Abstract
Summary
Heatmap is a powerful visualization method on two-dimensional data to reveal patterns shared by subsets of rows and columns. In this work, we introduce a new R package InteractiveComplexHeatmap that brings interactivity to the widely used ComplexHeatmap package. InteractiveComplexHeatmap is designed with an easy-to-use interface where static complex heatmaps can be directly exported to an interactive Shiny web application only with one additional line of code. InteractiveComplexHeatmap also provides flexible functionalities for integrating interactive heatmap widgets to build more complex and customized Shiny web applications.
Availability and implementation
The InteractiveComplexHeatmap package and documentations are freely available from the Bioconductor project: https://bioconductor.org/packages/InteractiveComplexHeatmap/. A complete and printer-friendly version of the documentation can also be found in Supplementary File S1.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Heatmap is a popular visualization method on two-dimensional (2D) matrices where colors are the major esthetic elements mapping to data (Wilkinson and Friendly, 2009). Matrices for heatmap visualization are normally accompanied with row and column reordering, e.g. by hierarchical clustering, so that features with similar patterns are closely grouped and they can be easily identified from the colors on heatmaps. We previously developed an advanced R package ComplexHeatmap (Gu et al., 2016) that provides a simple syntax to link parallel heatmaps and annotations for easily revealing relationships among different data types. The simplicity of its user interface and comprehensiveness of its functionalities make ComplexHeatmap widely used in biological data analysis to reveal interesting patterns.
After a specific pattern of a subset of rows and columns has been observed from a heatmap, the next step is to extract corresponding rows and columns for downstream analysis, which requires interactivity on heatmaps. There are several R packages that implement interactive heatmaps as web applications such as d3heatmap, heatmaply (Galili et al., 2018) and iheatmapr (Schep and Kummerfeld, 2017). The interactivity allows selecting a region from a heatmap to retrieve details associated with the selected heatmap cells. There are also other web-based and non-R implementations, such as Morpheus, Heatmapper (Babicki et al., 2016), Clustergrammer (Fernandez et al., 2017) and NG-CHM (Ryan et al., 2019) which allow users to directly upload data to a web server and manipulate heatmaps with no programming knowledge.
ComplexHeatmap is powerful for generating static complex heatmaps. Here, we developed a new R package InteractiveComplexHeatmap that brings interactivity to ComplexHeatmap. InteractiveComplexHeatmap provides a simple user interface with which any static complex heatmap generated by ComplexHeatmap can be directly exported to an interactive Shiny web application only with one additional line of code. The interactive application contains comprehensive and unique tools for manipulating heatmaps, e.g. selecting among multiple heatmaps or searching heatmaps via row or column labels. InteractiveComplexHeatmap also provides functionalities for integrating interactive heatmap widgets into general Shiny application development. InteractiveComplexHeatmap provides a user interface for self-defining response to the mouse events on heatmaps, e.g. by clicking or brushing, which greatly helps developing more complex Shiny web applications.
2 Implementation
For any heatmap generated by ComplexHeatmap or other packages that depend on ComplexHeatmap, the function htShiny() is simply applied to export it to an interactive Shiny web application. The Shiny application by default has three components: the original heatmap, the selected subheatmap and the output that prints information of the selected heatmap cells (Supplementary File S1 and Fig. S2). The original heatmap has several tools integrated, where the most useful one is to search matrix labels to obtain a subset of rows or columns of interest. The subheatmap also has several tools integrated, in particular supporting recursive exporting of subheatmaps to additional layers of interactive heatmaps for more specific inspection (Supplementary File S1 and Fig. S4) and viewing or exporting subheatmap as a text table or image files.
ComplexHeatmap supports implementing high-level visualizations for matrix-like data for specific purposes. More advanced plots based on ComplexHeatmap can be easily exported to interactive applications as well. Figure S6 in Supplementary File S1 demonstrates interactive visualizations of: (i) an UpSet plot which visualizes intersections in a large number of normal sets or genomic intervals; (ii) an oncoPrint which visualizes various types of genomic alterations for a list of genes in multiple patients; (iii) a density heatmap which visualizes a large number of distributions; and (iv) enriched heatmaps (Gu et al., 2018) which visualize the enrichment of certain genomic signals (e.g. DNA methylation) on specific genomic features (e.g. gene transcriptional start sites). To facilitate analysis for users who still use other heatmap functions such as heatmap(), heatmap.2() and pheatmap(), InteractiveComplexHeatmap supports a seamless way to convert their heatmaps to interactive ones without changing their original code of heatmap generation (Supplementary File S1 and Fig. S5). More details as well as the implementation of the interactivity can be found in Sections S3 and S4 of Supplementary File S1.
Besides htShiny() that directly exports static heatmaps to stand-alone Shiny applications, InteractiveComplexHeatmap also provides flexible solutions for integrating into general Shiny application development. In Section S5 of Supplementary File S1, we described in detail the methods of (i) integrating multiple interactive heatmap widgets into a single Shiny application, (ii) self-defining response to the mouse events on the original heatmap, (iii) adjusting the front-end user interface and (iv) dynamically generating the heatmap widgets. These functionalities allow users to build more customized and complex interactive applications for specific purposes.
3 Application
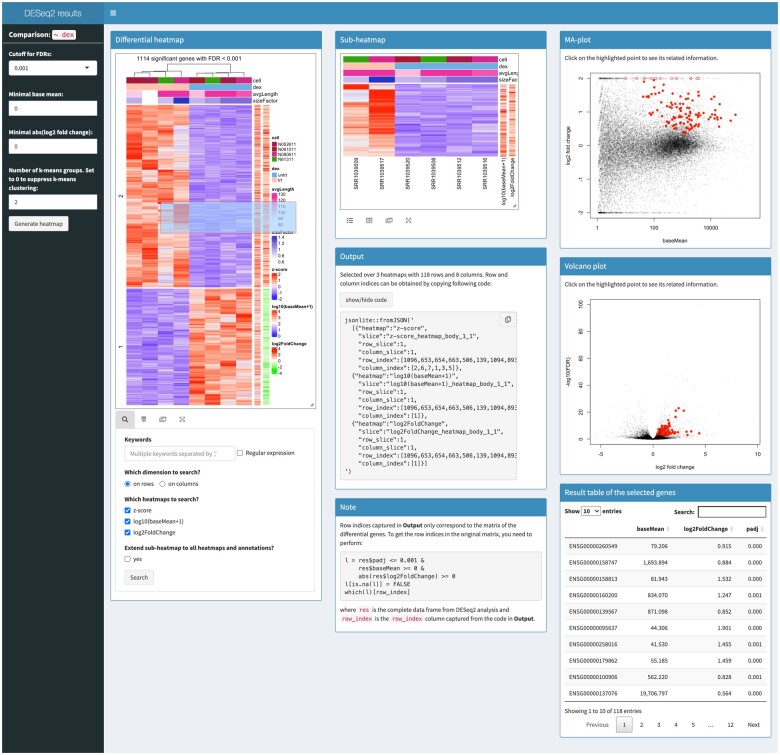
Figure 1 demonstrates an interactive heatmap application which visualizes results of differential gene expression analysis on the airway dataset performed by the package DESeq2 (Love et al., 2014). Based on a two-condition comparison, the heatmap visualizes genes that are significantly differently expressed. The application is arranged in a three-column layout where the original heatmap locates in the first column, the subheatmap and the default output locate in the second column and self-defined outputs are in the third column. In the original heatmap, there are two additional one-column heatmaps which visualize the absolute expression level of genes (‘baseMean’) and the fold change between the two conditions (‘-log10foldChange’). To get more comprehensive information of how genes are differentially expressed when selecting them from heatmap, we defined extra outputs for the click and brush events on the original heatmap. As shown in Figure 1, when a subset of genes are selected from the original heatmap, they are highlighted in an MA-plot which is a scatter plot of log2-fold change against base mean and in a volcano plot which is also a scatter plot of −log10(FDR) against log2-fold change. A table of statistics from DESeq2 analysis for the selected genes is printed below the two plots. Users can also search in the heatmap to obtain a subset of genes of interest to generate corresponding MA-plot, volcano plot and table.
Fig. 1.
An interactive heatmap application for visualizing results from a differential gene expression analysis. The interactive version of this example can be generated by the function interactivate()
Heatmap is a popular method for visualizing 2D density distributions. Nevertheless, when there are peaks that have large heights in the distribution, other peaks with smaller heights would be difficult to distinguish from each other and even not distinguishable from the background. Figure S16 in Supplementary File S1 demonstrates an interactive heatmap application where selecting a region from the original density heatmap triggers a new 2D density estimation but only on the selected subset of data. This approach helps to reveal more details in the regions which have relatively lower densities and are normally hidden in the complete dataset.
More examples with detailed explanations are available in Supplementary File S1 as well as in the vignettes of the package. InteractiveComplexHeatmap has also integrated 54 examples on both simple and advanced usages with runnable code.
4 Conclusion
Interactivity greatly facilitates to capture features which have specific patterns on heatmaps. In this work, we described a new R/Bioconductor package, InteractiveComplexHeatmap, which supports interactive visualization of complex heatmaps. It can easily deploy interactive Shiny applications from static heatmaps and it also provides flexible functionalities for implementing more comprehensive Shiny applications. We believe it will be a useful tool for efficiently interpreting data and developing new tools.
Funding
This work was supported by the NCT Molecular Precision Oncology Program.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Zuguang Gu, Molecular Precision Oncology Program, National Center for Tumor Diseases (NCT), Im Neuen-heimer Feld 280, 69120 Heidelberg, Germany.
Daniel Hübschmann, Molecular Precision Oncology Program, National Center for Tumor Diseases (NCT), Im Neuen-heimer Feld 280, 69120 Heidelberg, Germany; Heidelberg Institute of Stem Cell Technology and Experimental Medicine (HI-STEM), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; German Cancer Consortium (DKTK), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; Department of Pediatric Immunology, Hematology and Oncology, University Hospital Heidel-berg, 69120 Heidelberg, Germany.
References
- Babicki S. et al. (2016) Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res., 44, W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez N.F. et al. (2017) Clustergrammer: a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci. Data, 4, 170151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili T. et al. (2018) heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics, 34, 1600–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z. et al. (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics, 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Gu Z. et al. (2018) EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics, 19, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I. et al. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. et al. (2019) Interactive Clustered Heat Map Builder: an easy web-based tool for creating sophisticated clustered heat maps. F1000Res, 8, 1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep A.N., Kummerfeld S.K. (2017) iheatmapr: interactive complex heatmaps in R. J. Open Source Softw., 2, 359. [Google Scholar]
- Wilkinson L., Friendly M. (2009) The history of the cluster heat map. Am. Stat., 63, 179–184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.