Abstract
Summary
We have implemented the pypgatk package and the pgdb workflow to create proteogenomics databases based on ENSEMBL resources. The tools allow the generation of protein sequences from novel protein-coding transcripts by performing a three-frame translation of pseudogenes, lncRNAs and other non-canonical transcripts, such as those produced by alternative splicing events. It also includes exonic out-of-frame translation from otherwise canonical protein-coding mRNAs. Moreover, the tool enables the generation of variant protein sequences from multiple sources of genomic variants including COSMIC, cBioportal, gnomAD and mutations detected from sequencing of patient samples. pypgatk and pgdb provide multiple functionalities for database handling including optimized target/decoy generation by the algorithm DecoyPyrat. Finally, we have reanalyzed six public datasets in PRIDE by generating cell-type specific databases for 65 cell lines using the pypgatk and pgdb workflow, revealing a wealth of non-canonical or cryptic peptides amounting to >5% of the total number of peptides identified.
Availability and implementation
The software is freely available. pypgatk: https://github.com/bigbio/py-pgatk/ and pgdb: https://nf-co.re/pgdb.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Proteogenomics is a rapidly developing multiomics field that integrates genomics and transcriptomics information with proteomics to improve gene annotation, often uncovering novel or non-canonical protein-coding regions in the genome (Branca et al., 2014). One of the most important applications is in the study of cancer cells and tumors, where identifying cancer-specific proteins holds great potential in both elucidating cancer biology and in developing cancer therapies. However, the discovery of such proteins remains particularly challenging and is still largely linked to evidence from genome sequencing data, rather than directly from the protein data that have become abundant (Perez-Riverol et al., 2019). Recent applications of proteogenomics have enabled multiomics detection of novel peptide sequences that are not present in the canonical protein database. For instance, Ruiz Cuevas et al. (2021) recently identified a large number of non-canonical proteins in B cell lymphomas. However, customized protein databases are needed to enable the identification of such peptides. Recently, tools for generating sample-specific protein databases have been implemented using genomic sequencing data (Ruggles et al., 2016) and transcriptomics data (Cesnik et al., 2021; Cifani et al., 2018). Since matching sequencing data is not available for a large fraction of the currently available proteomics datasets, resources have been developed to provide protein databases generated from cancer somatic mutations and genomic variants (Zhang et al., 2017).
To make progress in high throughput proteogenomics analysis, we present a Python application integrated into a Nextflow workflow to facilitate the generation of proteogenomics databases from sample-specific and public resources under varying conditions (e.g. cancer type and transcript biotype). The aim is to enable the identification of variant proteins (derived from single nucleotide variant mutations) and non-canonical or cryptic proteins (from normally dormant regions of the genome).
2 Implementation
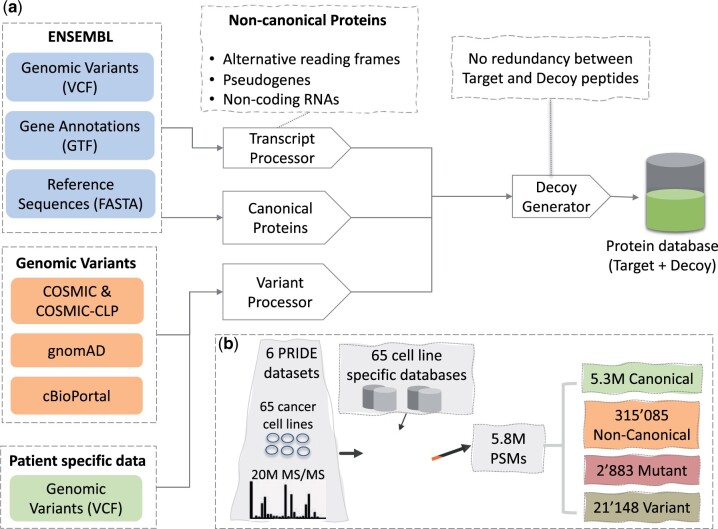
We implemented pypgatk, a Python package that provides tools to generate protein databases from non-canonical sequences as well as DNA variants and mutations from public resources and custom files (Fig. 1a).
Fig. 1.
(a) pypgatk and pgdb components to generate ENSEMBL-based proteogenomics databases. (b) Reanalyzed datasets (four human and two mice); number of identified canonical, non-canonical, variant and mutated peptides identified using cell-type specific proteogenomics databases
2.1 Non-canonical protein databases
Non-canonical proteins are a product of translation of transcripts that are not reported as protein coding in the reference protein databases, or a product of out-of-frame translation of canonical transcripts (Ruiz Cuevas et al., 2021). While many of the non-canonical proteins could be attributed to the yet incomplete reference databases, they might also be attributed to the activation of those genes under certain conditions such as genetic and epigenetic misregulation in cancer (Zhu et al., 2018). We have developed the dnaseq-to-proteindb tool to generate protein sequences from non-canonical transcripts such as pseudogenes and lncRNAs by performing three-frame translation. It also extracts alternative reading frames from canonical protein-coding genes to enable the detection of out-of-frame cryptic proteins. Furthermore, the ensemble-downloader tool enables automatic download of the latest ENSEMBL resources including gene annotations, the reference genome and canonical proteins for the species of interest.
2.2 Variant protein databases
Detection of altered proteins from proteomics data requires the inclusion of the mutated sequences in the target databases. However, due to a large number of potential DNA variants, only potentially relevant variant sequences should be included to keep the database size under control. Here, we implemented methods to automate generation of variant proteins from publicly available cancer mutations datasets, cancer cell lines and custom Variant Calling Format (VCF) files obtained from genome sequencing. cosmic-to-proteindb and cbioportal-to-proteindb enable the generation of cancer-type specific protein databases by generating mutated protein sequences based on genomic mutations identified in cancer samples. cosmic-to-proteindb curates mutations from the Catalogue Of Somatic Mutations In Cancer (COSMIC). It allows filtering the mutations based on cancer type or tissue of origin. Alternatively, cbioportal-to-proteindb translates genomics mutations reported by thousands of cancer studies through cBioPortal. pypgatk enables downloading and processing mutations from ENSEMBL and gnomAD resources. vcf-to-proteindb translates the genomic variants into variant protein sequences. The variants can be filtered based on functional consequences as well as allele frequency to enable a special focus on common variants. The vcf-to-proteindb command accepts a custom VCF file from any species or sample of interest and generates a database of altered protein-coding sequences, which is valuable when whole-exome or whole-genome sequencing data are available; for instance to detect cancer neoantigens from passenger mutations.
3 ENSEMBL-based proteogenomic databases
To enable the generation of ENSEMBL-based proteogenomic databases, we have also built the Proteomics-Genomics DataBase (pgdb—https://nf-co.re/pgdb) workflow in Nextflow using bioconda and BioContainers. The pipeline integrates the various commands of pypgatk allowing the user to generate protein databases by simple parameter selection without any additional input required from the user. Also, the pipeline can be used to generate protein databases for any ENSEMBL species, except for the processes that are dependent on data that are only available for Homo sapiens.
3.1 Identification of non-canonical peptides
We applied pgdb to generate cell-type specific databases for 64 human cell lines (Fig. 1b and Supplementary Note S1 and S2). Mutations from the COSMIC Cell Line project and the Broad CCLE project through cBioPortal were downloaded for each cell line to generate the respective set of variant protein sequences. Additionally, a database of non-canonical proteins was generated from the latest human genome assembly. The variant protein database from each cell line was appended to the non-canonical and canonical protein databases and the decoy sequences were generated to search MS/MS proteomics datasets from the corresponding cell lines. The proteomics data were obtained through the PRIDE database (PXD005946, PXD019263, PXD004452 and PXD014145). proteomicsLFQ (https://nf-co.re/proteomicslfq) was used to identify the novel peptides (Supplementary Note S3). Overall, 402 512 target peptide sequences were identified, including 43 501 non-canonical peptides and 786 variant peptide sequences (Table 1 and Supplementary Note S4 and S5). The majority of the non-canonical peptides were novel coding sequences in their entirety whereas only 16% matched canonical protein sequences with one amino acid mismatch.
Table 1.
Number of peptides identified per class
| Species | Class | #PSMs | #Peptide sequences | #Novel peptides |
|---|---|---|---|---|
| Homo sapiens | Canonical | 4 125 497 | 322 967 | NA |
| Non-canonical | 315 085 | 74 001 | 43 501 | |
| Mutated | 16 518 | 5544 | 786 | |
| Mus musculus | Canonical | 1 159 049 | 105 338 | NA |
| Variant | 4630 | 1928 | 374 | |
| Mutated | 2883 | 913 | 166 |
Additionally, we reanalyzed two mice datasets (PXD018891 and PXD006439) obtained from the B16 melanoma cell line. A proteogenomic database was generated using mice germline variants from the ENSEMBL variation database (release 104) and somatic mutations detected in mouse melanoma tumors. Overall, 374 variant peptides and 166 mutated peptides were identified. The identified peptides with the corresponding mass spectra and metadata annotations can be accessed via ProteomeXchange (PXD029360 and PXD029362).
4 Conclusions
The developed tools facilitate the creation of proteogenomics databases based on ENSEMBL genomes and other relevant sources of genome variation information. The pgdb is the first Nextflow workflow for proteogenomics database generation and its development within the nf-core community will ensure its stability, continued development and community support. pypgatk (https://pgatk.readthedocs.io/en/latest/pypgatk.html) and pgdb (https://nf-co.re/pgdb/1.0.0/usage) include extensive documentation to help researchers create their custom proteogenomics databases.
Funding
This work was supported by the Swedish Cancer Society [CAN 2017/685 and CAN 2020/1269 PjF], the Erling-Persson Family Foundation [12/12-2017 and 22/9-2020], DART and Rescuer EU-projects to H.U., J.L. and R.B.; the National Natural Science Foundation of China [32100505] and Guangdong Science and Technology Department [2020B1212060018, 2020B1212030004] to Y.Z.; the German Ministry of Research and Education [BMBF, project 031A535A] to T.S.; and the Wellcome Trust [208391/Z/17/Z] to Y.P.R.
Conflict of Interest: none declared.
Data availability:
We here explored proteomics datasets PXD005946, PXD019263, PXD004452 and PXD014145, which are from the public domain PRIDE database, at https://www.ebi.ac.uk/pride/. Further data underlying this article are available in its online supplementary material.
Supplementary Material
Contributor Information
Husen M Umer, Department of Oncology‐Pathology, Science for Life Laboratory, Karolinska Institutet, Stockholm 17165, Sweden.
Enrique Audain, Department of Congenital Heart Disease and Pediatric Cardiology, Universitätsklinikum Schleswig-Holstein Kiel, Kiel 24105, Germany.
Yafeng Zhu, Medical Research Center, Sun Yat-Sen Memorial Hospital, Sun Yat-sen University, Guangzhou 510120, China.
Julianus Pfeuffer, Algorithmic Bioinformatics, Freie Universität Berlin, Berlin 14195, Germany; Visualization and Data Analysis, Zuse Institute Berlin, Berlin 14195, Germany.
Timo Sachsenberg, Institute for Bioinformatics and Medical Informatics, University of Tübingen, 72076 Tübingen, Germany.
Janne Lehtiö, Department of Oncology‐Pathology, Science for Life Laboratory, Karolinska Institutet, Stockholm 17165, Sweden.
Rui M Branca, Department of Oncology‐Pathology, Science for Life Laboratory, Karolinska Institutet, Stockholm 17165, Sweden.
Yasset Perez-Riverol, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
References
- Branca R.M. et al. (2014) HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat. Methods, 11, 59–62. [DOI] [PubMed] [Google Scholar]
- Cesnik A.J. et al. (2021) Spritz: a proteogenomic database engine. J. Proteome Res., 20, 1826–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifani P. et al. (2018) ProteomeGenerator: a framework for comprehensive proteomics based on de novo transcriptome assembly and high-accuracy peptide mass spectral matching. J. Proteome Res., 17, 3681–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y. et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res., 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggles K.V. et al. (2016) An analysis of the sensitivity of proteogenomic mapping of somatic mutations and novel splicing events in cancer. Mol. Cell. Proteomics, 15, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Cuevas M.V. et al. (2021) Most non-canonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep., 34, 108815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. et al. (2017) CanProVar 2.0: an updated database of human cancer proteome variation. J. Proteome Res., 16, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. et al. (2018) Discovery of coding regions in the human genome by integrated proteogenomics analysis workflow. Nat. Commun., 9, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We here explored proteomics datasets PXD005946, PXD019263, PXD004452 and PXD014145, which are from the public domain PRIDE database, at https://www.ebi.ac.uk/pride/. Further data underlying this article are available in its online supplementary material.