Abstract
The prevalence, epidemiology, and genomovar status of Burkholderia cepacia complex strains recovered from Italian cystic fibrosis (CF) patients were investigated using genetic typing and species identification methods. Four CF treatment centers were examined: two in Sicily, one in central Italy, and one in northern Italy. B. cepacia complex bacteria were isolated from 59 out of 683 CF patients attending these centers (8.6%). For the two geographically related treatment centers in Sicily, there was a high incidence of infection caused by a single epidemic clone possessing the cblA gene and belonging to B. cepacia genomovar III, recA group III-A, closely related to the major North America-United Kingdom clone, ET12; instability of the cblA sequence was also demonstrated for clonal isolates. In summary, of all the strains of B. cepacia encountered in the Italian CF population, the genomovar III, recA group III-A strains were the most prevalent and transmissible. However, patient-to-patient spread was also observed with several other genomovars, including strains of novel taxonomic status within the B. cepacia complex. A combination of genetic identification and molecular typing analysis is recommended to fully define specific risks posed by the genomovar status of strains within the B. cepacia complex.
Bacteria of the Burkholderia cepacia complex have been increasingly isolated as pathogens from cystic fibrosis (CF) patient populations due to their capacity for spread between patients, and their potential role in declining lung function with necrotizing pneumonia and frequently fatal septicemia, the so-called B. cepacia syndrome, has also been noted. Published reports indicate that different Burkholderia strains, identified as B. cepacia by conventional laboratory procedures, may be associated with a poor clinical prognosis for some individuals and/or with enhanced person-to-person transmissibility (13). Cross-infection between CF patients and epidemic outbreaks have been documented both within and outside hospitals (8, 10). Various markers have been associated with transmissible strains of B. cepacia, such as extracellular appendages known as cable pili (7, 20, 23) and a conserved 1.4-kb open reading frame called esmR (B. cepacia epidemic strain marker [16]).
The taxonomic diversity and the peculiar genomic characteristics of these organisms present diagnostic laboratories with many problems (18, 28). The name “B. cepacia complex” was proposed to comprise a cluster of five closely related species, originally referred to as B. cepacia genomovars I through V (6, 29). Recent research has also defined genomovar VI as a new member of the B. cepacia complex which shares considerable similarity with Burkholderia multivorans (4), and the name Burkholderia ambifaria, for bacteria belonging to genomovar VII (5), has been proposed. Determination of the genomovar status of B. cepacia complex strains is based on a polyphasic taxonomic approach encompassing traditional phenotypic and genotypic tests (27, 28, 29). To simplify the identification process, recent genetic procedures based on nucleotide sequence polymorphisms of the 16S rRNA gene (1, 13, 14, 21) or the recA gene (18) have been developed. Rapid and precise identification of bacteria is essential to evaluate specific risks, in terms of clinical prognosis and epidemicity, posed by each genomovar within the B. cepacia complex. Our study was performed in order to (i) evaluate the prevalence of B. cepacia complex infection in CF patients attending four Italian treatment centers over a period of 10 months, (ii) identify the genomovar status of each isolate, (iii) study the epidemiological and genetic relatedness of Burkholderia isolates, and (iv) evaluate the frequency of transmissibility markers and their association with the epidemiological classification of the strains.
MATERIALS AND METHODS
Bacterial strains and culture.
From September 1998 to July 1999, 683 patients were screened for B. cepacia complex infection, by sputum cultures on oxidation-fermentation base polymyxin B agar (Becton Dickinson). Strains were presumptively identified by the API 20NE (Bio Merieux) system; positive cultures were then sent to our reference laboratory for further characterization. A total of 92 B. cepacia isolates were obtained from the respiratory tract of patients attending the following CF centers: 28 were from 9 CF patients from Catania, Sicily; 33 were from 33 CF patients hospitalized in Palermo, Sicily; 1 isolate was from 1 CF patient in Gualdo Tadino, central Italy; and 30 were from 22 CF patients in Milan, northern Italy.
Six control strains, all isolated from CF patients in Vancouver, British Columbia, Canada, were obtained from a previously published collection (11, 16, 19, 23). Additional control strains for each current genomovar and recA gene restriction fragment length polymorphism (RFLP) type were also included (17, 18). One reference isolate (ATCC 25608) was obtained from the American Type Culture Collection.
Genomovar status identification based on the rRNA genes.
Preliminary identification of genomovar status was performed on the basis of a previously published PCR-based procedure for the identification of sequence motifs within the 16S and 23S ribosomal DNAs (1). Three separate PCRs were run, including primers of different degrees of specificity, in order to achieve stepwise exclusion of single species: Ce-16-21028 excluded B. multivorans, Burkholderia vietnamiensis, and Burkholderia gladioli; Mu-Vi-16-21028 excluded B. cepacia; and Gl-16-2457 excluded B. cepacia, B. multivorans, and B. vietnamiensis.
Genomovar status identification based on the recA gene.
A total of 53 strains, representative of the genetic diversity within the collection (determined by pulsed-field gel electrophoresis [PFGE]; see below), were examined (18). Briefly, identification of B. cepacia complex was carried out using PCR with primers which amplify the entire recA gene of bacteria within the B. cepacia complex. Genomovar status was then identified by RFLP of the amplified recA gene and confirmed using PCR primers specific for each genomovar. Strains that produced novel RFLP types and that tested negative with the genomovar-specific primers were subjected to nucleotide sequence analysis of the upstream region of the recA gene using PCR primers. Phylogenetic analysis of the resulting sequences was then used to place these strains within the complex. In addition, RFLP analysis of the 16S rRNA operon was performed on these strains to confirm their subclassification within genomovar I, III, or IV, B. multivorans, or B. vietnamiensis.
PFGE.
DNA fingerprinting by PFGE was carried out by the method of Grothues et al. (9). In brief, isolates were grown overnight on nutrient agar and then suspended in 1 ml of SE buffer (75 mM NaCl, 25 mM EDTA, pH 7.5) and adjusted to 1010 CFU/ml. Cell suspensions were mixed with an equal volume of 1.6% low-melting-point agarose, molded into plugs at 4°C, and lysed at 56°C overnight; the DNA inserts were then digested with SpeI, according to the supplier's instructions (New England Biolabs). Macrorestriction fragments were separated using a Gene Navigator apparatus (Pharmacia Biotech) at 10°C for 19 h, with a start time of 5 s and an end-pulse time of 35 s, at a field strength of 6 V/cm. A concatemer ladder of lambda phage DNA was used as a size marker. Interpretation of genomic relatedness was performed using well-established criteria (22, 26).
PCR detection of cblA and esmR.
Whole-cell DNA was amplified in a reaction mixture consisting of 200 μM deoxynucleoside triphosphates, 1 μM (each) primer, 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2), 2.5 mM MgCl2, and 1 U of Taq DNA polymerase. Each mixture was overlaid with 50 μl of liquid paraffin and placed in a PT 100 thermal cycler. The primer sequences used to amplify the esmR gene were as previously described (16). DNA sequences were amplified as follows: 30 cycles of 1 min at 94°C, 1 min at 62°C, and 1 min at 72°C and a final extension step at 72°C for 5 min. For the amplification of the cblA gene, primers were as previously described (20). Amplification was done for 30 cycles, each consisting of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C.
RESULTS
Prevalence of B. cepacia complex bacteria.
Our study involved 683 out of 2,717 patients (25%) included in the National Italian CF Register up to the end of 1995 (2, 25), under microbiological surveillance in CF treatment centers. From the initial biochemical screening performed in the four centers, the prevalence of B. cepacia infection was 9.5% overall (65 out of 683 patients): 30% (9 out of 30 patients) in Catania, 16.5% (33 out of 200 patients) in Palermo, 4% (1 out of 23 patients) in Gualdo Tadino, and 5% (22 out of 430 patients) in Milan. Further molecular characterization based on the 16S rRNA and recA gene (see below) indicated that isolates from six of these patients were not of the B. cepacia complex, providing a final prevalence of infection of 8.6% (59 out of 683 patients). Misidentification of B. cepacia complex was associated with all four CF treatment centers participating in this survey (Catania, Palermo, and Gualdo Tadino each contributing one misidentified strain and Milan contributing three non-B. cepacia complex bacteria).
Identification of Burkholderia spp. based on the 16S rRNA gene.
A total of 92 isolates were identified as B. cepacia after routine biochemical tests. Preliminary molecular identification based on the 16S rRNA gene was performed on all 92 CF isolates. Briefly, 89 of these 92 isolates (97%) were found to belong to B. cepacia genomovar I, III, or IV, these genomovars not being discriminated. One strain (1%) was identified as B. multivorans or B. vietnamiensis, and two (2%) were B. gladioli (Table 1).
TABLE 1.
16S rRNA gene-based identification and characterization of Burkholderia isolates from the four Italian centers
| Center | No. of strains
|
|||||
|---|---|---|---|---|---|---|
| 16S rRNA-PCR identification
|
cblA+ | esmR+ | ||||
| Total no. | Burkholderia genomovars I, III, and IV | B. multivorans/ B. vietnamiensis | B. gladioli | |||
| Catania | 28 | 27 | 0 | 1 | 9 | 6 |
| Palermo | 33 | 33 | 0 | 0 | 24 | 15 |
| Gualdo Tadino | 1 | 1 | 0 | 0 | 0 | 1 |
| Milan | 30 | 28 | 1 | 1 | 0 | 18 |
| Total | 92 | 89 | 1 | 2 | 33 | 40 |
Identification of B. cepacia complex genomovars based on the recA gene.
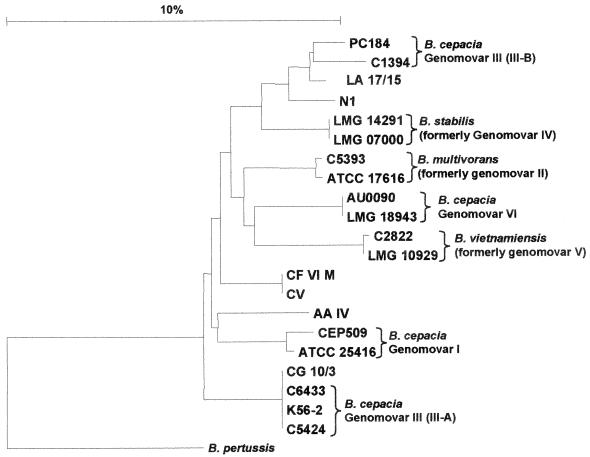
Due to the inability of the 16S rRNA gene PCR to distinguish all the current genomovars, the nucleotide sequence polymorphism in the recA gene was analyzed to determine specific genomovar status (Table 2). Examination of the PCR-RFLP patterns of the recA gene was performed first. Ten distinct HaeIII-derived RFLP patterns were found among 53 isolates that were representative of the genetic diversity of the B. cepacia complex strains present at the four CF centers (see PFGE results below). Six of these RFLP patterns correlated with those described previously, and their genomovar status was then confirmed using the genomovar-specific primers and was as follows: RFLP type E, genomovar I; RFLP type F, B. multivorans; RFLP type G, genomovar III (recA group III-A); RFLP types H and I, genomovar III (recA group III-B); and RFLP J, Burkholderia stabilis. Four recA gene RFLP types were not previously described and were designated RFLP types AZ, I3, S, and U. Apart from amplification of the entire recA gene with the B. cepacia complex-specific primers BCR1 and BCR2, strains possessing these novel RFLP types failed to react with any of the genomovar-specific recA PCR primers. To confirm the status of these strains as members of the B. cepacia complex, nucleotide sequence analysis of the 5′ 527-bp region of the recA gene was performed and phylogenetically analyzed. All strains possessing novel recA RFLP types were members of the B. cepacia complex based on recA phylogenetic analysis. The resulting phylogenetic tree is shown in Fig. 1. However, using current methods these strains have been found to have indeterminate genomovar status and may be novel taxonomic groups within the B. cepacia complex. To confirm this recA-based result, RFLP analysis of the 16S rRNA gene was also performed. All strains of recA RFLP types AZ, I3, S, and U possessed the 16S rRNA gene RFLP pattern 2 (data not shown). This 16S rRNA gene RFLP is shared by the control strains of genomovar I, genomovar III, B. ambifaria (genomovar VII), and B. stabilis. However, none of these novel strains possessed other phenotypic or genetic characteristics associated with B. ambifaria or B. stabilis; hence, they were identified as B. cepacia complex genomovar I-III indeterminate status (Fig. 1).
TABLE 2.
Comparison between 16S rRNA- and recA-based identification of Burkholderia isolates
| Genomovar or species | No. of strains
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA-based identification |
recA-based identification
|
||||||||
| Genomovar or species
|
Not B. cepacia complex | ||||||||
| I | B. multivorans | III-A | III-B | B. stabilis | B. vietnamiensis | I or III | |||
| Burkholderia genomovars I, III, and IV | 89 | 3 | 4 | 50 | 13 | 3 | 0 | 12 | 4 |
| B. multivorans/B. vietnamiensis | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| B. gladioli | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Total | 92 | 3 | 4 | 51 | 13 | 3 | 0 | 12 | 6 |
FIG. 1.
Phylogenetic tree of the B. cepacia complex based on the recA gene. The location of the Catania cable pilus-encoding strain within genomovar III-A is shown (represented by strain CG 10/3). Multiple sequence alignment was performed, and the tree was rooted with the published recA sequence from Bordetella pertussis. Genetic distance is indicated by the scale.
Final prevalence of each B. cepacia complex genomovar.
Final genomovar status identification was attributed based on the recA polymorphism; the results, in comparison with the 16S rRNA gene analysis, are summarized in Table 2. B. cepacia genomovar III was the dominant species present among the Italian isolates examined (69.5%, 64 out of 92 isolates), and of these, the majority (80%, 51 out of 64 isolates) belonged to B. cepacia genomovar recA group III-A.
Genome macrorestriction analysis.
On the basis of PFGE-based RFLP analysis, the epidemiology of B. cepacia strains was assessed. Serial isolates recovered from the same patients produced conserved macrorestriction patterns, demonstrating chronic persistence of individual strain types in 10 patients and recurrence of infection in 2 patients (data not shown).
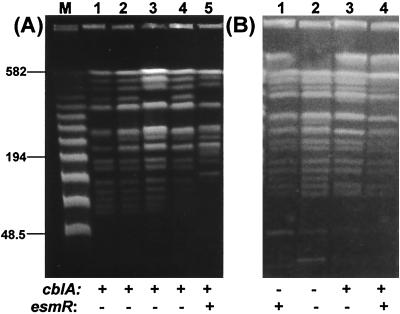
Macrorestriction analysis enabled the identification of at least 27 different clones responsible for the spread of B. cepacia infection. Different epidemiological features, showing cross-transmission or sporadicity of infection, were present in each center (Table 3). A predominant clone was identified in the two Sicilian centers. This “Sicilian epidemic clone” (PFGE strain type A) chronically infected 26 of the 42 B. cepacia complex-positive patients (62%) attending the two treatment centers. This highly transmissible strain was not found outside Sicily. Its PFGE profile was closely related (similarity coefficient, 78%) to the C5424 reference strain, a member of the ET12 North America-United Kingdom transatlantic clone (Fig. 2A).
TABLE 3.
Numbers of patients colonized with cross-transmitted or sporadic strains of different genomovars or species and PFGE types
| Center | No. of patients colonized with strain of genomovar or species and PFGE type
|
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cross-transmitted strains
|
Sporadic strains
|
||||||||||||||||||||||||||||
| I XB | III-A
|
III-B
|
B. stabilis, S | I-III
|
B. multivorans
|
III-A
|
III-B
|
B. stabilis, I | I-III
|
||||||||||||||||||||
| Ab | P | T | XE | UA | Q | BA | SB | L | NDa | Bb | Cb | H | SA | X | Y | Z | U | ND* | G | V | Dc | Ec | Fc | R | UB | ||||
| Catania | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||
| Palermo | 23 | 2 | 2 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| Milan | 3 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||
ND, not determined. DNA was shared in repeated samples.
Three strains, one each of types A, B, and C, were isolates from the same patient.
Three strains, one each of types D, E, and F, were isolates from the same patient.
FIG. 2.
PFGE-macrorestriction profiles of different cblA or esmR variants of the Sicilian epidemic clone. (A) Lanes 1 to 4, isolates from different Sicilian patients; lane 5, ET12 strain C5424 (16). (B) Lanes 1 to 4, cblA or esmR variants of the epidemic clone from different Sicilian patients. A λ ladder molecular size marker was run in the lane labeled M, and the size of relevant bands is indicated in kilobases. The presence (+) or absence (−) of the cblA and esmR markers is indicated below each lane.
PCR detection of cblA and esmR.
Within this study, the presence of published molecular markers of B. cepacia complex transmissibility was demonstrated for the first time within the Italian CF population (Table 1). Cable pilus-associated sequences were present in a total of 33 of the 92 isolates, all from the Sicilian centers, belonging to the B. cepacia genomovars III and I-III. Notably, the epidemic outbreak involving the two Sicilian centers was sustained by 26 different variants of the same genomovar III-A clone: 14 were cblA positive and esmR negative, 7 were cblA positive and esmR positive, 3 were cblA negative and esmR positive, and 2 were cblA negative and esmR negative (Fig. 2B). A total of 43 out of the 59 patients were colonized by B. cepacia complex strains as a result of epidemic spread or cross-transmission: for 23 of these patients, the strains involved were shown to bear the cblA gene, while the remaining 20 strains were cblA negative.
The presence of esmR sequences was shown for 40 of the 92 isolates examined, and all of these belonged to B. cepacia genomovar III: 22 of these esmR-positive strains were associated with epidemic spread, while the remaining 18 were not. Instability of both transmissibility markers within a single genomovar III-A clone was also observed (Fig. 2).
Correlation between bacterial genomovar status and the epidemicity of infection.
Using the genomovar status of each strain obtained by analysis of the recA gene, a correlation between the risk of patient-to-patient cross-infection and genomovar status was made. The highest correlation between cross-infection and genomovar was associated with strains of the B. cepacia complex genomovar III, recA group III-A. There were 43 patients sharing the same strain as a result of cross-infection: in 28 of these cases, the strain involved belonged to genomovar III, recA group III-A. A relative risk of cross-infection with genomovar III-A was calculated as 1.9 (28 cases associated with genomovar III-A compared to 15 cases associated with strains of the other genomovars). recA group III-A included epidemic CF strains from the cable pilus-encoding lineage (18, 20) and the Vancouver outbreaks (15, 17). Phylogenetic analysis of the recA gene sequence confirmed that the Catania cable pilus-carrying strain, responsible for the Sicilian epidemic, was within the same genetic cluster and closely related to these epidemic genomovar III-A strains (Fig. 1). A second genomovar III-A strain, of distinct PFGE type, was also responsible for cross-infection of two patients. Other instances of patients sharing the same strains were observed for B. cepacia of all the other genomovars except for B. multivorans (Table 3).
DISCUSSION
The worldwide increase of B. cepacia infection in CF patients suggests its epidemic spread, but the source and transmissibility of strains involved remain controversial. It has been suggested elsewhere that strains of the B. cepacia complex are not equally transmissible; rather, there exist highly transmissible lineages, presumably of a clonal nature, and heterogeneous lineages of negligible transmissibility (23). We investigated the hypothesis that transmissibility might be associated with a particular taxonomic group, genomovar or species, within the B. cepacia complex.
From our screening, the prevalence of infection of B. cepacia complex in CF patients in Italy was 8.6% overall, 18.3% in Sicily and 5% in Milan. The prevalence of B. cepacia infection, as reported in a survey published in 1997, was 3.8%, clearly underestimated, since at that time only a few laboratories used the selective culture media for isolation (24). A higher prevalence (20.5%) was reported from the Campania region, southern Italy, in 1999, when appropriate microbiological procedures were used, although identification was based on standard laboratory procedures (30).
Analysis of the recA gene of the B. cepacia complex is a more discriminatory molecular approach than the analysis of the 16S rRNA gene to identify isolates and evaluate the specific risk posed by infection with a given genomovar and the epidemic spread of infection. We evaluated this risk as being about twice that for infection sustained by microorganisms of genomovar III, recA group III-A, with respect to other strains of the complex. This group included the Sicilian epidemic clone, which was phylogenetically related to the ET12 North America-United Kingdom transatlantic clone.
Although infection of the majority of patients involved in the Sicilian outbreak was sustained by strains possessing the cblA gene, a genetic marker associated with transmissibility, 5 of the 26 patients involved were actually colonized with a cblA-negative variant of the same clone, suggesting that the region may be subject to some instability. The PFGE fingerprints of these cblA-negative variants of the type clone showed minor changes in their banding profile, which may be associated with genome rearrangement and loss of the cblA gene. Early studies of the B. cepacia genome showed pronounced genome plasticity due to its multiple replicon organization and large numbers of insertion sequences (12, 29). In light of the heavy use of genetic markers for the epidemiological management of B. cepacia complex infections in CF (3, 16, 20, 23), the stability of these markers must be better understood.
Our findings suggest that the esmR and the cblA sequences may be encoded on a chromosomal region which is unstable in some B. cepacia genomovar III epidemic strains. Genetic instability has been well documented for the esmR open reading frame (16, 18), while instability of the cblA gene had not been previously demonstrated.
In conclusion, we have demonstrated that genomovar III is the most prevalent genomovar of B. cepacia complex bacteria among a population of Italian CF patients as previously reported (28). We also observed CF infection and patient-to-patient spread of B. cepacia complex bacteria of novel taxonomic status. Genetic identification of bacteria recovered from clinical settings as well as from other sources is essential to further understanding of the transmissibility and pathogenic potential of different genomovars or species within the B. cepacia complex.
ACKNOWLEDGMENTS
This work was partially supported by a grant to A.A. and S.S. from the University of Catania (Progetti di Ricerca di Ateneo). E.M. is grateful to the UK Cystic Fibrosis Trust (project grant PJ472) for financial support.
We thank M. L. Garlaschi, Istituti Clinici di Perfezionamento, Milan; D. Natoli, A. Collura, and T. Pensabene, Ospedale G. Di Cristina, ARNAS, Palermo; C. Pasquarella, Dipartimento di Igiene di Perugia; and I. Collebrusco, Ospedale R. Calai, Gualdo Tadino, for sending strains included in the study. E.M. is grateful to Julie Fadden for her technical assistance. We are grateful to Antony Bridgewood for the critical English revision of the manuscript.
REFERENCES
- 1.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and V by PCR. J Clin Microbiol. 1999;37:1335–1339. doi: 10.1128/jcm.37.5.1335-1339.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossi A, Padoan R, Gagliardini R, Manca A, Zanda M, Miano A, Marianelli L, Romano L, Magazzù G, Raia V, Pardo F, Gazincich G, Salvatore D, Quattrucci S, de Candusio G, Lucidi V, Faraguna D, Mastella G, Giunta A. 20th European Cystic Fibrosis Conference, Brussels, Belgium, 18 to 21 June 1995. 1995. The Italian cystic fibrosis patients registry; p. 39. [Google Scholar]
- 3.Clode F E, Kaufmann M E, Malnick H, Pitt T L. Distribution of genes encoding putative transmissibility factors among epidemic and nonepidemic strains of Burkholderia cepacia from cystic fibrosis patients in the United Kingdom. J Clin Microbiol. 2000;38:1763–1766. doi: 10.1128/jcm.38.5.1763-1766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye T, LiPuma J J, Henry D, Vandemeulebroucke K, Gillis M, Speert D P, Vandamme P. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int J Syst Evol Microbiol. 2001;51(Part 2):271–279. doi: 10.1099/00207713-51-2-271. [DOI] [PubMed] [Google Scholar]
- 5.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme.Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex comprising biocontrol and cystic-fibrosis related isolates. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 6.Gillis M, Van T V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 7.Goldstein R, Sun L, Jiang R, Sajjan U, Forstner J, Campanelli C. Structurally variant classes of pilus appendage fibers coexpressed from Burkholderia (Pseudomonas) cepacia. J Bacteriol. 1995;177:1039–1052. doi: 10.1128/jb.177.4.1039-1052.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govan J R W, Brown P H, Maddison J, Doherty C J, Nelson J W, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 9.Grothues D, Koopmann U, Van der Hardt H, Tummler B. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol. 1988;26:115–123. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes A, Nolan R, Taylor R, Finley R, Riley M, Jiang R, Steinbach S, Goldstein R. An epidemic of Burkholderia cepacia transmitted between patients with and without cystic fibrosis. J Infect Dis. 1999;179:1197–1205. doi: 10.1086/314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson W, Tyler S, Rozee K. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J Clin Microbiol. 1994;32:924–930. doi: 10.1128/jcm.32.4.924-930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessie T G, Hendrickson W, Manning B D, Devereux R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol Lett. 1996;144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 13.LiPuma J J. Burkholderia cepacia. Management issues and new insights. Clin Chest Med. 1998;19:473–486. doi: 10.1016/s0272-5231(05)70094-0. [DOI] [PubMed] [Google Scholar]
- 14.LiPuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 2000;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahenthiralingam E, Simpson D, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahenthiralingam E, Coenye T, Chung J, Speert D P, Govan J R W, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahenthiralingam E, Bischof J, Byrne S K, Radomski C, Davies J E, Av-Gay Y, Vandamme P. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and II. J Clin Microbiol. 2000;38:3165–3173. doi: 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitt T L, Kaufmann M E, Patel P S, Benge L C A, Gaskin S, Livermore D M. Type characterization and antibiotic susceptibility of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J Med Microbiol. 1996;44:203–210. doi: 10.1099/00222615-44-3-203. [DOI] [PubMed] [Google Scholar]
- 20.Sajjan U, Sun L, Goldstein R, Forstner J. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J Bacteriol. 1995;177:1030–1038. doi: 10.1128/jb.177.4.1030-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Struelens M. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2:2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, Jiang R, Steinbach S, Holmes A, Campanelli C, Forstner J, Tan Y, Riley M, Goldstein R. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat Med. 1995;1:661–666. doi: 10.1038/nm0795-661. [DOI] [PubMed] [Google Scholar]
- 24.Taccetti G, Campana S. Microbiologic data of Italian cystic fibrosis patients. Eur J Epidemiol. 1997;13:323–327. doi: 10.1023/a:1007373700089. [DOI] [PubMed] [Google Scholar]
- 25.Taccetti G, Campana S, Marianelli L. Multiresistant non-fermentative Gram-negative bacteria in cystic fibrosis patients: the results of an Italian multicenter study. Eur J Epidemiol. 1999;15:85–88. doi: 10.1023/a:1007504524034. [DOI] [PubMed] [Google Scholar]
- 26.Tenover F, Arbeit R, Goering R, Mickelsen P, Murray B, Persing D, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandamme P, Holmes B, Vancanneyet M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 29.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, de Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villari P, Iacuzio L, Vozzella E A, Raia V, Torre I. Epidemiologia di Burkholderia cepacia in pazienti con fibrosi cistica: primi risultati di uno studio comprendente la tipizzazione genotipica dei ceppi isolati. Ann Ig. 1999;11:501–506. [PubMed] [Google Scholar]