Abstract
Background:
We aimed to prepare a nanofluid, containing f-MWCNTs, and investigate the antibacterial efficacy of f-MWCNTs+ ciprofloxacin (cip) on Klebsiella pneumoniae by evaluating the virulence gene expression.
Methods:
This study was carried out from 2019 to 2020, in the Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran. The nanofluid containing antibiotic and f-MWCNTs were prepared by the ultrasonic method. The minimum inhibitory concentrations (MICs) of ciprofloxacin and f-MWCNTs were determined using the broth micro dilution MIC tests. For examining the antibacterial effects, the expression level of virulence genes, under the influence of f-MWCNTs, was evaluated by a real-time PCR.
Results:
The effect of 8 μg/ml ciprofloxacin + 400 μg/ml f-MWCNTs, completely inhibited the growth of the resistant isolate of K. pneumoniae, while, in the ATCC 700,603 isolate, 2 μg/ml ciprofloxacin with 100 μg/ml f-MWCNT could inhibit a bacterial growth. In the resistant K. pneumoniae clinical isolate, after f-MWCNT+cip treatment, the expression of fimA, fimD, wza, and wzi genes was significantly downregulated, compared to the ciprofloxacin treatment, and upregulated, compared to the negative control. For the ATCC 700,603 isolate treated with f-MWCNT+cip, the expression of fimA, fimD and wza virulence genes showed upregulation, compared to the negative control and downregulated in comparison with the ciprofloxacin treatment.
Conclusion:
Simultaneous treatment of resistant isolate of K. pneumoniae with f-MWCNTs +antibiotic could improve the effectiveness of antibiotic at lower doses, due to the reduced expression of virulence genes in comparison with antibiotic treatment, besides the increased cell wall permeability to antibiotics.
Keywords: Carbon nanotube, Klebsiella pneumoniae, Virulence genes
Introduction
The importance of Klebsiella pneumoniae, as an opportunistic pathogen in humans, is due to the infections in the hospitalized patients and increased resistance to antibiotics (1). These bacterial species are the main reasons of nosocomial infections (NIs) including respiratory tract infections, urinary tract infections (UTIs), and blood stream-associated infections (BSIs) especially in patients confined in the intensive care unit (ICU) (2–5). K. pneumoniae is responsible for 44.22% of hospital-acquired infections in Iran (6). Pneumonia is one of the principal causes of childhood death and infection-related fatality in adults in low- and middle-income countries (7). The hospital-acquired pneumonia (HAP), as the second most common nosocomial infection, accounts for 30% of all nosocomial infections (8). A high percentage of patients with HAP, develops the infection after connecting to the ventilator (9). Capsular polysaccharides (CPS), are considered as the major determinant for K. pneumoniae involved in pathogenesis through resistance to phagocytosis and serum killing (10–13). Another group of virulence factors of K. pneumoniae are fimbrial adhesins (14, 15).
The majority of K. pneumoniae clinical isolates usually express type 1 fimbrial adhesins. FimA is the main structural protein of the type 1 fimbrial filament. Ciprofloxacin (cip), from the quinolone antibiotic family, is one of the most widely used antibiotics against K. pneumoniae. The major problem in the K. pneumoniae treatment is the antibiotic resistance (16). 75% of K. pneumoniae clinical strains isolated from HAP specimens in Iran showed resistant to this antibiotic (16, 17). Given the increasing resistance of pathogens to various antibiotics, multidrug-resistant (MDR) pathogens could lead to the death of 10 million people annually by 2050, and surprisingly more than any other life-threatening diseases, including cancer (18). An association between MDR and biofilm genes expression is highly approved in numerous studies (16). Bacteria, from the Enterobacteriaceae family, which are resistant to the third-generation cephalosporins, have been prioritized in the WHO list of pathogens for new research and development of antibiotics (19). According to the WHO announcement, the use of nano-materials in a post-antibiotic era has the potential to overcome the MDR (multi drug resistant) pathogen issue (20). Carbon nanotube (CNT), a hollow cylindrical shape with high strength and hardness, has unique biological, mechanical, thermal, and electronic properties that have many diverse applications in biomedicine. Nanofluid containing nanomaterials has been a good candidate in a wide range of applications, including drug delivery, antibacterial applications, cancer treatment, diagnosis, medical imaging (ex. MRI), and wound dressing (17, 21–23). Functionalization with a carboxyl group (−COOH) would make MWCNTs binding to bacteria more efficient. Overcoming the antibiotic resistance is one of the advantages of combination therapy. The studies showed that treatment of several tumor cell lines with functionalized multi-wall carbon nanotubes (f-MWCNT), led into high percentages of necrosis and apoptosis (24).
This study aimed to prepare nanofluid containing f-MWCNTs and evaluate the efficacy of f-MWCNTs+cip combination on virulence genes expression of both resistant and ATCC isolates of K. pneumoniae.
Methods
The present study was carried out from 2019 to 2020, in the Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran.
Nanofluid (NF) preparation
The MWCNT functionalized with −COOH in powder form, was prepared from United States Research Company. The ingredients including 0.2 g f-MWCNTs, 100 ml of deionized water, 6 ml of ethanol 96% and 0.06 g of Arabic gum were mixed and stirred for 20 minutes. Then, the suspension was placed in an ice bucket to the ultrasonic device (Ultrasonic Homogenizer 400 w, 200 kHz), with the power of 200 W for 20 minutes (25).
Bacteria identification
The clinical and ATCC 700,603 isolate of K. pneumoniae were prepared from the microbial bank of the mycobacterium and pulmonary research department, Pasteur institute of Iran. The ATCC 700,603 isolate of K. pneumoniae was used as a control in this study. We did the conventional biochemical and molecular validation tests according to the relevant protocols (26, 27).
Drug susceptibility tests
To determine the resistance of both clinical and standard isolates to the conventional antibiotics, the disk diffusion (Kirby–Bauer) testing on Mueller-Hinton agar was carried out and the growth inhibition zone diameters were compared to the clinical and laboratory standards institute (CLSI) guideline (Table 1) (28). The MICs of ciprofloxacin and f-MWCNTs, were determined using broth micro dilution MIC tests, according to CLSI 2020 instructions in triplicates (28) and was confirmed by MBC test (Fig. 1).
Table 1:
Antibiogram results of resistant and standard K. pneumoniae strains to antibiotic disks
| Antibiotic disk | Symbol | Disk Content (μg) | Strain | R/I /S | Zone Diameter (mm) | CLSI | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| S | I | R | ||||||
| Cefoxitin | CFX | 30 | ATCC | R | 0 | >16 | 13–15 | <12 |
| 133 | R | 0 | ||||||
| Ceftazidime | CAZ | 30 | ATCC | - | - | >21 | 18–20 | <17 |
| 133 | R | 0 | ||||||
| Cefepime | CPM | 30 | ATCC | S | 26 | >25 | 19–24 | <18 |
| 133 | R | 0 | ||||||
| Ciprofloxacin | CIP | 5 | ATCC | R | 20 | >26 | 22–25 | <21 |
| 133 | R | 0 | ||||||
| Imipenem | IMI | 10 | ATCC | - | - | >23 | 20–22 | <19 |
| 133 | R | 8 | ||||||
| Ertapenem | ETP | 10 | ATCC | R | 17 | >22 | 19–21 | <18 |
| 133 | R | 0 | ||||||
| Meropenem | MEM | 10 | ATCC | R | 0 | >23 | 20–22 | <19 |
| 133 | R | 0 | ||||||
| Doripenem | DOR | 10 | ATCC | - | - | >23 | 20–22 | <19 |
| 133 | R | 8 | ||||||
| Aztreonam | ATM | 30 | ATCC | - | - | >21 | 18–20 | <17 |
| 133 | R | 0 | ||||||
| Gentamicin | GM | 10 | ATCC | S | 17 | >15 | 13–14 | <12 |
| 133 | I | 14 | ||||||
| Chloramphenicol | C | 30 | ATCC | - | - | >18 | 13–17 | <12 |
| 133 | I | 13 | ||||||
| Colistin | CO | 20 | ATCC | - | 19 | - | - | - |
| 133 | - | 11 | ||||||
S: Sensitive, I: Intermediate, R: Resistant, -: Not determined
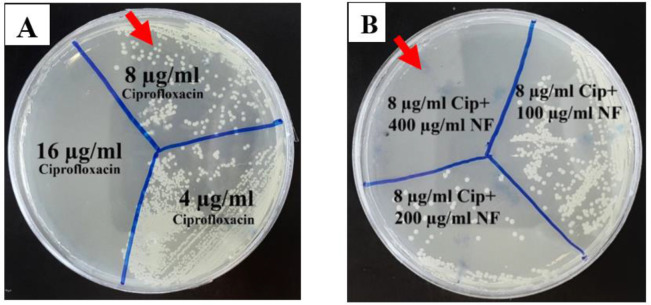
Fig. 1:
MBC results of the effect of ciprofloxacin and f-MWCNTs on K. pneumoniae. A) The effect of 8 μg/ml ciprofloxacin on the ES133 resistant strain did not inhibit the growth, but 16 μg/ml ciprofloxacin inhibited that. B) The effect of 8 μg/ml ciprofloxacin+ 400 μg/ml f- MWCNT completely killed the bacteria. As a conclusion, the effective dose of the antibiotic was reduced when combined with 400 μg/ml f-MWCNT
RNA extraction, cDNA synthesis, Real-Time-PCR
RNA extraction was performed according to RiboEX kit instructions (GeneAll Biotechnology Company, South Korea). The reverse transcriptase PCR (RT-PCR) method was used and the procedure were performed according to the Pars Tous company (Iran) instructions. Relevant primers were designed using the Gene runner software and the primer synthesis order was submitted by Pishgam Company in Tehran to Metabion Company in Germany. Moreover, a real-time PCR assay was performed using SYBR Green 2X master mix (Yekta Tajhiz company, Iran), following the manufacturer’s instructions.
TEM imaging
To confirm the internalization of f-MWCNTs, the treated and untreated samples were observed under a transmission electron microscope (TEM) (Fig. 2).
Fig. 2:
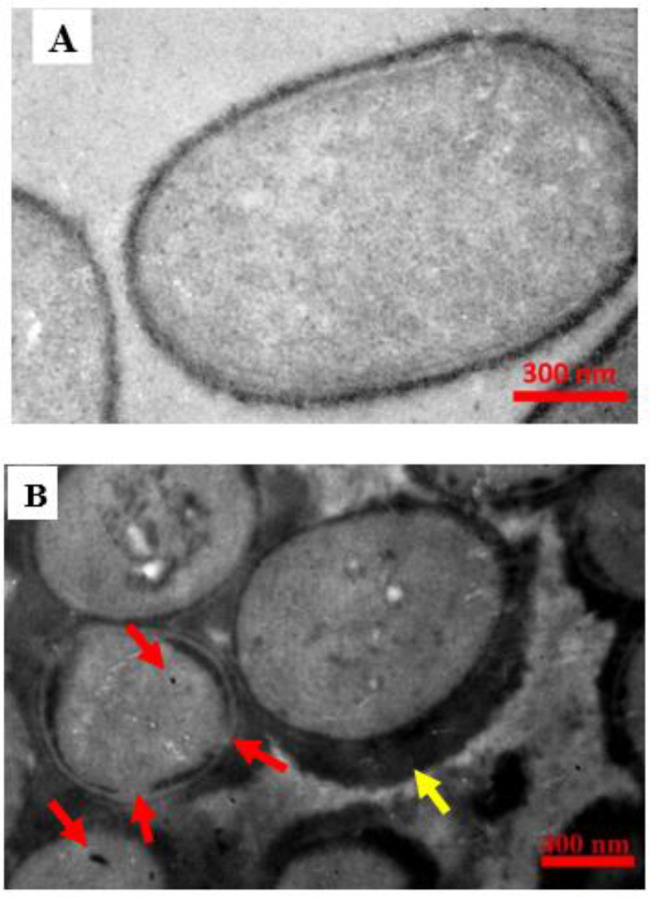
Transmission Electron Microscopy (TEM) image of K pneumoniae. A) Untreated resistant strain (control) B) The same strain treated with 8 μl/ml ciprofloxacin with 100 μg/ml f-MWCNTs. Arrows indicate the f-MWCNTs particles that have entered the bacteria, as well as the location of the wall destruction. The accumulation of f-MWCNTs on the bacterial wall, indicated by a yellow arrow
Statistical analysis
The data of the studied groups were collected in GraphPad Prism 7.0 and subjected to analysis. The expression levels were also normalized. Data are expressed as mean ± standard deviation (SD). The one-way ANOVA was used to determine significance. P-values<0.05 were considered significant.
Results
Nanofluid preparation
By functionalization, the compatibility of f-MWCNT improved and dispersion stability was increased up to 6 months with lower toxicity.
Results of microbial tests
The clinical and ATCC 700,603 isolates were identified by differential biochemical tests. They were also genetically confirmed by sequencing, followed by the BLAST technique.
Drug susceptibility tests results
By comparing the inhibition zone diameter, based on the CLSI guidelines 2020 (27), the ES133 clinical isolate of K. pneumonae showed a resistance to all studied antibiotics including, ciprofloxacin (5 μg), cefepime (30 μg), cefoxitin (30 μg), ceftazidime (30 μg), aztreonam (30 μg), imipenem (10 μg), meropenem (10 μg) ertapenem (10 μg), doripenem (10 μg) aztreonam (30 μg), and intermediate sensitivity to gentamicin (10 μg) and chloramphenicol (30 μg). The ATCC 700,603 isolate showed resistance to ciprofloxacin, cefoxitin, meropenem and sensitive to cefepime and gentamicin. Considering the inhibition zone diameter, the resistance of ATCC isolate to ciprofloxacin was less than that of the ES133 clinical isolate (Table 1).
MIC results
The broth micro dilution MIC breakpoints were read and compared with the CLSI 2020 guidelines (28). The lowest dose was considered as the minimum inhibitory concentration. By the effect of ciprofloxacin serial dilutions ranging 0.62–16 μg/ml on the resistant isolate of K. pneumoniae, no growth was observed from the dilution of 1 (16 μg/ml). In the cases of nanofluids containing f-MWCNTs and p-MWCNTs, the clearance (growth inhibition) began at a dose of 400 μg/ml and 600 μg/ml for f-MWCNTs and p-MWCNTs respectively.
MBC results
The ciprofloxacin resistant isolate grew under the influence of each 8 μg/μl ciprofloxacin and 400 μg/μl f-MWCNTs alone, but the effect of 8 μg/ml ciprofloxacin in combination with 400 μg/μl f-MWCNTs, completely inhibited the growth of the bacterium. Therefore, 8 μg/ml ciprofloxacin and 400 μg/ml f-MWCNTs were considered as the MICs (Fig. 2). In the ATCC 700,603 isolate of K. pneumoniae, doses of 2, 4, 16 μg/ml ciprofloxacin + 100 μg/ml f-MWCNT could inhibit bacterial growth. Therefore, 2 μg/ml ciprofloxacin + 100 μg/ml f-MWCNT were considered as the MICs.
Transmission Electron Microscopy (TEM)
To confirm the internalization of f-MWCNTs and visualize the wall degradation of bacteria, the treated and untreated samples were prepared and visualized under TEM. As a result of the accumulation of f-MWNCNTs, a thick layer surrounding the bacterium was observed (Fig. 2-B) and also the particles that managed to internalize into bacteria are illustrated (Fig 2-A, -B).
The virulence genes real time-PCR results
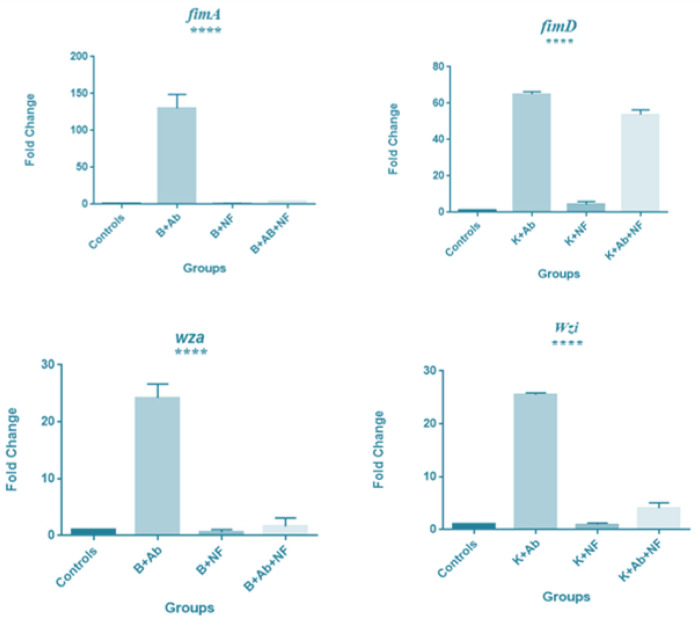
The Real Time-PCR data were analyzed by oneway ANOVA method and graphs were drawn with GraphPad prism 7.0 software. In the resistant K. pneumoniae isolate, after f-MWCNT+cip treatment, the expression of fimA, fimD, wza, and wzi genes was significantly down regulated compared to the ciprofloxacin treatment and upregulated compared to the control group (Fig. 3).
Fig. 3:
fimA, fimD, wza and wzi genes expression in K. pneumoniae resistant strain to antibiotic and f-MWCNT. From left to right respectively: the untreated K. pneumoniae (negative control), treated with 8 μg/ml antibiotic, 400 μg/ml f-MWCNT, 400 μg/ml f-MWCNT + 8 μg/ml antibiotics. After treatment with 400 μg/ml f-MWCNTs + 8 μg/ml ciprofloxacin in the resistant strain, fimA, fimD, wza and wzi genes expression significantly upregulated in comparison with the control, (P value <0.0001, <0.0001, <0.003, <0.0001 respectively) and downregulated compared to the antibiotic alone. Abbreviations: B; Bacteria, K: K. pneumoniae; Ab: Antibiotic; NF: Nanofluid. Expression levels were normalized against 16s rRNA as the reference housekeeping gene
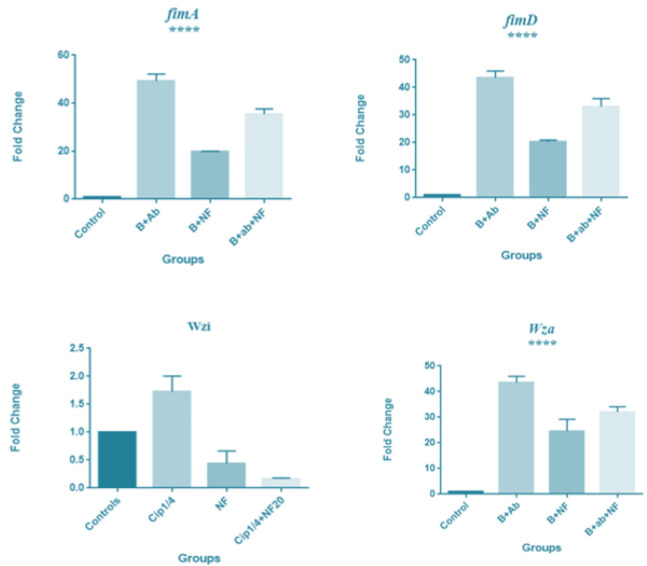
The Real Time-PCR analysis graph of the ATCC 700,603 isolate of K. pneumoniae treated with 100 μg/ml f-MWCNT with 2 μg/ml ciprofloxacin, showed that the fimA, fimD and wza virulence genes expression were upregulated compared to the negative control and downregulated in comparison with ciprofloxacin treatment (P values: 0.005, 0.005, 0.06). Though, wzi gene expression was downregulated compared to both the control and the ciprofloxacin treatment (P<0.37) (Fig. 4).
Fig. 4:
fimA, fimD, wza and wzi genes expression in K. pneumoniae ATCC strain to antibiotic and f-MWCNT. From left to right respectively: untreated K. pneumoniae (negative control), treated with 2 μg/ml ciprofloxacin, 100 μg/ml f-MWCNT, 2 μg/ml ciprofloxacin + 100 μg/ml f-MWCNT. In the ATCC strain treated with 2 μg/ml ciprofloxacin + 100 μg/ml f-MWCNT, the expression of fimA, fimD and wza genes showed an upregulation compared to the control and showed a downregulation in comparison with ciprofloxacin treatment (P value <0.005, 0.005, 0.06), while wzi gene expression was downregulation compared to both the control and the ciprofloxacin treatment, although the rate of reduction was lower than the significant level (P value <0.37). Abbreviations: B: Bacteria; Ab: Antibiotic; NF: Nanofluid. Expression levels were normalized against 16s rRNA as the reference housekeeping gene
Agarose gel electrophoresis
The electrophoresis of Real Time-PCR products showed sharp bands, formed exactly where they were expected on 2% agarose gel and matched the product length of the designed primers.
Discussion
Owing to the increasing emergence of antibiotic resistance as a public health concern, an efficient drug delivery system can improve the therapeutic effects of antibiotics against human pathogens. This study aimed to prepare a nanofluid containing f-MWCNTs and to evaluate the efficacy of f-MWCNT+cip co-administration on the virulence gene expression of both resistant and standard K. pneumoniae isolates. Since unique physicochemical properties, conjugation capabilities as well as targeting capacity, CNTs can facilitate antimicrobial drugs delivery and overcome some of the limitations of traditional antibiotic treatments. New methods using f-MWCNTs, such as combination of f-MWCNTs with antibiotics, can improve their effectiveness at lower doses (25).
The combination of AgNPs with various antibiotics were evaluated for antibacterial activity, and the greatest increase, obtained by the ciprofloxacin-AgNPs combination (29). After exposure to f- MWCNT+ antibiotic, the expression profile of each bacterial virulence gene, might change in a different pattern (24, 30). Similarly, in this study, the expression pattern of wzi gene was different from other studied genes. Thus, in f-MWCNT+cip treated bacteria compared to both the control and ciprofloxacin-treated, wzi expression showed a decrease, though it was below the significance level of < 0.05 for P-value (P<0.37) (Fig. 4).
The findings of the present study are consistent with our recent research, as f-MWCNT+ cefepime could inhibit bacterial growth in resistant isolate of K. pneumoniae at lower doses compared to cefepime alone. Our previous investigations on Pseudomonas aeruginosa, Acinetobacter baumannii, and Staphylococcus aureus antibiotic resistant isolates, showed that simultaneous treatment with f-MWCNTs and antibiotics could increase the permeability of the bacteria and improve the effectiveness of antibiotics at lower doses (25). Furthermore, based on the data obtained in our other research, f-MWCNT conjugated with isoniazid nano-drug, resulted in optimal drug delivery, at lower concentrations than in the free state (17). The remarkable synergistic effect of ciprofloxacin and f-MWCNTs offer their potential in nanomedicine for clinical application as a combined therapy in the future.
Conclusion
By accompanying the f-MWCNTs, the drug delivery efficiency of ciprofloxacin, that K. pneumoniae is resistant to that, was increased. Due to the high durability of K. pneumoniae on surfaces and also the antibacterial effects of carbon nano-tubes, the CNTs can be used as surface disinfect-ants to prevent infection, though our studies in this area are ongoing. For further comprehension the f-MWCNT's mechanism of action, and the immune system response to them, further cell signaling pathways and in vivo studies are also required. Although functionalization of MWCNTs with carboxyl group could reduce the toxicity and improve biocompatibility, the 3-(4,5-dimethylthiazol-2-yl) -2,5-diphenyl-2H-tetrazolium bromide (MTT) assay is proposed prior to the clinical usage of them. Conjugation of ciprofloxacin with f-MWCNT's is recommended in future studies for optimal drug delivery to K. pneumoniae as well.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
The authors sincerely appreciate the staff of the Mycobacteriology and Pulmonary Research Department of the Pasteur Institute of Iran for their kindly cooperation.
Footnotes
Conflict of interest
The authors have no conflict of interest.
References
- 1.Jiang W, Yang W, Zhao X, Wang N, Ren H. (2020). Klebsiella pneumoniae presents antimicrobial drug resistance for β-lactam through the ESBL/PBP signaling pathway. Exp Ther Med, 19(4): 2449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dosunmu E, Chaudhari AA, Singh Sh, et al. (2015). Silver-coated carbon nanotubes downregulate the expression of Pseudomonas aeruginosa virulence genes: a potential mechanism for their antimicrobial effect. Int J Nanomedicine, 10: 5025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths J, Maguire JH, Heggenhougen K, Quah S. (2010). Public Health and Infectious Diseases. 1st ed. Elsevier. USA. pp.119–124. [Google Scholar]
- 4.Marcoleta AE, Varas MA, Ortiz-Severín J, et al. (2018). Evaluating Different Virulence Traits of Klebsiella pneumoniae Using Dictyostelium discoideum and Zebrafish Larvae as Host Models. Front Cell Infect Microbiol, 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamlesh JRK, Mannheim W. (1974). Differentiation of the Oxytocum group from Klebsiella by deoxyribonucleic acid hybridization. Int J Syst Bacteriol, 24 (4): 402–7. [Google Scholar]
- 6.Ranjbar R, Fatahian Kelishadrokhi A, Chehelgerdi M. (2019). Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infect Drug Resist,12: 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herridge W, Shibu P, O'shea J, Brook TC, Hoyles L. (2020). Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J Med Microbiol, 69: 176–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jean Sh, Chang YCh, Lin WCh, et al. (2020). Epidemiology, Treatment, and Prevention of Nosocomial Bacterial Pneumonia. J Clin Med, 9:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu D, Liu B, Dijkshoorn L, Wang L, Reeves PR. (2013). Diversity in the Major Polysaccharide Antigen of Acinetobacter baumannii Assessed by DNA Sequencing, and Development of a Molecular Serotyping Scheme. PLoS One, 8(7): e70329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catalán-Nájera JC, Garza-Ramos U, Camacho HB. (2017). Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes. Virulence, 8(7): 1111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Follador R, Heinz E, Wyres KL. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom, 2(8): e000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doorduijn J D, Rooijakkers SHM, Schaik WV. (2016). Complement Resistance Mechanisms of Klebsiella pneumoniae. Immunobiology, 221(10): 1102–9. [DOI] [PubMed] [Google Scholar]
- 13.Gerlach G, Clegg S, Allen BL. (1989). Identification and Characterization of the Genes Encoding the Type 3 and Type 1 Fimbrial Adhesins of Klebsiella pneumoniae. J Bacteriol, 171(3): 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh YCh, Huang TH, Yang Sh, Chen Ch, Fang JY. (2020). Nano-Based Drug Delivery or Targeting to Eradicate Bacteria for Infection Mitigation: A Review of Recent Advances. Front Chem, 8:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray PR, Rosenthal KS, Pfaller MA. (2016). Medical Microbiology. 8th ed. Elsevier. Canada. [Google Scholar]
- 16.Mirzaie A, Ranjbar R. (2021). Antibiotic resistance, virulence-associated genes analysis and molecular typing of Klebsiella pneumoniae strains recovered from clinical samples. AMB Express, 11:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zomorodbakhsh Sh, Abbasian Y, Naghinejad M, Sheikhpour M. (2020). The Effects Study of Isoniazid Conjugated Multi-Wall Carbon Nanotubes Nanofluid on Mycobacterium tuberculosis. Int J Nanomedicine, 15: 5901–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill J. (2016). Tackling drug-resistant infections globally: final report and recommendations. Review on antimicrobial Resistance. https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf
- 19.Tacconelli E CE, Savoldi A. (2017). Global priority list (ppl) of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf
- 20.Rai M, Ingle AP, Pandit R, et al. (2017). Broadening the spectrum of small-molecule antibacterials by metallic nanoparticles to overcome microbial resistance. Int J Pharm, 532(1): 139–148. [DOI] [PubMed] [Google Scholar]
- 21.Sheikhpour M, Arabi M, Kasaeian A, et al. (2020). Role of Nanofluids in Drug Delivery and Biomedical Technology: Methods and Applications. Nanotechnol Sci Appl, 13: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheikhpour M, Barani L, Kasaeian A. (2017). Biomimetics in drug delivery systems: A critical review. J Control Release, 253: 97–109. [DOI] [PubMed] [Google Scholar]
- 23.Sheikhpour M, Golbabaeie A, Kasaeian A. (2017). Carbon Nanotubes: A review of novel strategies for cancer treatment and diagnosis. Mater Sci Eng C Mater Biol Appl, 76:1289–1304. [DOI] [PubMed] [Google Scholar]
- 24.Zardini HZ, Davarpanah M, Shanbedi M, et al. (2014). Microbial Toxicity of Ethanolamines-Multi Walled Carbon nanotubes. J Biomed Mater Res A, 102(6): 1774–1781. [DOI] [PubMed] [Google Scholar]
- 25.Yazdani MR, Sheikhpour M, Siadat SD, Safarian P. (2021). Overcoming the antibiotic resistance of Acinetobacter baumannii by using nanofluid containing functionalized carbon nanotubes. Nanomed Res J, 6(2): 179–187. [Google Scholar]
- 26.Brown C, Seidler RJ. (1973). Potential Pathogens in the Environment: Klebsiella pneumoniae, a Taxonomic and Ecological Enigma. Appl Microbiol, 25(6): 900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes B, Willcox WR, Lapage SP. (1978). Identification of Enterobacteriaceae by the API 20E system. J Clin Pathol, 31: 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstein MP, Lewis JS, Bobenchick AM. (2020). Clinical Labratory Standard Institute, performance standards for Antimicrobial Susceptibility Testing, M100 30th ed. pp. 31–37. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf
- 29.Namivandi Zangeneh R, Sadrearhami Z, Dutta D, et al. (2019). Synergy between Synthetic Antimicrobial Polymer and Antibiotics: A Promising Platform to Combat Multidrug-resistant Bacteria. ACS Infect Dis, 5(8): 1357–1365. [DOI] [PubMed] [Google Scholar]
- 30.Kon K, Rai M. (2016). Antibiotic Resistance Mechanisms and New Antimicrobial Approaches. 1st ed. Elsevier Inc,UK. pp.130–134. [Google Scholar]