Abstract
Exosomes, a subset of the extracellular vesicle (EV) group of organelles, hold great potential for biomarker detection, therapeutics, disease diagnosis, and personalized medicine applications. The promise and potential of these applications are hindered by the lack of an efficient means of isolation, characterization, and quantitation. Current methods for exosome and EV isolation (including ultracentrifugation, microfiltration, and affinity-based techniques) result in impure recoveries with regard to remnant matrix species (e.g., proteins, genetic material) and are performed on clinically irrelevant time and volume scales. To address these issues, a polyethylene terephthalate (PET) capillary-channeled polymer (C-CP) fiber stationary phase is employed for the solid-phase extraction (SPE) of EVs from various matrices using a micropipette tip-based format. The hydrophobic interaction chromatography (HIC) processing and a spin-down workflow are carried out using a table-top centrifuge. Capture and subsequent elution of intact, biologically active exosomes are verified via electron microscopy and bioassays. The performance of this method was evaluated by capture and elution of exosome standards from buffer solution and three biologically relevant matrices: mock urine, reconstituted non-fat milk, and exosome-depleted fetal bovine serum (FBS). Recoveries were evaluated using UV-Vis absorbance spectrophotometry and ELISA assay. The dynamic binding capacity (50%) for the 1-cm-long (~ 5 μL bed volume) tips was determined using a commercial exosome product, yielding a value of ~ 7 × 1011 particles. The novel C-CP fiber spin-down tip approach holds promise for the isolation of exosomes and other EVs from various matrices with high throughput, low cost, and high efficiency.
Graphical abstract
Electronic supplementary material
The online version of this article (10.1007/s00216-020-02728-z) contains supplementary material, which is available to authorized users.
Keywords: Exosomes, Extracellular vesicles (EVs), Capillary-channeled polymer (C-CP) fibers, Hydrophobic interaction chromatography (HIC), Isolation, Purification
Introduction
Exosomes are 30–130 nm-sized extracellular vesicles (EVs) containing genetic, proteomic, and intracellular content that reflect the biophysical characteristics of the cells of origin, and engage in diverse pathological and physiological roles [1, 2]. Exosomes are released from most cell types through multivesicular bodies (MVBs), which are distinctly created through the endosomal pathway [1, 3, 4], different from the biogenesis of many other extracellular vesicles [5, 6]. Exosomes carry intravesicular cargo, including DNA, RNA, miRNA, as well as surface biomarker proteins—all promising tools for unraveling the inner-workings of disease progression [7]. Exosomes mediate a plethora of inter- and intracellular processes, including cellular communication and signaling phenomena, and contain essential cargo for local and distal cargo transport processes [8]. Because the dysregulation of intercellular communication processes leads to cancers and immune-physical malfunctions, exosomes/EVs have become relevant to understanding many complex biochemical interactions [9–11]. Additionally, as the rate of exosome biogenesis differs based on the cell of origin, the simple ability to readily determine the concentration of exosomes is of high interest. An upregulation of exosome biogenesis is indicative of active disease progression [12, 13]. Increased exosome-mediated signaling is characteristic of invasive tumor phenotypes [12], so it is essential to be able to efficiently quantify exosomes in various types of samples. A pool of cells is well represented by a collection of exosomes, making them a promising analytical target for liquid biopsy applications.
Exosomes/EVs are expressed by most cells and, as such, can be collected from bodily fluids, including urine [14], saliva [15], blood (plasma [15] and serum [16]), breast milk [17, 18], and cerebrospinal fluids [19, 20], and are also released in vitro by cultured cells [21]. Furthermore, exosomes and other extracellular vesicles have been identified in all three physiological domains of life (archaea [22], bacteria [23, 24], and eukarya [22]) and are active agents of nutrient delivery by interspecies communication through intake of foods like raw vegetables [25]. A challenge in the progression of exosome/EV-based applications lies in the recovery of clean, stable, and biologically relevant vesicles for genetic profiling, bioengineering, and biomarker classification.
A large number of approaches have been used for exosome/EV isolation, including ultracentrifugation, differential centrifugation, density-gradient centrifugation, size exclusion chromatography, affinity chromatography, and several polymer-based precipitation techniques [8, 26, 27]. These separation techniques rely on either the size and density of the EVs, or the affinity of the exosomes for antibodies to specific surface marker proteins such as Alix, CD9, CD81, TSG101, and HSP70 [27]. Current exosome/EV isolation methods are tedious and are often used following multiple high-speed ultracentrifugation (> 100,000×g) steps to remove debris and pelletize the exosomes [26]. While the high-speed ultracentrifugation method is the most widely used technique for generic exosome isolation, it does not always efficiently isolate exosomes from large protein aggregates or other vesicular structures efficiently. In all, these techniques are time-consuming and sample-size burdensome to the point of limiting the use of exosomes on the clinical scale [27, 28]. Because biological sample matrices are extremely complex and varied, robust separation techniques are crucial for future clinical applications and fundamental research [29].
Marcus and co-workers have described the use of capillary-channeled polymer (C-CP) fibers as stationary phases for liquid chromatography (LC) separations of proteins via reversed-phase (RP), ion exchange (IEC), hydrophobic interaction (HIC), and affinity modalities [30–36]. The combination of high column permeability and low surface porosity provides high throughput and yield macromolecule separations [37–39]. Bruce, Marcus, and colleagues have also recently reported a method for exosome/EV isolation using an HIC mode on a poly (ethylene terephthalate) (PET) C-CP fiber phase [40–43]. The use of the HIC elution strategy allows for exosome isolations based on the vesicles’ inherent hydrophobicity (partially a function of their size), allowing non-destructive bulk recoveries of exosomes/EVs for further interrogation and applications. Capture/elution under HIC solvent conditions preserves the morphology of the vesicles isolated from various matrices, including cell culture milieu [40], urine [40, 42], and human plasma [41]. In terms of potential implementation scenarios, isolations are performed on < 100 μL sample volumes on time scales of < 10 min. The simple chromatographic method also shows promise for bulk recovery of EVs for fundamental biochemistry and preparative applications.
While a standard liquid chromatograph is not overly burdensome in the analytical chemistry laboratory, it is not practical in many biochemical and clinical situations. Solid-phase extraction (SPE) techniques are widely applied for sample preparation of biological specimens, as they allow for efficient separation of analytes from the originating complex matrices [44–46]. SPE is a form of step-wise chromatography that is designed to extract and adsorb components of interest from a liquid phase onto a stationary phase (similar to LC separations), thus serving as a means of pre-concentration and affecting a matrix modification. Many modalities influence the passage of sample solutions through an SPE bed, but the use of a table-top or microcentrifuge is particularly attractive in terms of very low operational complexity and overhead [47, 48]. In this regard, C-CP fibers can be employed either as the stationary phase column for HPLC or employed in 1-cm segments fit to a micropipette tip to affect SPE in a spin-down mode using a table-top centrifuge [49–51]. In these applications, fiber phases have also been used as a means of desalting proteins before MS characterization [52] and also to affect immunoaffinity capture [51].
Here, PET C-CP fiber micropipette tips are employed in a novel spin-down HIC protocol for the timely, efficient, and structurally preserving isolation and quantification of exosomes from various matrices (aqueous solution, mock urine, reconstituted non-fat milk, and an exosome-depleted fetal bovine serum). Mock matrices were used to normalize and control the exosome quantity input, while also presenting basic sample constituents. The goal is to quantify and characterize exosome recoveries of known spike concentrations from the various matrices without interference or introduced bias from native exosome-containing biofluids. The sequential aspects of immobilization and recovery are affected for multiple tips in parallel, in a total processing time of < 5 min. The capture of intact exosomes is verified via scanning electron microscopy (SEM) and confocal microscopy, with the efficacy of the elution confirmed via transmission electron microscopy (TEM) and dot blot analysis. The binding capacity of the 1 cm fiber tips is evaluated via sequential applications of sample aliquots until an observed breakthrough, with the recoveries determined spectrophotometrically. Finally, the ability to quantify EV recoveries via absorbance measurements is demonstrated and employed in the evaluation of recoveries of exosomes spiked into various mock-biofluid matrices. This simple and straightforward method for exosome isolation and quantification opens the door for future fiber platform optimization for selective EV-type isolations for clinical diagnostics and fundamental biochemistry research.
Materials and methods
Materials
HPLC-grade acetonitrile (ACN) was obtained from ThermoFisher Scientific (Pittsburgh, PA, USA). Deionized water (DI-H2O, 18.2 MΩ-cm) was obtained from a Milli-Q water purification system (Millipore Sigma, Merck, Germany, USA). Ultra-pure ammonium sulfate was obtained from VWR (Radnor, PA, USA). Biotechnology-grade glycerol was purchased from VWR (Sokon, OH, USA). Non-fat dry milk was purchased from Bio-Rad (Hercules, CA, USA). Phosphate-buffered saline (PBS, 0.067 M (PO4), pH 7.4) and exosome-depleted fetal bovine serum (FBS) were obtained from ThermoFisher Scientific (Waltham, MA, USA). Tris-buffered saline (TBS, 0.1 M, pH 8.0) was obtained from Sigma Aldrich (St. Louis, MO, USA). Uranyl acetate, 16% paraformaldehyde (formaldehyde) aqueous solution, and formvar/carbon film 10 nm/1 nm thick on square 200-mesh copper grids were obtained from Electron Microscopy Sciences (Hatfield, PA, USA). The 1-Step Ultra TMB-ELISA substrate was purchased from ThermoFisher Scientific (Pittsburgh, PA, USA). The Anti-rabbit IgG (H+L) 10 nm Silver Conjugate (OD 7.5) and Silver Enhancer Kit for membranes were obtained from Cytodiagnostics (Burlington, ON, Canada).
The mock urine was prepared based on the recipe as reported by Khan et al. [52], consisting of an aqueous solution of potassium chloride (0.2 g L−1), sodium chloride (8 g L−1), disodium hydrogen phosphate (1.14 g L−1), potassium dihydrogen phosphate (0.2 g L−1), 200 μL L−1 McCormick yellow food coloring (water, propylene glycol, FD&C yellow 5, and propylparaben), urea (114.1 g L−1), and DI-H2O up to 1 L. Hydrochloric acid and sodium hydroxide were used to adjust the pH to 7.5. The solution of 2% non-fat dry milk was dissolved in DI-H2O to create the milk matrix.
Instrumentation
Three absorbance spectrophotometers were employed in these studies, based on the required sample volume for measurement, the instrument’s sensitivity to changes in absorbance, and the sample introduction method. A NanoVue Plus UV-Vis spectrophotometer (GE Healthcare, Chicago, IL, USA) was used to measure the direct absorbance of the concentrated exosome eluates (1 μL) from the C-CP fiber tip. A GENESYS 10S UV-Vis spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA) was used to measure the absorbance of diluted exosome eluate. The Synergy H1 Hybrid Multi-Mode Reader (BioTek, Winooski, VT) was used to determine the UV-Vis absorbance (450 nm) of samples in the 96-well format as employed in an ELISA assay employing the 1-Step Ultra TMB Substrate.
Electron microscopy was employed as a confirmatory tool for the structural integrity of both immobilized and eluted EVs. SEM was performed using a Hitachi S-4800 (Chiyoda, Tokyo, Japan) to confirm the capture of intact EVs on the C-CP fiber surface. Transmission electron microscopy (TEM) was performed using a Hitachi HT7830 (Chiyoda, Tokyo, Japan) to confirm the release of intact EVs from the C-CP fiber surface. STEM imaging was performed using the Hitachi SU9000 CFE SEM/STEM to observe the integrity of eluted exosomes. The methods for fixing and imaging of these populations, which are not innovative in their own right, are described in the Electronic Supplementary Material (ESM).
Confocal microscopy was used to image the C-CP fiber tip-captured exosomes after undergoing immune-recognition procedures for the confirmation of the capture of exosomes exhibiting the CD81 tetraspanin marker protein. In preparation for this technique, the fiber-captured vesicles were stained using a mouse primary antibody to CD81 (Santa Cruz Biotechnology, Dallas, TX) followed by a goat anti-mouse secondary antibody labeled with AlexaFluor 647 before super-resolution confocal imaging with a Leica SP8 confocal microscope with Hyvolution super-resolution software (Leica Microsystems, Buffalo Grove, Illinois). A sandwich enzyme-linked immunosorbent assay (ELISA) was used to detect and quantify the expression of the tetraspanin exosomal marker protein, CD81, in exosome recoveries following elution from the C-CP fiber tip from various matrices. CD81 expression in the recoveries of exosomes isolated by the C-CP spin-down tip was further confirmed using an immunodot blot assay. Briefly, recovered exosomes were captured by the immobilized CD81 mouse antibody on a PVDF membrane, subsequently detected using rabbit primary antibodies to generic tetraspanin antibodies (CD9, CD81, CD63), and visualized using a goat anti-rabbit silver nanoparticle conjugate, followed by the use of a silver enhancement kit to amplify the resultant response (see the ESM).
Methods
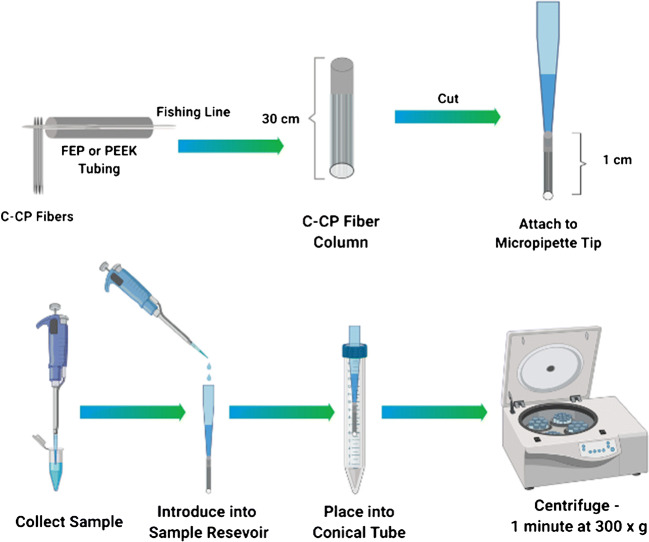
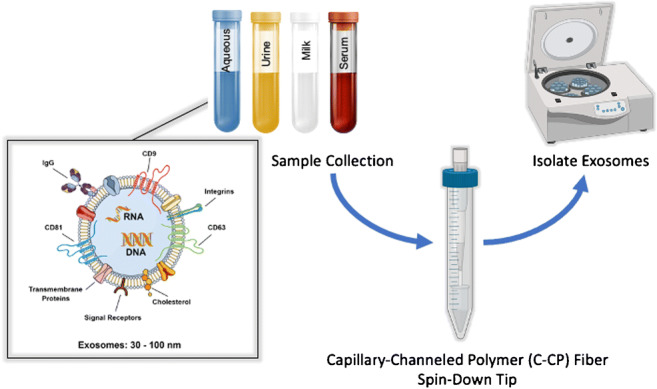
The C-CP fiber SPE tip assembly process is depicted in Fig. 1. Poly(ethylene terephthalate) (PET) capillary-channeled polymer fibers were extruded by the Clemson University School of Materials Science. The C-CP fiber tips were constructed as previously reported [48] (see the ESM). Ultimately, tips of 1 cm length, having an inner diameter of 0.8 mm, and an interstitial fraction of ~ 0.6, yielded bed volumes of ~ 3 μL. The method for mounting the spin-down tips for processing and collection of EV fractions has also been described previously [48] (see the ESM). The efficient reuse (n > 15) of the C-CP fiber stationary phase has been demonstrated in a column format used in HPLC isolation of exosomes from a mock urine matrix [42]. However, given the low consumable cost (< $0.5 USD per tip) and for the sake of convenience, new C-CP micropipette tips were employed for each exosome isolation here.
Fig. 1.
The practical steps of C-CP fiber tip fabrication and the spin-down approach to isolation and purification of EVs. (See ESM for details)
Lyophilized and purified exosomes from the urine of reportedly healthy donors were obtained from Galen Laboratory Supplies (North Haven, CT, USA) with a prepared suspension concentration of 2.27 × 1012 particles mL−1 (provider-determined by nanoparticle tracking analysis (NTA)). For HIC-based processing, EVs in the mock sample matrices were mixed 1:1 with 2 M ammonium sulfate at pH = 7.5. Aliquots of 100 μL per trial were passed through the C-CP fiber tips under 300×g centrifugal force for 1 min each. Under the high salt conditions, the target vesicles and latent proteins (from the original sample) were retained on-fiber. After the capture of the vesicles, the fiber surfaces were washed with 100 μL of DI-H2O. Protein elution was induced by passage of 50 μL of 25% glycerol in PBS under the same centrifugation conditions, with the final elution of the captured EVs induced using 50 μL of 50% glycerol in PBS. The elution of proteins by 25% glycerol and exosomes by 50% glycerol has been confirmed by SEM imaging of the fiber surfaces after the various steps in this workflow as well as in the use of acetonitrile as the mobile phase modifier [42, 43], as it is here. The eluted EVs were quantified by diluting a 3-μL aliquot to 1.5 mL with DI-H2O. Absorbance measurements were performed using a GENESYS 10S UV-Vis spectrophotometer. Additionally, an ELISA was used to confirm the presence of CD81-expressing EVs in the spin-down tip recoveries.
To determine dynamic binding capacity, breakthrough experiments were performed using 21 successive 50 μL aliquots of the diluted exosome standard (4.65 × 107 particles per 50 μL aliquot in 1 M ammonium sulfate with 25% glycerol), spun through the tips (300×g, 1 min each). Use of the glycerol modifier inhibits the adsorption of adventitious proteins. The fiber surfaces were then washed five times with 50 μL aliquots of diH2O.
Results and discussion
Capture and elution fidelity
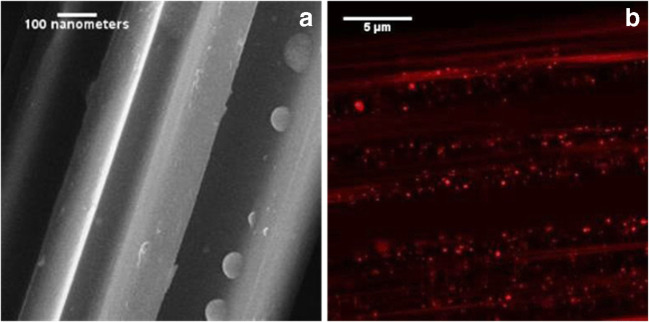
As with all forms of biomolecule/particle isolation, a successful SPE spin-down methodology for exosome/EV isolation and recovery must provide not only for separation, but must do so without compromising the physical and biological attributes of the EVs. In this case, EVs must be isolated with respect to the components of the sample matrix, including salts, small molecules, such as amino acids, sugars, proteins, and genetic material. Previous reports have illustrated this capability via HIC separation of exosomes from diverse media [40–42]. In the case of the spin-down tip processing, the integrity of the physical and biological attributes of the exosomes was evaluated via SEM and immunofluorescence, respectively, following the elution steps to remove salts and adventitious proteins. In Fig. 2a (for the case of the commercial exosomes dispersed in water), the surface of the C-CP fibers at this stage is pristine, as indicated by the presence of globular vesicles without any remnants of salt crystals or the like.
Fig. 2.
Physical and biologic imaging of exosomes adsorbed to PET C-CP fiber surface via a scanning electron microscopy and b super-resolution confocal fluorescence microscopy
To further illustrate the integrity of the captured exosomes, super-resolution confocal microscopy imaging was performed. Exosomes captured on C-CP fiber surfaces were immunolabeled using a primary antibody to the tetraspanin surface marker protein, CD81, and a fluorescent secondary antibody (AlexaFluor 647 goat anti-mouse). As seen in Fig. 2b, there are dispersed nanobodies (of the size range expected for the target exosomes) within the ~ 25 × 25 μm2 viewing region. Due to the resolution limits of the confocal microscope (~ 140 nm), it is important to note that fluorescent particles observed here are not necessarily individual exosomes, but perhaps small aggregates producing a more intense fluorescent response. Nevertheless, with regard to capture, the target exosomes are well dispersed on the fiber surface (without substantial debris), while maintaining their basic physical morphology and surface protein makeup. Indeed, the characteristics depicted in Fig. 2 are the first steps towards affecting a practical exosome diagnostic platform.
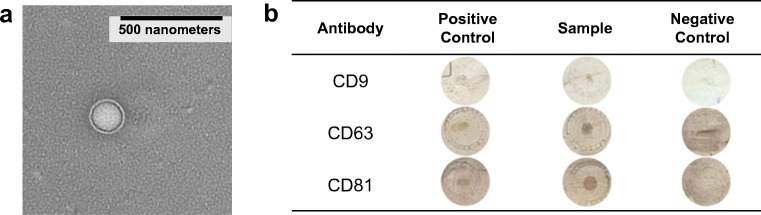
In those cases where further exosome characterization is required, such as in the search for surface biomarkers or genetic analysis (e.g., RNA-Seq) of the vesicular cargo, the organelles must be recovered (eluted) while maintaining their physical integrity and biological function. The most common method for assessment of the morphology of individual exomes is transmission electron microscopy, where both the size and vesicular structure are revealed. The TEM micrograph of an HIC-eluted EV (Fig. 3a) illustrates the successful maintenance of the physical structure through the isolation process. The biological fidelity of exosome populations is readily assessed through the use of dot blot assays (Fig. 3b), wherein a positive immune-response is obtained for the CD9, CD81, and CD63 antibodies in the post-tip isolation eluates. As seen in the various exposures, the recovered exosomes do indeed retain the surface markers of the three tetraspanin proteins confirming the presence and viability of the exosomes. While the dot blots do not reflect the retention of the encapsulated genetic materials, they suggest that the expected membrane-bound proteins remain intact.
Fig. 3.
Physical and biological characterization of exosomes eluted from PET C-CP fiber spin-down tips via a transmission electron microscopy and b dot blot immunoassay
Dynamic binding capacity (DBC)
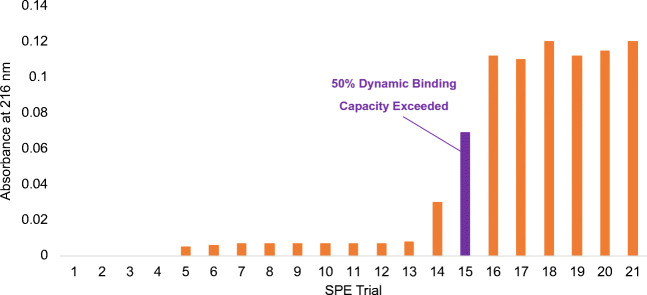
The ability to effectively isolate and purify EVs is only relevant to the extent that it yields the required density of EVs necessary to provide meaningful sample data. As a general rule, most RNA-sequencing analyses require 109–10 exosomes for accurate profiling, while LC/MS proteomic studies require on the order of 1010–11 exosomes [53–59] To this end, the dynamic binding capacity of the 1 cm C-CP fiber spin-down tips was determined (Fig. 4). Unlike in the case of continuous processes [38], a frontal analysis was required. This was performed using discrete 50 μL aliquots of test solutions (exosomes in 25% glycerol/1 M (NH4)2 SO4), with the pass-through exosome content used to assess breakthrough/overload. Figure 4 shows the determined absorbance values, obtained by diluting 3 μL of the eluate in DI-H2O in a 1.5-mL cuvette. The absorbance response is not significant until aliquot #14, wherein the pass-through content increases rapidly, and a plateau is reached beyond aliquot #16, suggestive of surface saturation. Based on the general response, the eluate absorbance reaches one half of the steady maximum value (a measure of reaching DBC) with aliquot #15. At this point, based on per-aliquot particle densities of 4.65 × 1010, a DBC value on the order of ~ 7 × 1011 is achieved for a total volume of 750 μL. Though there is no consensus regarding a “healthy range” for exosome concentration, this value is in line with that expected in many native biofluids, including urine, milk, serum, and plasma. The capacity demonstrated at this early stage is on-par for what would be desired in the clinical and biochemical laboratory arenas.
Fig. 4.
Breakthrough analysis of 50 μL aliquot additions of 4.65 × 1010 particles per dose. The 50% dynamic binding capacity is surpassed during trial 15 at 6.98 × 1011 particles
Quantification
In previous EV separations employing PET C-CP fiber columns in a Dionex Ultimate 3000 HPLC system (ThermoFisher Scientific, Waltham, MA, USA), UV-Vis absorbance at 203, 216, and 280 nm was used as a method for EV detection [40]. Even with the well-known optical absorbance of some buffer/matrix components at these wavelengths, a successful method of exosome isolation should alleviate their contributions and allow ready quantification. The absorbance response observed in this instance is not due to the molecular absorption of an innate biomolecule, but rather corresponds to the light scattering due to the presence of the particles. Ultimately, the absorbance response was found to be directly proportional to the exosome content, for particles of different sources. As most methods of EV isolation carry along remnant proteins, there is a potential that the absorbance (scattering)-based measurement could be affected by their presence.
Based on the fact that the extent of scattering would be (nominally) inversely related to the incident wavelength and that proteins (being composed of aromatic amino acids) absorb at 280 nm, response functions were prepared at 203, 216, and 280 nm. Lyophilized exosome standards from the urine of reportedly healthy donors (previously shown to have latent proteins present) were used to create standard curves. Here, 1–35 μL of the exosome standards (2.3 × 1012 particles mL−1) were diluted to 1.0 mL in DI-H2O, presenting a concentration range of ~ 2.3 × 109–8.0 × 1010 particles mL−1 (Table 1). The slope of the 280 nm function is approximately 40% higher than the lower wavelengths. The stronger absorbance at 280 nm reflects the inevitable presence of proteins (which contain aromatic amino acids) in the commercial exosome material. Indeed, the characteristics for the lower wavelengths are virtually identical, with much better regression statistics than at the higher wavelength. Based on these figures of merit, and fewer contributions from background proteins, the shorter wavelengths are preferred. While the limits of detection and quantification are not as low as with other methods (e.g., immunoassays) [60–65], the values are relevant for most biological and clinical systems of interest, particularly in consideration of the total sample volume required (< 50 μL) and ease of determination.
Table 1.
Absorbance response characteristics for exosome standards in aqueous solution at 203, 216, and 280 nm
| λ (nm) | Response function | R2 | LOD (particles mL−1) | LOQ (particles mL−1) |
|---|---|---|---|---|
| 203 | y = 5E−16x + 0.0076 | 0.9958 | 6.05 × 109 | 2.02 × 1010 |
| 216 | y = 5E−16x + 0.0077 | 0.9938 | 7.58 × 109 | 2.53 × 1010 |
| 280 | y = 7E−16x + 0.0069 | 0.9808 | 1.22 × 1010 | 4.08 × 1010 |
Isolation and quantification of EVs/exosomes in diverse media
As proof of concept towards the efficacy of the HIC spin-down tip approach to exosome isolation and quantification, the commercial exosome standards (2.73 × 1012 particles mL−1) were spiked into DI-H2O, mock urine, reconstituted non-fat milk, and exosome-depleted fetal bovine serum (FBS) matrices. Two dilution factors were employed (1/100; 1/1000), as a quantitative test of the response, as well as tolerance towards the challenges of the matrices themselves. The matrices were mixed (50:50 v/v) with the HIC loading solvent (2 M (NH4)2SO4) in PBS. While diH2O presents a pristine environment, the mock urine matrix presents high salinity and is small molecule-heavy, the milk has high protein content, and the FBS contains fat and high-protein content. These model biofluids are obvious target matrices from which exosome/EVs may be extracted for diagnostic purposes. In terms of loading and elution, the procedure involved a spin-down under high-salt conditions, followed by elution of proteins with 50 μL of 25% glycerol and 1 M (NH4)2SO4 in PBS. This fraction was collected for absorbance measurements of protein/exosome content. Finally, the EV fraction was eluted in 50 μL of 50% glycerol in PBS and collected for the determination of vesicle content. Though glycerol has been used as a biological preservative [66], it is not ideal for all downstream analyses (i.e., proteomic analysis) where necessary vesicle lysing may be hindered. In these cases, acetonitrile may be used as a substitute elution phase, as previously reported [42, 43].
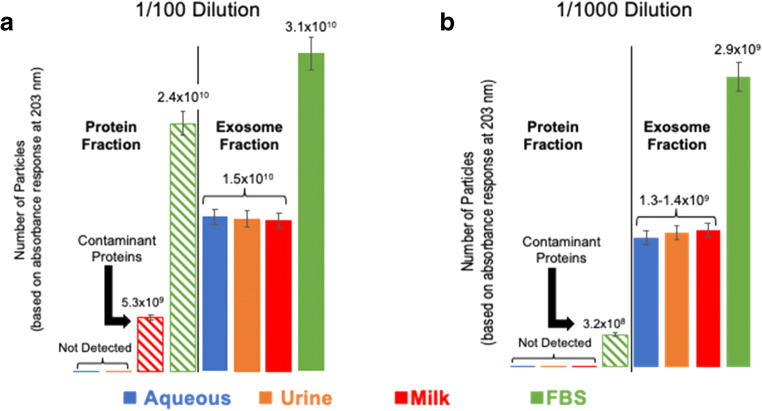
Essential to the quantification process of EVs in different matrices is the assumption that EVs may be quantitatively immobilized and recovered from the fiber surfaces. The latter point has been evaluated in recent studies using the chromatographic (column) platform, wherein recoveries of adsorbed EVs were greater than 80% [43]. Parallel evaluation of the recoveries was performed here via UV absorbance (using the previously generated aqueous matrix calibration functions) and an ELISA assay. The determined numbers of EV particles for the two dilution factors, as determined via optical absorbance (203 nm), are presented in Fig. 5. Aliquots (50 μL) for both the protein and exosome elution fractions were diluted to 1 mL for the absorbance measurements. Starting with the lowest (1/100) dilution factor, no absorbance response is seen in the protein fractions for aqueous and mock urine phases, but there is a measurable absorbance, equivalent to 5.3 × 109 and 2.4 × 1010 EVs, for the milk and exosome-depleted FBS matrices, respectively. These respective responses are not surprising, as the latter matrices have appreciable protein content and corresponding appreciable absorbance, while the aqueous and urine matrices do not. On the other hand, absorbance measurements taken of the presumed EV fraction yield statistically identical values for the aqueous, mock urine, and non-fat milk matrices, as they would be expected. Interestingly, a much higher (~ 2×) recovery of EVs was observed in the exosome-depleted FBS exosome elution fraction. The precision of triplicate measurements for each of the matrices was better than 8.4% RSD.
Fig. 5.
Post-isolation of exosome standards spiked into various matrices (50 μL) using the PET C-CP fiber tip spin-down method. The concentrations of exosomes recovered were determined based on absorbance response (1 μL) when compared to the standard curve of Table 1. a Quantified recovery of exosomes from mock matrices of 1/100 concentration and b quantified recovery of exosomes from mock matrices of 1/1000 concentration. The loaded 1/100 and 1/1000 solutions theoretically contain 2E10 and 2E9 exosomes, respectively
For the case of the higher dilution factor (1/1000), it would be expected that the recoveries would be proportionally (~ 10×) less, but potential matrix effects would be lessened as well. Here, the responses for the protein elution fractions for the aqueous, mock urine, and non-fat milk matrices fall below the level for accurate determination. The FBS protein elution still shows a measurable absorbance response, equivalent to 3.2 × 108 EVs. This is to be expected with the high concentration of total protein in the original matrix. The greater than expected decrease in apparent concentration is due to lessened amounts of protein aggregation in the more dilute solution. That noted, the determined concentrations in the respective EV fractions are indeed ~ 10× less than the more concentrated case for all matrices. Here again, a high level of precision in the EV recovery is obtained (< 6.9% RSD), with the determined particle numbers across the first three matrices being virtually the same, and a significantly higher exosome recovery again for the exosome-depleted FBS matrix. Thus, based on the absorbance-based quantification method, there is no significant difference in EV recoveries across the diverse aqueous, mock urine, and non-fat dry milk matrices. More importantly, the fractional recoveries for the two dilutions are approximately 75% versus the initial number of EVs applied to each tip for these matrices. This value reflects a significantly more efficient recovery of exosomes when compared to the fractional recoveries of other methods, such as ultracentrifugation, which results in equal or lesser concentrated recoveries of exosomes, though requiring nearly 90 times the starting sample volume. As previously mentioned, a significant increase in recovery was observed from the FBS matrix. Marketed as an “exosome-depleted” FBS source, the manufacturer claims the depletion of 90% or more of native endogenous exosomes. The increase in EV recovery for the FBS matrix may be due to remnant exosomes from the native FBS matrix (known to contain high concentrations of EVs).
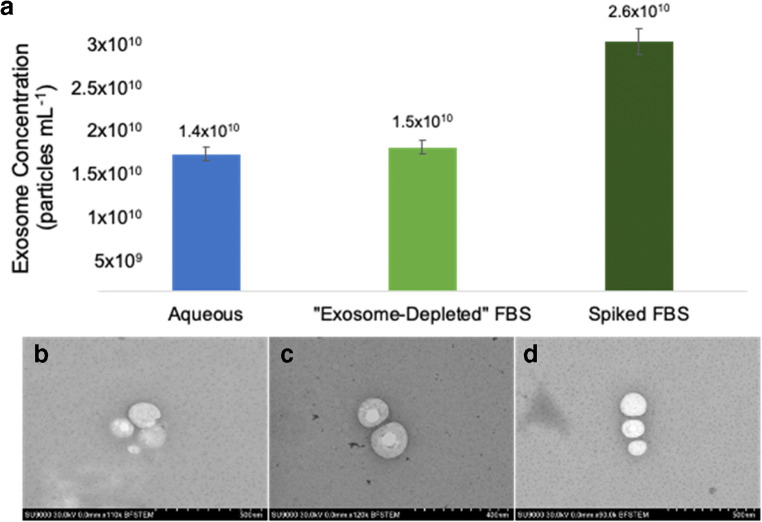
To verify and quantify the presence of remnant (native) extracellular vesicles in the exosome-depleted FBS matrix, the tip isolation of exosomes was performed on an exosome-spiked aqueous solution, the exosome-depleted FBS, and the FBS spiked with exosome standard. In the spiked-solution cases, the primary stock solution was added at a 1:100-μL ratio to the matrix. The optical absorbance of the eluate was detected at the 203 nm wavelength and used to quantify the exosomes based on the previous aqueous solution calibration function. Figure 6a shows the resulting exosome concentrations, where approximately the same number of exosomes were quantified in the eluates from the aqueous and native exosome-depleted FBS solutions. Addition of the spike to the FBS yields an ~ 73% increase in the determined density, a value in line with a combination of the responses for the aqueous solution and the FBS sample, as would be expected as the spike values are the same for the first and third cases. Importantly, the levels of precision are very uniform ranging from 5.4–8.2% RSD for triplicate isolation and measurement sets. Based on the determinations performed here, the exosome concentration in the “depleted” FBS is approximately 1.5 × 1010 particles mL−1. This value is less than recently published values of 2.27–2.93 × 1011 particles mL−1 [67]. Based on those values, the material employed here meets the stated 90% clearance target stated by the manufacturer, with ~ 6.6% remaining based on the published values.
Fig. 6.
a Quantification of exosomes in the eluates from aqueous solution (1/100), exosome-depleted FBS, and a spiked exosome-depleted FBS matrix (1/100). b–d STEM images of eluted exosomes, all containing their characteristic spherical structure post isolation using the C-CP tip method
The presence of exosomes in the depleted FBS was further confirmed physically using STEM and nanoparticle tracking analysis. Figures 6b–d are micrographs of the exosome eluted fractions for the same three cases, exosomes spiked (1:100) in aqueous solution, the native FBS, and exosomes spiked into FBS. In all three cases, the typical halo-structure objects are clearly revealed, having diameters on the order of 80–120 nm. NTA analysis was performed to analyze the size distribution of the eluted exosome populations. The graphical size distributions of the eluted exosomes are presented in ESM. Statistically, larger numbers of exosomes are observed in the case of the spiked-FBS (as suggested in the data of Fig. 5), though the means (~ 96 nm) and modes (74.3 and 77.7 nm) of the distributions are very similar. What are quite different are the broader distribution aspects, where the spiked-FBS displays a D90 (upper limit inclusive of 90% of the population) of 155.4 nm while the spiked aqueous population exhibits a D90 of 130.1 nm. This relationship is not surprising as the FBS is a far more diverse matrix than the human urine originating spike matrix.
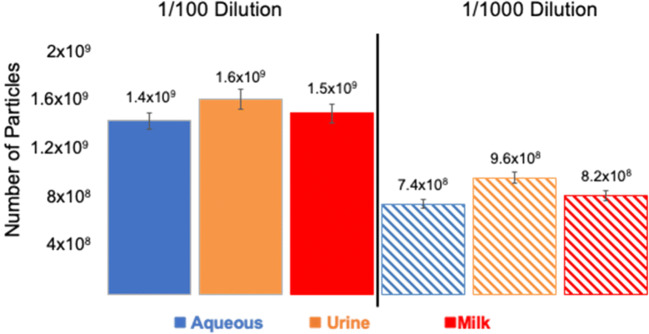
As a complement to the use of absorbance spectrophotometry to perform quantification, spin-down tip recoveries were also assessed via a standard ELISA assay for the antibody response to the CD81 tetraspanin surface protein. Presented in Fig. 7 are the determined number of particles is reported for the same two dilution values (1/100 and 1/1000) as depicted in Fig. 5 aqueous, mock urine, and non-fat milk test matrices. (The FBS material was received after the University ELISA facilities were closed due to COVID-19 protocols and so were not part of this assay.) The determinations were made on the same collected EV elution fractions as used in the absorbance measurements. As reported for the 1/100 dilution samples, the numbers of collected EVs are statistically identical for the three different matrices. As expected, the level of precision of the bioassay is somewhat degraded from the absorbance measurements, but with a variability of < 9.1% RSD, the results are in line with what would be expected. With increased dilution, the number of particles is statistically lower, with similar repeatability, but not in the direct proportion seen in the absorbance case. Again, the recoveries across the matrices are similar, maintaining the same relative responses among each. The measured CD81 expression reflects the fact that the exosome biogenesis process, and therefore, surface protein expression is due to many stochastic processes. Though exosomes from identical cells may be produced via the same mechanisms, exosome populations are heterogeneous, and differences in protein expression are expected. Also, while glycerol in the elution buffer was used to increase exosome stability and prevent aggregation, the presence of glycerol may also have an effect on the conformation of exosome surface proteins in the eluate. Changes in protein conformation due to the presence of glycerol has been previously reported [68, 69], where proteins are altered to more compact states. This has been found to affect antigen–antibody binding interactions, specifically in ELISA applications [70]. The observation of non-linear quantification of exosomes seen in Fig. 7, when compared to absorbance-based results in Fig. 5, is most likely due to these effects.
Fig. 7.
Post-isolation of exosome standards spiked into various matrices (50 μL) using the PET C-CP fiber tip spin-down method; the concentration of exosomes recovered were determined based on ELISA readout to an exosome standard curve of linear response was performed to quantitatively detect the expression of the exosomal tetraspanin protein-CD81 (n = 3) employing a capture antibody of 1:250 concentration. Quantified recovery of exosomes from mock matrices of 1/100 and 1/1000 concentration. The loaded 1/100 and 1/1000 solutions theoretically contain 2 × 1010 and 2 × 109 exosomes, respectively
Conclusions
Presented here is an efficient, timely, and vesicle-preserving method for exosome/EV isolation using a simple PET C-CP fiber tip workflow followed by quantitation via absorbance and ELISA assay quantification. There is a high demand for clean and reproducible EV recoveries from complex matrices for potential uses as targets of clinical, diagnostic, and therapeutic relevance. The isolation of exosomes using hydrophobicity-based chemical separation allows for the gentle and effective capture and subsequent release of exosomes despite the complexity of the matrix of origin. It should be pointed out that the process is not directly related to, nor is it impacted by, traditional size exclusion effects as the fibers are non-porous and the interfiber channels have widths of 1–4 μm. That said, there may be some size-based effects in terms of elution characteristics as size will affect the extent of hydrophobic interactions with the fiber surface. The combination of low-volume, high-throughput processing, high recoveries, and practical simplicity of the method bodes well in comparison to other approaches, particularly for clinical situations.
The HIC mode C-CP fiber tip workflow introduces a plethora of potential capabilities as modes of fiber capture selectivity are explored and optimized. The present method would be classified as a generic exosome/EV capture approach, but previously demonstrated methods of fiber surface modification could be implemented for selective capture based on the presence of target surface proteins [36, 50]. Likewise, as shown here, protein-specific immunofluorescent labeling could be affected for on-fiber detection. Continued optimization of this technique and characterization of the purity (freedom from matrix species) and the proteomic and genetic cargo are essential to the future implementation of this technique to complex biofluid samples. To this end, there indeed may be instances, such as mass spectrometry-based proteomics, where the use of acetonitrile will be the preferred elution phase modifier, allowing more efficient processing to recover surface and sequestered proteins. Ultimately, the use of other C-CP fiber platforms could be implemented to affect point-of-care (POC) assays. Importantly, each of these aspects could be scaled up to volumes necessary for the isolation and purification of exosomes/EVs for various biotherapeutic applications.
Electronic supplementary material
(PDF 254 kb)
Acknowledgments
Financial support for the chromatography development efforts from the National Science Foundation, Division of Chemistry under grant CHE-1608663, is gratefully acknowledged. Financial support for the EV and exosome isolation efforts from the Eppley Foundation for Scientific Research is gratefully acknowledged. The Gibson Foundation, the Prisma Health ITOR Biorepository, and the Greenville Hospital System are gratefully acknowledged. Special thanks to George Wetzel, Clemson University Electron Microscopy Facility, for assistance with EM. The content of this material and any opinions, findings, conclusions, or recommendations expressed in this material are solely the responsibility of the author(s) and do not necessarily represent the official views of the National Science Foundation. The research in this publication was conducted using a Leica SP8X Confocal multiphoton/HyVolution microscope system, housed in the Clemson Light Imaging Facility (CLIF). CLIF gratefully acknowledges the support of the Clemson University Division of Research, NIH EPIC COBRE Award #P20GM109094, NIH SCBiocraft COBRE Award #5P20RR021949-03, and NSF MRI Award #1126407.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 2.Rashed MH, Bayraktar E, Helal GK, Abd-Ellah MF, Amero P, Chavez-Reyes A, Rodriguez-Aguayo C. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci. 2017;18(3):538. doi: 10.3390/ijms18030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107(2):102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 5.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Rai AJ, DeCastro GJ, Zeringer E, Barta T, Magdaleno S, Setterquist R, Vlassov AV. An optimized procedure for exosome isolation and analysis using serum samples: application to cancer biomarker discovery. Methods. 2015;87:26–30. doi: 10.1016/j.ymeth.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Schageman J, Zeringer E, Li M, Barta T, Lea K, Gu J, Magdaleno S, Setterquist R, Vlassov AV. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int. 2013;2013:253957. doi: 10.1155/2013/253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harischandra DS, Ghaisas S, Rokad D, Kanthasamy AG. Exosomes in toxicology: relevance to chemical exposure and pathogenesis of environmentally linked diseases. Toxicol Sci. 2017;158(1):3–13. doi: 10.1093/toxsci/kfx074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendler F, Bota-Rabassedas N, Franch-Marro X. Cancer becomes wasteful: emerging roles of exosomes(dagger) in cell-fate determination. J Extracell Vesicles. 2013;2:22390. doi: 10.3402/jev.v2i0.22390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucharzewska P, Belting M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J Extracell Vesicles. 2013;2(1):20304. [DOI] [PMC free article] [PubMed]
- 12.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koritzinsky EH, Street JM, Star RA, Yuen PS. Quantification of exosomes. J Cell Physiol. 2017;232(7):1587–1590. doi: 10.1002/jcp.25387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, Bossios A, Sjostrand M, Gabrielsson S, Lotvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halvaei S, Daryani S, Eslami SZ, Samadi T, Jafarbeik-Iravani N, Bakhshayesh TO. Majidzadeh AK, Esmaeili R. Exosomes in cancer liquid biopsy: a focus on breast cancer. Mol Ther Nucleic Acids. 2018;10:131–141. doi: 10.1016/j.omtn.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A, Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179(3):1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 19.Yoo YK, Lee J, Kim H, Hwang KS, Yoon DS, Lee JH. Toward exosome-based neuronal diagnostic devices. Micromachines (Basel). 2018;9(12):634. [DOI] [PMC free article] [PubMed]
- 20.Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, Jackson B, McKee AC, Alvarez VE, Lee NC, Hall GF. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287(6):3842–3849. doi: 10.1074/jbc.M111.277061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thery C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3:Unit 3 22. [DOI] [PubMed]
- 22.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11(2):240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heusermann W, Hean J, Trojer D, Steib E, von Bueren S, Graff-Meyer A, Genoud C, Martin K, Pizzato N, Voshol J, Morrissey DV, Andaloussi SE, Wood MJ, Meisner-Kober NC. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J Cell Biol. 2016;213(2):173–184. doi: 10.1083/jcb.201506084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J Biol Chem. 2007;282(35):25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, Wang B, Zhang L, Kakar S, Jun Y, Miller D, Zhang HG. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol Nutr Food Res. 2014;58(7):1561–1573. doi: 10.1002/mnfr.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, Soria CE, Oquin S, Bonebreak CM, Saracoglu E, Skog J, Kuo WP. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394:1253–1262. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Jeppesen DK, Hvam ML, Primdahl-Bengtson B, Boysen AT, Whitehead B, Dyrskjot L, Orntoft TF, Howard KA, Ostenfeld MS. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J Extracell Vesicles. 2014;3:25011. doi: 10.3402/jev.v3.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, Salumets A, Peters M. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47(1–2):135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 30.Nelson DM, Marcus RK. Characterization of capillary-channeled polymer fiber stationary phases for high-performance liquid chromatography protein separations: comparative analysis with a packed-bed column. Anal Chem. 2006;78(24):8462–8471. doi: 10.1021/ac061223w. [DOI] [PubMed] [Google Scholar]
- 31.Stanelle RD, Straut CM, Marcus RK. Nylon-6 capillary-channeled polymer fibers as a stationary phase for the mixed-mode ion exchange/reversed-phase chromatography separation of proteins. J Chromatogr Sci. 2007;45(7):415–421. doi: 10.1093/chromsci/45.7.415. [DOI] [PubMed] [Google Scholar]
- 32.Jiang L, Jin Y, Marcus RK. Polyethylenimine modified poly(ethylene terephthalate) capillary channeled-polymer (C-CP) fibers for anion exchange chromatography of proteins. J Chromatogr A. 2015;1410:200–209. doi: 10.1016/j.chroma.2015.07.102. [DOI] [PubMed] [Google Scholar]
- 33.Stanelle RD, Marcus RK. Nylon-6 capillary-channeled polymer (C-CP) fibers as a hydrophobic interaction chromatography stationary phase for the separation of proteins. Anal Bioanal Chem. 2009;393(1):273–281. doi: 10.1007/s00216-008-2457-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Marcus RK. Evaluation of protein separations based on hydrophobic interaction chromatography using polyethylene terephthalate capillary-channeled polymer (C-CP) fiber phases. J Chromatogr A. 2019;1585:161–171. doi: 10.1016/j.chroma.2018.11.068. [DOI] [PubMed] [Google Scholar]
- 35.Trang HK, Marcus RK. Application of protein A-modified capillary-channeled polymer polypropylene fibers to the quantitation of IgG in complex matrices. J Pharm Biomed Anal. 2017;142:49–58. doi: 10.1016/j.jpba.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 36.Jiang L, Marcus RK. Biotin-functionalized poly(ethylene terephthalate) capillary-channeled polymer fibers as HPLC stationary phase for affinity chromatography. Anal Bioanal Chem. 2015;407(3):939–951. doi: 10.1007/s00216-014-8235-4. [DOI] [PubMed] [Google Scholar]
- 37.Randunu KM, Marcus RK. Microbore polypropylene capillary channeled polymer (C-CP) fiber columns for rapid reversed-phase HPLC of proteins. Anal Bioanal Chem. 2012;404(3):721–729. doi: 10.1007/s00216-012-6163-8. [DOI] [PubMed] [Google Scholar]
- 38.Randunu KM, Dimartino S, Marcus RK. Dynamic evaluation of polypropylene capillary-channeled fibers as a stationary phase in high-performance liquid chromatography. J Sep Sci. 2012;35(23):3270–3280. doi: 10.1002/jssc.201200523. [DOI] [PubMed] [Google Scholar]
- 39.Randunu KM, Marcus RK. Initial evaluation of protein throughput and yield characteristics on nylon 6 capillary-channeled polymer (C-CP) fiber stationary phases by frontal analysis. Biotechnol Prog. 2013;29(5):1222–1229. doi: 10.1002/btpr.1773. [DOI] [PubMed] [Google Scholar]
- 40.Bruce TF, Slonecki TJ, Wang L, Huang S, Powell RR, Marcus RK. Exosome isolation and purification via hydrophobic interaction chromatography using a polyester, capillary-channeled polymer fiber phase. Electrophoresis. 2019;40(4):571–581. doi: 10.1002/elps.201800417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Bruce TF, Huang S, Marcus RK. Isolation and quantitation of exosomes isolated from human plasma via hydrophobic interaction chromatography using a polyester, capillary-channeled polymer fiber phase. Anal Chim Acta. 2019;1082:186–193. doi: 10.1016/j.aca.2019.07.035. [DOI] [PubMed] [Google Scholar]
- 42.Huang S, Wang L, Bruce TF, Marcus RK. Isolation and quantification of human urinary exosomes by hydrophobic interaction chromatography on a polyester capillary-channeled polymer fiber stationary phase. Anal Bioanal Chem. 2019;411(25):6591–6601. doi: 10.1007/s00216-019-02022-7. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Wang L, Bruce TF, Marcus RK (2020) Evaluation of exosome loading characteristics in their purification via a glycerol-assisted hydrophobic interaction chromatography method on a polyester, capillary-channeled polymer fiber phase. Biotechnol Prog in press. [DOI] [PubMed]
- 44.Snow NH. Solid-phase micro-extraction of drugs from biological matrices. J Chromatogr A. 2000;885(1–2):445–455. doi: 10.1016/S0021-9673(00)00192-8. [DOI] [PubMed] [Google Scholar]
- 45.Hennion MC. Solid-phase extraction: method development, sorbents, and coupling with liquid chromatography. J Chromatogr A. 1999;856(1–2):3–54. doi: 10.1016/S0021-9673(99)00832-8. [DOI] [PubMed] [Google Scholar]
- 46.Xu H, Wang S, Zhang G, Huang S, Song D, Zhou Y, Long G. A novel solid-phase microextraction method based on polymer monolith frit combining with high-performance liquid chromatography for determination of aldehydes in biological samples. Anal Chim Acta. 2011;690(1):86–93. doi: 10.1016/j.aca.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Zarei M, Sprenger A, Gretzmeier C, Dengjel J. Rapid combinatorial ERLIC-SCX solid-phase extraction for in-depth phosphoproteome analysis. J Proteome Res. 2013;12(12):5989–5995. doi: 10.1021/pr4007969. [DOI] [PubMed] [Google Scholar]
- 48.Brambilla G, Fiori M, Rizzo B, Crescenzi V, Masci G. Use of molecularly imprinted polymers in the solid-phase extraction of clenbuterol from animal feeds and biological matrices. J Chromatogr B. 2001;759(1):27–32. doi: 10.1016/S0378-4347(01)00199-2. [DOI] [PubMed] [Google Scholar]
- 49.Burdette CQ, Marcus RK. Solid phase extraction of proteins from buffer solutions employing capillary-channeled polymer (C-CP) fibers as the stationary phase. Analyst. 2013;138(4):1098–1106. doi: 10.1039/C2AN36126D. [DOI] [PubMed] [Google Scholar]
- 50.Manard BT, Jones SM, Marcus RK. Capillary-channeled polymer (C-CP) fibers for the rapid extraction of proteins from urine matrices prior to detection with MALDI-MS. Proteomics Clin Appl. 2015;9(5–6):522–530. doi: 10.1002/prca.201400081. [DOI] [PubMed] [Google Scholar]
- 51.Schadock-Hewitt AJ, Marcus RK. Initial evaluation of protein a modified capillary-channeled polymer fibers for the capture and recovery of immunoglobulin G. J Sep Sci. 2014;37(5):495–504. doi: 10.1002/jssc.201301205. [DOI] [PubMed] [Google Scholar]
- 52.Fornea DS, Wu Y, Marcus RK. Capillary-channeled polymer fibers as a stationary phase for desalting of protein solutions for electrospray ionization mass spectrometry analysis. Anal Chem. 2006;78(15):5617–5621. doi: 10.1021/ac060447b. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Zeringer E, Barta T, Schageman J, Cheng AG, Vlassov AV. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos T R Soc B. 2014;369(1652):20130502. [DOI] [PMC free article] [PubMed]
- 54.Guidi L, Felice C, Procoli A, Bonanno G, Martinelli E, Marzo M, Mocci G, Pugliese D, Andrisani G, Danese S, De Vitis I, Papa A, Armuzzi A, Rutella S. FOXP3(+) T regulatory cell modifications in inflammatory bowel disease patients treated with anti-TNFalpha agents. Biomed Res Int. 2013;2013:286368. doi: 10.1155/2013/286368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao X, Wu Y, Duan J, Ma Y, Shen Z, Wei L, Cui X, Zhang J, Xie Y, Liu J. Quantitative proteomic analysis of exosome protein content changes induced by hepatitis B virus in Huh-7 cells using SILAC labeling and LC-MS/MS. J Proteome Res. 2014;13(12):5391–5402. doi: 10.1021/pr5008703. [DOI] [PubMed] [Google Scholar]
- 57.Kruger S, Abd Elmageed ZY, Hawke DH, Worner PM, Jansen DA, Abdel-Mageed AB, Alt EU, Izadpanah R. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer. 2014;14:44. doi: 10.1186/1471-2407-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffiths SG, Cormier MT, Clayton A, Doucette AA. Differential proteome analysis of extracellular vesicles from breast cancer cell lines by chaperone affinity enrichment. Proteomes. 2017;5(4):16. doi: 10.3390/proteomes5040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.An M, Wu J, Zhu J, Lubman DM. Comparison of an optimized ultracentrifugation method versus size-exclusion chromatography for isolation of exosomes from human serum. J Proteome Res. 2018;17:3599–3605. doi: 10.1021/acs.jproteome.8b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He M, Zeng Y. Microfluidic exosome analysis toward liquid biopsy for cancer. J Lab Autom. 2016;21(4):599–608. doi: 10.1177/2211068216651035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ko J, Hemphill MA, Gabrieli D, Wu L, Yelleswarapu V, Lawrence G, Pennycooke W, Singh A, Meaney DF, Issadore D. Smartphone-enabled optofluidic exosome diagnostic for concussion recovery. Sci Rep. 2016;6:31215. doi: 10.1038/srep31215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14(19):3773–3780. doi: 10.1039/C4LC00662C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17(21):3558–3577. doi: 10.1039/C7LC00592J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He F, Liu H, Guo X, Yin BC, Ye BC. Direct exosome quantification via bivalent-cholesterol-labeled DNA anchor for signal amplification. Anal Chem. 2017;89(23):12968–12975. doi: 10.1021/acs.analchem.7b03919. [DOI] [PubMed] [Google Scholar]
- 65.van der Pol E, Coumans FA, Grootemaat AE, Gardiner C, Sargent IL, Harrison P, Sturk A, van Leeuwen TG, Nieuwland R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12(7):1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 66.Sumida S. Transfusion and transplantation of cryopreserved cells and tissues. Cell Tissue Bank. 2006;7(4):265–305. doi: 10.1007/s10561-006-9005-0. [DOI] [PubMed] [Google Scholar]
- 67.Lehrich BM, Liang YX, Khosravi P, Federoff HJ, Fiandaca MS. Fetal bovine serum-derived extracellular vesicles persist within vesicle-depleted culture media. Int J Mol Sci. 2018;19(11):11. doi: 10.3390/ijms19113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vagenende V, Yap MGS, Trout BL. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry. 2009;48(46):11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
- 69.Vagenende V, Han AX, Pek HB, Loo BLW. Quantifying the molecular origins of opposite solvent effects on protein-protein interactions. PLoS Comput Biol. 2013;9(5):9. doi: 10.1371/journal.pcbi.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kjaer S, Stausbol-Gron B, Wind T, Ravn P, Jensen KP, Kahns L, Clark BFC. Glycerol diversifies phage repertoire selections and lowers non-specific phage absorption. FEBS Lett. 1998;431(3):448–452. doi: 10.1016/S0014-5793(98)00789-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 254 kb)