Abstract
We report the emergence of non-susceptibility to cefiderocol from a subpopulation of Pseudomonas aeruginosa recovered from a patient without history of cefiderocol exposure. Whole genome sequencing identified mutations in major iron transport pathways previously associated with cefiderocol uptake. Susceptibility testing should be performed before therapy with siderophore cephalosporins.
Keywords: Pseudomonas aeruginosa, cefiderocol, antibiotic resistance
The emergence of antimicrobial resistance has led to a global public health emergency, particularly for healthcare-associated infections caused by carbapenem-resistant gram-negative pathogens. There is an urgent need for new therapeutic options and drug delivery systems [1]. Cefiderocol is a novel siderophore cephalosporin that shows activity against multidrug-resistant bacteria, including carbapenem-resistant Pseudomonas aeruginosa isolates [2]. Cefiderocol possesses a cathecol moiety, which binds iron and promotes active transport across the outer membrane of bacteria via specialized iron transporter channels. The β-lactam structure of cefiderocol is similar to both ceftazidime and cefepime, providing stability in the presence of some β-lactamases [3]. Overall rates of resistance to cefiderocol in clinical isolates is low, and has been primarily associated with Acinetobacter baumannii producing Pseudomonas extended resistant (PER) enzymes or Enterobacterales with metallo-β-lactamases [4]. Resistance to this drug in P. aeruginosa has been demonstrated in vitro, and was linked to alterations of the iron uptake pathways, but the clinical implications of this phenotype are uncertain [5]. We report the emergence of resistance to cefiderocol in a P. aeruginosa clinical isolate in the absence of prior exposure to this antibiotic, raising major concerns on the effectiveness of this drug without susceptibility testing.
METHODS
Clinical P. aeruginosa isolates P-3614 and P-3615 were recovered from blood cultures by the microbiology laboratory at the Banner—University Medical Center Tucson (B-UMCT). Isolate P-3616 was purified from one of the colonies recovered inside the zone of cefiderocol inhibition on the P-3615 disk diffusion test. Antimicrobial susceptibility testing was performed by broth microdilution or by gradient diffusion strip in accordance with Clinical and Laboratory Standards Institute guidelines. Cefiderocol susceptibility testing of P. aeruginosa isolates was initially performed by disk diffusion at B-UMCT clinical microbiology laboratory, and confirmed by broth microdilution at International Health Management Associates, Inc. (the reference microbiology laboratory of the Shionogi Sidero compassionate use program). Genomic DNA was isolated from strain P-3614, P-3615, and P-3616 and sequencing was performed on the MiSeq platform (Illumina, Inc.) with 2x 300-bp paired-end reads. Genomic data was processed using a custom analysis pipeline. A detailed description of the genomic analysis is available in the supplementary information. The mutations identified in the piuD and pirR genes on whole genome sequencing were confirmed by Sanger sequencing in P-3616. Genome sequences were deposited in the NCBI database, accession numbers JACSDX000000000 (P-3614), JADWTC000000 (P-3615), and JACSDW000000000 (P-3616).
RESULTS AND DISCUSSION
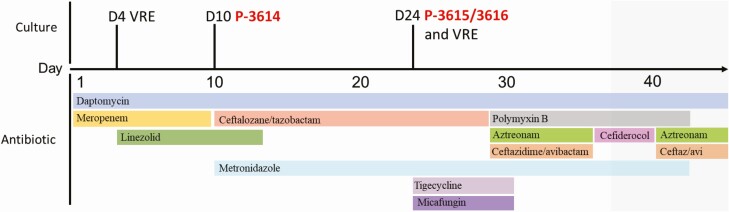
A 30-year-old woman with a history of orthotopic liver transplant (OLT) was admitted to the hospital a month after OLT because of an episode of acute cellular rejection and cholangitis. On the day of admission, the patient was started on empiric antibiotic therapy with intravenous daptomycin and meropenem for the treatment sepsis based on prior microbiological results from previous admissions and underlying OLT infectious risk factors (Figure 1). Four days after starting daptomycin, she developed vancomycin-resistant Enterococcus faecium (VRE) bacteremia. Thus, linezolid was added for 9 days with clearance of subsequent blood cultures. On the 9th day of hospitalization, the patient underwent endoscopic retrograde cholangiopancreatography (ERCP) with placement of a common bile duct stent. The next day she developed P. aeruginosa (PSA, isolate P-3614) bacteremia that was resistant to meropenem (Supplementary Table 1), resulting in a change of her antibiotic regimen from meropenem to ceftolozane-tazobactam and metronidazole. She underwent repeat ERCP on day 24 of hospitalization with exchange of the biliary stent. After the procedure she developed a fever, therefore tigecycline and micafungin were added to the regimen for 1 week. One day after the ERCP attempt, she developed a new episode of VRE and PSA bacteremia (isolate P-3615) and 8 days later multiple small hepatic abscesses were identified on MRI of the abdomen.
Figure 1.
Clinical timeline and antimicrobial regimens used in this patient.
The PSA recovered from the second episode of bacteremia was resistant to ceftolozane-tazobactam and ceftazidime-avibactam. The antibiotic regimen was changed to ceftazidime-avibactam, aztreonam, and polymyxin B on hospital day 30, and a decision was made to request cefiderocol from Shionogi, Inc., on a compassionate use basis. Cefiderocol was started on day 8 of polymyxin-based therapy (in place of ceftazidime-avibactam and aztreonam), but was stopped after 4 days due to the elevation of liver enzymes and susceptibility testing results. The P-3614 isolate recovered in the first bacteremia episode was susceptible to cefiderocol. However, the second isolate (P-3615, recovered before starting cefiderocol therapy) was reported as intermediate by disk diffusion at B-UMCT with the presence of a subpopulation growing within the inner border of the zone of inhibition on disk diffusion. From the latter isolate, an inner zone colony was purified (P-3616), and isolate P-3614 (cefiderocol-susceptible) and P-3616 (cefiderocol non-susceptible) MICs to cefiderocol were determined by broth microdilution. The organisms were reported as susceptible and intermediate by CLSI interpretive criteria (MICs of 2 and 8 μg/mL), respectively. The patient continued on polymyxin for an additional 4 weeks, and ceftazidime-avibactam combined with aztreonam for a total of 12 weeks, with hepatic abscesses showing improvement on repeated MRI imaging.
According to the antimicrobial susceptibility testing (Supplementary Table 1), the first P. aeruginosa isolate recovered (P-3614) was susceptible to ceftalozane/tazobactam (2 μg/mL) and cefiderocol (2 μg/mL), and resistant to meropenem (>16 μg/mL). After 14 days of ceftalozane/tazobactam treatment, the subsequent isolate (P-3615) displayed elevated MICs to ceftalozane/tazobactam (64 μg/mL). Using the genome sequence of the three P. aeruginosa isolates, we determined that all strains belonged to sequence type (ST) 639 (https://pubmlst.org/paeruginosa) [6]. No acquired β-lactamases were found in any strain. The alignment of core genome single nucleotide polymorphisms (SNPs) between the strains showed 126 SNPs in P-3616 that were not present in P-3614,indicating that the isolates were related but that significant population diversity existed in this patient (Supplementary Table 2). In fact, there was a 30 SNP difference between isolates P-3615 and P-3616, suggesting that several subpopulations may have coexisited in the mileu of the hepatic abscess. Interestingly, there were SNPs identified in genes belonging to TonB dependent receptors (TBDRs), associated with iron acquisition, and in the chromosomal ampC β-lactamase gene. Mutations and genes associated with resistance to other antibiotics are shown in Supplementary Table 3. When compared to the ancestor isolate, the AmpC β-lactamase present in P-3615 and P-3616 (PDC-191) exhibited a substitution of leucine for phenylalanine at Ambler amino acid position 147. This substitution lies near the YSN loop where it is predicted to result in an enlarged substrate binding pocket, and has been associated with elevated ceftolozane MICs in vitro [7, 8]. A 2-amino acid deletion in the R2 loop of the AmpC of Enterobacter cloacae was recently associated with a >32-fold increase in cefiderocol MICs, and it is possible AmpC mutations may contribute in part to the phenotype observed in this isolate [9].
The core genome analysis also identified mutations in two TBDRs, transmembrane proteins with a β-barrel structure involved in siderophore import. A total of 34 TBDRs have been identified in P. aeruginosa, with PirA, PiuA, and its ortholog PiuD experimentally implicated in susceptibility to siderophore-antibiotic combinations in vitro [10]. In P. aeruginosa PA01, inactivation of piuA (encoding the drug import channel) led to a 16-fold increase in cefiderocol MIC (0.5 to 8 μg/mL). Expression of the TBDR pirA and piuD on plasmids restored susceptibility to cefiderocol, with a 4-fold and 64-fold increase in susceptibility, respectively [10]. The expression of TBDRs is regulated by sigma/antisigma factors (PiuC) or two-component systems (PirRS) and can be induced by the presence of cefiderocol [11]. As compared to the susceptible P-3614, isolate P-3616 was found to have a deletion of an A nucleotide in the piuD gene, leading to mistranslation starting at predicted amino acid 42 with a premature stop codon at position 89, and an insertion of a G nucleotide in the pirR gene leading to a premature stop codon at predicted amino acid 201 (Supplementary Table 4). The presence of these premature stop codons would be predicted to lead to increased cefiderocol MICs in the P-3616 isolate, due to loss of cefiderocol import via PiuD and PirA.
To investigate whether these mutations were present in the P-3615 population, we examined the individual short reads covering the regions of interest in pirR and piuD. In the pirR gene, the insertion occurred at a repeat region of 7 bases that was covered at a sequencing depth of 85–92x. Of these reads, 2 definitively showed the insertion, and an additional 16 reads did not cross the entire sequence and thus could not be definitively assessed for presence or absence of the mutation. The piuD gene had a depth of coverage of 79x, and all reads were wild type. Thus, this mutation was present at a frequency of less than approximately 1 in 100 genomes, or it arose on exposure to cefiderocol during the disk diffusion assay. Given that there were 29 additonal SNPs between P-3615 and P-3616, it is likely that a small subpopulation harboring the mutation was present rather than selection on the disk diffusion plate. Nonetheless, if multiple spontaneous mutants arose after 24 hours of exposure to cefiderocol on susceptibility testing, this would have important clinical implications for the use of this antibiotic.
To the best of our knowledge, the in vivo evolution of non-susceptibility to cefiderocol in P. aeruginosa clinical isolates has not been reported before. Whole genome sequencing suggests the phenotype was driven by a subpopulation of bacteria harboring mutations leading to a loss of function of TonB-dependent receptors needed for cefiderocol import. The finding of the development of resistance to cefiderocol without prior exposure to this compound is concerning, especially because this drug represents a new option for the treatment of a variety of MDR pathogens. Further studies are needed to understand if the emergence of resistance in this manner is a more widespread phenomenon.
Supplementary Material
Nonstandard Abbreviations: ERCP, endoscopic retrograde cholangiopancreatiography; OLT, orthotopic liver transplant; SNP, single nucleotide polymorphism; TBDR, TonB-dependent receptors; VRE, vancomycin-resistant Enterococcus.
Notes
Acknowledgments. This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases NIH/NIAID grants R01 AI134637, R01 AI148342, K01AI148593-01, and K24 AI121296, UTHealth Presidential Award, and University of Texas System STARS Award to C. A. A. W. R. M. is supported by NIH/NIAID K08 AI135093. The funding agency had no role in experimental design, data collection, or interpretation of this work.
Potential conflicts of interest. C. A. A. has received grants from Merck, MeMed Diagnostics and Entasis Pharmaceuticals. C. A. A. has also received Chapters royalties from UptoDate, Harrison Principles of Internal Medicine, Mandell Principles and Practice of Infectious Diseases; Study section member/Grant reviewer fees from NIH/NIAID; travel fees from Infectious Diseases Society of America and American Society for Microbiology; and Antimicrobial Agents and Chemotherapy Editor’s stipend from American Society for Microbiology, outside the submitted work. W. R. M. has received grants and/or honoraria from NIH/NIAID, Merck, Entasis, Achaogen, IDSA, and Shionogi. A. P. S., M. M. A. O., W. D. L., A. K., B. H., and A. Q. D., report no conflict.
Contributor Information
Ana Paula Streling, Center for Antimicrobial Resistance and Microbial Genomics, UTHealth McGovern School of Medicine, Houston, Texas, USA; Universidade Federal de São Paulo - UNIFESP, Laboratório Alerta, Division of Infectious Diseases, Department of Internal Medicine. Escola Paulista de Medicina–EPM, São Paulo–SP, Brazil.
Mohanad M Al Obaidi, Division of Infectious Diseases, University of Arizona College of Medicine, Tucson, Arizona, USA; Department of Medicine, University of Arizona College of Medicine, Tucson, Arizona, USA.
William D Lainhart, Division of Infectious Diseases, University of Arizona College of Medicine, Tucson, Arizona, USA; Department of Medicine, University of Arizona College of Medicine, Tucson, Arizona, USA; Department of Pathology, University of Arizona College of Medicine, Tucson, Arizona, USA.
Tirdad Zangeneh, Division of Infectious Diseases, University of Arizona College of Medicine, Tucson, Arizona, USA; Department of Medicine, University of Arizona College of Medicine, Tucson, Arizona, USA.
Ayesha Khan, Center for Antimicrobial Resistance and Microbial Genomics, UTHealth McGovern School of Medicine, Houston, Texas, USA; Department of Microbiology and Molecular Genetics, UTHealth McGovern School of Medicine, Houston, Texas, USA; MD Anderson Cancer Center and UT Health Graduate School of Biomedical Sciences, Houston, Texas, USA.
An Q Dinh, Center for Antimicrobial Resistance and Microbial Genomics, UTHealth McGovern School of Medicine, Houston, Texas, USA; Division of Infectious Diseases, UTHealth McGovern School of Medicine, Houston, TX, USA; Center for Infectious Diseases, UTHealth, School of Public Health, Houston, TX, USA.
Blake Hanson, Center for Antimicrobial Resistance and Microbial Genomics, UTHealth McGovern School of Medicine, Houston, Texas, USA; Division of Infectious Diseases, UTHealth McGovern School of Medicine, Houston, TX, USA; Center for Infectious Diseases, UTHealth, School of Public Health, Houston, TX, USA.
Cesar A Arias, Center for Antimicrobial Resistance and Microbial Genomics, UTHealth McGovern School of Medicine, Houston, Texas, USA; MD Anderson Cancer Center and UT Health Graduate School of Biomedical Sciences, Houston, Texas, USA; Division of Infectious Diseases, UTHealth McGovern School of Medicine, Houston, TX, USA; Center for Infectious Diseases, UTHealth, School of Public Health, Houston, TX, USA; Molecular Genetics and Antimicrobial Resistance Unit, International Center for Microbial Genomics, Universidad El Bosque, Bogotá, Colombia.
William R Miller, Center for Antimicrobial Resistance and Microbial Genomics, UTHealth McGovern School of Medicine, Houston, Texas, USA; Division of Infectious Diseases, UTHealth McGovern School of Medicine, Houston, TX, USA.
References
- 1. Christaki E, Marcou M, Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol 2020; 88:26–40. [DOI] [PubMed] [Google Scholar]
- 2. O’Donnell JN, Bidell MR, Lodise TP. Approach to the treatment of patients with serious multidrug-resistant Pseudomonas aeruginosa infections. Pharmacotherapy 2020; 40:952–69. [DOI] [PubMed] [Google Scholar]
- 3. Zingg S, Nicoletti GJ, Kuster S, et al. Cefiderocol for extensively drug-resistant gram-negative bacterial infections: real-world experience from a case series and review of the literature. Open Forum Infect Dis 2020; 7:ofaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamano Y. In vitro activity of cefiderocol against a broad range of clinically important gram-negative bacteria. Clin Infect Dis 2019; 69:544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ito A, Nishikawa T, Ishii R, et al. Mechanism of cefiderocol high MIC mutants obtained in non-clinical FoR studies. Poster presented at: IDWeek 2018, San Francisco, CA, 3–7 October 2018. Poster 69.
- 6. Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 2012; 50:1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berrazeg M, Jeannot K, Ntsogo Enguéné VY, et al. Mutations in β-Lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 2015; 59:6248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cabot G, Bruchmann S, Mulet X, et al. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 2014; 58:3091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shields RK, Iovleva A, Kline EG, Kawai A, McElheny CL, Doi Y. Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in Enterobacter cloacae complex following exposure to cefepime. Clin Infect Dis 2020:ciaa355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luscher A, Moynié L, Auguste PS, et al. TonB-dependent receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugates. Antimicrob Agents Chemother 2018;62:e00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moynié L, Luscher A, Rolo D, et al. Structure and function of the PiuA and PirA Siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob Agents Chemother 2017; 61:e02531-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.