Synopsis
Information, energy, and matter are fundamental properties of all levels of biological organization, and life emerges from the continuous flux of matter, energy, and information. This perspective piece defines and explains each of the three pillars of this nexus. We propose that a quantitative characterization of the complex interconversions between matter, energy, and information that comprise this nexus will help us derive biological insights that connect phenomena across different levels of biological organization. We articulate examples from multiple biological scales that highlight how this nexus approach leads to a more complete understanding of the biological system. Metrics of energy, information, and matter can provide a common currency that helps link phenomena across levels of biological organization. The propagation of energy and information through levels of biological organization can result in emergent properties and system-wide changes that impact other hierarchical levels. Deeper consideration of measured imbalances in energy, information, and matter can help researchers identify key factors that influence system function at one scale, highlighting avenues to link phenomena across levels of biological organization and develop predictive models of biological systems.
Introduction
Our current understanding of Biology has been built primarily through reductionism. This reductionist approach has resulted in highly specialized knowledge; however, developing better ways to identify, characterize, and predict phenomena across spatial and temporal scales is becoming increasingly important. Better understanding and engineering of biological systems will undoubtedly require new conceptual and analytical frameworks that transcend traditional disciplinary boundaries in order to discover universal rules of life. Here, we describe one possible conceptual framework: that biological systems in their broadest definition are anti-entropy systems that emerge from the nexus of energy, information, and matter. We propose that one avenue for reintegrating biological disciplines is through studying this nexus across systems and scales in order to develop quantitative and, ultimately, predictive models of biological phenomena. This perspective piece explains each of the three pillars of this nexus. We will articulate examples from multiple biological scales that highlight how this nexus approach leads to a more complete understanding of the biological system. Finally, we provide some opportunities and challenges to employing this nexus approach in practice. Our vision is that characterizing the nexus of energy, information, and matter will help researchers identify system components currently missing from biological studies and thereby reshape our experiments and interpretations.
How do energy, information, and matter interact to produce outcomes in biological systems?
Information, energy, and matter are fundamental properties of all levels of biological organization (Schroedinger 1944; Prigogine 1967; Beck and Schlögl 1993; Murphy and O'Neill 1997; Smith 1999, 2000; Brewer and Smith 2011), and life emerges from the continuous fluxes of matter, energy, and information. We use the term energy to refer to a property of physical or chemical resources that enables work to support life functions such as moving, feeding, reproducing, and growing. The practical definition of information for the biological sciences that we use here is anything that has the potential to reduce uncertainty for a biological entity (e.g., DNA, RNA, communication signals between cells or organisms, and presence or absence of participants in a community). This definition of information is based primarily in information theory and is necessarily broad because of our focus on a diversity of biological systems (i.e., from atomic to ecological). However, different types of information may be more relevant to certain biological questions (summarized recently by O'Connor et al. 2019). We propose that a quantitative characterization of the complex interconversions between matter, energy, and information that constitute this nexus will help us derive biological insights that connect phenomena across different levels of biological organization and that could improve our ability to predict responses of biological systems to disturbance.
Matter can be quantified as mass, making it relatively straightforward to relate across levels of biological organization. Although experimentally determining mass may be technically challenging, particularly at the molecular or ecosystem scales, the same units apply across scales. However, the form that matter takes will have a dramatic impact on both energy and information. For example, biological units with the same mass may differ in elemental composition, molecular complement, cell number or cell size, organ sizes, or species composition. Determining which mass components or forms are most relevant for information and energy flow is essential to formulating a predictive theory of the energy–information–matter nexus.
Energy can be quantified at all levels of biological organization as well. For example, at the molecular scale, chemical reactions such as ATP hydrolysis can release energy, such that production or breakdown of ATP or other molecules are common measures of energetic flux. At the cellular, tissue, or organismal levels, energy use is typically estimated by measuring respiration rate. At the ecosystem scale, energy is quantified by using energy balance equations to estimate radiation inputs and outputs and storage of energy in biomass (e.g., photosynthetic carbon fixation) and its mobilization and transformation as it moves through an ecosystem (e.g., Kooijman 2010). Radiative balance regulates heat exchange and temperature, both of which are crucial in determining critical biological processes ranging from metabolism to survival and reproduction. Quantifying energy necessarily depends on measuring energy change, and experimental techniques often measure only a subset of the energetic flux, challenging a complete consideration of energy conversion.
Information can also be quantified, in principle, at all levels of biological organization. Entropy is a measure of uncertainty in a system (Box 1), and information is negatively related to uncertainty in a quantifiable way. The field of information theory, first developed by Claude Shannon (1948), measures information as statistical entropy and provides well-defined methods for quantifying information. The term “entropy” is used both in information theory and in thermodynamics, and entropy in these two cases does not represent equivalent concepts (Wicken 1987; Box 1). Because biological systems are composed of structural units (e.g., atoms, molecules, cells, and organisms), information is inherent to the structure of the biological system; this syntactic information results from the non-randomness of biological systems in time and space (O'Connor et al. 2019). To the extent that biological systems are ordered and in disequilibrium with their surroundings, they require energy to overcome the natural tendency of physical systems to move towards increasing entropy. Information theoretic approaches have been applied across scales, from the molecular scale (e.g., in the genetic code; Vetsigian et al. 2006) to the ecological scale (e.g., to quantify species diversity using Shannon's Diversity Index and Maximum Entropy methods (Barnes et al. 1998; Spellerberg and Fedor 2003; Haegeman and Etienne 2010). Information theory is a central tenet both of sensory neuroscience and of signal detection theory in psychology and animal behavior, and has recently been applied to biochemical and social networks. However, despite the formal statistical definition and its demonstrated application to quantifying information, actually measuring information in biological contexts is quite difficult. One reason is that biological systems include different types of information, and the unification of these types of information remains challenging (O'Connor et al. 2019). While syntactic information describes the nonrandom spatiotemporal structure in a system, semiotic information refers to what signals (e.g., between cells or between organisms) or sensory cues from the environment represent about the state of the system (O'Connor et al. 2019; see also structural and dynamic information; Morowitz 1968). The distinction between these types of information is useful because different subfields of biology may use the same term (i.e., “information”) to describe vastly different concepts. As our case studies below illustrate, information within a molecule, cell, or ecosystem can take multiple forms, so a comprehensive accounting of information requires a deep understanding of the biology. Moreover, biological systems do not process and act upon all available information, such that quantifying information content using information theory may overestimate the amount of information that is biologically relevant.
Box 1.
We outline here fundamental physical principles that demonstrate some of the different ways that information, energy, and matter can be mathematically conceptualized and related quantitatively. We note that these equations are not directly applied to most biological empirical studies, in that few biologists fully account for energy and information flow. We introduce these equations and relationships to illustrate the mathematical underpinning of the nexus and to clarify physics terminology that is foundational to the framework proposed here.
The thermodynamic entropy S of a system with N possible microstates (i) is given by:
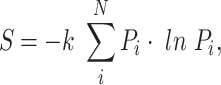 |
(1) |
where Pi denotes the probability that the system is in microstate i, and k represents the Boltzmann's constant. Thermodynamic entropy of the system represents the uncertainty about which microstate the system occupies. Empirical measurements can reveal the system macrostate, but each macrostate has multiple possible microstates that are indistinguishable to experimenters (see Wicken (1987) for additional discussion of unique features of thermodynamic entropy).
Similarly, the Shannon entropy (designated “H(SE)” for clarity) is defined as:
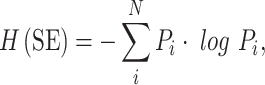 |
(2) |
where Pi is the probability of occurrence of state i given the N possible states of the system. H(SE) is a measure of the information content of an observed event, which in a biological context could refer to an experiment, population, or signal. Shannon entropy differs from thermodynamic entropy in the constant k and the base of the logarithm, which typically would be two for a binary system or four in the case of DNA, representing the four nucleotides. Moreover, Shannon entropy calculations vary depending on the choice of the possible set of states to which the observed state is compared (see Wicken (1987) for extensive comparison between thermodynamic entropy and Shannon entropy). The set of reference states for this calculation represent those considered possible before the event is observed. For example, prior knowledge might reduce the set of possible states of the system under consideration, hence altering the measure of H(SE) for a particular event.
To further explore Shannon entropy, we consider the information in a DNA molecule. For one nucleotide, there are four possible states: A, C, G, or T. For a strand with two nucleotides, there are 16 possible states (AA, AC, AG, AT, CA, CC, CG, CT, GA, GC, GG, GT, TA, TC, TG, and TT) because the information in DNA depends on order. A strand with three nucleotides has 64 possible states. The Shannon entropy, like the thermodynamic entropy, depends on the size of the system; entropy is higher for DNA strands with more nucleotides, as the system contains more possible states. Note that both S and H(SE) are maximized at a value of log(N) when all of the N system (micro)states have equal probability (Pi = 1/N). DNA sequences within conserved protein coding genes thus have lower entropy than maximal, as all arrangements of nucleotides are not equally likely. Both S and H(SE) are zero for the case where one (micro)state j has probability Pj = 1 and all other (micro)states have zero probability Pi = 0 for i ≠ j. The event or outcome from the vantage point of information theory contains zero information. For example, if you know the genome sequence of a person, the sequence of their monozygotic twin has very low entropy (only non-zero because of somatic mutations). The Shannon entropy thus is linked to the amount of uncertainty or surprise in an experimental outcome or observation. We note that despite the generally negative relationship between entropy and information, a variety of equations relating information and entropy apply, reflecting the diversity of uses of the word "information" (e.g., Morowitz 1968; Xu and Jiang 2010).
A higher-order probabilistic representation of a biological system can be described that theoretically relates the entropy and energy of the system. Reducing thermodynamic entropy S costs free energy:
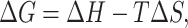 |
(3) |
where G is the Gibbs free energy, H is the enthalpy, and T is the Kelvin temperature.
A derivation of the Gibbs equation that is useful in biological systems on a small scale is:
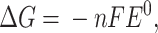 |
(4) |
where the change in Gibbs free energy is related to the number of electrons n, the Faraday constant F, and the electrical potential of the cell E⁰, which can be interpreted as any electron donor/acceptor pair.
Many equations relate energy and matter (e.g., Kooijman 2010 at organismal–ecological scales), as interconversions between energy and matter are a widely studied part of the Energy–Information–Matter nexus in some fields. Small biological scales permit the adaptation of Einstein's Theory of Relativity to biology:
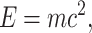 |
(5) |
where E is the total energy of the system, m is the mass of the system, and c is the speed of light. The Theory of Relativity has been used to estimate the energy needed for electrical conductance, or rates of proton or carbon tunneling in biochemical catalysis (e.g., Kohen and Klinman 1999; Chowdhury and Banerjee 2000; Reece et al. 2006).
These physical laws have been directly applied to few biological systems. Even the examples of biological nexuses we discuss in this paper have mathematical formulations that describe only a subset of the nexus. Comprehensive application of physical laws to biological questions is most feasible at sub-micrometer scale (often in dilute aqueous buffers), under special cases (e.g., sealed anaerobic microbial cultures), or at very short timescales; however, quantitative treatment of energy, information, and matter interconversions that span scales of biological organization has great promise to provide biological insights.
Although living systems use energy and matter to create order and hence are anti-entropy systems, noise itself can be advantageous. For example, random mutation is necessary for evolution to occur. New enzyme functions (Näsvall et al. 2012), strain variants (Woods et al. 2011), and metabolic potential that can be accessed to gain a competitive advantage (Vemuri et al. 2006; Catlett et al. 2015) all require some amount of entropy as the creative canvas. On more rapid time scales, stochasticity in neural systems can also improve information processing performance (McDonnell and Ward 2011). These examples illustrate that living systems do not universally move towards reducing uncertainty or stochasticity by maximizing information transmission and processing.
In biological scenarios, energy, matter, and information are constantly being interconverted, so the nexus between them is a dynamic system. Thermodynamics uses straightforward equations that relate energy, information, and matter (Box 1). Because living systems are not closed systems, but rather open systems that exist far from equilibrium (Schneider and Kay 1994), applying physical principles to biology is inherently complex. Our typical goal as biologists is not to completely account for the changes in statistical entropy, enthalpy, and Gibbs free energy in cells, organisms, or ecosystems. Practitioners of biological subdisciplines typically study a part of any given system and account for only a subset of all inputs, outputs, and system components in their studies. Explicit consideration of the energy–information–matter nexus could help identify overlooked parts of complex systems. For example, a physiologist might measure oxygen consumption as a measure of energy consumption through respiration but not quantify heat loss or gain. Similarly, a molecular biologist might measure DNA content without characterizing the genetic sequence that determines information content or potential (but see, Jiang and Xu (2010) for one approach to quantify the information content of DNA). We propose that explicitly considering the nexus of energy, information, and matter can highlight critical inputs, outputs, and components currently missing from biological studies and thereby reshape our experiments and interpretations.
To illustrate insights derived from considering the energy–information–matter nexus, we highlight the ribosome, a highly conserved ribonucleoprotein complex—a molecular machine—that generates the polypeptides essential to all organisms (Schmeing and Ramakrishnan 2009). By iteratively unravelling the many ways energy and information interact in the context of ribosome function, we have learned not only how cells engage with, develop in, and respond to their environment (Ecker and Schaechter 1963; Maitra and Dill 2015); we have also gained a tool to describe the evolutionary trajectory of life on earth (Woese and Fox 1977). Ribosomes integrate environmental and cellular information to translate the ribonucleotide triplet code from messenger RNA into a polypeptide of amino acid residues (Nirenberg and Matthaei 1961; Nirenberg and Leder 1964), thereby using information and energy to reconfigure matter into proteins necessary for essential cellular chemical and physical work. The resulting proteins both use and produce energy. Concepts from information theory and thermodynamics have been key to understanding ribosome function (Gamow et al. 1956, Sievers et al. 2004). Ribosomes reduce thermodynamic entropy by binding substrates and using energy stored in bonds of guanosine triphosphate (GTP) to catalyze polypeptide chain elongation and termination. GTP availability largely determines the rate of protein synthesis as guided by information from the triplet code (Savelsbergh et al. 2000; Åqvist and Kamerlin 2015). The physical structure of ribosomes depends on RNA and protein sequences of ribosomal components and on environmental factors such as temperature (VanBogelen and Neidhardt 1990), such that information and energy external to the ribosome influence its material form. Characterizing information, energy, and matter within the ribosome and its environment has revealed ribosomes to be sophisticated sensors of metabolism that gate protein production and cellular behavior (reviewed in Chubukov et al. 2014; Maitra and Dill 2015; Dai and Zhu 2020).
Fundamentally, insights into the nexus of energy, information, and matter on biological systems require an understanding across multiple levels of biological organization at multiple spatial and temporal scales. Even the relatively straightforward function of the ribosome depends on a broader cellular environment, as previously described. One challenge mentioned above is finding metrics of energy, information, and matter that can be quantified in a common way across levels of biological organization. Another challenge is that the propagation of information and energy through levels of biological organization can result in emergent properties and system-wide changes that impact other hierarchical levels. Defining the spatial and temporal scale of study necessarily draws boundaries that often preclude fully accounting for the flows of energy, information, and matter that are relevant to biology. Thus, defining the boundaries of a biological system typically results in imbalanced flows of energy, information, and matter. For example, an individual cell exists in relationship to other cells that influence its physiological processes. While we can quantify the conversion between information and matter that occurs during transcription, the information, energy, or matter that motivate that transcription may originate in the extracellular environment. Additionally, the genomic DNA that stores the information of the cell is itself produced by and the result of a long series of prior events and experiences of abiotic and biotic interactions that have shaped through natural selection the genome sequence. Similarly, if the scale is a multicellular organism, such as an animal or plant, then its mere movement or dispersal across space moves information, energy, and matter and exposes the organism to new energy, information, and matter in the environment. Deeper consideration of measured imbalances in energy, information, and matter can help researchers identify key factors that influence system function at one scale, highlighting avenues to link phenomena across levels of biological organization.
In summary, the energy–information–matter nexus is relevant at all scales of biological organization and may provide a common framework that helps link phenomena at different levels of biological organization. This framework offers great potential to highlight emergent phenomena and missing key regulators of biological systems. The next section illustrates applications of this framework at different levels of organization from molecules to ecosystems.
Energy–information–matter nexuses: challenges and examples
We outline below four examples of applications of how considering the nexus of energy, information, and matter can facilitate new insights. Examples are ordered from the smallest to largest spatial scales, and each highlights distinct features of the energy–information–matter nexus. We explain throughout how prior research has focused on complex relationships between energy, information, and matter, and we identify challenges and opportunities in applying the energy–information–matter nexus framework.
-
Microbes as a nexus: integrating matter, energy, and information to predict and design biological systems
One area where the concept of the energy–information–matter nexus is already being applied is in microbiology. The ease with which laboratory strains of Escherichia coli and Saccharomyces cerevisiae can be manipulated has led to the emerging field of synthetic biology where genetic elements are recombined to design new traits and behaviors. Synthetic biology has contributed to, and continues to benefit from, efforts to understand the interplay between metabolic pathways and energy conservation mechanisms of the organism as a whole. Synthetic biologists are developing increasingly sophisticated approaches to study microbial growth grounded in energy and information (via physics and information theory) with the aim toward predicting and designing microbial behaviors to benefit society.
An essential concept in understanding the inherent potential of a microbial system is the use of Gibbs free energy to describe the energy that is available to do chemical or mechanical work within the cell (Shapiro and Shapley 1965). Microbiologists explore the nexus of energy, information, and matter in living cells when they use the Gibbs equation (Box 1) to explain, predict, and successfully cultivate new organisms. For example, the application of Gibbs free energy to microbe-catalyzed biochemical reactions has allowed us to estimate key thermodynamic properties of ATP and the enzyme ATPase that allows release of bioavailable energy by ATP hydrolysis (Friedl and Shairer 1981; Müller and Hess 2017). The Gibbs equation predicts the environmental conditions that can support life such as in extreme Earth habitats or in theoretical or extraterrestrial habitats (as in the sub-field of Astrobiology; Decker et al. 1970; Tijhuis et al. 1993). It has also been used to predict the electrical current required to support growth of microbes on cathodes (Rabaey and Rozendal 2010). Finally, thermodynamic calculations involving Gibbs free energy were essential to guiding the laborious, expensive, and time-consuming culturing experiments leading to discovery of new Orders of microorganisms.
However, the spontaneity of a chemical reaction as determined from Gibbs free energy change is not sufficient to indicate whether an organism can grow, as the Gibbs equation does not consider information transmission or kinetics. Organisms transmit hereditary (syntactic) information that specifies the molecules and resulting chemical processes that are necessary to harness available free energy to maintain homeostasis and create progeny. The biochemistry to support life must also occur on a timescale faster than the entropic decay of cellular structures. The physical environment (temperature, pH, oxidation/reduction potentials, and so on) determines the physical stability of biological macromolecules and directly influences the free energy required to maintain homeostasis. Additionally, the nutrient requirements of the organism (as defined by the genetic information) must be obtained on a timescale that maintains sufficient intracellular chemical fluxes to maintain homeostasis and allow reproduction.
Nevertheless, applying the Gibbs equation approach systematically across the broader field of microbiology offers a promising avenue to extend descriptions of microbial genetic diversity so as to characterize functional roles of species in complex communities. The Gibbs equation is a useful starting point to formulate hypotheses about the function of known or unknown microbes that can guide experiments. Sequencing microbial metagenomes reveals a vast unexplored DNA information space. Amongst that cacophony of sequences, we can discern patterns of energy conservation strategies that relate to the Gibbs equation. For instance, the enzymes and cofactors underpinning growth are biochemically coupled in cultured microbes. Enzymes such as those of the TCA cycle, Wolfe cycle, and Wood–Ljungdahl metabolic pathways are not only often coupled in how they assemble inside cells (Beeckmans and Kanarek 1981; Förster and Staib 1990; Costa et al. 2010, Lieber et al. 2014; Adam et al. 2018), but also at the informational level of gene expression when organized into co-transcribed operons or co-regulated modules (Teichmann and Babu 2002; Grahame et al. 2005; Müller et al. 2013). Furthermore, the presence and relative abundance of enzymes in different metabolic pathways roughly relate to the nutrient environment experienced by the organisms. However, without full knowledge of the energetic inputs and outputs, enzyme functions, and regulatory information processing, we struggle to confidently ascribe functions of metabolic enzymes and hence ecological roles of organisms (Friedberg 2006; Widder et al. 2016). We cannot determine which patterns in metabolic pathway diversity are due to chance, the extent of undiscovered pathways, or whether and how physical and environmental factors constrain metabolic evolution (Bordbar et al. 2014; Crona et al. 2020). This lack of knowledge limits our ability to design novel pathways, to infer soil greenhouse gas emissions using environmental microbial community metagenome data, or to develop synthetic gut microbiomes as therapeutics. To produce technologies that fully harness microbial biodiversity, the applications must account for conservation of mass, bioenergetics, and both intracellular signaling and heritable information.
-
Morphogenesis: integration of intrinsic and extrinsic information, energy, and matter for optimum phenotypes
Morphogenesis is the formation of organismal structures through development. During morphogenesis, information guides the transformation of energy and matter to produce mature cells, tissues, organs, and organisms capable of living in their environment. Because morphogenesis is fundamental to producing phenotypes that can function in selective regimes, explicitly considering the energy–information–matter nexus would help identify missing inputs and components that shape phenotypic diversity. The rise of molecular genetics and genome sequencing promised to decode all the information needed to produce a living organism. Acquisition of this “code” was thought to be a clear step towards predicting and manipulating organismal structure and function based on the underlying assumption that all the information needed to form an organism is contained in the genome. Under this model, genetic information guides conversions between energy and matter that ultimately result in a biological system. This metaphor of “genes as blueprints” is pervasive in developmental biology but woefully inadequate (Nijhout 1990). In fact, developmental processes are elaborate, context-dependent temporal and spatial processes, and morphogenesis occurs when gene products interact with their environment (Nijhout 1990). Indeed, the external environment provides more than just energy (e.g., sunlight) and matter (e.g., water). It also contains informational inputs that must be integrated successfully during development. The information stored in genomic matter must be decoded and converted into chemical materials that are used in developmental processes, which are largely self-organizing (Nijhout 1990; Moczek 2012). Self-organizing processes use a wealth of extrinsic information, in addition to that stored in the genome (Vinogradov 2004), to influence developmental trajectories. In this way self-organization can be viewed as an informationally efficient way of producing phenotypes that naturally meet the physical demands of the biotic and abiotic environments in which they occur.
One example of how the information–energy–matter nexus is important in characterizing morphogenesis is in the development of plant cells and tissues. Plant cells expand when internal turgor pressure exceeds the strength of the cell wall, resulting in plastic deformation of the cell wall (Kutschera 1991). Turgor pressure is controlled partially by biological processes (e.g., gene expression influencing osmotic accumulation) but also by abiotic conditions such as water availability, which influences the Gibbs free energy in the system and, thus, the amount of work that can be done (Box 1). Plastic deformation of the cell wall is opposed by deposition of additional cellulose fibrils that strengthen the cell wall. Interestingly, the orientation of the microtubules that direct fibril deposition is controlled by mechanical stresses on the cell (Green 1962; Panteris et al. 1993; Wernicke et al. 1993; Panteris and Galatis 2005; Paradez 2006; Hamant et al. 2008; Sampathkumar et al. 2014; Mirabet et al. 2018). Thus, genetic information is combined with external and internal resources (energy and matter) to produce biochemical building blocks (e.g., microtubules and cellulose fibrils), but the resulting cell and tissue phenotypes result from physical interactions with the external environment. Insofar as the extracellular environment influences cell wall properties and cell shape morphogenesis, cell sizes and shapes may themselves store information about the external environment. The cell types for which this process has been well-elucidated are leaf epidermal “puzzle” cells. In a given leaf, these cells all possess the same genome yet are highly variable in shape (Sapala et al. 2018; Vőfély et al. 2019). The variation in puzzle cell shape within an organ can be modeled strictly by incorporating the mechanical feedback process described above, in which cell wall reinforcement responds to local stresses to oppose turgor-driven cell wall expansion and deformation. Additionally, much of the large diversity of epidermal cell shapes apparent among species (Vőfély et al. 2019) can be recapitulated by modifying just a few parameters in this model, such as growth anisotropy and overall organ growth rate (Sapala et al. 2018). While we typically consider that energy and matter are provided by the environment of the cell (e.g., sunlight and water), the cell's interactions with its physical environment also provide a wealth of information. Understanding the developmental origins of phenotypes, therefore, requires explicit consideration of information, energy, and matter both inside the cell and outside the cell. Furthermore, using environmental information—rather than employing rigid developmental programs based solely on genetic information—allows biological structures to be built with less encoded information, and to be both physically robust and also variable and tunable to the environment.
Another aspect of genomic information is worth noting. Because the genome is composed of matter (nucleic acids), it occupies physical space inside the cell. Therefore, the size of the genome limits minimum cell size, and because smaller cells have a higher ratio of surface area-to-volume, smaller cells enable higher rates of diffusion and photosynthesis (Roddy et al. 2019; Théroux-Rancourt et al. 2021). While smaller genomes, in principle, contain less syntactic information, in plants small genomes are composed of a higher fraction of gene-coding regions than are large genomes (Novák et al. 2020). Additionally, genome downsizing enables greater cell size variation and, thus, greater anatomical plasticity. This allows species with smaller genomes to better fine-tune their anatomy to the environment (Simonin and Roddy 2018; Roddy et al. 2019). Thus, genome downsizing (i.e., potential loss of syntactic information) may be associated with greater reliance on environmental information (i.e., semiotic information) to drive cell and tissue development, thereby matching phenotypes to their ecological setting better than if the developmental program were strictly and entirely encoded in the genome. The “genes as blueprints” metaphor ignores the role of the environment in providing information, but by embracing the nexus of energy, information, and matter, we can improve our understanding of morphogenesis in order to better predict and synthesize phenotypes and functions.
-
The energy-information tradeoff in neural systems depends on neuron size, cell types, and coding strategies
Neural systems exhibit two related aspects of the energy–information–matter nexus that remain open questions. One is the influence of qualities and quantities of matter in defining energy and information tradeoffs, and the other is that all available information is not necessarily used by the biological system.
Neurons use action potentials and other changes in membrane potential to encode semiotic information such as signals from other organisms and cues from the environment. Every temporary membrane depolarization initiates neuron repolarization. This repolarization requires energy, as the enzyme sodium–potassium ATPase hydrolyzes one ATP molecule to move two potassium ions into the cell and three sodium ions out of the cell. Investigators studying the photoreceptor cells that convert light information to neural signals have been remarkably successful in defining these energy and information intersections. Analyses of photoreceptor ion channel function and Shannon information (a syntactic information measure) have identified a direct tradeoff. Calculated estimates of sodium-potassium ATPase activity approximate single-cell biophysical measurements of the energetic costs of photoreceptor function (Laughlin et al. 1998). Further, photoreceptor cells that can transmit information at higher rates are more energetically costly to maintain (Niven et al. 2007). The latter conclusion indicates that features that can be considered qualities of matter, such as cell size and ion channel content, alter the quantitative relationship between energy and information. Although the information–energy tradeoff in photoreceptors can be precisely quantified in ATP molecules consumed per bit, even these precise experiments have limitations. For instance, this energetic accounting evaluates immediate ion flow but not broader costs of signaling over time, such as those of neurotransmitter packaging, protein production and transport, or photoreceptor development. Moreover, the measure of information and calculated reduction in Shannon entropy is restricted to the experimental stimulus set used (as in reference states in Box 1). In contrast, real world stimuli are multiple and constantly changing.
These limitations aside, these studies illustrate relationships between energetic costs and information processing in sensory systems. However, extensions of these same approaches in other neural systems have been limited by our incomplete understanding of neural coding principles. We have an incomplete understanding of how information inherent in neurophysiology is used. In classic neural coding studies, a sensory neuron's rate of action potentials represents properties of the sensory input, enabling a simple calculation of energy consumption per bit based on the number of action potentials used in representing a stimulus. Ion channel complements and placement establish varying costs of action potentials, so this calculation would vary based on both cell type (the form of matter) and firing patterns in response to a stimulus (Sengupta et al. 2010; Lewis et al. 2014; Niven 2016). However, neurons employ other dynamic types of coding as well. Examples include shifts in relative timing of action potentials or phase of action potentials relative to large-scale oscillations (Panzeri et al. 2010). In these examples, links between energetic costs and information content are less direct or understood. Likely, neural firing multiplexes different types of codes, and neural dynamics reflect multiple features of natural sensory stimuli on different time scales (Panzeri et al. 2010). The efficiency possible in multiplexed coding is in line with other features of neural systems that reduce energetic costs of information for individual cells. Understanding the implications of neural multiplexing would require looking at the neural energy–information–matter nexus with orders of magnitude higher complexity than is done currently, and methods of doing so remain to be developed.
Other factors complicate the idea of an energy-information tradeoff in neural systems. Populations of neurons can carry redundant information. Moreover, the presence of syntactic information in neural firing patterns does not imply the animal uses that information to direct its behavior (Panzeri et al. 2017). Hence, the energetic cost of information actually used by the organism is quite challenging to define. Related to matter, nervous systems have static or fixed costs, such as nonzero baseline firing rates and neurotransmitter turnover and cell maintenance, that constrain information capacity. These fixed costs, all in some way related to either quantity or quality of the biological matter, can vary widely among neuronal subtypes. Additionally, maintaining more neurons increases system information capacity but in a nonlinear fashion, typically with diminishing returns at some point. Hence there is no simple relationship between information capacity, energetic demand, and either total number of neurons or mass of neural tissue. This situation represents a tremendous opportunity for theoretical and empirical biological scientists to come together to identify general principles about how animals balance these energetic and material costs with the benefits of reduced uncertainty about the abiotic and biotic environment.
-
Ecological stoichiometry: phosphorus mediates information and energy across scales—from genes to ecosystems
A form of matter, the element phosphorus, is an important mediator of the relationship between energy and information, and phosphorus illustrates the potential for the energy–information–matter nexus to bridge levels of biological organization. Bioavailable phosphorus in the form of phosphate is a key factor limiting growth of species in many ecosystems. It is an essential component of fertilizer in most agroecosystems that provide the world's food supply. Along with nitrogen, phosphorous often limits primary productivity in terrestrial, inland, and oceanic ecosystems (Elser et al. 2000,2007).
We see the likely origins of this phenomenon on a much smaller scale. Phosphorous is also particularly abundant in ribosomal RNA, such that phosphate limitation in ecosystems is due to the high demand for phosphorous by ribosomal RNA during periods of rapid growth (Elser et al. 1996). Additionally, phosphorous plays an important role in energy storage and mobilization. Energy flow through biological systems is associated with the formation and hydrolysis of phosphate bonds, most commonly in ATP, and the free energy change associated with ATP hydrolysis enables enzymatic work in cells. Paradoxically, a phosphate group from ATP is also frequently transferred to proteins, where it mediates information processing by changing the functional characteristics of the protein. If the enzyme in question is a phosphate-adding kinase or a phosphate-removing phosphatase, phosphorylation or dephosphorylation enhances or suppresses enzyme activity. Cascades of kinases and phosphatases are the signature signaling mechanisms within a cell, turning processes on and off. Phosphate is thus also essential to the management of information processing by protein phosphorylation. Finally, at the molecular level, phosphate makes up the backbone of the critical information transmission molecules DNA and RNA.
The competition is so fierce for phosphate that some organisms have found ways to access other forms of phosphorus (phosphite, hypophosphite, phosphonate, and phosphine) and some are thought to specifically decrease the bioavailability of phosphorus for competing organisms by converting phosphate to phosphonate or phosphinate molecules (Yu et al. 2013; Pasek et al. 2014). Altogether, phosphorus illustrates the energy–information–matter nexus at the molecular level (as syntactic information in nucleic acid, as a mechanism of releasing energy during ATP hydrolysis, and as phosphate transfer altering protein function), at the cellular level (within signaling cascades serving as switches for cellular processes), and even at the ecosystem level (as a key limiting nutrient). On a global scale, phosphorus demand is expected to exceed supply in the coming decades, leading to a world-wide scarcity and increase in the commercial cost and need for recovery programs to obtain this essential element (Desmidt et al. 2015). Expanding our analysis of the energy–information–matter nexus would open new research questions. For example, do phosphorus limitation and consequent mediation of information and energy fluxes vary across taxa, at multiple trophic levels, or in organisms of different motility and energy demands (e.g., endotherms vs. exotherms)?
The above are just a few examples of why we view the pursuit of the energy–information–matter nexus as critical to uniting phenomena across scales in biological systems. In each case, the benefits and challenges of examining systems from the energy–information–matter nexus across scales demonstrates the potential power of this framework. These are:
A baseline of research into direct energy, information, and matter measurements and tradeoffs using principles described in Box 1 already exists, to differing degrees.
Practical and applied benefits exist, such as being able to accurately anticipate effects of global warming on the biosphere at all scales, to address a key globally limiting element, and to generate life that is possible but does not yet exist.
A universal conceptual framework to view the energy–information–matter nexus within and between scales is elusive.
Matter must be considered both quantitatively (mass) and qualitatively (substance and form).
An expanded concept of information sources and types must be considered.
Theoretical treatments of information capacity and information use must be developed.
Colloquial uses of “information” must be aligned with the mathematical formulations.
Carrying the vision forward: next steps
The largest hurdle to successful application of the nexus concept is determining practical implementation strategies within our scientific enterprise. Success will be possible only through a concerted effort from multiple angles. Here we identify broad challenges as starting points.
Educational
As a discipline, we can certainly improve the training of biologists in a common vocabulary related to energy, information, and matter at the curricular and continuing education levels (Brewer and Smith 2011). The ultimate goal is to train biologists at every stage who are capable of communicating their work using this vocabulary. While many undergraduate biology curricula include physics and mathematics coursework, these required courses rarely, if ever, focus on biological applications of information and energetic approaches or, even more critically, their nexus. We also propose a coordinated effort to disseminate examples quantifying energy, information, and matter at the nexus that can be readily adapted for use in undergraduate courses. Another concrete resource could be a primer of relevant physics for biologists at any career stage. As our efforts to reach across our own disciplines have taught us, such a written resource will be most accessible if informed by physicists and informaticists but written by biologists using approachable examples. In sum, theoretical approaches must be integrated into our current empirical approaches.
Experimental
We propose to identify specific nexuses as research targets for larger scale collaborative funding. Interdisciplinary research teams would invent or identify new scientific methodologies, quantitative measures, and system components necessary to evaluate changes in the balance of energy, information, and matter at all biological levels. Effective ways to facilitate teams with the essential collaborative skills have been addressed recently by the National Research Council and others, including an explicit focus on the importance of diversity and interpersonal skills (Cheruvelil et al. 2014; National Research Council 2015). For this and other efforts to achieve scientific summits previously considered unconquerable, we rely on the continuing efforts to remove systemic barriers in scientific communities and their funders that limit interdisciplinary work (Bromham et al. 2016).
In conclusion, we propose that studying the nexuses of energy, information, and matter can reintegrate biological subdisciplines. The stakes for reintegration are increasingly high, as robust predictive models and biological technologies may form essential responses to climate change, may contribute to sustainable food and fuel production, and may mitigate the associated risks to social order. Employing energy, information, and matter as common currencies that apply across spatial and temporal scales provides unique opportunities to integrate across levels of biological organization. Expanding ongoing efforts to support team-based work and novel approaches to train young investigators will be at the forefront of these efforts to unite the plurality of approaches and reduce sub-disciplinary boundaries in the biological sciences.
Acknowledgments
This paper was initiated at, and supported by, the US National Science Foundation (NSF) Reintegrating Biology Jumpstart Workshop held in Austin, Texas in December 2019. We acknowledge Amadee des Georges for extensive contributions to discussions that led to the ideas presented here as well as anonymous reviewers who provided feedback.
Notes
Authors contributed equally and are listed in randomized order
Based on a Jumpstart-Reintegrating Biology Vision Paper, developed during Town Hall meetings funded by The National Science Foundation in 2019–2020.
Contributor Information
Kim L Hoke, Department of Biology, Colorado State University, Fort Collins, CO 80523-1878, USA.
Sara L Zimmer, Department of Biomedical Sciences, University of Minnesota Medical School, Duluth campus, Duluth, MN 55812, USA.
Adam B Roddy, Department of Biological Sciences, Institute of Environment, Florida International University, Miami, FL 33199, USA.
Mary Jo Ondrechen, Department of Chemistry and Chemical Biology, Northeastern University, Boston, MA 02115, USA.
Craig E Williamson, Department of Biology, Miami University, Oxford, OH 45056, USA.
Nicole R Buan, Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE 68588-0662, USA.
Author contributions
All authors contributed to the development of the ideas, the drafting of the manuscript, and editing of the final version. KLH coordinated the group efforts and synthesized editorial suggestions of group members.
Funding
This work was supported by the following: CEW acknowledges support from an NSF OPUS grant (number DEB-1950170). MJO acknowledges support from NSF (grant numbers CHE-1905214 and CHE-2030180). ABR acknowledges support from NSF grant (numbers DEB-1838327 and CMMI-2029756). NRB was supported by NSF (grant number IOS-1938948), USDA Hatch Multistate (NC1200 Photosynthetic Processes, grant number NEB-30–133), and Nebraska Center for Energy Sciences Research (Cycle 15) grants. KLH acknowledges support from an NSF OPUS grant (number DEB-1911619). SLZ was supported by NIH (grant number 1R15AI135885-01).
References
- Adam PS, Borrel G, Gribaldo S. 2018. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc Natl Acad Sci. 115:E1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åqvist J, Kamerlin SCL. 2015. Exceptionally large entropy contributions enable the high rates of GTP hydrolysis on the ribosome. Sci Rep. 5:15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes BV, Zak DR, Denton SR, Spurr SH. 1998. Forest ecology, New York (NY): Wiley. [Google Scholar]
- Beck C, Schögl F. 1993. Thermodynamics of chaotic systems. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Beeckmans S, Kanarek L. 1981. Demonstration of physical interactions between consecutive enzymes of the citric acid cycle and of the aspartate-malate shuttle: a study involving fumarase, malate dehydrogenase, citrate synthase and aspartate aminotransferase. Eur J Biochem. 117:527–35. [DOI] [PubMed] [Google Scholar]
- Bordbar A, Monk JM, King ZA, Palsson BO. 2014. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 15:107–20. [DOI] [PubMed] [Google Scholar]
- Brewer CA, Smith D, eds. 2011. Vision and change in undergraduate biology education: a call to action. Washington (DC): American Association for the Advancement of Science. (http://visionandchange.org/finalreport). [Google Scholar]
- Bromham L, Dinnage R, Hua X. 2016. Interdisciplinary research has consistently lower funding success. Nature. 534:684–7. [DOI] [PubMed] [Google Scholar]
- Catlett JL, Ortiz AM, Buan NR. 2015. Rerouting cellular electron flux to increase the rate of biological methane production. Appl Environ Microbiol. 81:6528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheruvelil KS, Soranno PA, Weathers KC, Hanson PC, Goring SJ, Filstrup CT, Read EK. 2014. Creating and maintaining high-performing collaborative research teams: the importance of diversity and interpersonal skills. Front Ecol Environ. 12:31–8. [Google Scholar]
- Chowdhury S, Banerjee R. 2000. Evidence for quantum mechanical tunneling in the coupled cobalt− carbon bond homolysis− substrate radical generation reaction catalyzed by methylmalonyl-CoA mutase. J Am Chem Soc. 122:5417–8. [Google Scholar]
- Chubukov V, Gerosa L, Kochanowski K, Sauer U. 2014. Coordination of microbial metabolism. Nat Rev Microbiol. 12:327–40. [DOI] [PubMed] [Google Scholar]
- Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, Burn JA, Hackett M, Leigh JA. 2010. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci. 107:11050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crona K, Luo M, Greene D. 2020. An uncertainty law for microbial evolution. J Theor Biol. 489:110155. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhu M. 2020. Coupling of ribosome synthesis and translational capacity with cell growth. Trends Biochem Sci. 45:681–92. [DOI] [PubMed] [Google Scholar]
- Decker K, Jungermann K, Thauer RK. 1970. Energy production in anaerobic organisms. Angewandte Chemie International Edition in English. 9:138–58. [DOI] [PubMed] [Google Scholar]
- Desmidt E, Ghyselbrecht K, Zhang Y, Pinoy L, Bruggen BV der, Verstraete W, Rabaey K, Meesschaert B. 2015. Global phosphorus scarcity and full-scale P-recovery techniques: a review. Crit Rev Environ Sci Technol. 45:336–84. [Google Scholar]
- Ecker RE, Schaechter M. 1963. Ribosome content and the rate of growth of Salmonella typhimurium. Biochimica et Biophysica Acta (BBA) Special Sec Nucleic Acids Rel Sub. 76:275–9. [PubMed] [Google Scholar]
- Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE. 2007. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett. 10:1135–42. [DOI] [PubMed] [Google Scholar]
- Elser JJ, Dobberfuhl DR, MacKay NA, Schampel JH. 1996. Organism size, life history, and N:P stoichiometry: Toward a unified view of cellular and ecosystem processes. Bioscience. 46:674–84. [Google Scholar]
- Elser JJ, Fagan WF, Denno RF, Dobberfuhl DR, Folarin A, Huberty A, Interlandi S, Kilham SS, McCauley E, Schulz KL, et al. 2000. Nutritional constraints in terrestrial and freshwater food webs. Nature. 408:578–80. [DOI] [PubMed] [Google Scholar]
- Förster M, Staib W. 1990. Further evidence for direct interaction of pyruvate dehydrogenase multienzyme complex with citric acid cycle based on nonlinearity of hill plots in presence of C2 and C4 substrate analogues. Int J Biochem. 22:773–8. [DOI] [PubMed] [Google Scholar]
- Friedberg I. 2006. Automated protein function prediction—the genomic challenge. Brief Bioinform. 7:225–42. [DOI] [PubMed] [Google Scholar]
- Friedl P, Schairer HU. 1981. The isolated of F0 of Escherichia coli ATP-synthase is reconstitutively active in H+-conduction and ATP-dependent energy-transduction. FEBS Lett. 128:261–4. [DOI] [PubMed] [Google Scholar]
- Gamow G, Rich A, Yčas M. 1956. The problem of information transfer from the nucleic acids to proteins. Adv Biol Med Phys. 4:23–68. [DOI] [PubMed] [Google Scholar]
- Grahame DA, Gencic S, DeMoll E. 2005. A single operon-encoded form of the acetyl-CoA decarbonylase/synthase multienzyme complex responsible for synthesis and cleavage of acetyl-CoA in Methanosarcina thermophila. Arch Microbiol. 184:32–40. [DOI] [PubMed] [Google Scholar]
- Green PB. 1962. Mechanism for plant cellular morphogenesis. Science. 138:1404–5. [DOI] [PubMed] [Google Scholar]
- Haegeman B, Etienne RS. 2010. Entropy maximization and the spatial distribution of species. Am Nat. 175:E74–90. [DOI] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. 2008. Developmental patterning by mechanical signals in Arabidopsis. Science. 322:1650–5. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xu C. 2010. The calculation of information and organismal complexity. Biol Direct. 5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohen A, Klinman JP. 1999. Hydrogen tunneling in biology. Chem Biol. 6:R191–8. [DOI] [PubMed] [Google Scholar]
- Kooijman SALM. 2010. Dynamic energy and mass budgets in biological systems. Cambridge: Cambridge University Press. [Google Scholar]
- Kutschera U. 1991. Regulation of cell expansion. The cytoskeletal basis of plant growth and form. Cambridge (MA): Academic Press, The University of Michigan, 85–99. [Google Scholar]
- Laughlin SB, de Ruyter van Steveninck RR, Anderson JC. 1998. The metabolic cost of neural information. Nat Neurosci. 1:36–41. [DOI] [PubMed] [Google Scholar]
- Lewis JE, Gilmour KM, Moorhead MJ, Perry SF, Markham MR. 2014. Action potential energetics at the organismal level reveal a trade-off in efficiency at high firing rates. J Neurosci. 34:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber DJ, Catlett J, Madayiputhiya N, Nandakumar R, Lopez MM, Metcalf WW, Buan NR. 2014. A multienzyme complex channels substrates and electrons through acetyl-CoA and methane biosynthesis pathways in Methanosarcina. PLoS ONE. 9:e107563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Dill KA. 2015. Bacterial growth laws reflect the evolutionary importance of energy efficiency. Proc Natl Acad Sci. 112:406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell MD, Ward LM. 2011. The benefits of noise in neural systems: bridging theory and experiment. Nat Rev Neurosci. 12:415–25. [DOI] [PubMed] [Google Scholar]
- Mirabet V, Krupinski P, Hamant O, Meyerowitz EM, Jönsson H, Boudaoud A. 2018. The self-organization of plant microtubules inside the cell volume yields their cortical localization, stable alignment, and sensitivity to external cues. PLoS Comput Biol. 14:e1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP. 2012. The nature of nurture and the future of evodevo: toward a theory of developmental evolution. Integr Comp Biol. 52:108–19. [DOI] [PubMed] [Google Scholar]
- Morowitz HJ. 1968. Energy flow in biology; biological organization as a problem in thermal physics. New York (NY): Academic Press. [Google Scholar]
- Müller B, Sun L, Schnürer A. 2013. First insights into the syntrophic acetate-oxidizing bacteria–a genetic study. MicrobiologyOpen. 2:35–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller V, Hess V. 2017. The minimum biological energy quantum. Front Microbiol. 8:2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP, O'Neill LAJ, Eds. 1997. What is life?. In: The Next Fifty Years. Speculations on the Future of Biology. Cambridge: Cambridge University Press. [Google Scholar]
- Näsvall J, Sun L, Roth JR, Andersson DI. 2012. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 338:384–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council, Committee on the Science of Team Science, Board on Behavioral, Cognitive, and Sensory Sciences, Division of Behavioral and Social Sciences and Education. 2015. Enhancing the Effectiveness of Team Science. Washington (DC): National Academies Press. [PubMed] [Google Scholar]
- Nijhout HF. 1990. Problems and paradigms: metaphors and the role of genes in development. Bioessays. 12:441–6. [DOI] [PubMed] [Google Scholar]
- Nirenberg M, Leder P. 1964. RNA codewords and protein synthesis: the effect of trinucleotides upon the binding of sRNA to ribosomes. Science. 145:1399–407. [DOI] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei JH. 1961. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci. 47:1588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven JE, Anderson JC, Laughlin SB. 2007. Fly photoreceptors demonstrate energy-information trade-offs in neural coding. PLoS Biol. 5:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven JE. 2016. Neuronal energy consumption: biophysics, efficiency and evolution. Curr Opin Neurobiol. 41:129–35. [DOI] [PubMed] [Google Scholar]
- Novák P, Guignard MS, Neumann P, Kelly LJ, Mlinarec J, Koblížková A, Dodsworth S, Kovařík A, Pellicer J, Wang W, et al. 2020. Repeat-sequence turnover shifts fundamentally in species with large genomes. Nat Plants. 6:1325–9. [DOI] [PubMed] [Google Scholar]
- O'Connor MI, Pennell MW, Altermatt F, Matthews B, Melián CJ, Gonzalez A. 2019. Principles of ecology revisited: integrating information and ecological theories for a more unified science. Front Ecol Evolut. 7:219. [Google Scholar]
- Panteris E, Apostolakos P, Galatis B. 1993. Microbutule organization, mesophyll cell morphogenesis, and intercellular space formation in Adiantum capillus veneris leaflets. Protoplasma. 172:97–110. [Google Scholar]
- Panteris E, Galatis B. 2005. The morphogenesis of lobed plant cells in the mesophyll and epidermis: organization and distinct roles of cortical microtubules and actin filaments. New Phytol. 167:721–32. [DOI] [PubMed] [Google Scholar]
- Panzeri S, Brunel N, Logothetis NK, Kayser C. 2010. Sensory neural codes using multiplexed temporal scales. Trends Neurosci. 33:111–20. [DOI] [PubMed] [Google Scholar]
- Panzeri S, Harvey CD, Piasini E, Latham PE, Fellin T. 2017. Cracking the neural code for sensory perception by combining statistics, intervention, and behavior. Neuron. 93:491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. 2006. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 312:1491–5. [DOI] [PubMed] [Google Scholar]
- Pasek MA, Sampson JM, Atlas Z. 2014. Redox chemistry in the phosphorus biogeochemical cycle. Proc Natl Acad Sci. 111:15468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigogine I. 1967. Introduction to thermodynamics of irreversible processes. 3rd ed. New York (NY): Interscience. [Google Scholar]
- Rabaey K, Rozendal RA. 2010. Microbial electrosynthesis — revisiting the electrical route for microbial production. Nat Rev Microbiol. 8:706–16. [DOI] [PubMed] [Google Scholar]
- Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. 2006. Proton-coupled electron transfer: the mechanistic underpinning for radical transport and catalysis in biology. Philos Trans R Soc B Biol Sci. 361:1351–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddy AB, Théroux-Rancourt G, Abbo T, Benedetti JW, Brodersen CR, Castro M, Castro S, Gilbride AB, Jensen B, Jiang G-F, et al. 2020. The scaling of genome size and cell size limits maximum rates of photosynthesis with implications for ecological strategies. Int J Plant Sci. 181:75–87. [Google Scholar]
- Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jönsson H, Meyerowitz EM. 2014. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife. 3:e01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapala A, Runions A, Routier-Kierzkowska A-L, Das Gupta M, Hong L, Hofhuis H, Verger S, Mosca G, Li C-B, Hay A, et al. 2018. Why plants make puzzle cells, and how their shape emerges. eLife. 7:e32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Mohr D, Wilden B, Wintermeyer W, Rodnina MV. 2000. Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. J Biol Chem. 275:890–4. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature. 461:1234–42. [DOI] [PubMed] [Google Scholar]
- Schneider ED, Kay JJ. 1994. Life as a manifestation of the second law of thermodynamics. Math Comput Modell. 19:25–48. [Google Scholar]
- Schrödinger E. 1944. What is life? The physical aspect of the living cell. Cambridge: Cambridge University Press. [Google Scholar]
- Sengupta B, Stemmler M, Laughlin SB, Niven JE. 2010. Action potential energy efficiency varies among neuron types in vertebrates and invertebrates. PLoS Comput Biol. 6:e1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J. 27:379–423. [Google Scholar]
- Shapiro NZ, Shapley LS. 1965. Mass action laws and the Gibbs free energy function. J Soc Ind Appl Math. 13:353–75. [Google Scholar]
- Sievers A, Beringer M, Rodnina MV, Wolfenden R. 2004. The ribosome as an entropy trap. Proc Natl Acad Sci. 101:7897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin KA, Roddy AB. 2018. Genome downsizing, physiological novelty, and the global dominance of flowering plants. PLoS Biol. 16:e2003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ. 1999. The idea of information in biology. Q Rev Biol. 74:395–400. [DOI] [PubMed] [Google Scholar]
- Smith MJ. 2000. The concept of information in biology. Philos Sci. 67:177–94. [Google Scholar]
- Spellerberg IF, Fedor PJ. 2003. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ Index. Glob Ecol Biogeogr. 12:177–9. [Google Scholar]
- Teichmann SA, Babu MM. 2002. Conservation of gene co-regulation in prokaryotes and eukaryotes. Trends Biotechnol. 20:407–10. [DOI] [PubMed] [Google Scholar]
- Théroux-Rancourt G, Roddy AB, Earles JM, Gilbert ME, Zwieniecki MA, Boyce CK, Tholen D, McElrone AJ, Simonin KA, Brodersen CR. 2021. Maximum CO2 diffusion inside leaves is limited by the scaling of cell size and genome size. Proc R Soc B. 288: 20203145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijhuis L, Loosdrecht MCMV, Heijnen JJ. 1993. A thermodynamically based correlation for maintenance Gibbs energy requirements in aerobic and anaerobic chemotrophic growth. Biotechnol Bioeng. 42:509–19. [DOI] [PubMed] [Google Scholar]
- VanBogelen RA, Neidhardt FC. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci. 87:5589–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri GN, Eiteman MA, Altman E. 2006. Increased recombinant protein production in Escherichia coli strains with overexpressed water-forming NADH oxidase and a deleted ArcA regulatory protein. Biotechnol Bioeng. 94:538–42. [DOI] [PubMed] [Google Scholar]
- Vetsigian K, Woese C, Goldenfeld N. 2006. Collective evolution and the genetic code. Proc Natl Acad Sci. 103:10696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. 2004. Evolution of genome size: multilevel selection, mutation bias or dynamical chaos?. Curr Opin Genet Dev. 14:620–6. [DOI] [PubMed] [Google Scholar]
- Vőfély RV, Gallagher J, Pisano GD, Bartlett M, Braybrook SA. 2019. Of puzzles and pavements: a quantitative exploration of leaf epidermal cell shape. New Phytol. 221:540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernicke W, Günther P, Jung G. 1993. Microtubules and cell shaping in the mesophyll of Nigella damascena L. Protoplasma. 173:8–12. [Google Scholar]
- Wicken JS. 1987. Entropy and information: suggestions for common language. Philos Sci. 54:176–93. [Google Scholar]
- Widder S, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, Cordero OX, Brown SP, Momeni B, Shou W, et al. 2016. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10:2557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci. 74:5088–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE. 2011. Second-order selection for evolvability in a large Escherichia coli population. Science. 331:1433–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Doroghazi JR, Janga SC, Zhang JK, Circello B, Griffin BM, Labeda DP, Metcalf WW. 2013. Diversity and abundance of phosphonate biosynthetic genes in nature. Proc Natl Acad Sci. 110:20759–64. [DOI] [PMC free article] [PubMed] [Google Scholar]


