse series; n > 55 cells). Scale bars, 100 µm in (A–C).
Anther growth and differentiation
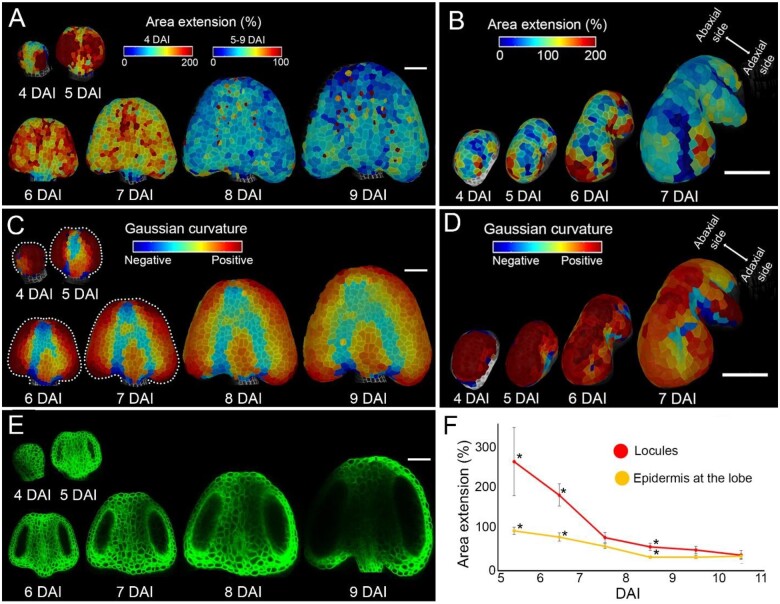
Mature anthers present an elaborate shape with four lobes encapsulating locules in the internal tissues (Sanders et al., 1999; Ma, 2005; Figure 1, B and C). Our data show that early stages of anther development (4–6 DAI) are characterized by very fast, mostly isotropic growth on the abaxial side of the anther (Figure 2, B and C; Figure 4, D and E). To get further insight into the formation of lobes, we closely observed local differences in growth rates at the anther surface on both abaxial and adaxial sides (Figure 5, A and B). Between 4 and 6 DAI, cells located in the lateral parts of the abaxial side of the anther increased their size three times, while cells in the median part doubled in size (Figure 5A). During this early time frame, cellular growth rates were high at the location of all future lobes and slower at all future boundaries (Figure 5, A and B).
Figure 5.
Fast growth at the anther lobes correlates with early stages of locule formation. A and B, Heat maps of area extension of the abaxial (A) and adaxial (B) sides of anthers displayed on the digitally extracted organ surface. C and D, Gaussian curvature (red, positive; blue, negative) displayed on the digitally extracted abaxial (C) and adaxial (D) anther surface. Dotted lines in (C) mark stamen outlines for clarity. Heat maps generated between consecutive time points are displayed on the first time point. E, Digital longitudinal sections through the confocal stacks located in the middle of developing locules. Plasma membrane marker in green. F, Quantification of area extension of the longitudinal sections through the locules (red, n > 6 locules) and of the surface expansion of cells located at the lobe (yellow, n > 8 lobes). Error bars indicate sem. Asterisks mark the means significantly different between locules and lobe at the same time point according to Student’s t test (P < 0.05). Scale bars, 50 µm in (A–E).
These regional growth differences coincided with the gradual change of the anther surface geometry. The early primordium at 4 DAI had an oval shape with positive (convex) Gaussian curvature on the entire abaxial surface (Figure 5C), and was relatively flat in the center of the adaxial side (Figure 5D). At 5 DAI, a concavity started to be apparent in the central region of the abaxial surface, consistent with an increased growth at the lobes (Figure 5, A and C). As the growth started to equalize in the entire abaxial side from 6 DAI (Figures 2B and 5A), this concave region broadened and started to split into two shallow boundaries separated by emerging connective tissue (Figure 5C). In contrast, from 5 DAI, the slow growing, concave region on the adaxial side deepened and became the central notch separating the two symmetrical sides of the anther (Figure 5D). The boundary region between the abaxial and adaxial lobes (stomium) started to be geometrically distinguishable at 6 DAI, and became clearly visible at 7 DAI (Figure 5D). This is consistent with lower cellular growth at the future stomium and intensive growth at the incipient lobes at 5–7 DAI (Figure 5B). Curiously, high cellular growth rates were maintained for longer at the adaxial lobes (until at least 8 DAI) while growth in the abaxial lobes already slowed down (Figure 5, A and B), indicating a temporal difference between abaxial and adaxial lobe formation.
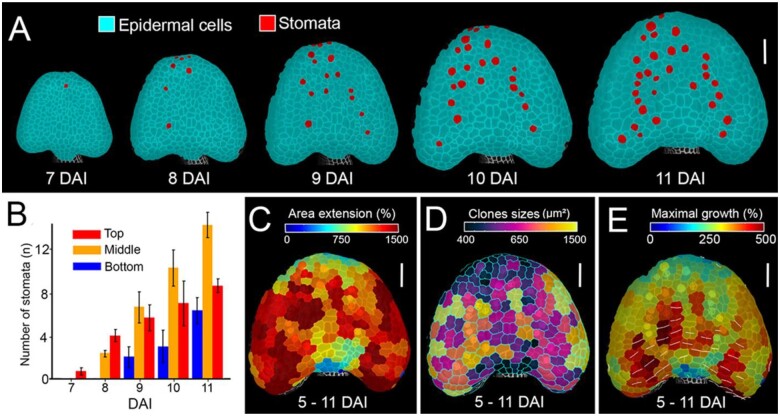
In most aerial organs in Arabidopsis, cell differentiation progresses basipetally (from the tip to the base of the organ; Kang and Dengler, 2002; Andriankaja et al., 2012; Hervieux et al., 2016; Fox et al., 2018; Kierzkowski et al., 2019). Such a gradient is also present in the anther, as indicated by the pattern of stomata differentiation (Wei et al., 2018). Consistently, we observed a clear basipetal gradient in stomata emergence starting from 7 DAI on the abaxial side (Figure 6, A and B). Cells located at the tip of the anther tended to grow slower compared with more proximal locations on both the abaxial (from 9 DAI; Figure 5A) and adaxial sides (from 7 DAI; Figure 5B). Slower growth at the tip is especially apparent in maps showing cumulative growth and sector size over several days (from 5 to 11 DAI; Figure 6, C–E). Our data indicate that differentiation-dependent, basipetal gradient of growth seems to be established in the Arabidopsis anther. It is however partially obstructed by another gradient in the mediolateral direction, most probably associated with the developing lobes.
Figure 6.
Anther displays a basipetal gradient of growth and differentiation. A, Cell-type tracing in the developing anther. Stomata marked in red displayed on the digitally extracted organ surface. B, Quantification of stomata distribution from 7 to 11 DAI as a function of distance from the anther tip. Error bars indicate sem (n = 3 independent time-lapse series; n > 2 cells). C–E, Heat maps of area extension (C), sizes (D), and maximal growth (E) of clones developing from single cells at 5 DAI. White lines in (E) indicate orientation of maximum growth for clones where growth anisotropy was >60%. Scale bars, 50 µm in (A) and (C–E).
We next investigated how high epidermal growth rates and geometric changes at the developing lobes correlate with the differentiation of internal anther tissues. We could observe the outline of developing locules from optical sections, since our plasma membrane fluorescent marker shows a very weak signal in the germ lines. Locules were clearly visible starting from 5 DAI (Figure 5E). Between 5 and 7 DAI, locule growth (measured as the increase of area of its longitudinal section) was around two times faster than the epidermal growth at the abaxial lobes (Figure 5F). Starting from 7 DAI, growth in both the locules and the epidermis significantly decreased and equalized (Figure 5F). These results suggest that rapid early growth of cells located in internal tissues could drive the formation of the lobes.
Filament elongation
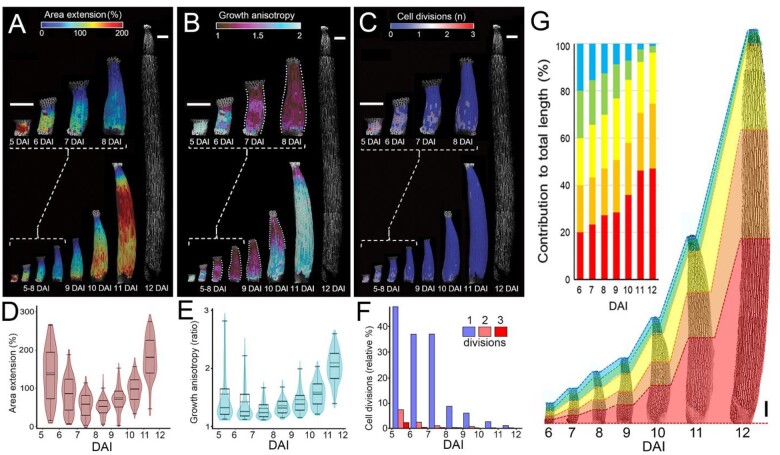
Filament elongation is critical for successful pollination. To understand the cellular dynamics during this process, we first quantified growth parameters from filament specification at 5 DAI until the final length of the stamen is reached at 12 DAI (Figure 7, A–F; Figure 8; Supplemental Figure S2). Right after its specification (5–6 DAI), growth in the entire filament was homogenous, very fast, and anisotropic. From 6 DAI, cellular growth rates progressively decreased, and growth became restricted to the proximal region of the filament (Figure 7, A, B, D, and E; Figure 8A). High cell division rates coincided with the first rapid phase of growth (5–7 DAI). Similar to growth patterns, cell proliferation decreased from 7 DAI and became restricted to the filament base (Figure 7, C and F; Figure 8, A and B). From 9 DAI, growth started to accelerate again, reaching its maximum just before anthesis (Figure 7, A and D). This resumption of growth first occurred at the base of the filament and was later displaced distally (Figure 7, A and D; Figure 8A). Fast elongation of the filament in later stages happened mainly without cell divisions (Figure 7, C and F; Figure 8D), leading to a nearly three-fold increase in average cell size between 11 and 12 DAI (Figure 8B;Supplemental Figure S2, A and D). Sections of the early filament contributed unequally to its final length, due to the nonuniform distribution of growth (Figure 7G). The most proximal section (red in Figure 7G) gave rise to nearly half of the filament length, while the two top sections (blue and green in Figure 7G) produced only 4% of the total length.
Figure 7.
Filament elongation occurs in three phases. A–C, Heat maps of area extension (A), growth anisotropy (B), and cell proliferation (C) displayed on the digitally extracted filament surface. Dotted lines in (B) mark filament outlines for clarity. Heat maps generated between consecutive time points are displayed on the first time point. White signals in (A–C) at 12 DAI indicate cell outlines. D–F, Quantification of area extension (D), growth anisotropy (E), and relative proportion of cell divisions (F). Violin plots contain 95% and boxplots 90% of the values, respectively. The boxes indicate the range between the first and the third quartile and the whiskers include 90% of the values. Lines represent median and dashed lines represent the mean (nine independent time-lapse series, n > 127 cells). G, Contribution of different portions of the early filament to the final length of the fully elongated filament. Graph represents relative contribution of equal segments marked at 6 DAI to the final filament length at 12 DAI. Time series of the developing filament divided at 6 DAI into five equal portions marked with different colors. Scale bars, 100 µm in (A–C) and (G).
Figure 8.
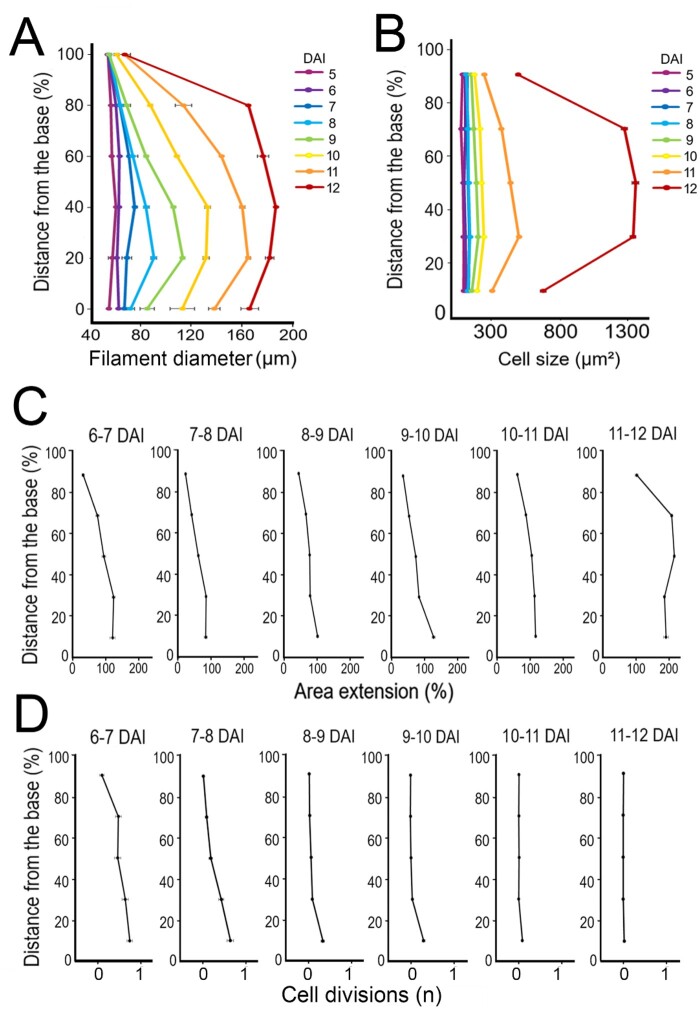
Proximodistal distribution of cellular growth and proliferation during filament elongation. A and B, Alignment graphs of mean filament diameter (A), and cell size (B), from 5 to 12 DAI as a function of distance from the filament base. C and D, Quantification of area extension (C) and cell divisions (D), from 5 to 12 DAI as a function of distance from the filament base. Error bars indicate standard error of the mean (nine independent time-lapse series; n > 127 cells).
Although growth occurred predominantly along the main axis of the organ (Figure 7, A and B; Supplemental Figure S2, B and E), we observed that the filament also grew in width (Supplemental Figure S2, C and F), leading to its progressive thickening (Figure 8A). The increase of the filament diameter followed a proximodistal gradient, with an initial broadening close to the base from 8 DAI (Figure 8A). Filament width progressively equalized before anthesis (mainly between 10 and 12 DAI; Figure 8D). The topmost, nongrowing part of the filament remained very narrow throughout its development (Figure 8D).
Altogether, these data suggest that filament elongation occurs in three phases: (1) fast growth along the entire length of the organ associated with high cell proliferation; (2) restriction of growth and proliferation to the broadening filament base; and (3) resumption of fast and anisotropic growth (without cell proliferation) that progressively moves distally and leads to uniformization of cell size and equalization of the filament width.
Discussion
Quantitative live-imaging of entire development of internal floral organs of Arabidopsis has never been achieved so far, due to the technical challenges presented by such experiments. The method we developed makes it possible to access the hidden stamen primordia and image them for several days at cellular resolution, without major perturbation of developmental processes. Our time-lapse data combined with quantitative image analysis provide a full characterization of the entire organogenesis of stamen at unprecedented spatial and temporal resolution.
Lineage tracing analysis revealed that around 28 epidermal cells of the floral meristem contribute to stamen development (Figure 3B). This number is much higher than the four epidermal founder cells that were previously inferred from genetic sector analysis (Bossinger and Smyth, 1996) or from the expression of a stamen primordia genetic marker (Chandler et al., 2011). Interestingly, the number of stamen initial cells found here is comparable with estimations from live-imaging experiments for Arabidopsis leaf, sepal, and flower primordia (Grandjean et al., 2004; Kwiatkowska, 2006; Schiessl et al., 2012; Burian et al., 2016). In contrast, a very low number of founder cells (approximately three cells) contribute to Arabidopsis stipules, small, leaf-associated appendages that lack vascular tissue and emerge exclusively from the epidermis (Vuolo et al., 2018). When stipules are transformed into leaves in the lmi1 (late meristem identity 1) mutant, the number of epidermal founder cells increases, and the internal tissue starts to be recruited into the primordium (Vuolo et al., 2018). This suggests that to generate stamen, as well as other typical aerial organs containing vascular tissue, several dozen epidermal cells need to be recruited at the incipient primordium.
Stamens, as all other aerial organs, are believed to evolutionarily derive from a leaf-like ancestral structure (Pelaz et al., 2001). As in leaves, basipetal gradients of growth and proliferation are present in the elongated anthers of easter lily (Lilium longiflorum; Gould and Lord, 1988). In Arabidopsis, cell differentiation appears to progress basipetally throughout the anther (Wei et al., 2018). Here we confirmed this gradient of differentiation and showed that it corresponds with a decrease of growth at the tip of the anther (Figure 6, C and D). Such differentiation-related gradients of growth are important for organ shaping. In leaves, delaying cell differentiation causes a redistribution of growth toward the distal part of the organ that normally stops growing first (Alvarez et al., 2016; Kierzkowski et al., 2019). Consistently, accelerating cell differentiation produces narrower petals by reducing growth in its distal regions (Sauret-Güeto et al., 2013). A similar mechanism seems to be conserved in anthers, as delaying differentiation via the overexpression of NUBBIN transcription factor mainly affects the morphology of the most distal tip of the anther, causing the tissue to extend apically beyond the normal limits of the anther (Dinneny et al., 2006).
We showed that the specification of the filament happened early in development (Supplemental Figure S1), and the entire filament originated only from two to three rows of cells along the main axis of the organ (Figure 4A). In several species, filament elongation was reported to occur predominantly at its base, resulting in the establishment of the basipetal gradient of cell sizes in the mature filament (Greyson and Tepfer, 1966; Hess and Morré, 1978; Lobello et al., 2000). Here we revealed that after initial restriction of growth to the filament base, a wave of growth moves acropetally (from the base to the tip) during the final stages of filament elongation (Figures 7 and 8). Such an acropetal wave of growth is characteristic for the elongation of the dark-grown hypocotyl (Gendreau et al., 1997; Refrégier et al., 2004; Pelletier et al., 2010; Peaucelle et al., 2015; Bou Daher et al., 2018), pointing to some common mechanisms underlying the elongation of both organs. The filament could become a model system to study cell growth and organ elongation as an alternative to the commonly used hypocotyl (Gendreau et al., 1997; Peaucelle et al., 2015; Reed et al., 2018). Hypocotyl elongation is sensitive to both gravity and light, making it difficult to live-image with standard microscopy setups without inducing a gravitropic response or a light-induced growth inhibition. Here we demonstrate that the stamen filament has comparable growth characteristics to the hypocotyl, while its development is neither affected by light exposure nor gravity, which makes it easier to handle. Furthermore, filament growth can be followed from its very initiation, while the hypocotyl is already established before germination (Bassel et al., 2014). Filament anatomy is also simpler, potentially providing a more practical system for biomechanical modeling.
The anther in itself could be used as a model system to study the establishment of complex 3D shapes, since it quickly develops a clear abaxial–adaxial asymmetry starting from a relatively symmetric primordium, forms sharp boundaries between growing lobes, and presents local differences in cell growth dynamics. Strikingly, our results showed a correlation between locule initiation and growth at the anther surface, suggesting that growth of internal tissues could be a driving force for the formation of the lobes. Previous studies described that division of cells derived from the L2 is responsible for internal anther patterning, resulting in concentric subepidermal layers surrounding the pollen mother cells (Sanders et al., 1999; Zhao, 2009; Huang et al., 2016; Xue et al., 2021; Zheng et al., 2021). Following lobe development in 3D would shed light on the biophysical interactions between internal and epidermal layers and their role in morphogenesis.
The quantitative atlas of cellular growth dynamics developed here, when combined with genetics and computational modeling, will provide a powerful tool to investigate the links between molecular mechanisms, cellular behavior, and tissue biomechanics during stamen morphogenesis. We believe that our approach can be further extended to investigate internal tissues of the stamen by employing advanced imaging methods, such as light sheet microscopy (Valuchova et al., 2020) in combination with modern 3D segmentation methods using convolutional neural networks (Eschweiler et al., 2019) to understand how different tissues control anther morphogenesis and filament elongation.
Materials and methods
Plant material and growth conditions
Wild-type A. thaliana (Col-0) carrying plasma membrane-localized fluorescent marker pUBQ10::myr:YFP was described previously (Yang et al., 2016). Plants were grown on soil in a growth chamber under long-day conditions (16 h of illumination, 95 µmol m−2 s−1), with 60%–70% relative humidity at 22°C ± 1°C. Dissected flowers were cultivated in vitro on Ø60 mm Petri dishes with one-half Murashige and Skoog (MS) medium plus vitamins (Murashige and Skoog, 1962) supplemented with 1.5% agar (w/v), 1% sucrose (w/v), and 0.1% Plant Protective Medium (Plant Cell Technologies; v/v) in long-day conditions (16 h illumination, 80 µmol m−2 s−1) with 60%–70% relative humidity at 22°C ± 1°C.
Live-imaging microscopy
Inflorescences were removed from 4-week-old soil-grown plants. Flower buds at stage 5 or 7 (Smyth et al., 1990) were dissected under a stereomicroscope on a wet paper tissue to prevent drying. Using fine tweezers and injection needles, the abaxial sepal was removed to expose the medial (long) stamens. Petals and lateral sepals were dissected when needed to avoid coverage of the developing stamen. To image late stages of filament elongation, the dissection was performed in flower buds at stage 10 (Smyth et al., 1990) by removing the abaxial and lateral sepals. Dissected stems were placed horizontally in a cavity cut out in the culture medium to image an abaxial stamen surface (Figure 1E). To follow the adaxial side, the stem was placed vertically in the medium and was imaged from the top.
Confocal imaging was performed with a Zeiss LSM800 upright confocal microscope, equipped with a long working-distance water immersion ×40 objective (1 NA, Apochromat). Excitation was performed using a diode laser with 488 nm for yellow flouresent protein YFP, and the signal was collected at 500–600 nm. Images were acquired at 512 × 512 resolution with 0.5–1 µm distance in z-dimension. Depending on the zoom used for image acquisition, the voxel size varied from 0.28 to 0.62 µm in the xy dimensions. For samples that were larger than the microscope field of view, multiple overlapping stacks were acquired and stitched. The abaxial surface of the stamen was imaged every 24 h for up to 11 days. For late stages of filament elongation, anthers were not imaged. To acquire data for the adaxial surface, the time-lapse series were acquired by imaging the flower buds from the top and gradually increasing laser power as we imaged deeper into the z-stack.
The images of anther development shown in Figure 5 are derived from two independent overlapping time-lapse series (4–5 and 5–10 DAI). The images of filament elongation shown in Figure 7 and Supplemental Figure S2 are derived from three independent overlapping time-lapse series (5–7, 7–10, and 10–12 DAI). Three to nine independent time-lapse series were acquired for each experiment. Images of dissected inflorescences and flower buds (Figure 1) were acquired using digital microscope VHX-970F (Keyence).
Image analysis and growth tracking
Time-lapse series were processed and analyzed using the MorphoGraphX software (Barbier de Reuille et al., 2015; Strauss et al., 2019). Stacks were manually stitched into a single file using the “combine stack” tool. Surface detection was performed with the “edge detect” tool with a threshold from 8,000–10,000 followed by “edge detect angle” (threshold 4,000–6,000). Initial meshes of 5-µm cube size were created and subdivided three times before projecting the membrane signal (2–4 µm from the mesh). Segmentation was performed with the “auto-segmentation” tool, followed by manual curation and segmentation of additional cells at the periphery of the mesh. Parent relations between cells of successive time points were attributed manually.
For lineage tracking analysis, the parent relations between each of the consecutive time points were combined to compute corresponding cell lineages over multiple days. The lineage data were later used to compute area extension, cell growth anisotropy, and cell proliferation. Area extension is calculated as the relative increase between the surface area of a mother cell and the area of its daughter cell(s) at the next time point. It is expressed in percentage of area increase ([{Total area of daughter cells at T1/Area of mother cell T0}−1] × 100). Heat maps of growth, growth anisotropy, and cell divisions generated between consecutive time points were displayed in the first time point. Heat maps of clone sector size, accumulated growth, and maximal growth were displayed in the last analyzed time point. Cell shape analysis was performed by calculating the lobeyness using a custom plugin in MorphoGraphX described previously (Sapala et al., 2018; Figure 4C).
Tracing the origin of the anther and filament was performed by following cell lineages from different early time points and finding the time point when resulting sectors contributed exclusively to anther or filament development (Supplemental Figure S1).
Anther regions were defined as three equal size zones (top, middle, and base) at 5 DAI based on the heat map of distance from the anther tip. The three zones were then identified in each of the following time points by following the clone sectors derived from them (based on lineage relations). Cell-type attribution was performed by manually labeling differentiated stomata (two guard cells) at each time point based on their stereotypical bean-shape.
Stamen primordia curvature heat maps were obtained by calculating the Gaussian curvature (neighborhood value 6). Anther surface curvature heat maps were computed on a cellular mesh (radius 40 µm).
Growth rates along the proximodistal and mediolateral axis of the filament were calculated using a custom Bezier grid that was manually placed so that it closely followed the geometry of the organ at each time point. Distance from the filament base was calculated using the “Cell distance” tool, which measures the shortest path between cells following the organ surface geometry.
To measure the contribution of different portions of the early filament to its final length, the early filament at 6 DAI was divided into five equal parts. Those parts were then identified at later time points using lineage sectors. To find corresponding landmarks between overlapping time lapse series, the length of different parts of the last filament of the first series was transferred to the first filament of the similar length in the following series.
Diameter measurements were performed using digital transverse cross-sections obtained from confocal stacks (n = 9 series) for each time point at six different positions corresponding to the top and bottom of five equal portions of the filament. Maximal locule area measurements were performed using digital longitudinal cross-sections for each time point (n = 6 locules from 3 series). All measurements were carried out in Fiji software (Schindelin et al., 2012). Cell surface extension in the lobes was measured by calculating area extension of the abaxial epidermal sector corresponding to the lobes, which was determined using data for growth and curvature in the anthers.
All figures were assembled using Adobe Photoshop software.
Quantification and statistical analysis
Plots in Figure 4, Figure 7, D–F, and Supplemental Figure S2 were generated using Python scripts, and in Figures 5, 6, 7G, and 8 using Microsoft Excel. For box plots, the lines represent median and dashed lines represent the mean of at least three independent time-lapse series (n > 55 cells). The violin plot outlines represent the proportion of the data located in specific positions; violin plots contain 95% and boxplots 90% of the values, respectively. Series starting from different stages were aligned according to the developmental stages and size prior to plotting. Parameters quantified as a function of distance from the filament base were plotted in line charts with data markers representing the mean value calculated for each one of the five regions representing 20% of filament length. The alignment graph of filament diameter was plotted per region for each of the six regions measured. Error bars in line charts represent the standard error of the mean (n = 9 for Figure 8; n = 6 for Figure 5) per region per time point. Relative contributions of different segments of the early filament to the final length were represented in a stacked bar plot per time point for the representative sample. The precise number of how many samples were analyzed (n) are indicated in figure legends.
Data availability
The data used to extract growth are available to download from the Open Science Framework repository (https://osf.io/95bn7/?view_only=64a5d538c1584c558004c86d2ab5cd81).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The specification of anther and filament occurs 4 days after stamen primordia initiation.
Supplemental Figure S2. Increase in cell size along the proximodistal axis underlies late filament elongation.
Supplementary Material
Acknowledgments
We thank George Bassel for discussions and Dorota Kwiatkowska, Richard Smith, and Viraj Alimchandani for critical reading of the manuscript.
Funding
This work was supported by the New Frontiers in Research Fund Exploration grant (no. NFRFE-2018-00953) from the Government of Canada, and Discovery grants (nos. RGPIN-2018-05762 and RGPIN-2018-04897) from the Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. Authors declare no conflict of interest.
S.R.S. performed all experiments with the help from A.G.F.; S.R.S. and D.K. extracted data from time-lapse series with the help from A.-L.R.-K.; S.R.S. and C.L.G. analyzed the data; D.K. conceived the study and supervised the project. S.R.S., D.K., and A.-L.R.-K. wrote the manuscript with input from A.G.F. and C.L.G.; D.K. and A.-L.R.-K. provided the funding.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Daniel Kierzkowski (daniel.kierzkowski@umontreal.ca).
References
- Alvarez JP, Furumizu C, Efroni I, Eshed Y, Bowman JL (2016) Active suppression of a leaf meristem orchestrates determinate leaf growth. Elife 5: e15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, DeBodt S, Vanhaeren H, Coppens F, DeMilde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78. [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Routier-Kierzkowska A-L, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, et al. (2015) MorphoGraphX: a platform for quantifying morphogenesis in 4D. Elife 4: 05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Smith RS (2016) Quantifying morphogenesis in plants in 4D. Curr Opin Plant Biol 29: 87–94 [DOI] [PubMed] [Google Scholar]
- Bassel GW, Stamm P, Mosca G, De Reuille PB, Gibbs DJ, Winter R, Janka A, Holdsworth MJ, Smith RS (2014) Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc Natl Acad Sci U S A 111: 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger G, Smyth DR (1996) Initiation patterns of flower and floral organ development in Arabidopsis thaliana. Development 122: 1093–1102 [DOI] [PubMed] [Google Scholar]
- Bou Daher F, Chen Y, Bozorg B, Clough J, Jönsson H, Braybrook SA (2018) Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. Elife 7: e38161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian A, Barbier de Reuille P, Kuhlemeier C (2016) Patterns of stem cell divisions contribute to plant longevity. Curr Biol 26: 1385–1394 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Jacobs B, Cole M, Comelli P, Werr W (2011) DORNRÖSCHEN-LIKE expression marks Arabidopsis floral organ founder cells and precedes auxin response maxima. Plant Mol Biol 76: 171–185 [DOI] [PubMed] [Google Scholar]
- Coen E, Kennaway R, Whitewoods C (2017) On genes and form. Development 144: 4203–4213 [DOI] [PubMed] [Google Scholar]
- Coen E, Rolland-Lagan A-G, Matthews M, Bangham JA, Prusinkiewicz P (2004) The genetics of geometry. Proc Natl Acad Sci U S A 101: 4728–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF (2006) NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133: 1645–55 [DOI] [PubMed] [Google Scholar]
- Dong Y, Majda M, Šimura J, Horvath R, Srivastava AK, Łangowski Ł, Eldridge T, Stacey N, Slotte T, Sadanandom A, et al. (2020) HEARTBREAK controls post-translational modification of INDEHISCENT to regulate fruit morphology in Capsella. Curr Biol 30: 3880–3888.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevin E, Le Gloanec C, Skowrońska N, Routier-Kierzkowska A-L, Burian A, Kierzkowski D (2019) Growth and biomechanics of shoot organs. J Exp Bot 70: 3573–3585 [DOI] [PubMed] [Google Scholar]
- Eldridge T, Łangowski Ł, Stacey N, Jantzen F, Moubayidin L, Sicard A, Southam P, Kennaway R, Lenhard M, Coen ES, et al. (2016) Fruit shape diversity in the Brassicaceae is generated by varying patterns of anisotropy. Development 143: 3394–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschweiler D, Spina TV, Choudhury RC, Meyerowitz E, Cunha A, Stegmaier J (2019) CNN-based preprocessing to optimize watershed-based cell segmentation in 3D confocal microscopy images. In Proceedings of International Symposium on Biomedical Imaging (ISBI 2019). IEEE Computer Society, Piscataway, NJ, pp. 223–227
- Fox S, Southam P, Pantin F, Kennaway R, Robinson S, Castorina G, Sánchez-Corrales YE, Sablowski R, Chan J, Grieneisen V, et al. (2018) Spatiotemporal coordination of cell division and growth during organ morphogenesis. PLoS Biol 16: e2005952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders SM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KS, Lord EM (1988) Growth of anthers in Lilium longiflorum—a kinematic analysis. Planta 173: 161–171 [DOI] [PubMed] [Google Scholar]
- Grandjean O, Vernoux T, Laufs P, Belcram K, Mizukami Y, Traas J (2004) In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16: 74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PB, Erickson RO, Richmond PA (1970) On the physical basis of cell morphogenesis. Ann NY Acad Sci 175: 712–731 [Google Scholar]
- Greyson RI, Tepfer SS (1966) An analysis of stamen filament growth of Nigella hispanica. Am J Bot 53: 485–490 [Google Scholar]
- Hervieux N, Dumond M, Sapala A, Routier-Kierzkowska A-L, Kierzkowski D, Roeder AHK, Smith RS, Boudaoud A, Hamant O (2016) A mechanical feedback restricts sepal growth and shape in Arabidopsis. Curr Biol 26: 1–10 [DOI] [PubMed] [Google Scholar]
- Hess K, Morré DJ (1978) Fine structural analysis of the elongation zone of Easter Lily (Lilium longiflorum) staminal filaments. Bot Gaz 139: 312–321 [Google Scholar]
- Hong L, Dumond M, Tsugawa S, Sapala A, Routier-Kierzkowska AL, Zhou Y, Chen C, Kiss A, Zhu M, Hamant O, et al. (2016) Variable cell growth yields reproducible organ development through spatiotemporal averaging. Dev Cell 38: 15–32 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang T, Linstroth L, Tillman Z, Otegui MS, Owen HA, Zhao D (2016) Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet 12: e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Dengler N (2002) Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216: 212–219 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D, Runions A, Vuolo F, Strauss S, Lymbouridou R, Routier-Kierzkowska A-L, Wilson-Sánchez D, Jenke H, Galinha C, Mosca G. et al. (2019) A Growth-Based Framework for Leaf Shape Development and Diversity. Cell 177: 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska D (2006) Flower primordium formation at the Arabidopsis shoot apex: quantitative analysis of surface geometry and growth. J Exp Bot 57: 571–580 [DOI] [PubMed] [Google Scholar]
- Lobello G, Fambrini M, Baraldi R, Lercari B, Pugliesi C (2000) Hormonal influence on photocontrol of the protandry in the genus Helianthus. J Exp Bot 51: 1403–1412 [PubMed] [Google Scholar]
- Ma H (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Peaucelle A, Wightman R, Höfte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol 25: 1746–1752 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-López R, Alvarez-Buylla ER, Yanofsky MF (2001) Conversion of leaves into petals in Arabidopsis. Curr Biol 11: 182–184 [DOI] [PubMed] [Google Scholar]
- Pelletier S, Van Orden J, Wolf S, Vissenberg K, Delacourt J, Ndong YA, Pelloux J, Bischoff V, Urbain A, Mouille G, et al. (2010) A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol 188: 726–739 [DOI] [PubMed] [Google Scholar]
- Rebocho AB, Kennaway JR, Bangham JA, Coen E (2017) Formation and shaping of the Antirrhinum flower through modulation of the CUP boundary gene. Curr Biol 27: 2610–2622.e3 [DOI] [PubMed] [Google Scholar]
- Reed JW, Wu MF, Reeves PH, Hodgens C, Yadav V, Hayes S, Pierik R (2018) Three auxin response factors promote hypocotyl elongation. Plant Physiol 178: 864–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refrégier G, Pelletier S, Jaillard D, Höfte H (2004) Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol 135: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll JJ, Zhu M, Brocke S, Hon CT, Yanofsky MF, Boudaoud A, Roeder AHK (2019) Growth dynamics of the Arabidopsis fruit is mediated by cell expansion. Proc Natl Acad Sci U S A 116: 25333–25342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland-Lagan A-G, Bangham JA, Coen E (2003) Growth dynamics underlying petal shape and asymmetry. Nature 422: 161–163 [DOI] [PubMed] [Google Scholar]
- Sablowski R (2015) Control of patterning, growth, and differentiation by floral organ identity genes. J Exp Bot 66: 1065–1073 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Sapala A, Runions A, Routier-Kierzkowska AL, Das Gupta M, Hong L, Hofhuis H, Verger S, Mosca G, Li CB, Hay A, et al. (2018) Why plants make puzzle cells, and how their shape emerges. Elife 7: e32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E (2013) JAGGED controls arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol 11: e1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K, Kausika S, Southam P, Bush M, Sablowski R (2012) JAGGED controls growth anisotropy and coordination between cell size and cell cycle during plant organogenesis. Curr Biol 22: 1739–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk WK, Lord EM, Eckard KJ (1989) Growth patterns inferred from anatomical records. Plant Physiol 90: 708–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solly JE, Cunniffe NJ, Harrison CJ (2017) Regional growth rate differences specified by apical notch activities regulate liverwort thallus shape. Curr Biol 27: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss S, Sapala A, Kierzkowski D, Smith RS (2019) Quantifying plant growth and cell proliferation with MorphoGraphX. Methods Mol Biol 1992: 269–290 [DOI] [PubMed] [Google Scholar]
- Tauriello G, Meyer HM, Smith RS, Koumoutsakos P, Roeder AHK (2015) Variability and constancy in cellular growth of Arabidopsis sepals. Plant Physiol 169: 2342–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa S, Hervieux N, Kierzkowski D, Routier-Kierzkoswska A-L, Sapala A, Hamant O, Smith RS, Roeder AHK, Boudaoud A, Li C-B (2017) Cells from the same lineage switch from reduction to enhancement of size variability in Arabidopsis sepals. Development 144: 4398–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valuchova S, Mikulkova P, Pecinkova J, Klimova J, Krumnikl M, Bainar P, Heckmann S, Tomancak P, Riha K (2020) Imaging plant germline differentiation within Arabidopsis flowers by light sheet microscopy. Elife 9: e52546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan A, Tofanelli R, Strauss S, Cerrone L, Wolny A, Strohmeier J, Kreshuk A, Hamprecht FA, Smith RS, Schneitz K (2021) A digital 3D reference atlas reveals cellular growth patterns shaping the Arabidopsis ovule. Elife 10: e63262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuolo F, Kierzkowski D, Runions A, Hajheidari M, Mentink RA, Gupta MD, Zhang Z, Vlad D, Wang Y, Pecinka A, et al. (2018) LMI1 homeodomain protein regulates organ proportions by spatial modulation of endoreduplication. Genes Dev 32: 1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V, Egger RL (2016) Pre-Meiotic anther development: cell fate specification and differentiation. Annu Rev Plant Biol 67: 365–395 [DOI] [PubMed] [Google Scholar]
- Wei D, Liu M, Chen H, Zheng Y, Liu Y, Wang X, Yang S, Zhou M, Lin J (2018) INDUCER OF CBF EXPRESSION 1 is a male fertility regulator impacting anther dehydration in Arabidopsis. PLoS Genet 14: e1007695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitewoods CD, Gonçalves B, Cheng J, Cui M, Kennaway R, Lee K, Bushell C, Yu M, Piao C, Coen E (2020) Evolution of carnivorous traps from planar leaves through simple shifts in gene expression. Science 367: 91–96 [DOI] [PubMed] [Google Scholar]
- Xue JS, Yao C, Xu QL, Sui CX, Jia XL, Hu WJ, Lv YL, Feng YF, Peng YJ, Shen SY, et al. (2021) Development of the middle layer in the anther of Arabidopsis. Front Plant Sci 12: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Schuster C, Beahan CT, Charoensawan V, Peaucelle A, Bacic A, Doblin MS, Wightman R, Meyerowitz EM (2016) Regulation of meristem morphogenesis by cell wall synthases in Arabidopsis. Curr Biol 26: 1404–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, McEvoy D, Le Y, Ambrose C (2021) Live imaging of microtubule organization, cell expansion, and intercellular space formation in Arabidopsis leaf spongy mesophyll cells. Plant Cell 33: 623–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Runions A, Mentink RA, Kierzkowski D, Karady M, Hashemi B, Huijser P, Strauss S, Gan X, Ljung K, et al. (2020) A WOX/Auxin biosynthesis module controls growth to shape leaf form. Curr Biol 30: 4857–4868. e6 [DOI] [PubMed] [Google Scholar]
- Zhao D (2009) Control of anther cell differentiation: a teamwork of receptor-like kinases. Sex Plant Reprod 22: 221–228 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wang D, Ye S, Chen W, Li G, Xu Z, Bai S, Zhao F (2021) Auxin guides germ cell specification in Arabidopsis anthers. Proc Natl Acad Sci U S A 118: e2101492118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen W, Mirabet V, Hong L, Bovio S, Strauss S, Schwarz EM, Tsugawa S, Wang Z, Smith RS, et al. (2020) Robust organ size requires robust timing of initiation orchestrated by focused auxin and cytokinin signalling. Nat Plants 6: 686–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to extract growth are available to download from the Open Science Framework repository (https://osf.io/95bn7/?view_only=64a5d538c1584c558004c86d2ab5cd81).