Abstract
In Angiosperms, the development of the vascular system is controlled by a complex network of transcription factors. However, how nutrient availability in the vascular cells affects their development remains to be addressed. At the cellular level, cytosolic sugar availability is regulated mainly by sugar exchanges at the tonoplast through active and/or facilitated transport. In Arabidopsis (Arabidopsis thaliana), among the genes encoding tonoplastic transporters, SUGAR WILL EVENTUALLY BE EXPORTED TRANSPORTER 16 (SWEET16) and SWEET17 expression has been previously detected in the vascular system. Here, using a reverse genetics approach, we propose that sugar exchanges at the tonoplast, regulated by SWEET16, are important for xylem cell division as revealed in particular by the decreased number of xylem cells in the swt16 mutant and the accumulation of SWEET16 at the procambium–xylem boundary. In addition, we demonstrate that transport of hexoses mediated by SWEET16 and/or SWEET17 is required to sustain the formation of the xylem secondary cell wall. This result is in line with a defect in the xylem cell wall composition as measured by Fourier-transformed infrared spectroscopy in the swt16swt17 double mutant and by upregulation of several genes involved in secondary cell wall synthesis. Our work therefore supports a model in which xylem development partially depends on the exchange of hexoses at the tonoplast of xylem-forming cells.
ylem development and secondary cell wall formation partially depend on the control of cytosolic sugar availability, which is regulated by tonoplastic sugar transporters.
Introduction
The plant vasculature, composed of phloem, procambium/cambium, and xylem, is an elaborate system responsible for the transport of most biological compounds throughout the plant (Lucas et al., 2013). At the molecular level, vasculature development is governed by a complex network of transcription factors that are under the control of several signals, including hormones, peptides, and microRNAs (Fukuda and Ohashi-Ito, 2019; Smit et al., 2019). However, within this well-organized framework, a certain plasticity is required to adjust to cellular variations in terms of the availability of nutrients (i.e. sugars and amino acids).
Sugars, which represent the main source of energy, are required for metabolic activities, and they serve as a carbon reserve, and as intermediates for the synthesis of cell wall polysaccharides. Additionally, they have morphogenetic activity and act as primary messengers in signal transduction pathways (Sakr et al., 2018). It is therefore logical that modifications of sugar metabolism, transport or signaling can lead to multiple defects in plant growth and development (Eveland and Jackson, 2012). However, despite this central role, the role of sugar availability in the development of the vascular system in general and more specifically in heterotrophic tissues such as cambium and xylem is still elusive.
In these tissues, it has been suggested that lateral transport of sugars, coming from leakages from phloem sieve tubes, provides the sugars needed for vascular cell development (Minchin and McNaughton, 1987; Sibout et al., 2008; Spicer, 2014; Furze et al., 2018). Lateral transport is especially crucial for xylem secondary cell wall formation, since sugars are intermediate compounds in the synthesis of the cell wall polysaccharides, which represent 80% of the secondary cell wall (Marriott et al., 2016; Verbančič et al., 2018). The xylem tissue thus represents a strong sink for sugars that must be imported from surrounding tissues to serve as the source of carbon and energy. This is supported by the fact that perturbations in sugar transport at the plasma membrane of vascular cells, via SUGAR WILL EVENTUALLY BE EXPORTED TRANSPORTERS (SWEET) or SUCROSE TRANSPORTERS (SUC/SUT), affect the composition of the xylem secondary cell wall both in aspen (Populus tremuloides) and in Arabidopsis (Arabidopsis thaliana) inflorescence stems (Mahboubi et al., 2013; Le Hir et al., 2015). Furthermore, in the Arabidopsis inflorescence stem, it has been suggested that movements of sucrose and/or hexoses towards the apoplast, mediated by SWEET11 and SWEET12, occur between the vascular parenchyma cells and the developing conducting cells to drive cell wall formation in a cell-specific manner (Dinant et al., 2019). Intercellular sugar availability seems, therefore, to play an important role in xylem development. However, the question remains open as to whether the modification of sugar partitioning within the vasculature cells is also of importance.
The vacuole represents the main storage compartment for numerous primary and specialized metabolites including sugars (Martinoia, 2018). In tobacco (Nicotiana tabacum) leaves, up to 98% of hexoses are found in the vacuole (Heineke et al., 1994). In Arabidopsis leaves compartmentation of sugars is different. Sucrose is mostly present in the cytosol and the plastids while glucose and fructose are mostly found in the vacuole (Weiszmann et al., 2018). Sugar exchanges between the vacuole and the cytosol are therefore required for dynamic adjustment of the quantity of sugar needed for metabolic and signaling pathways. In herbaceous and ligneous plants, few sugar transporters have been functionally characterized at the tonoplast (Hedrich et al., 2015), and localization in the cells of the vascular system has been shown only for SUCROSE TRANSPORTER 4 (SUC4/SUT4), EARLY RESPONSE TO DEHYDRATION SIX-LIKE 1 (ESL1), SWEET16, and SWEET17 (Yamada et al., 2010; Payyavula et al., 2011; Chardon et al., 2013; Klemens et al., 2013). In Populus, sugar export mediated by the tonoplastic sucrose symporter PtaSUT4 is required for carbon partitioning between the source leaves and the lateral sinks (e.g. xylem; Payyavula et al., 2011). In Arabidopsis, SWEET16 and SWEET17 transporters were localized in the roots (Guo et al., 2014) and we showed that the SWEET16 promoter is active in the xylem parenchyma cells (Klemens et al., 2013), while the SWEET17 promoter is active in the xylem parenchyma cells and young xylem cells of the Arabidopsis inflorescence stem (Chardon et al., 2013). Moreover high levels of SWEET17 transcripts have been measured in the inflorescence stem, compared to other organs including roots, after 7 to 8 weeks of growth (Guo et al., 2014). SWEET16 and SWEET17 are therefore good candidates with which to assess whether the maintenance of sugar homeostasis between the cytosol and the vacuole influences xylem development in Arabidopsis.
In the present work, through a reverse genetic approach, we demonstrate that SWEET16 and SWEET17 have specific and overlapping roles during xylem development. In particular, we suggest that tonoplastic sugar exchanges across the procambium–xylem boundary regulated by SWEET16 are important for xylem cell proliferation. By using infrared spectroscopy and gene expression analysis, we also show that both SWEET16 and SWEET17 are required for correct development of the secondary cell wall of xylem cells. Finally, since glucose and fructose accumulation are observed in the inflorescence stem of the double mutant, we suggest that maintenance of hexose homeostasis through the action of SWEET16 and/or SWEET17 is important at different stages of xylem development.
Results
Radial growth of the inflorescence stem is altered in the swt16swt17 double mutant
To explore to what extent mutations in SWEET16 and/or SWEET17 impact inflorescence stem development in Arabidopsis, we used the previously described sweet17-1 (hereafter called swt17) mutant line (Chardon et al., 2013) and identified two new T-DNA insertion lines in the SWEET16 gene. The mutants sweet16-3 (SM_3.1827) and sweet16-4 (SM_3.30075) possess an insertion in the promoter region and in the first exon of the SWEET16 gene respectively (Supplemental Figure S1, A). They were named after the previous alleles already published (Guo et al., 2014). A full-length SWEET16 transcript was detected by reverse transcription polymerase chain reaction (RT-PCR) in the sweet16-3 mutant, while no full-length transcript could be detected in the sweet16-4 mutant (Supplemental Figure S1, B and C). The sweet16-4 allele (hereafter called swt16) was therefore deemed to be a null allele. We generated the double mutant sweet16sweet17 (hereafter called swt16swt17) and confirmed by reverse transcription quantitative polymerase chain reaction (RT-qPCR) that both full-length genes were absent in this double mutant (Supplemental Figure S1, C).
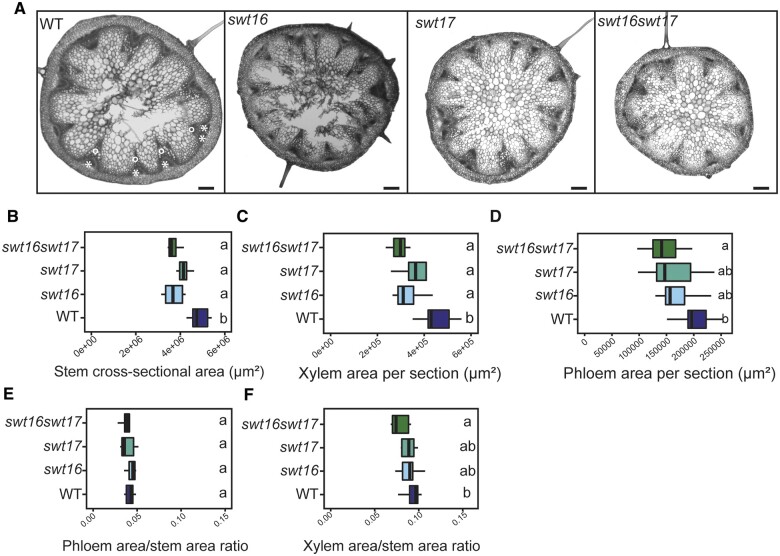
Analysis of the inflorescence stems of the swt16, swt17, and swt16swt17 mutants showed that the area of stem cross-sections was significantly smaller compared to that of the wild-type (Figure 1, A and B). More precisely, the stem of all mutant lines contained less xylem tissue compared with the wild-type, while only the stem of swt16swt17 displayed significantly less phloem tissue (Figure 1, C and D). Additionally, the proportion of xylem or phloem per stem was calculated (Figure 1, E and F). While no change in the proportion of phloem was observed in the mutants compared to the wild-type (Figure 1, E), a significant reduction in the xylem proportion was observed in the double swt16swt17 mutant (Figure 1, F).
Figure 1.
Altered development of the inflorescence stem in the swt16swt17 double mutant. A, Transverse sections of the basal part of the inflorescence stem of 7-week-old plants stained with FASGA solution. Stars indicate the phloem tissue while circles indicate the xylem tissue. Bars = 200 µm. B–F, Boxplots showing the inflorescence stem cross-sectional area (B), the area occupied by xylem tissue (C) or phloem tissue (D) within a stem section, the ratio of xylem area to stem area (E), and the ratio of phloem area to stem area (F). The box and whisker plots represent values from seven to eight independent plants. The lines represent median values, the tops and bottoms of the boxes represent the first and third quartiles respectively, and whisker extremities represent maximum and minimum data points. A one-way analysis of variance combined with the Tukey’s comparison post hoc test were performed. The values marked with the same letter were not significantly different from each other, whereas different letters indicate significant differences (P < 0.05).
To further assess the involvement of SWEET16 and/or SWEET17 in the radial growth, constructs containing N-ter GFP-fused SWEET16 and/or SWEET17 genomic DNA driven by their native promoter were introduced into the swt16, swt17, and swt16swt17 mutants. The GFP-SWEET17 construct successfully complemented the phenotype of the swt17 mutant (Supplemental Figure S2, A), while GFP-SWEET16 only partially complemented the stem phenotype of the swt16 mutant (Supplemental Figure S2, A). However, full complementation of the double mutant swt16swt17 was achieved when both translational GFP fusions were expressed (Supplemental Figure S2, A).
Altogether, our results show that both SWEET16 and SWEET17 transporters are required for proper radial growth of the stem by affecting the vascular system development.
SWEET16 and SWEET17 proteins interact physically and are expressed in the xylem during its early development
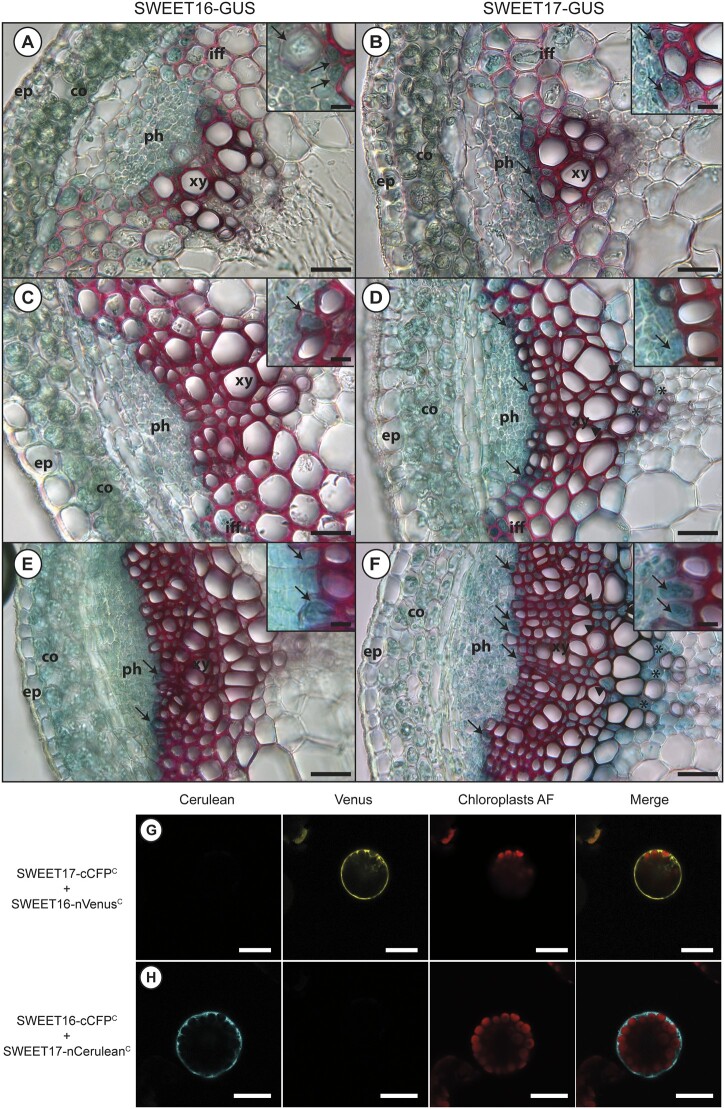
Previously, using lines expressing pSWT:GUS transcriptional fusions, we showed that SWEET16 and SWEET17 were expressed in the xylem tissue of petioles and inflorescence stems (Chardon et al., 2013; Klemens et al., 2013). To confirm the presence of SWEET16 and SWEET17 proteins in the inflorescence stem we analyzed the N-ter translational GFP fusions that complemented the mutant’s phenotype (Supplemental Figure S2, A). Unfortunately, despite the phenotype complementation, we could not detect any GFP signal in these lines. Nonetheless, we obtained lines from Guo et al. (2014) expressing translational fusions between GUS and the C-terminus of SWEET16 or SWEET17 coding sequences under the control of their respective native promoter in a wild-type background. We analyzed the expression in three different zones: (1) a stem region where the growth was rapid, (2) a stem region where elongation growth had finished but where further thickening of the secondary cell wall was still ongoing, and (3) the base of the stem, which corresponds to a mature zone (Hall and Ellis, 2013; Figure 2).
Figure 2.
SWEET16 and SWEET17 are expressed in xylem cells throughout inflorescence stem development and form heterodimers. A, C, and E, Histochemical analysis of GUS activity in lines expressing SWEET16-GUS fusion proteins driven by the SWEET16 native promoter in sections taken at different positions in the inflorescence stem of 7-week-old plants. B, D, and F, Histochemical analysis of GUS activity in lines expressing SWEET17-GUS fusion proteins driven by the SWEET17 native promoter in sections taken at different positions in the inflorescence stem section of 7-week-old plants. Sections were taken in a stem region where the growth was still rapid (A, B, and insets), in a stem region where elongation growth had finished but where thickening of the secondary cell wall was still ongoing (C, D, and insets), and at the bottom of the stem, a region that corresponds to a mature stem (E, F, and insets). Arrows point to cells showing blue GUS staining in developing xylem cells and arrow heads point to axial parenchyma cells and asterisks indicate xylary parenchyma cells. Lignin is colored pink after phloroglucinol staining. The intensity of the pink color indicates the level of lignification of the xylary vessels. ep, epidermis; co, cortex; iff, interfascicular fibers; ph, phloem; xy, xylem. G, Arabidopsis mesophyll protoplast expressing SWEET17-cCFPC and SWEET16-nVenusC interaction revealed by false color yellow, chloroplast auto-fluorescence is in false color red. H, Arabidopsis mesophyll protoplast expressing SWEET16-cCFPC and SWEET17-nCeruleanC interaction revealed by false color cyan, chloroplast auto-fluorescence is in false color red. Invaginations around the chloroplasts in (G) and (H) indicate that SWEET16 and SWEET17 interact at the vacuolar membrane. Scale bar = 50 µm (A–F) or 25 µm (insets in A–F) or 20 µm (G and H).
In the region where the stem was growing rapidly SWEET16 and SWEET17 proteins are expressed in the cortex, the phloem cells, the interfascicular fibers, and in the developing xylem cells (Figure 2, A and B and insets). In the phloem tissue whether SWEET16 and SWEET17 were expressed in the phloem parenchyma cells and/or the companion cells will need to be further addressed by using for instance translational GFP fusions (Guo et al., 2014). In the region where secondary cell wall thickening was still ongoing and in the mature stem, SWEET16 and SWEET17 were found to be expressed in the cortex and across the phloem–procambium–xylem region (Figure 2, C and F). The expression of SWEET16 and SWEET17 was also observed in young xylem cells, that could be developing xylary fibers and/or in developing xylem vessels, before extensive cell wall lignification had occurred, as shown by the weaker phloroglucinol cell wall staining (insets in Figure 2, C–F). Additionally, expression of SWEET17 was observed in xylem cells showing a lignified cell wall and situated close to the xylem vessels (Figure 2, D and F). Based on their localization in the middle of the vascular bundle, this cell type is more likely to be axial parenchyma cells (Turner and Sieburth, 2002; Baghdady et al., 2006) than developing xylary fibers. Finally, SWEET17 was also expressed in xylary parenchyma cells situated at the bottom of the vascular bundle (Berthet et al., 2011; Figure 2, D and F). In addition, we also verified the expression pattern of pSWEET16:GUS and pSWEET17:GUS, used to generate the translational GFP fusions, which contain shorter promoters (1,295 bp for SWEET16 and 2,004 bp for SWEET17) than the ones used for the translational GUS fusions (1,521 bp for SWEET16 and 2,746 bp for SWEET17; Supplemental Figure S2, B–G). Overall we observed that the expression pattern obtained with the pSWT:GUS transcriptional fusions is included in that of the translational SWT:GUS fusions (Supplemental Figure S2, B–G). In conclusion, SWEET16 and SWEET17 expression patterns overlap in cortex cells as well as in the phloem–procambium–xylem region and in the young xylem cells, whereas SWEET17 is specifically expressed in the xylem parenchyma cells.
It has been established that plant sugar SWEET transporters require homo or hetero-oligomerization to gain functionality (Xuan et al., 2013). Because SWEET16 and SWEET17 expression patterns overlap in several cells types of the inflorescence stem, we investigated whether these proteins could interact. Xuan et al. (2013) previously showed that SWEET16 and SWEET17 can form homodimers as well as heterodimers in a split ubiquitin yeast two-hybrid assay. We confirmed that SWEET16 and SWEET17 can form a heterodimer at the vacuolar membrane in a bimolecular fluorescence complementation assay in Arabidopsis mesophyll protoplasts (Figure 2, G–H; Supplemental Figure S3).
SWEET16 but not SWEET17 is required for proliferation of xylem cells
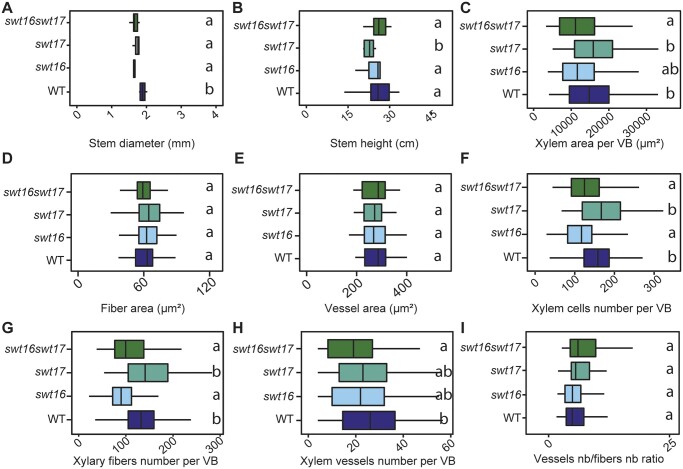
The localization of SWEET16 and SWEET17 in the young xylem cells prompted us to further analyzed the phenotype of the xylem tissue (Figure 3). On an independent set of plants, we first checked the robustness of the inflorescence stem phenotype and confirmed that the swt16 and swt17 single mutants and the swt16swt17 double mutant consistently displayed a significantly thinner inflorescence stem compared to the wild-type (Figure 3, A). In addition, we confirmed our previous results that showed a significantly shorter inflorescence stem in the swt17 mutant compared to the wild-type (Figure 3, B;Chardon et al., 2013). Interestingly, we did not observed any alteration in the inflorescence stem height in swt16 or swt16swt17 plants compared to the wild-type (Figure 3, B). This suggests a compensation by other sugar transporters yet to be identified in the absence of SWEET16.
Figure 3.
Knockout of SWEET16 gene expression impacts the proliferation of xylem cells. A–I, Boxplots showing the inflorescence stem height (A) and diameter (B), the cross-sectional area occupied by xylem tissue per vascular bundle (C), the average cross-sectional area of a xylary fiber (D) or of a xylem vessel (E), and the average number of xylem cells (F), of xylary fiber vessels (G), of xylem vessels (H) per vascular bundle and the ratio of vessel number to fiber number (I). The box and whisker plots represent values from five to seven independent plants (A and B) or from 71, 53, 41, and 50 individual vascular bundles from the wild-type, swt16, swt17, and swt16swt17 plants, respectively, coming from five to seven independent plants for each genotype (C–I). The lines represent median values, the tops and bottoms of the boxes represent the first and third quartiles, respectively, and whisker extremities represent maximum and minimum data points. A one-way analysis of variance combined with the Tukey’s comparison post hoc test was performed. The values marked with the same letter were not significantly different from each other, whereas different letters indicate significant differences (P < 0.05).
The xylem phenotype was then studied in more detail by counting the number of xylary fibers (cells with an area of between 5 and 150 µm2) and xylem vessels (cells with an area greater than 150 µm2) as well as measuring the individual cross-sectional areas within each vascular bundle (Figure 3, C–I). In the vascular bundles of the swt16 and swt16swt17 mutants, but not swt17, the area occupied by the xylem tissue was significantly smaller than in the wild-type (Figure 3, C). These changes could result from modification of either cell size or cell number. While no changes in the size of the xylary fibers or the xylem vessels were observed in any of the genotypes analyzed (Figure 3, D and E), the total number of xylem cells per vascular bundle was significantly reduced, by about 20%, in the single mutant swt16 and the double mutant swt16swt17 but not in swt17 (Figure 3, F). The numbers of xylary fibers and xylem vessels per vascular bundle were significantly reduced in the stem of the swt16 single and swt16swt17 double mutant but not in the swt17 mutant (Figure 3, G and H). The decreased number of xylary fibers was proportional to that of xylem vessels since the vessels-to-fibers ratio was the same in the wild-type and the swt16, swt17, and swt16swt17 mutant lines (Figure 3, I).
Overall, these results show that the single swt16 mutant and the swt16swt17 double mutant have the same phenotype (Figure 3 andSupplemental Table S1) and suggest that the expression of SWEET16, but not that of SWEET17, is required for correct division of xylem cells.
SWEET16 and SWEET17 are required for normal secondary cell wall composition and development in xylem cells
To explore whether modifications in the vacuolar transport of sugars impact the formation of the xylem cell wall, we first exploited the transcriptomic dataset obtained from plants overexpressing a dexamethasone (DEX)-inducible version of the VASCULAR RELATED NAC-DOMAIN PROTEIN 7 (VND7) gene, the master secondary wall-inducing transcription factor (Li et al., 2016). The VND7-VP16-GR plants allow the transcriptional and metabolic changes occurring during secondary cell wall formation to be studied. From the RNA-sequencing data set, we extracted information related to the expression of the family of SWEET genes at different time points after induction of VND7 expression (Supplemental Figure S4). Out of the 17 SWEET genes identified in Arabidopsis, 7 were differentially expressed during secondary cell wall formation (Supplemental Figure S4). Most interestingly, the vacuolar SWEET2 and SWEET17 were upregulated 3 h after DEX induction while SWEET16 expression was upregulated 12 hours after DEX induction (Supplemental Figure S4). In contrast, the expression of genes encoding the plasma membrane localized SWEET transporters (e.g. SWEET1, SWEET3, SWEET11, and SWEET12) was downregulated during secondary cell wall formation (Supplemental Figure S4). Additional analysis of the dataset showed that SWEET2 and SWEET17 were co-regulated with genes related to cell wall synthesis, namely CELLULOSE SYNTHASE (CESA), SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 2 (SND2), SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 (SND3), and MYB DOMAIN PROTEIN 46 (MYB46) as well as those encoding other sugar transporters localized at the tonoplast (ESL1) or at the plasma membrane SUGAR TRANSPORTER 1 (STP1), SUGAR TRANSPORTER 13 (STP13), and POLYOL/MONOSACCHARIDE TRANSPORTER 4 (PMT4) (Supplemental Table S2). These results support the fact that, in Arabidopsis seedlings, sugar export from the vacuole to the cytosol is ongoing during secondary cell wall formation, most probably to provide sugars to be used as intermediates for cell wall formation. Since VND7 is also expressed in xylem vessels in Arabidopsis inflorescence stems (Shi et al., 2021), we can postulate that similar sugar exchanges involving tonoplastic sugar transporters take place during secondary cell wall formation in this organ.
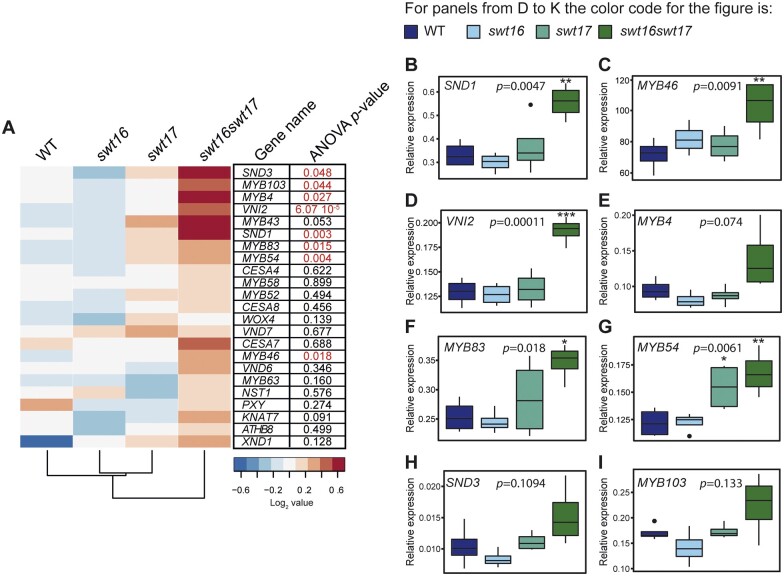
To assess whether SWEET16 and SWEET17 are indeed functionally involved in xylem secondary cell wall formation, we performed a targeted gene expression analysis including genes that are known to be part of the transcriptional network involved in stem cell proliferation/organization: PHLOEM INTERCALATED WITH XYLEM (PXY) or WUSCHEL RELATED HOMEOBOX 4 (WOX4) (Etchells et al., 2013); xylem cell identity: HOMEOBOX GENE 8 (ATHB8) (Smetana et al., 2019); and secondary cell wall biosynthesis in vessels/fibers: CELLULOSE SYNTHASE 4 (CESA4), CESA7, CESA8, KNOTTED-LIKE HOMEOBOX OF ARABIDOPSIS THALIANA 7 (KNAT7), MYB DOMAIN PROTEIN 4 (MYB4), MYB43, MYB46, MYB52, MYB54, MYB58, MYB63, MYB83, MYB103, NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 (NST1), VND6, VND7, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 1 (SND1/NST3), SND3, VND-INTERACTING 2 (VNI2), and XYLEM NAC DOMAIN 1 (XND1) (Hussey et al., 2013; Figure 4, A–I). When looking at the overall transcriptional profile of the wild-type and the swt16, swt17, and swt16swt17 mutants, two clusters can be identified (Figure 4, A). The first cluster contains the wild-type, the swt16 and swt17 single mutants, whereas the second includes only the swt16swt17 double mutant. Only a subset of genes shows significantly increased expression among the different genotypes, namely SND3, MYB103, MYB4, VNI2, SND1, MYB83, MYB54, and MYB46 (Figure 4, A), though a tendency, albeit not significant, is observed for MYB43 (P = 0.053) and KNAT7 (P = 0.091) (Figure 4, A). Interestingly, all these genes belong to the transcriptional network involved in secondary cell wall biosynthesis in xylem vessels and/or in xylary fibers (for review, Hussey et al., 2013). A Student’s t test was then performed to compare each mutant line with the wild-type plants (Figure 4, B–I). On average, a two-fold increase in expression was measured for the genes SND1, MYB46, VNI2, MYB83, and MYB54 in the swt16swt17 double mutant compared to the wild-type (Figure 4, B–D, F and G), while a similar tendency was observed for MYB4, SND3, and MYB103 expression (Figure 4, E, H, and I). Overall, these results show that in the swt16swt17 double mutant neither stem cell maintenance nor xylem identity genes are affected, whereas secondary cell wall biosynthesis genes are deregulated.
Figure 4.
Genes involved in the development of xylem secondary cell walls are differentially regulated in the swt16swt17 double mutant. A–I, mRNAs were extracted from total inflorescence stems collected from 7-week-old wild-type, swt16, swt17, and swt16swt17 plants. The mRNA contents are expressed relative to those of the reference gene UBQ5. A, Heatmap of expression of candidate genes involved in xylem development and secondary cell wall biosynthesis in the inflorescence stem of the wild-type, and swt16, swt17, and swt16swt17 mutants. The values used to build the heatmap are the mean accumulation of transcripts (n=4) normalized by the median value of each gene and expressed as log2 values. For each gene, the result of one-way ANOVA is displayed beside the heatmap. Those with P-values below the significance threshold (P<0.05) are in red. B–I, Boxplots showing the relative expression of SND1 (B), MYB46 (C), VNI2 (D), MYB4 (E), MYB83 (F), MYB54 (G), SND3 (H), and MYB103 (I). The box-and-whisker plots represent values from four biological replicates. The lines represent median values, the tops and bottoms of the boxes represent the first and third quartiles, respectively, and the ends of the whiskers represent maximum and minimum data points. Black dots are outliers. Stars denote significant differences between the mutant line compared to the wild-type according to a one-way ANOVA followed by a Dunnett post hoc test (*P<0.05; **P<0.01; ***P<0.001). The P-values for the comparison between the wild-type and swt16swt17 plants are indicated on each graph. The experiment was repeated twice and gave similar results.
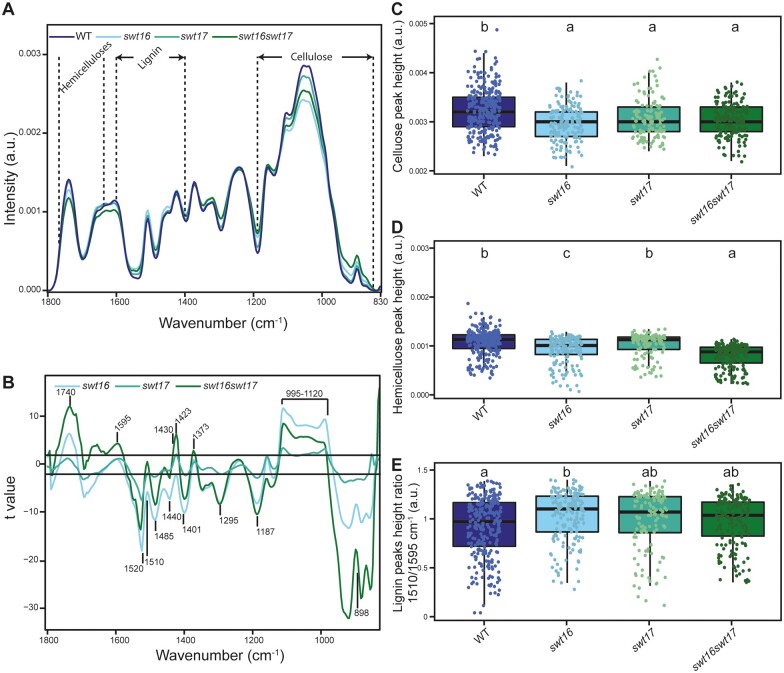
Next, we tested whether this transcriptional deregulation was accompanied by modifications in the cell wall composition. We used Fourier-transformed infrared spectroscopy (FTIR) on inflorescence stem cross-sections to analyze the xylem secondary cell wall composition as previously described in Le Hir et al. (2015; Figure 5). The average spectra for all three mutants showed several differences compared to the wild-type spectra in fingerprint regions associated with cellulose, hemicelluloses, and lignin (Figure 5, A). The t values, plotted against each wavenumber of the spectrum, showed that the mutant lines exhibited several significant negative and positive peaks (higher or lower absorbance than in the wild-type) at wavenumbers associated with cellulosic and hemicellulosic polysaccharides (898 cm−1, 995–1,120 cm−1, 1,187 cm−1, 1,295 cm−1, 1,373 cm−1, 1,401 cm−1, 1,423 cm−1, 1,430 cm−1, 1,440 cm−1, and 1,485 cm−1) (Åkerholm and Salmén, 2001; Kačuráková et al., 2002; Lahlali et al., 2015; Figure 5, A and B). More precisely, wavenumbers at 898 cm−1, associated with the amorphous region of cellulose (Kačuráková et al., 2002), and at 1,430 cm−1, associated with crystalline cellulose (Åkerholm and Salmén, 2001), showed opposite and significant differences (Figure 5, B). This suggests a potential defect in cellulose organization in the xylem secondary cell wall. Measurements of the cellulose C–O vibrations at a peak height at 1,050 cm−1 (Lahlali et al., 2015) further indicate modifications of the cellulose composition in the cell wall of all the mutant lines (Figure 5, C).
Figure 5.
The composition of the xylem secondary cell wall is altered in the swt16swt17 double mutant. FTIR spectra were acquired on xylem tissue from sections of the basal part of the inflorescence stem. All spectra were baseline-corrected and area-normalized in the range 1800–800 cm−1. A, Average FTIR spectra were generated from 266, 170, 123, and 170 spectra for the wild-type, swt16, swt17, and swt16swt17 plants, respectively, obtained using three independent plants for each genotype. B, Comparison of FTIR spectra obtained from xylem cells of the swt16, swt17, and swt16swt17 mutants. A Student’s t test was performed to compare the absorbances for the wild-type, single, and double mutants. The results were plotted against the corresponding wavenumbers. t-Values (vertical axis) between −2 and +2 correspond to nonsignificant differences (P < 0.05) between the genotypes tested (n = 3). t-Values above +2 or below −2 correspond to, respectively, significantly weaker or stronger absorbances in the mutant spectra relative to the wild-type. C–E, Boxplots of the cellulose (C–O vibration band at 1,050 cm−1) (C), hemicellulose (C–O and C–C bond stretching at 1,740 cm−1) (D) peak height and lignin peak height ratio (1,510/1,595 cm−1) (E). The lines represent median values, the tops and bottoms of the boxes represent the first and third quartiles, respectively, and the ends of the whiskers represent maximum and minimum data points. The boxplots represent values (shown as colored dots) from 266, 170, 123, and 170 spectra from the wild-type, swt16, swt17, and swt16swt17 plants, respectively, obtained from three independent plants for each genotype. A one-way analysis of variance combined with the Tukey’s comparison post hoc test was performed. Values marked with the same letter were not significantly different from each other, whereas different letters indicate significant differences (P < 0.05).
The swt16 and swt16swt17 mutants also displayed significant variations compared with the wild-type at 1,740 cm−1 (band specific for acetylated xylan; Gou et al., 2008) and 1,369 cm−1 (deformation of C–H linkages in the methyl group O-acetyl moieties; Mohebby, 2008) suggesting modifications in xylan acetylation (Figure 5, A and B). Furthermore, the hemicellulose peak height at 1740 cm−1 (C–O and C–C bond stretching) was significantly smaller in the swt16 mutant suggesting less acetylated xylan (Figure 5, D). Although the swt17 single mutant was not distinguishable from the wild-type, the swt16swt17 double mutant had significantly fewer acetylated xylans than the wild-type and the swt16 single mutant (Figure 5, D). The lignin-associated bands at 1,510 cm−1 (Faix, 1991), 1,520 cm−1 (Faix, 1991; Gou et al., 2008), and 1,595 cm−1 also exhibited significant differences in the single and double mutants compared to the wild-type plants (Figure 5, B). In addition we measured the lignin height peak ratio (1,510/1,595 cm−1) that can be used as a proxy for G-type lignins, to have a more detailed analysis of FTIR spectra of lignins. G-type lignins are mainly present in the cell wall of xylem vessels while G-type and S-type lignins are present in cell wall of xylary fibers (Schuetz et al., 2012; Öhman et al., 2013). We showed that the secondary cell wall of the swt16 single mutant contains significantly more G-type lignin than that of the wild-type (Figure 5, E). On the other hand, only a tendency for more G-type lignin was measured for the swt17 and the swt16swt17 mutants which suggest that S-type lignin, also present in fibers with G-type lignin (Gorzsás et al., 2011), are responsible for the changes observed in the FTIR profiles (Figure 5, A and B).
Overall, these results suggest that sugar export between the cytosol and the vacuole regulated by SWEET16 and/or SWEET17 is required to provide the intermediates needed for the synthesis of cellulosic and hemicellulosic polysaccharides.
The hexose content is modified in the inflorescence stem of the swt16swt17 double mutant
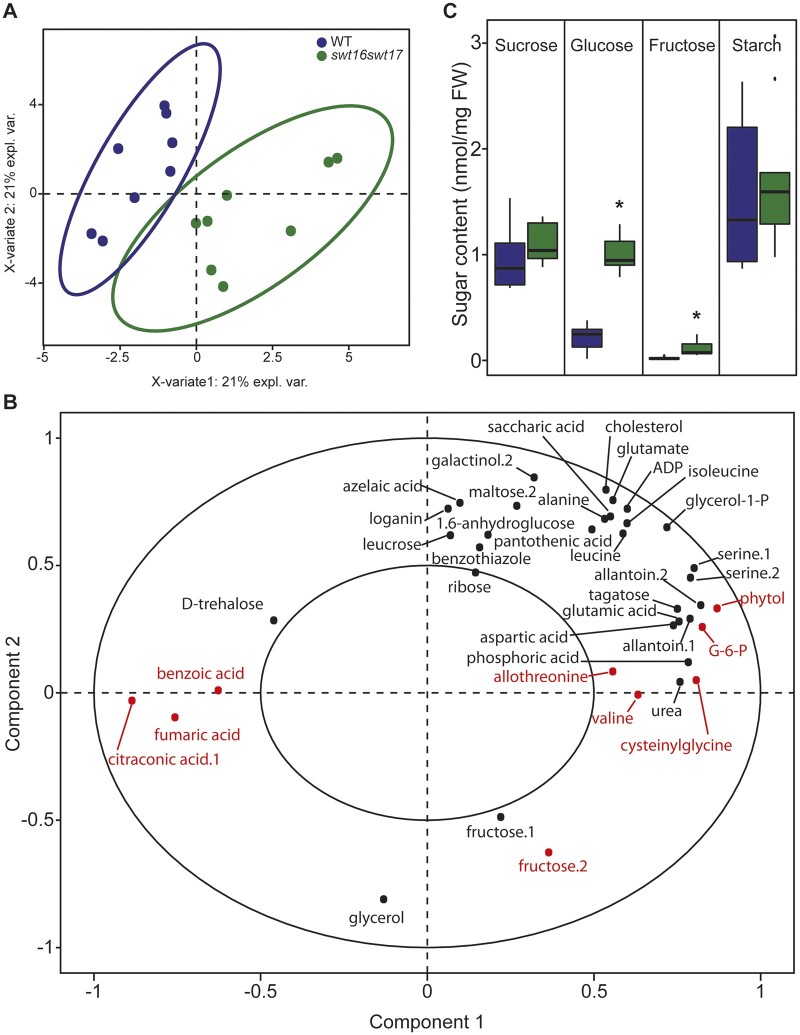
Assuming that SWEET16 and SWEET17 are sugar carriers, we wondered what would be the metabolic status of the inflorescence stem in the swt16swt17 double mutant. We therefore used gas chromatography–mass spectrometry (GC–MS) to explore the global metabolomic profiles of the wild-type and the double swt16swt17 mutant, identifying a total of 158 metabolites. In order to identify the subset of metabolites that best discriminate between the genotypes, we performed a sparse partial-least-squares discriminant analysis (sPLS-DA; Figure 6, A). The resulting score plot clearly allows the two genotypes to be separated by the first dimension (Figure 6, A). Among the metabolites selected by the sPLS-DA analysis, a subsequent t test identified nine that were significantly different between the wild-type and the swt16swt17 double mutant: allothreonine, benzoic acid, citraconic acid, cysteinylglycine, fructose, fumaric acid, glucose-6-phosphate, phytol, and valine (Figure 6, B andSupplemental Table S3). The relative quantities of benzoic acid, citraconic acid, and fumaric acid (a component of the tricarboxylic cycle) were significantly reduced in the double mutant compared with the wild-type (Supplemental Figure S5, A, B, and F). On the other hand, significant accumulation of cysteinylglycine (an intermediate in glutathione biosynthesis (Hasanuzzaman et al., 2017), hexoses and hexose-phosphates (e.g. glucose-6-phosphate and fructose), amino acids (e.g. allothreonine and valine), and phytol (a chlorophyll component; Gutbrod et al., 2019) was measured in the swt16swt17 mutant compared to the wild-type stems (Supplemental Figure S5, C–E, G–I). We further quantified the soluble sugars and starch content in both genotypes by enzymatic methods. Consistent with the metabolomics results, a six-fold significant increase of fructose in the double mutant was confirmed (Figure 6, C). In addition, the glucose content was significantly increased by four-fold in the stem of the double mutant (Figure 6, C), while no variation in the sucrose and starch contents was observed (Figure 6, C). Interestingly, the inflorescence stem of the swt16swt17 double mutant accumulated mostly hexoses while no significant changes in glucose or sucrose were observed in the stem of the swt16 and swt17 single mutants (Supplemental Figure S6, A). Although it was not significant, a tendency to accumulate glucose was observed in the single mutants (Supplemental Figure S6, B). A significant increase in fructose content was measured only in the swt17 mutant compared to the wild-type (Supplemental Figure S6, C).
Figure 6.
Hexoses accumulate in the inflorescence stem of the swt16swt17 double mutant. A, B, Multivariate analysis of the metabolomic data sets obtained from the wild-type and swt16swt17 inflorescence stems. Metabolites were extracted and analyzed by GC–MS from eight individual plants for each genotype. Plants were grown under long-day conditions for seven weeks. A, sPLS-DA score plot for the wild-type (purple) and swt16swt17 (green) samples. The variable plot for the sPLS-DA is presented in (B) and metabolites in red are significantly different between the wild-type and the swt16swt17 mutant according to Student’s t test (P<0.05) (Supplemental Table S3 and Supplemental Figure S3). ADP, adenosine-5-diphosphate; G-6-P, glucose-6-phosphate. C, Boxplots showing the sucrose, glucose, fructose, and starch contents of the inflorescence stems of the wild-type (in purple) and the swt16swt17 (in green) mutant grown under long-day conditions for 7 weeks. The box-and-whisker plots represent values from nine biological replicates coming from plants grown at two separate times The lines represent median values, the tops and bottoms of the boxes represent the first and third quartiles, respectively, and the ends of the whiskers represent maximum and minimum data points. Black dots are outliers. Asterisks above the boxes indicate statistical differences between genotypes according to Student’s t test (P<0.05).
Discussion
To efficiently control carbon homeostasis within a cell and to fuel the different metabolic and signaling pathways, dynamic sugar storage in the plant vacuole is critical. Over the past years, several vacuolar transporters have been identified at the molecular level (for review see Hedrich et al., 2015). Among them, SWEET16 and SWEET17 have been characterized as bidirectional tonoplastic sugar facilitators and shown to be involved in seed germination, root growth, and stress tolerance (Chardon et al., 2013; Klemens et al., 2013; Guo et al., 2014; Valifard et al., 2021). In addition, the expression of both genes has been shown in the inflorescence stem’s vascular parenchyma cells, but this had not previously been explored further. In this work, we ask whether facilitated sugar transport (via SWEET16 and SWEET17) across the vacuolar membrane limits vascular tissue development in the inflorescence stem of Arabidopsis.
Our data show that vascular tissue development in the inflorescence stem is regulated by both SWEET16 and SWEET17 transporters. This conclusion is supported by the fact that mutations in both genes impact the inflorescence stem diameter along with the quantity of phloem and xylem tissues. These phenotypes are consistent with our analysis of lines expressing translational CDS–GUS, fusions which confirmed the presence of both transporters in phloem and xylem tissues of the flower stem. Nonetheless, because SWEET16 and SWEET17 are also expressed in the root (Guo et al., 2014), the defects observed in the flower stem could also result from a disruption of sugar allocation between roots and shoot. Further experiment will be required to address the relative role of SWEET16 and SWEET17 in shoot and root parts and the consequences on the development of the vascular system in the inflorescence stem.
Our data also highlight modifications of hexose homeostasis in the inflorescence stem of the different mutant lines. Although a tendency to accumulate glucose was observed in the swt16 mutant, a significant increase in fructose was measured in the swt17 mutant stem. Furthermore, mutations in both SWEET16 and SWEET17 induced somewhat specific accumulation of glucose, glucose-6-phosphate, and fructose in the inflorescence stem. It has been previously shown that defects in the expression of vacuolar sugar transporters alter carbon partitioning and allocation in different organs, which is in line with our findings for the inflorescence stem (Wingenter et al., 2010; Yamada et al., 2010; Poschet et al., 2011; Chardon et al., 2013; Klemens et al., 2013; Guo et al., 2014; Klemens et al., 2014). Knowing that SWEET proteins are sugar facilitators working along the concentration gradient (Chen et al., 2010) and that at least half of the hexoses are present in the plant vacuole (Heineke et al., 1994; Weiszmann et al., 2018), we can reasonably propose that some of the hexoses are trapped inside the vacuole in the single swt16 and swt17 mutants and the swt16swt17 double mutant. As a consequence, modifications in the distribution of hexose concentrations between the vacuole and the cytosol, which would impact the availability of hexoses for subsequent metabolic and signaling purposes, could be expected. Hexoses are known to favor cell division and expansion, while sucrose favors differentiation and maturation (Koch, 2004). In addition, after metabolization, hexoses and hexoses-phosphates constitute the building blocks for the synthesis of cell wall polysaccharides (Verbančič et al., 2018). Since SWEET16 and/or SWEET17 are expressed in the xylem initials, in young xylem cells and in xylem parenchyma cells, we propose that enhanced storage of vacuolar hexoses in these cells will affect different stages of xylem development.
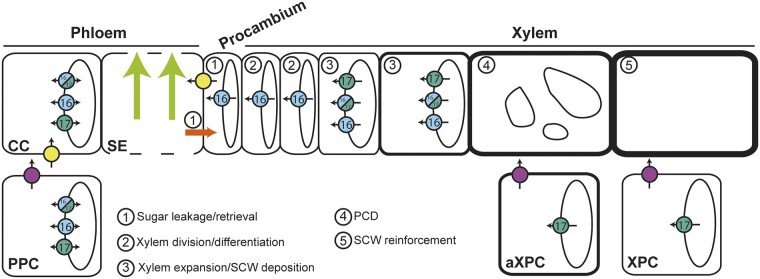
(Pro)cambium and xylem tissues can be regarded as sinks because they rely mostly on the supply of carbohydrates from the surrounding cells to sustain their development (Sibout et al., 2008; Spicer, 2014). In aspen stems, a gradual increase in sucrose and reducing sugars, together with a rise in the activities of sugar metabolism enzymes, are observed across the cambium–xylem tissues (Roach et al., 2017). In addition, in tomato (Solanum lycopersicum), the modification of fructose phosphorylation by the FRUCTOKINASE 2 (SlFRK2) leads to a defect in cambium activity (Damari-Weissler et al., 2009). Taken together, these results support the need for maintenance of sugar homeostasis in the (pro)cambium to respond to the high metabolic activity required during cell division. Our work identified SWEET16 as a player in the dividing xylem cells, acting to balance the tradeoffs between the need for sugars in the cytosol and their storage in the vacuole (Figure 7). This conclusion is supported by the fact that SWEET16 is expressed across the procambium–xylem boundary and that a mutation in SWEET16 leads to defects in the number of xylem cells and in radial growth of the inflorescence stem. Furthermore, the expression of the gene coding for the WUSCHEL RELATED HOMEOBOX 4 (WOX4) transcription factor (Etchells et al., 2013), which is involved in cellular proliferation, was unchanged in both swt16 and sw16swt17 mutants. These results suggest that the defects in xylem cell division could result from reduced availability of energy and matter resources due to a reduction in sugar transport and/or from a defect in sugar signaling.
Figure 7.
Model for the role of SWEET transporters during xylem development in Arabidopsis inflorescence stems. This model is based on the results presented in this work on SWEET16 and SWEET17 and those previously published on SWEET11, SWEET12, and SUCROSE-PROTON SYMPORTER 2 (SUC2) transporters (Truernit and Sauer, 1995; Chen et al., 2012; Gould et al., 2012; Le Hir et al., 2015). In the phloem tissue, sugar exchanges between cytosol and vacuole in companion cells and/or parenchyma cells are regulated by SWEET16 and SWEET17. In addition, cytosolic sucrose and hexoses present in the phloem parenchyma cells (PPC) are exported into the apoplastic space between PPC and companion cells (CC) by the sugar transporters SWEET11 and SWEET12 (fuchsia circles). Apoplastic sucrose is then imported into the CC cytosol via the SUC2 transporter (yellow circles) before entering the phloem sieve tubes (SE) and being transported over long distances (light green arrows). A part of these sugars leak from the SE, most probably through plasmodesmata (orange arrow), and reach axial sinks (e.g. procambium and xylem) while another part of the sugars is reimported inside the SE, mostly through the action of SUC2 (1). In the cells at the cambium–xylem boundary, soluble sugars are probably exported by SWEET16 (light blue) into the cytosol in order to sustain the division of xylem cells (2). Given the high cytosolic sugar demand required to sustain the secondary cell wall (SCW) deposition process (3), sugars stored in the vacuole are likely exported into the cytosol through the action of SWEET16 and/or SWEET17. Interaction between SWEET16 and SWEET17 is shown as bicolor circles. After the completion of programmed cell death (PCD) and the disintegration of the vacuole (4), the SCW is still being reinforced (5) and we can assume that the sugar demand is still high. At this stage, the sugars stored in the vacuole of the xylary parenchyma cells (XPC) and the axial xylem parenchyma cells (aXPC) are likely released by SWEET17 and then exported into the apoplastic space by SWEET11 and SWEET12. Whether it is the sugars themselves or more complex cell wall sugar-derived molecules that reach the dead xylem cells remains an open question.
The reduced number of xylem vessels in the swt16swt17 double mutant’s vascular bundle could be explained by the upregulation of VND-INTERACTING 2 (VNI2), which is a repressor of the activity of the master regulator of xylem vessel differentiation VND7 (Zhong et al., 2008; Yamaguchi et al., 2010). On the other hand, the overexpression in the double mutant swt16swt17 of SECONDARY WALL-ASSOCIATED NAC DOMAIN 1 (SND1), the master switch for fiber differentiation, would be expected to result in a shift towards increased differentiation of xylary fibers (Zhong et al., 2006), which is not consistent with the fewer fibers observed in the double mutant. Based on these results, we can assume that the increase in storage of vacuolar hexoses in the double mutant also affects xylem cell differentiation. Against this hypothesis, our results show that both xylary fiber and xylem vessel numbers decreased proportionately, since no change in the xylem vessels/xylary fibers ratio was measured. Consistent with this observation, the expression of the gene coding for the PHLOEM INTERCALATED WITH XYLEM (PXY) receptor, which is involved in xylem cell differentiation (Etchells et al., 2016), was not modified. Despite the upregulation of VNI2 and SND1 expression, which could be due to a feedback mechanism yet to be identified, these results therefore tend to suggest that no disequilibrium is occurring in xylem cell differentiation in the swt16swt17 mutant stem. The enhanced storage of hexoses in the vacuole of the double mutant is therefore affecting the overall pattern of xylem cell division rather than xylem cell differentiation.
After cell division and differentiation, xylem cells undergo a developmental program that includes secondary cell wall formation, lignification and programmed cell death, to produce functional xylem fibers and vessels (Schuetz et al., 2012; Figure 7). Along with the overexpression of SND1, the overexpression of genes encoding its downstream targets, namely MYB DOMAIN PROTEIN 46 and 83 (MYB46 and MYB83), was observed in the swt16swt17 double mutant. Furthermore, the targets of the MYB46/MYB83 node, which positively regulates SND3 (SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3), KNAT7 (KNOTTED-LIKE HOMEOBOX OF ARABIDOPSIS THALIANA 7), MYB43, and MYB54 or/and negatively regulates MYB103, KNAT7, and MYB4, all of which are involved in the formation of the xylem secondary cell wall, are also upregulated (Hussey et al., 2013). KNAT7 directly or indirectly represses cellulose, hemicellulose, and lignin biosynthetic genes (Li et al., 2012), while MYB54 is related to cellulose synthesis (Zheng et al., 2019). In Arabidopsis, MYB43 along with other MYB transcription factors regulates lignin biosynthesis (Geng et al., 2020), while its putative ortholog in rice (Oryza sativa) is involved in the regulation of cellulose deposition (Ambavaram et al., 2011). Finally, the upregulation of MYB4 in Arabidopsis results in downregulation of the lignin pathway (Jin et al., 2000), while a role for MYB103 in lignin biosynthesis has been shown in Arabidopsis stems (Öhman et al., 2013). In the swt16wt17 double mutant, these transcriptional changes were accompanied by modifications of the secondary cell wall in terms of cellulose and hemicellulose composition. However, a single mutation in SWEET16 or SWEET17 was sufficient to modify the composition of the xylem cell wall without any alteration in the expression of genes involved in secondary cell wall synthesis. Our data further show that the SWEET16 and SWEET17 expression patterns overlap in xylem cells that are building a secondary cell wall and that they form a heterodimer in Arabidopsis mesophyll protoplasts. We therefore postulate that the intermediate sugars required for the synthesis of cell wall polysaccharides come in part from the vacuole unloading mediated by SWEET16 and SWEET17 homo- and heterodimers (Figure 7). Previously, it has been shown that genes encoding vacuolar sugar facilitators are upregulated during secondary cell wall formation in xylem vessels, while sugar facilitators expressed at the plasma membrane are downregulated (Supplemental Figure S2; Li et al., 2016), supporting the idea that secondary cell wall formation relies on sugar export from the vacuole. In the current model of cell wall synthesis, the cytosolic catabolism of sucrose is thought to be the main source of nucleotide sugars (e.g. UDP-glucose, UDP-galactose, GDP-mannose) that act as precursors for cellulose and hemicellulose synthesis (Verbančič et al., 2018). Our data support the existence of a more complex system in which the export of vacuolar hexoses also represents a source for the synthesis of nucleotide sugars and subsequent cell wall formation (Figure 7).
Because SWEET17, a fructose-specific transporter (Chardon et al., 2013), is also expressed in the xylem parenchyma cells (including axial xylem parenchyma cells and xylary parenchyma cells), we postulate that the maintenance of fructose homeostasis within this cell type, is important and could contribute to the provision of carbon skeletons for secondary cell wall synthesis after the disappearance of the vacuole from the maturing xylem cells (Figure 7). Previously, the Arabidopsis fructokinases (FRKs), which allow the fructose phosphorylation before its metabolization by the cells, has been shown to play an important role in the vascular tissue development in hypocotyls (Stein et al., 2017). Furthermore, based on abnormal vascular cell shapes, Stein et al. (2017) proposed that FRKs may also contribute to the strength of the cell walls. This study and our results concur, therefore, to propose a link between fructose transport/metabolism and secondary wall formation that should be further explored. Within this scheme, the export of sugars in the apoplastic space between parenchyma cells and developing conducting cells could be carried out by the plasmalemmal SWEET11 and SWEET12 transporters which also expressed in the xylem parenchyma cells (Figure 7;Le Hir et al., 2015). Such cooperation between parenchyma cells and developing conducting cells was previously described as the “good neighbor” hypothesis in the context of H2O2 and monolignol transport (Barcelo, 2005; Smith et al., 2013; Smith et al., 2017). To further explore the importance of sugar transport between xylary parenchyma cells and developing xylem cells, more experiments, such as cell-specific complementation of the sweet mutant lines, will be needed in order to better comprehend the role of xylary parenchyma cells in xylem development. Additionally, it would be interesting to explore, with similar techniques, whether or not the vascular system development is impaired in the leaf petiole where both SWEET16 and SWEET17 genes are expressed (Chardon et al., 2013; Klemens et al., 2013). If this is the case, this would suggest a more general role of SWEET16 and SWEET17 in xylem development.
As sink tissues, cambium, and xylem rely on the lateral escape of sugars along the phloem pathway to nourish them (Sibout et al., 2008; van Bel, 2021). The localization of SWEET16 and SWEET17 at the phloem–cambium–xylem interface in the inflorescence stem along with our previous results on the localization of SWEET11 and SWEET12 (Le Hir et al., 2015) suggests that these SWEET transporters play a role in the radial transport (lateral transport) of sugars in order to provide energy and substrate to sustain cell division and xylem formation. However, such hypothesis needs to be tested by using for instance tissue-specific complementation to address the importance of their expression in either phloem or xylem on the global vascular system phenotype.
In conclusion, our results propose that the hexose exchanges regulated by SWEET16 are important for the division of the vascular cells while both SWEET16 and SWEET17 transporters, working as homo and/or heterodimers, are important for the secondary cell wall synthesis before the vacuole disruption. Finally, we identified SWEET17 as specifically involved in the maintenance of fructose homeostasis within the xylem parenchyma cells in order to sustain the latest phases of secondary cell wall formation. Overall, our work shows that the exchange of intracellular hexoses, regulated by SWEET16 and/or SWEET17 at the tonoplast, contributes to xylem development by modulating the amounts of sugars that will be made available to the different cellular processes. However, how the cell is prioritizing the distribution of sugars among the different processes remains an open question. Although these technologies are challenging, the use of nonaqueous fractionation and metabolomics approaches (Fürtauer et al., 2019) could help in resolving subcellular sugar metabolism in a cell-specific context in mutant lines affected in sugar metabolism, transport and signaling.
Materials and methods
Plant material and growth conditions
Seeds of Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines homozygous for SWEET17 (sweet17-1) and SWEET16 (sweet16-3 and sweet16-4) mutations in the Col-0 background were gifts from Dr F. Chardon and Pr. E. Neuhaus, respectively. The sweet17-1 line was previously reported to be a knock-out by Chardon et al. (2013). The sweet16-3 (SM_3_1827) and sweet16-4 (SM_3_30075) lines were numbered following the sweet16-1 and sweet16-2 lines already published by Guo et al. (2014). To verify whether sweet16-3 and sweet16-4 were knock-out mutants we performed RT-PCR with specific primers to amplify the full-length SWEET16 cDNA (Supplemental Figure S1 and Supplemental Table S4). Since only the sweet16-4 mutant turned out to be a knock-out (Supplemental Figure S1, B), we crossed it with swt17-1 to obtain the double mutant sweet16-4sweet17-1 (hereafter referred to as swt16swt17). Homozygous plants were genotyped using gene-specific primers in combination with a specific primer for the left border of the T-DNA insertion (Supplemental Table S4).
To synchronize germination, seeds were stratified at 4°C for 48 h and sown in soil in a growth chamber in long-day conditions (16-h day/8-h night and 150 µE m−2 s−1) at 22°C/18°C (day/night temperature) with 35% relative humidity. Plants were watered with Plant-Prod nutrient solution twice a week (Fertil, https://www.fertil.fr/). For all experiments, the main inflorescence stems (after the removal of lateral inflorescence stems, flowers, and siliques) were harvested from 7-week-old plants.
Inflorescence stem sample preparation
For each plant, the main inflorescence stem height was measured with a ruler before harvesting a 1- to 2-cm segment taken at the bottom part of the stem. The stem segments were embedded in 8% (w/v) agarose solution and sectioned with a VT100 S vibratome (Leica, https://www.leica-microsystems.com/). Some of the cross-sections were used for FT-IR analysis and the others were stained with a FASGA staining solution prepared as described in Tolivia and Tolivia (1987) for morphometric analysis of the xylem.
Morphometric analysis of the xylem
Previously stained inflorescence stem cross-sections were imaged under an Axio Zoom V16 microscope equipped with a Plan-Neofluar Z 2.3/0.57 FWD 10.6 objective (Zeiss, https://www.zeiss.fr/microscopie/). For each section, the diameter of the inflorescence stem was measured using the Image J software package (https://imagej.nih.gov/ij/). For the same sections all the vascular bundles were photographed individually using a confocal laser scanning microscope and morphological analysis of the xylem were performed as described in Le Hir et al. (2015). For each vascular bundle, the morphological segmentation made it possible to find the number of xylem cells (xylary fibers and xylem vessels) as well as their cross-sectional areas. Cells with a cross-sectional area of between 5 and 150 µm2 were considered to be xylary fibers and cells with a cross-sectional area greater than 150 µm2 were considered to be xylem vessels. The sum of all xylem cell cross-sectional areas was then calculated to give the total xylem cross-sectional area. The average xylary fiber and xylem vessel area was calculated by dividing the total xylem cross-sectional area by the number of each cell type.
GUS staining
The lines expressing pSWEET16:SWEET16-GUS and pSWEET17:SWEET17-GUS were kindly provided by Dr Woei-Jiun Guo (National Cheng Kung University, Tainan, Taiwan) and were used to assess the SWEET16 and SWEET17 expression pattern on seven-week-old plants grown under greenhouse conditions. The histochemical GUS staining was performed according to Sorin et al. (2005). Inflorescence stems subjected to GUS staining were then embedded in 8% (w/v) agarose and sectioned with a Leica VT100S vibratome (Leica, https://www.leica-microsystems.com/). Sections were counterstained for lignin by phloroglucinol staining (Pradhan Mitra and Loqué, 2014). Pictures were taken using a Leitz Diaplan microscope equipped with an AxioCam MRc camera and the ZEN (blue edition) software package (Zeiss, https://www.zeiss.com/).
FTIR analysis of the xylem secondary cell wall
The composition of the secondary cell wall of the xylem tissue was determined by Fourier Transformed Infra-red spectroscopy using an FT-IR Nicolet iN (Thermo Fisher Scientific, https://www.thermofisher.com). Spectral acquisitions were done in transmission mode on a 30 µm × 30 µm acquisition area targeting the xylem tissue (xylem vessels and xylary fibers) as described in Le Hir et al. (2015). Between 10 and 15 acquisition points sweeping the xylem tissue homogeneously were performed on each vascular bundle within a stem section. Three individual inflorescence stems were analyzed for each genotype. After sorting the spectra and correcting the baseline, the spectra were area-normalized and the different genotypes were compared as described in Le Hir et al. (2015). The absorbance values (maximum height) of the major cellulose, lignin and hemicellulose bands in the fingerprint region (1,800–800 cm−1) were collected using TQ Analyst EZ edition (Thermo Fisher Scientific, https://www.thermofisher.com).
Metabolomic analysis
The inflorescence stems of the wild-type and the swt16swt17 double mutant were harvested in the middle of the day (8 h after the beginning of the light period). Metabolites were extracted from 4.5 mg of lyophilized stem powder from eight individual plants and analyzed by GC–MS as described in Cañas et al. (2020). Relative concentrations of metabolites were determined relative to the internal standard ribitol, which was added after grinding the lyophilized material. Differential accumulation of metabolites was determined by one-way analysis of variance (ANOVA) and post hoc Tukey’s tests (P < 0.05).
Quantification of soluble sugars and starch
The main inflorescence stems of the wild-type, and the swt16, swt17, and swt16swt17 mutants, were harvested in the middle of the day (8 h after the beginning of the light period), frozen in liquid nitrogen, and ground with a mortar and a pestle. Soluble sugars and starch were extracted from 50 mg of powder from an individual stem as described in Sellami et al. (2019). Depending on the experiment, four to nine biological replicates were analyzed.
RNA isolation and cDNA synthesis
RNAs were prepared from the main inflorescence stem from four 7-week-old individual plants grown as described above. Samples were frozen in liquid nitrogen before being ground with a mortar and a pestle. Powders were stored at −80°C until use. Total RNA was extracted from frozen tissue using TRIzol reagent (Thermo Fisher Scientific, 15595-026, https://www.thermofisher.com) and treated with DNase I, RNase-free (Thermo Fisher Scientific, EN0521, https://www.thermofisher.com). cDNA was synthetized by reverse transcribing 1 µg of total RNA using RevertAid H minus reverse transcriptase (Thermo Fisher Scientific, EP0452, https://www.thermofisher.com) with 1 µL of oligo(dT)18 primer (100 pmoles) according to the manufacturer’s instructions. The reaction was stopped by incubation at 70°C for 10 min.
RT-qPCR experiment
Transcript levels were assessed for four independent biological replicates in assays with triplicate reaction mixtures by using specific primers either designed with the Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) or taken from the literature (Supplemental Table S5). RT-qPCR reactions were performed in a 96-well transparent plate on a Bio-Rad CFX96 Real-Time PCR machine (Bio-Rad) in 10-µL mixtures each containing 5-µL of Takyon ROX SYBR MasterMix dTTP Blue (Eurogentec, UF-RSMT-B0710, https://www.eurogentec.com/), 0.3-µL forward and reverse primer (30 µM each), 2.2-µL sterile water, and 2.5 µL of a 1/30 dilution of cDNA. The following RT-qPCR program was applied: initial denaturation at 95°C for 5 min, followed by 39 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 30 s. Melting curves were derived after each amplification by increasing the temperature in 0.5°C increments from 65°C to 95°C. The Cq values for each sample were acquired using the Bio-Rad CFX Manager 3.0 software package. The specificity of amplification was assessed for each gene, using dissociation curve analysis, by the precision of a unique dissociation peak. If one of the Cq values differed from the other two replicates by >0.5, it was removed from the analysis. The amplification efficiencies of each primer pair were calculated from a 10-fold serial dilution series of cDNA (Supplemental Table S5). Four genes were tested as potential reference genes: ADENINE PHOSPHORIBOSYL TRANSFERASE 1 (APT1, At1g27450), TAP42 INTERACTING PROTEIN OF 41 KDA (TIP41, At4g34270), ELONGATION FACTOR 1 ALPHA (EF1α, At5g60390), and UBIQUITIN 5 (UBQ5, At3g62250). The geNorm algorithm (Vandesompele et al., 2002) was used to determine the gene most stably expressed among the different genotypes analyzed, namely UBQ5 in this study. The relative expression level for each genotype was calculated according to the ΔCt method using the following formula: average Et−Cq(of target gene in A)/Er−Cq(of reference gene in A), where Et is the amplification efficiency of the target gene primers, Er is the reference gene primer efficiency, A represents one of the genotypes analyzed.
Production of complementation lines
For complementation of the single sweet16-4 and sweet17-1 and the double sweet16-4sweet17-1 mutants, N terminal with GFP were constructed as follow. First, the coding sequence of eGFP was amplified from the pKGWFS7 plasmid (Karimi et al., 2002) with or without a stop codon and then introduced into a modified donor pENTR vector to produce pENT-GFP (w/o stop). To make the N terminal translational GFP fusions (pSWEET16:GFP-SWEET16 and pSWEET17:GFP-SWEET17), the promoters (1,295 bp for SWEET16 and 2,004 bp for SWEET17) and genomic sequences (1,863 bp for SWEET16 and 2,601 bp for SWEET17) were amplified separately and then cloned on each side of the GFP gene in the intermediary vector pENT-GFP by taking advantage of the restriction sites generated by PCR. All the PCR reactions were performed using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, F-530S, https://www.thermofisher.com) with the primers listed in Supplemental Table S4. Donor vectors created in this way were analyzed by sequencing in order to check the reading frame of the translational fusions and the integrity of the whole genomic sequences. Destination binary vectors were then obtained by recombination, using Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific, 11791-100, https://www.thermofisher.com), between pENTR donor vectors and pMDC99 (for pSWEET16:GFP-SWEET16) or pMDC123 (for pSWEET17:GFP-SWEET17) (Curtis and Grossniklaus, 2003). All binary vectors were introduced into Agrobacterium tumefaciens C58pMP90 (Koncz and Schell, 1986) by electroporation. Arabidopsis single mutants swt16-4 and swt17-1 as well as the double mutant sweet16-4sweet17-1 plants were transformed by the floral dip method (Clough and Bent, 1998). Transformants were selected on hygromycin (15 mg·L-1) for pMDC99 constructs and/or Basta (7.5 mg·L-1) for pMDC123 constructs. For all constructs, three independent transgenic lines were analyzed and one representative line was selected for subsequent studies.
Bimolecular fluorescence complementation assay
For the bimolecular fluorescence complementation assay, the full-length ORFs of SWEET16 and SWEET17 were amplified from cDNA with the primers given in Supplemental Table S4, either with or without their stop codons, depending on the final vector used. The ORFs were further sub-cloned into pBlueScript II SK, blunt end cut with EcoRV. The resulting vectors were checked for errors and orientation of the insert by sequencing with T3 and T7 primers. Subsequently, positive clones in the T7 orientation and the corresponding pSAT1 vectors (Lee et al., 2008) were cut with EcoRI and XhoI. SWEET16 including the stop codon was ligated into pSAT1-cCFP-C, and SWEET17 without the stop codon was ligated into pSAT1-nCerulean-N. Plasmid DNA of the final constructs was isolated with a PureLink HiPure Plasmid Filter Midiprep Kit (Invitrogen/Thermo Fisher Scientific) according to the manufacturer’s manual. Isolation and transfection of Arabidopsis mesophyll protoplasts were performed as described by Yoo et al. (2007). For imaging protoplasts, a Leica TCS SP5 II confocal laser scanning microscope (http://www.leica-microsystems.com) was used. All pictures were taken using a Leica HCX PL APO 63/1.20 w motCORR CS objective with a VIS‐Argon laser suitable for constructs with CFP (Cerulean, 458 nm excitation/460–490 nm collection bandwidth, laser power 90% and gain at 735 V) or YFP (Venus, 514 nm excitation/520–540 nm collection bandwidth, laser power 70% and gain at 700 V) derivates. The chloroplast autofluorescence was imaged between 620 and 700 nm after excitation with the 514 nm laser line (laser power 70% and gain at 690 V).
Statistical analysis
Differences between genotypes were assessed by a Student’s t test for comparison between wild-type plants and mutant lines or by using one-way ANOVA with a Tukey’s HSD post hoc test or a Dunnett post hoc test (for analysis of the qPCR data set). The sPLS-DA analysis was performed according to Jiang et al. (2014) and Lê Cao et al. (2011). Irrelevant variables were removed using lasso (least absolute shrinkage and selection operator) penalizations and 20 variables were selected in each dimension. The “mixOmics” package (Rohart et al., 2017) was used to perform sPLS-DA. All the statistical analysis and graph production were done in RStudio (version 1.1.456) (RStudio Team, 2015), which incorporates the R software package (version 3.5.1) (R Core Team, 2017) using “ggplot2” (Wickham, 2016), “ggthemes” (Arnold, 2019), “cowplot” (Wilke CO, 2019), “hyperSpec” (Beleites and Sergo, 2020), and “multcompView”(Graves et al., 2015).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SWEET16 (AT3G16690), SWEET17 (AT4G15920). Metabolomic data can be found at https://www.ebi.ac.uk/metabolights/MTBLS2179.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of SWEET16 insertion lines and characterization of the sweet16-4sweet17-1 mutant line.
Supplemental Figure S2. Phenotypic complementation of the swt mutant lines.
Supplemental Figure S3. Negative controls for bimolecular fluorescence complementation experiment.
Supplemental Figure S4. Differential expression of SWEET genes during the secondary cell wall formation of the xylem vessels.
Supplemental Figure S5. Nine out of 158 metabolites identified by GC–MS differ significantly between the wild-type and the swt16swt17 double mutant.
Supplemental Figure S6. Fructose accumulation in the inflorescence stem of the swt17 mutant.
Supplemental Table S1. P-values from pairwise comparisons (Tukey’s post hoc test) between genotypes of anatomical parameters measured in the inflorescence stem xylem tissue.
Supplemental Table S2. Genes co-regulated during the secondary cell wall formation.
Supplemental Table S3. P-values from t test of metabolites identified from the GC–MS analysis.
Supplemental Table S4. Primers used for characterizing mutant lines.
Supplemental Table S5. Primers used for quantifying genes by RT-qPCR.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Fabien Chardon and Dr Anne Krapp (IJPB, INRAE Versailles, France) for providing seeds of the sweet17-1 mutant as well as Dr Woei-Jiun Guo for kindly providing us the SWEET16 and SWEET17 translational GUS fusions. They also thank Dr Grégory Mouille (IJPB, INRAE Versailles, France) for advice on FTIR data set analysis and anonymous reviewers who helped us to improve the manuscript.
E.A. performed most of the research and the first analysis of the data. P.K. performed the initial bimolecular fluorescence complementation experiment under the supervision of H.E.N. and R.L.H. performed the complementary bimolecular fluorescence complementation experiment. P.K. and H.E.N. provided seeds of the sweet16 mutant. F.Gi., F.Gu., and B.G. supervised and helped E.A. with the GC–MS analysis. F.V. cloned the complemented lines. B.H. provided technical assistance to E.A.; S.D., C.B., and R.L.H. reviewed and edited the manuscript. R.L.H. conceived the project, designed the research, and wrote the article with contributions of all the authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: rozenn.le-hir@inrae.fr.
Funding
This work has benefited from the support of IJPB’s Plant Observatory technological platforms and from a French State grant (Saclay Plant Sciences, reference ANR-17-EUR-0007, EUR SPS-GSR) managed by the French National Research Agency under an Investments for the Future program (reference ANR-11-IDEX-0003-02) through PhD funding to E.A.
Conflict of interest statement. The authors declare no conflict of interest.
References
- Åkerholm M, Salmén L (2001) Interactions between wood polymers studied by dynamic FT-IR spectroscopy. Polymer (Guildf) 42:963–969 [Google Scholar]
- Ambavaram MMR, Krishnan A, Trijatmiko KR, Pereira A (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 155:916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JB (2019) Ggthemes: extra themes, scales and geoms for “ggplot2.” https://cran.r-project.org/web/packages/ggthemes/index.html
- Baghdady A, Blervacq AS, Jouanin L, Grima-Pettenati J, Sivadon P, Hawkins S (2006) Eucalyptus gunnii CCR and CAD2 promoters are active in lignifying cells during primary and secondary xylem formation in Arabidopsis thaliana. Plant Physiol Biochem 44:674–683 [DOI] [PubMed] [Google Scholar]
- Barcelo RA (2005) Xylem parenchyma cells deliver the H2O2 necessary for lignification in differentiating xylem vessels. Planta 220:747–756 [DOI] [PubMed] [Google Scholar]
- van Bel AJE (2021) The plant axis as the command centre for (re)distribution of sucrose and amino acids. J Plant Physiol 265:153488. [DOI] [PubMed] [Google Scholar]
- Beleites C, Sergo V (2020) hyperSpec: a package to handle hyperspectral data sets in R. CRAN Package. https://cran.r-project.org/web/packages/hyperSpec/index.html
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, le Bris P, Herve J, Blondet E, Balzergue S, Lapierre C, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23:1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas RA, Yesbergenova-Cuny Z, Belanger L, Rouster J, Brulé L, Gilard F, Quilleré I, Sallaud C, Hirel B (2020) NADH-GOGAT overexpression does not improve maize (Zea mays L.) performance even when pyramiding with NAD-IDH, GDH and GS. Plants 9: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F, Bedu M, Calenge F, Klemens PAW, Spinner L, Clement G, Chietera G, Léran S, Ferrand M, Lacombe B, et al. (2013) Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr Biol 23: 697–702 [DOI] [PubMed] [Google Scholar]
- Chen L-Q, Hou B-H, Lalonde S, Takanaga H, Hartung ML, Qu X-Q, Guo W-J, Kim J-G, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-Q, Qu X-Q, Hou B-H, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335:207–211 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vectors set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damari-Weissler H, Rachamilevitch S, Aloni R, German MA, Cohen S, Zwieniecki MA, Holbrook NM, Granot D (2009) LeFRK2 is required for phloem and xylem differentiation and the transport of both sugar and water. Planta 230:795–805 [DOI] [PubMed] [Google Scholar]
- Dinant S, Wolff N, De Marco F, Vilaine F, Gissot L, Aubry E, Sandt C, Bellini C, Le Hir R (2019) Synchrotron FTIR and Raman spectroscopy provide unique spectral fingerprints for Arabidopsis floral stem vascular tissues. J Exp Bot 70:871–884 [DOI] [PubMed] [Google Scholar]
- Etchells JP, Provost CM, Mishr L, Turner SR (2013) WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Dev 140:2224–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Smit ME, Gaudinier A, Williams CJ, Brady SM (2016) A brief history of the TDIF-PXY signalling module: balancing meristem identity and differentiation during vascular development. New Phytol 209:474–484 [DOI] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP (2012) Sugars, signalling, and plant development. J Exp Bot 63:3367–3377 [DOI] [PubMed] [Google Scholar]
- Faix O (1991) Classification of lignins from different botanical origins by FT-IR spectroscopy. Holzforschung 45: 21–27 [Google Scholar]
- Fukuda H, Ohashi-Ito K (2019) Vascular tissue development in plants. Curr Top Dev Biol 131:141–160 [DOI] [PubMed] [Google Scholar]
- Fürtauer L, Küstner L, Weckwerth W, Heyer AG, Nägele T (2019) Resolving subcellular plant metabolism. Plant J 100:438–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furze ME, Trumbore S, Hartmann H (2018) Detours on the phloem sugar highway: stem carbon storage and remobilization. Curr Opin Plant Biol 43:89–95 [DOI] [PubMed] [Google Scholar]
- Geng P, Zhang S, Liu J, Zhao C, Wu J, Cao Y, Fu C, Han X, He H, Zhao Q (2020) MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol 182:1272–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzsás A, Stenlund H, Persson P, Trygg J, Sundberg B (2011) Cell-specific chemotyping and multivariate imaging by combined FT-IR microspectroscopy and orthogonal projections to latent structures (OPLS) analysis reveals the chemical landscape of secondary xylem. Plant J 66:903–914 [DOI] [PubMed] [Google Scholar]
- Gou JY, Park S, Yu XH, Miller LM, Liu CJ (2008) Compositional characterization and imaging of “wall-bound” acylesters of Populus trichocarpa reveal differential accumulation of acyl molecules in normal and reactive woods. Planta 229:15–24 [DOI] [PubMed] [Google Scholar]
- Gould N, Thorpe MRR, Pritchard J, Christeller JTT, Williams LEE, Roeb G, Schurr U, Minchin PEH (2012) AtSUC2 has a role for sucrose retrieval along the phloem pathway: evidence from carbon-11 tracer studies. Plant Sci 188–189:97–101 [DOI] [PubMed] [Google Scholar]
- Graves S, Piepho H-P, Selzer L, Dorai-Raj S (2015) multcompView: visualizations of paired comparisons.
- Guo W-J, Nagy R, Chen H-Y, Pfrunder S, Yu Y-C, Santelia D, Frommer WB, Martinoia E (2014) SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol 164:777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutbrod K, Romer J, Dörmann P (2019) Phytol metabolism in plants. Prog Lipid Res 74:1–17 [DOI] [PubMed] [Google Scholar]
- Hall H, Ellis B (2013) Transcriptional programming during cell wall maturation in the expanding Arabidopsis stem. BMC Plant Biol 13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Nahar K, Anee TI, Fujita M (2017) Glutathione in plants: biosynthesis and physiological role in environmental stress tolerance. Physiol Mol Biol Plants 23:249–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R, Sauer N, Neuhaus HE (2015) Sugar transport across the plant vacuolar membrane: nature and regulation of carrier proteins. Curr Opin Plant Biol 25:63–70 [DOI] [PubMed] [Google Scholar]
- Heineke D, Wildenberger K, Sonnewald U, Willmitzer L, Heldt HW (1994) Accumulation of hexoses in leaf vacuoles: studies with transgenic tobacco plants expressing yeast-derived invertase in the cytosol, vacuole or apoplasm. Planta 194:29–33 [Google Scholar]
- Le Hir R, Spinner L, Klemens PAW, Chakraborti D, De Marco F, Vilaine F, Wolff N, Lemoine R, Porcheron B, Géry C, et al. (2015) Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol Plant 8:1687–1690 [DOI] [PubMed] [Google Scholar]
- Hussey SG, Mizrachi E, Creux NM, Myburg AA (2013) Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front Plant Sci 4:1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Wang C, Zhang Y, Feng Y, Wang Y, Zhu Y (2014) Sparse partial-least-squares discriminant analysis for different geographical origins of salvia miltiorrhiza by 1 h-NMR-based metabolomics. Phytochem Anal 25:50–58 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Wiesshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kačuráková M, Smith AC, Gidley MJ, Wilson RH (2002) Molecular interactions in bacterial cellulose composites studied by 1D FT-IR and dynamic 2D FT-IR spectroscopy. Carbohydr Res 337:1145–1153 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195 [DOI] [PubMed] [Google Scholar]
- Klemens PAW, Patzke K, Deitmer J, Spinner L, Le Hir R, Bellini C, Bedu M, Chardon F, Krapp A, Neuhaus HE (2013) Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol 163:1338–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemens PAW, Patzke K, Trentmann O, Poschet G, Büttner M, Schulz A, Marten I, Hedrich R, Neuhaus HE (2014) Overexpression of a proton-coupled vacuolar glucose exporter impairs freezing tolerance and seed germination. New Phytol 202:188–197 [DOI] [PubMed] [Google Scholar]
- Koch KE (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7:235–246 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396 [Google Scholar]
- Lahlali R, Karunakaran C, Wang L, Willick I, Schmidt M, Liu X, Borondics F, Forseille L, Fobert PR, Tanino K, et al. (2015) Synchrotron based phase contrast X-ray imaging combined with FTIR spectroscopy reveals structural and biomolecular differences in spikelets play a significant role in resistance to Fusarium in wheat. BMC Plant Biol 15:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao KA, Boitard S, Besse P (2011) Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 12:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bhargava A, Qiang W, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ (2012) The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 194:102–115 [DOI] [PubMed] [Google Scholar]
- Li Z, Omranian N, Neumetzler L, Wang T, Herter T, Usadel B, Demura T, Giavalisco P, Nikoloski Z, Persson S (2016) A transcriptional and metabolic framework for secondary wall formation in Arabidopsis. Plant Physiol 172:1334–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Groover A, Lichtenberger R, Furuta K, Yadav SR, Helariutta Y, He XQ, Fukuda H, Kang J, Brady SM, et al. (2013) The plant vascular system: evolution, development and functions. J Integr Plant Biol 55:294–388 [DOI] [PubMed] [Google Scholar]
- Mahboubi A, Ratke C, Gorzsás A, Kumar M, Mellerowicz EJ, Niittylä T (2013) Aspen SUCROSE TRANSPORTER3 allocates carbon into wood fibers. Plant Physiol 163:1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott PE, Gómez LD, Mcqueen-Mason SJ (2016) Unlocking the potential of lignocellulosic biomass through plant science. New Phytol 209:1366–1381 [DOI] [PubMed] [Google Scholar]
- Martinoia E (2018) Vacuolar transporters - companions on a longtime journey. Plant Physiol 176:1384–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin PEH, McNaughton GS (1987) Xylem transport of recently fixed carbon within lupin. Aust J Plant Physiol 14:325–329 [Google Scholar]
- Mohebby B (2008) Application of ATR infrared spectroscopy in wood acetylation. J Agric Sci Technol 10:253–259 [Google Scholar]
- Öhman D, Demedts B, Kumar M, Gerber L, Gorzsás A, Goeminne G, Hedenström M, Ellis B, Boerjan W, Sundberg B (2013) MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J 73:63–76 [DOI] [PubMed] [Google Scholar]
- Payyavula RS, Tay KHC, Tsai C-J, Harding SA (2011) The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J 65:757–770 [DOI] [PubMed] [Google Scholar]
- Poschet G, Hannich B, Raab S, Jungkunz I, Klemens PAW, Krueger S, Wic S, Neuhaus HE, Buttner M (2011) A novel Arabidopsis vacuolar glucose exporter is involved in cellular sugar homeostasis and affects the composition of seed storage compounds. Plant Physiol 157:1664–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan Mitra P, Loqué D (2014) Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J Vis Exp (87):51381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017) R: A Language and Environment for Statistical Computing. R Core Team, Vienna, Austria
- Roach M, Arrivault S, Mahboubi A, Krohn N, Sulpice R, Stitt M, Niittylä T (2017) Spatially resolved metabolic analysis reveals a central role for transcriptional control in carbon allocation to wood. J Exp Bot 68:3529–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F, Gautier B, Singh A, Lê Cao KA (2017) mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 13:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team (2015) RStudio: Integrated Development for R. RStudio Team, Boston, MA
- Sakr S, Wang M, Dédaldéchamp F, Perez-Garcia M-D, Ogé L, Hamama L, Atanassova R (2018) The sugar-signaling hub: overview of regulators and interaction with the hormonal and metabolic network. Int J Mol Sci 19: 2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M, Smith R, Ellis B (2012) Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot 64:11–31 [DOI] [PubMed] [Google Scholar]
- Sellami S, Le Hir R, Thorpe MR, Vilaine F, Wolff N, Brini F, Dinant S (2019) Salinity effects on sugar homeostasis and vascular anatomy in the stem of the Arabidopsis thaliana inflorescence. Int J Mol Sci 20:3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Jouannet V, Agustí J, Kaul V, Levitsky V, Sanchez P, Mironova V V, Greb T (2021) Tissue-specific transcriptome profiling of the Arabidopsis inflorescence stem reveals local cellular signatures. Plant Cell 33:200–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Plantegenet S, Hardtke CS (2008) Flowering as a condition for xylem expansion in Arabidopsis hypocotyl and root. Curr Biol 18:458–463 [DOI] [PubMed] [Google Scholar]
- Smetana O, Mäkilä R, Lyu M, Amiryousefi A, Rodríguez FS, Wu M, Solé-gil A, Gavarrón ML, Siligato R, Miyashima S, et al. (2019) High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565:485–491 [DOI] [PubMed] [Google Scholar]
- Smit M, McGregor S, Sun H, Gough C, Bågman A-M, Soyars CL, Kroon JTM, Gaudinier A, Williams CJ, Yang X, et al. (2019) A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell 32:319–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Karlen SD, Bird D, Tokunaga N, Sato Y, Mansfield SD, Ralph J, Samuels AL (2017) Defining the diverse cell populations contributing to lignification in Arabidopsis thaliana stems. Plant Physiol 174:1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L (2013) Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25:3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, McKhann H, Garcion C, Vaucheret H, Sandberg G, et al. (2005) Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell Online 17:1343–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer R (2014) Symplasmic networks in secondary vascular tissues: parenchyma distribution and activity supporting long-distance transport. J Exp Bot 65:1829–1848 [DOI] [PubMed] [Google Scholar]
- Stein O, Avin-Wittenberg T, Krahnert I, Zemach H, Bogol V, Daron O, Aloni R, Fernie AR, Granot D (2017) Arabidopsis fructokinases are important for seed oil accumulation and vascular development. Front Plant Sci 7:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolivia D, Tolivia J (1987) Fasga: a new polychromatic method for simultaneous and differential staining of plant tissues. J Microsc 148:113–117 [Google Scholar]
- Truernit E, Sauer N (1995) The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of B-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196:564–570 [DOI] [PubMed] [Google Scholar]
- Turner S, Sieburth LE (2002) Vascular patterning. Arabidopsis Book 2:e0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valifard M, Le Hir R, Müller J, Scheuring D, Neuhaus HE, Pommerrenig B (2021) Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol 187: 2716–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, F. S (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1-0034.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbančič J, Lunn JE, Stitt M, Persson S (2018) Carbon supply and the regulation of cell wall synthesis. Mol Plant 11:75–94 [DOI] [PubMed] [Google Scholar]
- Weiszmann J, Fürtauer L, Weckwerth W, Nägele T (2018) Vacuolar sucrose cleavage prevents limitation of cytosolic carbohydrate metabolism and stabilizes photosynthesis under abiotic stress. FEBS J 285:4082–4098 [DOI] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. Springer, New York [Google Scholar]
- Wilke CO (2019) Cowplot: Streamlined Plot Theme and Plot Annotation for “ggplot2.” R package version 0.9.4.
- Wingenter K, Schulz A, Wormit A, Wic S, Trentmann O, Hoermiller II, Heyer AG, Marten I, Hedrich R, Neuhaus HE (2010) Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol 154:665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan YH, Hu YB, Chen L-Q, Sosso D, Ducat DC, Hou B-H, Frommer WB (2013) Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci USA 110:E3685–E3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Osakabe Y, Mizoi J, Nakashima K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K (2010) Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. J Biol Chem 285:1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22:1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572 [DOI] [PubMed] [Google Scholar]
- Zheng M, Liu X, Lin J, Liu X, Wang Z, Xin M, Yao Y, Peng H, Zhou DX, Ni Z, et al. (2019) Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J 97:587–602 [DOI] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye Z-H (2006) SND1, a NAC domain franscription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell Online 18:3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.