Abstract
We have developed an automated multiplex system for simultaneously screening hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus type 1 (HIV-1) in blood donations. The assay, designated AMPLINAT MPX HBV/HCV/HIV-1 Test (AMPLINAT MPX), consists of virus extraction and target sequence-specific probe capture on specimen preparation workstation GT-X (Roche Diagnostics K.K., Tokyo, Japan) and amplification and detection by TaqMan PCR on the ABI PRISM 7700 Analyzer (Perkin-Elmer Applied Biosystems, Foster City, Calif.). An internal control (IC) is incorporated in the assay to monitor the extraction, target amplification, and detection processes. The assay yields qualitative results without discrimination of the three targets. Detection limits (95% confidence interval) are 22 to 60 copies/ml for HBV, 61 to 112 IU/ml for HCV, and 33 to 66 copies/ml for HIV-1, using a specimen input volume of 0.2 ml. The AMPLINAT MPX assay detects a broad range of genotypes or subtypes for all three viruses and has a specificity of 99.6% for all three viruses with seronegative specimens. In an evaluation of seroconversion panels, the AMPLINAT MPX assay detects HBV infection an average of 24 days before the detection of HBsAg by enzyme immunoassay. HCV RNA was detected an average of 31 days before HCV antibody. HIV-1 RNA was detected an average of 14 days before HIV-1 antibody and an average of 9 days before p24 antigen. The Japanese Red Cross has been evaluating the AMPLINAT MPX system since October 1999. The clinical performance indicates that the AMPLINAT MPX system is robust, sensitive, and reproducible, with a high percentage of valid assay runs (96.8%), a low false-positive rate (0.34%), and a low IC failure rate (0.24%).
A small but significant transfusion risk of pathogenic viruses exists due to the inability of current serologic screening tests either to identify recently infected donors in the preseroconversion window phase of infection or to detect antigenic variants of these viruses. In recent years, applications of nucleic acid amplification tests (NATs) have significantly reduced the preseroconversion “window period” (M. P. Busch, Program Abstr. 52nd Annu. Meet. Am. Assoc. Blood Bank, p. 354–363, 1999).
A typical NAT involves sample preparation, target-specific amplification, and detection. Over the last several years, several methods for sequence-specific probe capture of viral nucleic acids to specific particles have been developed (5, 16). Recently, several amplification-based multiplex assays (1, 5, 8, 15, 21) have been developed by different laboratories and companies. Although a multiplex assay for detecting hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus type 1 (HIV-1) has been reported (5), the sample preparation process is labor-intensive and time-consuming. Thus, there is a need in testing centers for a system providing automated high-throughput sample preparation, along with automated amplification and detection. The main criteria for such a system are (i) complete automation with high-throughput sample processing, (ii) simultaneous detection of major blood-borne viruses, and (iii) amplification and detection methodologies that retain high sensitivity and specificity.
Currently, commercially available assay systems use one of the following four amplification methods (Busch, Program Abstr. 52nd Annu. Meet. Am. Assoc. Blood Bank): PCR (6), transcription-mediated amplification/nucleic acid sequence-based amplification (TMA/NASBA) (2), ligase chain reaction (LCR), and branched DNA signal amplification assay (bDNA assay). Although the feasibility of the above applications has been demonstrated and some of the platforms are under commercial evaluation, testing centers still face difficulties in implementing NATs due to the complexity of target selection, low throughput, and inadequate sensitivity and specificity.
Here, we describe an automated and sensitive test for simultaneously screening HBV, HCV, and HIV-1. This test, the AMPLINAT MPX HBV/HCV/HIV-1 (AMPLINAT MPX) Test, involves the multiplexed extraction and purification of viral RNA and DNA targets by probe capture technology on an automated sample preparation workstation (GT-X), followed by multiplexed amplification and detection using TaqMan technology on the PRISM 7700 Analyzer. An internal control (IC) is incorporated to monitor target extraction, amplification, and detection.
MATERIALS AND METHODS
Clinical specimens.
Sensitivity panels for HBV (genotype A; Consolidated Laboratory Services, Van Nuys, Alameda, Calif.), HCV (genotype 1b; Roche Molecular Systems), and HIV-1 (subtype B; Roche Molecular Systems) were prepared by diluting clinical isolate stocks in acid citrate dextrose (ACD)-treated negative human plasma (Interstate Blood Bank, Memphis, Tenn.). The stocks were quantified by AMPLICOR HIV-1 MONITOR version 1.5 for HIV-1, AMPLICOR HCV MONITOR version 2.0 for HCV, and AMPLICOR HBV MONITOR for HBV. Plasma units found to be PCR negative for all three viruses were pooled and used to dilute the stocks from 0 to 400 IU/ml or from 0 to 400 copies/ml. Additional HCV panels were prepared using the Roche Molecular Systems HCV Secondary Standard (HCV genotype 1a; 14). All members in the sensitivity panel were tested in eight replicates by using three different instrument combinations on 3 days, for a total of 24 replicates for each concentration. The assay sensitivity (95% confidence interval [CI]) for each target was determined by PROBIT analysis.
A seven-member genotype panel for HBV (HBV panel A; Roche Diagnostics, K.K., Tokyo, Japan) was quantitated by the Roche AMPLICOR HBV MONITOR assay and genotyped by sequence analysis. Another HBV genotype panel (HBV panel B; Millennium Biotech, Miami, Fla.) was also quantitated by the Roche AMPLICOR HBV MONITOR assay and genotyped by the supplier. A 25-member HCV genotype panel, lot no. HCV-G001b (Millennium Biotech), was quantitated by the COBAS AMPLICOR HCV Test, version 2.0, and genotyped by the supplier using the INNO LiPA HCV II method. A 30-member HIV-1 group M subtype panel (Walter Reed Army Institute of Research and Henry M. Jackson Foundation, Seattle, Wash.) was quantitated by electron microscopic counting of viral particles. Dilutions containing from 30 to 500 copies/ml (or IU/ml) in pooled PCR-negative human plasma were prepared for each panel member. Six to eight replicates were tested for each concentration, and the frequency of positive results was determined for each dilution.
Thirty-six seroconversion panels for HBV, HCV, and HIV-1 (Boston Biomedica Inc., Boston, Mass., and BioClinical Partners Inc., Franklin, Mass.) were tested by the AMPLINAT MPX assay. Each panel member was tested in duplicate. The AMPLINAT MPX assay results were compared with enzyme immunoassay (EIA) or antigen test results to determine the window period reduction that was achieved by the detection of RNA or DNA prior to seroconversion.
IC.
The IC is a noninfectious 142-bp in vitro-transcribed RNA molecule with primer binding regions identical to those of the HIV-1 target sequence (17). The IC amplicon is the same length (142 bases) and contains the same base composition as the HIV-1 target amplicon. The probe-binding region of the IC is modified to differentiate the IC-specific amplicon.
The IC is introduced at the lysis step for each specimen and carried through the specimen preparation, amplification, and detection steps along with the viral targets. Thus, it serves as both an extraction control and an amplification and detection control for each individually processed specimen.
External controls.
Individual HBV, HCV, and HIV-1 positive controls were made from human plasma specimens representing the most prevalent genotype of each virus as appropriate. The titer of each virus stock was determined by AMPLICOR HBV MONITOR, AMPLICOR HCV MONITOR version 2.0, and AMPLICOR HIV-1 MONITOR version 1.5. Each stock was diluted with seronegative, PCR-negative human plasma to 200 to 300 copies/ml (or IU/ml). A buffer solution was used as a negative control (non-template control [NTC]). External controls, including one positive virus control for each target and three replicates of a negative control (NTC), were used in each reaction plate.
Specimen and control preparation.
Specimens and controls (samples) were extracted on an automated specimen preparation workstation, GT-X. After manually loading the samples on the instrument, the lysis, hybridization, capture, and resuspension steps were performed by the GT-X without user intervention. For each extraction, 0.2 ml of sample was treated with 0.4 ml of lysis solution containing the IC, followed by incubation at 60°C for 11 min. This process results in efficient release of both DNA and RNA while inactivating RNases and maintaining the integrity of RNA. Released RNA or DNA was hybridized with a set of biotinylated capture probes that are specific for the 5′ untranslated region of the HCV genome, the pre-core region of the HBV genome, and the gag region of the HIV-1 genome. Four capture probes (two for HIV-1, one for HBV, and one for HCV) were designed to be complementary to target sequences that are highly conserved and present in most viral genotypes and subtypes. The hybridized RNA or DNA was then captured with streptavidin-coated magnetic microparticles (Dynalbead; Dynal A.S., Oslo, Norway). The particles were washed to remove nonspecifically bound materials, resuspended in 50 μl of specimen diluent, transferred into a MicroAmp Optical Reaction Plate (Perkin-Elmer Applied Biosystems, Foster City, Calif.), and mixed with 50 μl of working master mix.
Amplification and detection (TaqMan PCR assay).
The TaqMan methodology uses a real-time PCR technique (9, 10) to measure PCR product accumulation through a dual-labeled fluorogenic probe (TaqMan probe). The fluorescent signal is generated by means of 5′-nuclease activity that separates a fluorescent reporter dye and quencher dye (7).
Amplifying and detecting HBV DNA and HCV and HIV-1 RNA with equal efficiency in this multiplex assay proved to be challenging. Since both HIV-1 and HCV amplifications involve a reverse transcription (RT) step, RT primer concentrations and thermocycling conditions were adjusted so the amplification efficiencies for each of the four types of amplicons (three viral targets and one IC) were approximately equal during the exponential phase of the reaction (data not shown). Four fluorogenic detection probes, which are conserved among most viral genotypes and subtypes, were designed to hybridize to target sequences within their respective target amplicons. Both the PCR primers and fluorogenic probes hybridized to their respective complementary strands during the amplification step. Primer-template hybrids are stabilized when the thermal-stable enzyme extends the primer in the polymerization step. Since the fluorogenic probes can't be extended, the detection probes were designed to be longer than the primers to achieve a stable probe-template hybrid. Because efficient probe cleavage (and, consequently, the TaqMan 5′-nuclease fluorogenic signal) requires maximum probe-template hybridization, the probe annealing temperature was lowered to be less than the Tm for the fluorogenic probe-template hybrid. Three detection probes for HBV, HCV, and HIV-1 were labeled with the same fluorogenic reporter and quencher dyes. Since this assay is a qualitative assay without discrimination of each target due to the three probes having the same reporter dye, the fluorescent signals generated from all three targets have the same wavelength. The IC probe was labeled with a different fluorogenic reporter dye but the same quencher dye as the target. Target and IC probes generated one composite fluorescent spectrum contributed by individual overlapping component dye spectra. The multicomponent algorithm on the Sequence Detection System application used matrix calculation to determine the contributions of each component dye (PRISM 7700 Analyzer user manual).
Amplification and detection were carried out using the PRISM 7700 analyzer (Perkin-Elmer Applied Biosystems). The amplification and detection working master mix consisted of 1× PCR buffer; 300 to 500 μM concentrations of dATP, dGTP, dCTP, and dUTP; primer pairs at a concentration of 0.15 to 0.6 μM for HBV, HCV, and HIV-1; four detection probes specific for the HBV, HCV, HIV-1, and IC amplicons; 200 U of AmpErase uracil-N-glycosylase (UNG) (Perkin-Elmer)/ml; 800 U of a thermostable enzyme (ZO5) that has both reverse transcriptase and DNA polymerase activity (Roche Molecular Systems)/ml; and 3.0 mM manganese. The PCR thermocycling conditions were optimized to increase the PCR amplification efficiency, to increase the fluorogenic probe cleavage efficiency, and to reduce primer dimer formation. The thermocycling parameters were as follows: 10 min at 45°C for UNG to cleave any carryover amplicon and primer dimers (S. Kwok, S. Kinard, J. Spadoro, and J. J. Sninsky, Program Abstr. 8th Int. AIDS Conf., abstr. A2388 67, 1992); 30 min at 60°C; 5 cycles of 95°C for 15 s and 60°C for 40 s; and 45 cycles of 91°C for 15 s and 52°C for 40 s. The amplification products were detected by continuously monitoring the release of the fluorescent reporter during the TaqMan 5′-nuclease PCR assay.
Data analysis.
The raw data were initially analyzed by the ABI PRISM 7700 Analyzer's sequence detection system software (Perkin-Elmer Applied Biosystems), including multicomponent analysis. The software calculates a ΔRn value by using the normalized reporter signal minus the baseline signal that is established in the first few cycles of PCR. The ΔRn increases during PCR as the target is amplified to the point at which the reaction approaches a plateau. The ΔRn measurements were taken at the end of the annealing phase at 52°C. A cut-off algorithm was developed based on the distribution of the ΔRn values of the NTC. The ΔRn value for a positive sample is greater than the average ΔRn value of the NTC plus a constant for each individual run. A sample was considered inhibitory if the ΔRn value for the IC was at least 20% lower than the average ΔRn value for the IC in NTCs.
RESULTS
Assay sensitivity.
The limit of detection (LOD) of the AMPLINAT MPX assay for each viral target was evaluated using sensitivity panels described in Materials and Methods. Each member was tested in replicates of 24 using three different instrument combinations, with each combination performed on a different day. The frequency of positive reactions was calculated for two separate runs, each consisting of 24 replicates. The final LOD was determined by PROBIT, a statistical method. Overall, the LOD (with 95% CI) of the MPX assay was 22 to 60 copies/ml for HBV, 61 to 112 IU/ml for HCV, and 33 to 66 copies/ml for HIV-1 (Table 1). For low-level samples that yielded both positive and negative results for multiple replicates of that sample, the number of copies added to the reaction was calculated from the Poisson distribution by using the following formula (Z. Wang and J. Spadoro, Abstr. 94th Gen. Meet. Am. Soc. Microbiol., p. 141, 1994):
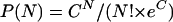 |
1 |
where P(N) is the probability that a unit volume contains N copies, N is the actual number of molecules in a unit volume, and C is the average copy number of molecules in a unit volume. The probability that a unit volume contains zero copies is as follows:
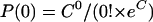 |
2 |
That is,
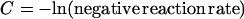 |
3 |
The assay recovery rate was calculated by dividing the calculated number of copies by the expected number of copies based on the sample titer. The average recovery rate was about 54% for HBV, 21% for HCV, and 33% for HIV-1. Since the equivalent of 0.2 ml of sample was added to each reaction mixture, a 50-copy/ml titer required to achieve a 100% positivity rate corresponds to approximately 5 copies of HBV target per reaction mixture (50 copies/ml × 0.2 ml × 54%). Similarly, we calculated that approximately 8 IU per reaction mixture and 5 copies per reaction mixture are required to achieve 100% positivity for HCV and HIV-1, respectively (Table 1).
TABLE 1.
LODs (95% CI) using PROBIT analysis
| Virus | Titera | Positivity rate (no. positive/total)
|
% Positive | LOD (95% CI) | Avg recovery rate (%)b | |
|---|---|---|---|---|---|---|
| Run no. 1 | Run no. 2 | |||||
| HBV type A adw | 0 | 0/24 | 0/24 | 0 | 30 copies/ml (22–60) | 54 |
| 12 | 19/24 | 19/24 | 79.1 | |||
| 25 | 22/24 | 21/23 | 89.6 | |||
| 50c | 24/24 | 24/24 | 100.0 | |||
| 100 | 24/24 | 24/24 | 100.0 | |||
| 200 | 24/24 | 24/24 | 100.0 | |||
| HCV type 1b | 0 | 0/24 | 0/23 | 0.0 | 77 IU/ml (61–112) | 21 |
| 20 | 13/24 | 12/18 | 59.5 | |||
| 30 | 11/24 | 23/23 | 72.3 | |||
| 50 | 20/24 | 21/22 | 89.1 | |||
| 70 | 22/24 | 22/23 | 93.6 | |||
| 100 | 23/24 | 24/24 | 97.9 | |||
| 140 | 23/24 | 24/24 | 97.9 | |||
| 200 | 24/24 | 24/24 | 100.0 | |||
| HIV-1 type B | 0 | 0/24 | 0/24 | 0 | 42 copies/ml (33–66) | 33 |
| 16 | 16/23 | 15/24 | 66.0 | |||
| 32 | 21/24 | 22/24 | 89.6 | |||
| 64 | 24/24 | 23/24 | 97.9 | |||
| 80 | 24/24 | 23/23 | 100.0 | |||
| 190 | 24/24 | 24/24 | 100.0 | |||
| 260 | 24/24 | 24/24 | 100.0 | |||
Units: HBV and HIV-1, copies/milliliter; HCV, international units/milliliter.
Average recovery rate was calculated as follows: calculated copy number/input copy number × 100%, where input copy number was adjusted (titer × 0.2 ml) since the equivalent of 0.2 ml of sample was added to each reaction mixture.
Bold numbers indicate the lowest titers that yielded 100% positivity.
Genotype and subtype detection.
This study was intended to assess the ability of the AMPLINAT MPX assay to detect the most prevalent HBV, HCV, and HIV-1 genotypes and subtypes. Specimens were diluted to concentrations ranging from 30 to 500 copies/ml (or IU/ml) using pooled seronegative and PCR-negative human plasma. The overall performance of the AMPLINAT MPX assay for genotype detection is summarized in Table 2. For HBV panel A, 100% of the isolates representing HBV genotypes A, B, and C were detected at 30 copies/ml (one isolate from genotype C was detected at 300 copies/ml with a 100% positivity rate). For HBV panel B, all genotypes were detected at 100 copies/ml with a 100% positivity rate. For the HCV genotype panel, genotypes 1a, 1b, 2b, and 3a yielded a 100% positivity rate at 100 IU/ml. Genotype 4 yielded a 100% positivity rate at 100 to 300 IU/ml, and genotype 5 yielded a 100% positivity rate at 100 to 500 IU/ml. For HIV-1, all isolates from subtypes A, C, and D yielded a 100% positivity rate at 100 copies/ml; isolates from subtypes B, E, F, and G yielded a 100% positivity rate at 100 to 300 copies/ml (Table 2).
TABLE 2.
Summary of detection of HBV, HCV, and HIV-1 genetic variants using AMPLINAT MPX assay
| ID no. | Genotype | % Positivity
|
|||||
|---|---|---|---|---|---|---|---|
| 30 copies/ml | 100 copies/ml | 300 copies/ml | 100 IU/ml | 300 IU/ml | 500 IU/ml | ||
| HBV ID no. | |||||||
| RDKK 131 | A | 100 | 100 | ||||
| RDKK 17 | B | 100 | 100 | ||||
| RDKK 94 | B | 100 | 100 | ||||
| RDKK 85 | B | 100 | 100 | ||||
| RDKK 46 | C | 100 | 100 | ||||
| RDKK 245 | C | 83 | 100 | ||||
| RDKK 73 | C | 100 | 100 | ||||
| MBID 11090 | A | 100 | |||||
| MBID 11158 | B | 100 | |||||
| MBID 11102 | C | 100 | |||||
| MBID 8908 | D | 100 | |||||
| MBID 7859 | E | 100 | |||||
| HCV member no. | |||||||
| 1 | 1a | 100 | |||||
| 2 | 1a | 100 | |||||
| 3 | 1a | 100 | |||||
| 4 | 1a | 100 | |||||
| 5 | 1a | 100 | |||||
| 19 | 1a | 100 | |||||
| 6 | 1b | 100 | |||||
| 7 | 1b | 100 | |||||
| 8 | 1b | 100 | |||||
| 9 | 1b | 100 | |||||
| 10 | 1b | 100 | |||||
| 11 | 2b | 100 | |||||
| 12 | 2b | 100 | |||||
| 13 | 2b | 100 | |||||
| 14 | 2b | 100 | |||||
| 15 | 2b | 100 | |||||
| 16 | 3a | 100 | |||||
| 17 | 3a | 100 | |||||
| 18 | 3a | 100 | |||||
| 22 | 3a | 100 | |||||
| 20 | 4 | 100 | |||||
| 21 | 4c/4d | 83 | 100 | ||||
| 23 | 4h | 67 | 100 | ||||
| 24 | 5a | 67 | 83 | 100 | |||
| 25 | 5a | 100 | |||||
| HIV isolate no. | |||||||
| GS001 | A | 100 | NAa | ||||
| GS002 | A | 100 | NA | ||||
| GS003 | A | 100 | NA | ||||
| GS004 | B | 100 | NA | ||||
| GS005 | B | 100 | NA | ||||
| GS006 | B | 83 | 100 | ||||
| GS007 | B | 83 | 100 | ||||
| GS008 | B | 83 | 100 | ||||
| GS009 | B | 100 | NA | ||||
| GS010 | B | 100 | NA | ||||
| GS011 | C | 100 | NA | ||||
| GS012 | C | 100 | NA | ||||
| GS013 | C | 100 | NA | ||||
| GS014 | C | 100 | NA | ||||
| GS016 | C | 100 | NA | ||||
| GS017 | D | 100 | |||||
| GS018 | D | 100 | |||||
| GS019 | D | 100 | |||||
| GS020 | E | 100 | |||||
| GS021 | E | 100 | |||||
| GS022 | E | 100 | |||||
| GS023 | E | 100 | |||||
| GS024 | E | 100 | |||||
| GS025 | E | 100 | |||||
| GS026 | E | 100 | |||||
| GS027 | E | 83 | 100 | ||||
| GS030 | F | 100 | |||||
| GS031 | F | 100 | |||||
| GS032 | F | 83 | 100 | ||||
| GS029 | G | 50 | 100 | ||||
NA, not available.
Detection of HBV, HCV, and HIV-1 in seroconversion panels.
The performance of the AMPLINAT MPX assay was further evaluated with 12 HBV, 9 HCV, and 15 HIV-1 seroconversion panels to assess the ability of the assay to close the preseroconversion window period for all three viruses. The predicted HBV window closure time is summarized in Tables 3 and 4. For these seroconversion panels, the average window closure time of AMPLINAT MPX assay for HBV was 24 days (range, 9 to 94 days). Four of the 12 panels (panels PHM915, PHM909, 6290, and 6272) were HBV DNA positive at day 0 (Table 3).
TABLE 3.
Performance of AMPLINAT MPX assay on HBV seroconversion panels
| Specimen ID | Day of bleed | EIA HBsAg | AMPLINAT MPX assay |
|---|---|---|---|
| PHM919-1 | 0 | 0.1 | − |
| PHM919-2 | 5 | 0.1 | +*b |
| PHM919-3 | 14 | 0.7 | + |
| PHM919-4 | 19 | 2.6a | + |
| PHM915-1 | 0 | 0.3 | +* |
| PHM915-2 | 7 | 0.2 | + |
| PHM915-3 | 21 | 0.5 | + |
| PHM915-4 | 33 | 1.0 | + |
| PHM911-1 | 58 | 0.1 | − |
| PHM911-2 | 63 | 0.1 | +* |
| PHM911-3 | 77 | 0.7 | + |
| PHM911-4 | 79 | 1.3 | + |
| PHM909-01 | 0 | 0.4 | +* |
| PHM909-02 | 4 | 0.2 | + |
| PHM909-03 | 9 | 1.2 | + |
| PHM909-04 | 14 | 4.2 | + |
| PHM922-01 | 0 | 0.1 | − |
| PHM922-02 | 2 | 0.1 | +* |
| PHM922-03 | 7 | 0.3 | + |
| PHM922-04 | 16 | 2.1 | + |
| 6284-01 | 0 | 0.16 | − |
| 6284-02 | 40 | 0.25 | +* |
| 6284-03 | 47 | 0.66 | + |
| 6284-04 | 49 | 2.46 | + |
| 6284-05 | 53 | 8.99 | + |
| 6290-01 | 0 | 0.44 | +* |
| 6290-02 | 2 | 0.55 | + |
| 6290-03 | 16 | 0.71 | + |
| 6290-04 | 21 | 1.92 | + |
| 6289-01 | 11 | 0.50 | − |
| 6289-02 | 16 | 0.21 | +* |
| 6289-03 | 18 | 0.22 | + |
| 6289-04 | 30 | 2.15 | + |
| 6287-01 | 44 | 0.32 | − |
| 6287-03 | 51 | 0.30 | +* |
| 6287-09 | 60 | 0.67 | + |
| 6287-11 | 77 | 57.24 | + |
| 6286-01 | 0 | 0.23 | − |
| 6286-02 | 22 | 0.24 | +* |
| 6286-03 | 29 | 0.76 | + |
| 6286-04 | 33 | 1.82 | + |
| 6292–01 | 2 | 0.27 | − |
| 6292-02 | 21 | 0.31 | +* |
| 6292-03 | 23 | 0.35 | + |
| 6292-04 | 29 | 0.51 | + |
| 6292-05 | 42 | 7.87 | + |
| 6272-01 | 0 | 0.49 | +* |
| 6272-18 | 72 | 0.64 | + |
| 6272-19 | 74 | 0.61 | + |
| 6272-20 | 94 | 1.60 | + |
| 6272-21 | 97 | 2.23 | + |
Boldface numbers represent the first positive result by serological test.
∗, the first positive result by AMPLINAT MPX assay.
TABLE 4.
HBV seroconversion window closure by AMPLINAT MPX assay
| Seroconversion panel ID | Days of closure |
|---|---|
| BBI PHM 919 | 14 |
| BBI PHM 915 | 33 |
| BBI PHM 911 | 16 |
| BBI PHM 922 | 14 |
| BBI PHM 909 | 9 |
| BCP 6290 | 21 |
| BCP 6284 | 9 |
| BCP 6289 | 14 |
| BCP 6287 | 26 |
| BCP 6286 | 11 |
| BCP 6292 | 21 |
| BCP 6272 | 94 |
| Avg | 24 |
The nine HCV seroconversion panels demonstrated a relatively consistent window closure time (Table 5). Two panels (PHV 908 and 9047) were positive at day 0. All samples in panel 9057 were negative for HCV EIA testing; therefore, the predicted window closure time is longer than 24 days (Table 5). Overall, the average window closure for these HCV seroconversion panels by AMPLINAT MPX assay was about 31 days (Table 6).
TABLE 5.
Performance of AMPLINAT MPX assay on HCV seroconversion panels
| Specimen ID | Day of bleed | EIA anti-HCV 2.0c | AMPLINAT MPX assay |
|---|---|---|---|
| PHV 905-1 | 0 | 0.1 | − |
| PHV 905-2 | 4 | 0.1 | +*b |
| PHV 905-3 | 25 | 1.6a | + |
| PHV 905-4 | 28 | 1.7 | + |
| PHV 908-1 | 0 | 0.1 | +* |
| PHV 908-2 | 3 | 0.1 | + |
| PHV 908-3 | 35 | 0.7 | + |
| PHV 908-4 | 41 | 1.0 | + |
| 6227-01 | 0 | 0.029 | − |
| 6227-02 | 22 | 0.067 | − |
| 6227-03 | 42 | 0.042 | +* |
| 6227-04 | 74 | 3.686 | + |
| 6227-05 | 76 | 3.730 | + |
| 6225-01 | 35 | 0.003 | − |
| 6225-02 | 39 | 0.003 | +* |
| 6225-03 | 52 | 0.003 | + |
| 6225-04 | 78 | 1.734 | + |
| 6225-05 | 80 | 2.110 | + |
| 9041-01 | 0 | 0.016 | +/− |
| 9041-02 | 24 | 0.013 | +* |
| 9041-03 | 27 | 0.016 | + |
| 9041-04 | 62 | 4.092 | + |
| 9041-05 | 64 | 4.092 | + |
| 9054-01 | 46 | 0.06 | − |
| 9054-02 | 51 | 0.06 | +* |
| 9054-03 | 73 | 0.09 | + |
| 9054-04 | 76 | 0.25 | + |
| 9054-05 | 81 | 1.41 | + |
| 9055-01 | 17 | 0.23 | − |
| 9055-02 | 21 | 0.19 | +* |
| 9055-03 | 24 | 0.24 | + |
| 9055-04 | 44 | 1.39 | + |
| 9047-01 | 0 | 0.008 | +* |
| 9047-02 | 2 | 0.012 | + |
| 9047-03 | 21 | 0.012 | + |
| 9047-04 | 28 | 1.286 | + |
| 9057-01 | 0 | 0.07 | − |
| 9057-02 | 17 | 0.06 | +* |
| 9057-03 | 19 | 0.07 | + |
| 9057-04 | 41 | 0.07 | + |
Boldface numbers represent the first positive result by serological test.
∗, the first positive result by AMPLINAT MPX assay.
Values are signal-to-cutoff ratios. A value of ≥1 was considered positive.
TABLE 6.
HCV seroconversion window closure by AMPLINAT MPX assay
| Seroconversion panel ID | Days of closure |
|---|---|
| BBI PHV 905 | 21 |
| BBI PHV 908 | 41 |
| BCP 6227 | 32 |
| BCP 6225 | 39 |
| BCP 9057 | >24 |
| BCP 9047 | 28 |
| BCP 9055 | 23 |
| BCP 9054 | 30 |
| BCP 9041 | 38 |
| Avg | 31 |
For the HIV-1 seroconversion panels, we assessed the AMPLINAT MPX window period closure compared to both an HIV-1 and -2 antibody assay and an HIV-1 p24 antigen assay. In most cases, RNA detection preceded antigen detection, which in turn preceded antibody detection. For 13 of the 15 panels, HIV-1 RNA detection preceded antigen detection, and for 12 of the 15 panels, antigen detection preceded antibody detection. In order to obtain an accurate estimation of the window closure time, panels containing samples obtained over infrequent intervals were excluded. For example, with panel PRB932, the last sample, which was positive for all markers, was obtained 14 days after the previous sample, which was negative for all markers (Table 7). Also, panel PBR939(E) had an 80-day interval between the day 23 antigen-positive sample and the day 103 antibody-positive sample. These two panels were not used in estimating the window closure. Another factor we considered was that if the panel member failed to have either antigen or antibody detection or if antibody detection preceded antigen detection, such panels were not included in the window period calculation. For example, in panel 9032, the antigen test was negative for all samples (Table 7). In panel 3031, the antibody detection was 7 days earlier than the antigen detection (Table 7). These data were also not included in the calculation. Overall, the average window closure for HIV-1 was 14 days when compared with antibody testing and 9 days when compared with antigen testing (Table 8).
TABLE 7.
Performance of AMPLINAT MPX assay on HIV seroconversion panels
| Specimen ID | Day of bleed | EIA anti-HIV-1/2 | Antigen test | AMPLINAT assay | |
|---|---|---|---|---|---|
| PRB 932-01 | 0 | 0.1 | 0.4 | − | |
| PRB 932-02 | 3 | 0.1 | 0.4 | − | |
| PRB 932-03 | 13 | 0.2 | 0.4 | − | |
| PRB 932-04 | 27 | 1.1a | 4.6 | +*b | |
| PRB 923-01 | 30 | 0.1 | 0.4 | − | |
| PRB 923-02 | 35 | 0.1 | 0.4 | +* | |
| PRB 923-03 | 37 | 0.1 | 1.0 | + | |
| PRB 923-04 | 47 | 1.4 | >23.3 | + | |
| PRB 929-01 | 0 | 0.2 | 0.5 | +* | |
| PRB 929-02 | 4 | 0.2 | 0.5 | + | |
| PRB 929-03 | 14 | 0.2 | 0.9 | + | |
| PRB 929-04 | 18 | 0.2 | 13.4 | + | |
| PRB 929-05 | 21 | 0.9 | >22.7 | + | |
| PRB 929-06 | 25 | >16.3 | >22.7 | + | |
| PRB 936-01 | 0 | 0.1 | 0.3 | − | |
| PRB 936-02 | 5 | 0.1 | 0.3 | +* | |
| PRB 936-03 | 7 | 0.1 | 0.4 | + | |
| PRB 936-04 | 12 | 0.1 | 13.0 | + | |
| PRB 936-05 | 19 | 1.5 | >31.7 | + | |
| PRB 937-01 | 0 | 0.1 | 0.3 | − | |
| PRB 937-02 | 7 | 0.1 | 0.3 | +* | |
| PRB 937-03 | 9 | 0.1 | 0.3 | + | |
| PRB 937-04 | 14 | 0.1 | 2.1 | + | |
| PRB 937-05 | 21 | 1.0 | 13.6 | + | |
| PRB 939(E)-01 | 2 | 0.2 | 0.4 | − | |
| PRB 939(E)-02 | 9 | 0.2 | 0.4 | +* | |
| PRB 939(E)-03 | 14 | 0.2 | 0.4 | + | |
| PRB 939(E)-04 | 21 | 0.1 | 13.9 | + | |
| PRB 939(E)-05 | 23 | 0.1 | 27.9 | + | |
| PRB 939(E)-06 | 103 | >14.7 | 0.3 | + | |
| PRB 943-01 | 0 | 0.1 | 0.4 | − | |
| PRB 943-02 | 5 | 0.1 | 0.4 | +* | |
| PRB 943-03 | 7 | 0.1 | 0.7 | + | |
| PRB 943-04 | 12 | 0.2 | 10.6 | + | |
| PRB 943-05 | 14 | 2.5 | 25.6 | + | |
| PRB 943-06 | 21 | >18.2 | 5.1 | + | |
| PRB 949-01 | 0 | 0.2 | 0.4 | − | |
| PRB 949-02 | 6 | 0.2 | 0.5 | +* | |
| PRB 949-03 | 9 | 0.2 | 0.4 | + |
| PRB 949-04 | 18 | 0.8 | 3.7 | + |
| PRB 949-05 | 20 | 10.9 | 7.1 | + |
| 6247-01 | 9 | 0.076 | 0.387 | − |
| 6247-02 | 14 | 0.119 | 0.339 | +* |
| 6247-03 | 21 | 0.110 | 0.290 | + |
| 6247-04 | 23 | 0.153 | 1.113 | + |
| 6247-05 | 30 | 3.534 | 11.823 | + |
| 9010-01 | 20 | 0.234 | 0.42 | − |
| 9010-02 | 25 | 0.234 | 0.28 | +* |
| 9010-03 | 34 | 0.234 | 0.32 | + |
| 9010-04 | 41 | 0.313 | 2.88 | + |
| 9010-05 | 45 | 1.211 | 0.77 | + |
| 6240-01 | 14 | 0.082 | 0.277 | − |
| 6240-02 | 21 | 0.055 | 0.538 | +* |
| 6240-03 | 23 | 0.045 | 2.785 | + |
| 6240-04 | 28 | 1.490 | >30.769 | + |
| 6240-05 | 44 | 7.110 | 1.169 | + |
| 6240-06 | 52 | 11.073 | 0.954 | + |
| 9032-01 | 10 | 0.091 | 0.370 | +/− |
| 9032-02 | 14 | 0.109 | 0.356 | +* |
| 9032-03 | 17 | 0.091 | 0.342 | + |
| 9032-04 | 24 | 1.691 | 0.658 | + |
| 9032-05 | 51 | 6.236 | 0.370 | + |
| 9032-06 | 56 | 7.128 | 0.342 | + |
| 9031-01 | 127 | 0.090 | 0.384 | − |
| 9031-02 | 131 | 0.081 | 0.370 | +* |
| 9031-03 | 134 | 0.063 | 0.397 | + |
| 9031-04 | 146 | 2.730 | 0.356 | + |
| 9031-05 | 153 | 1.333 | 1.534 | + |
| 9020-01 | 80 | 0.081 | 0.425 | − |
| 9020-02 | 83 | 0.072 | 0.918 | +* |
| 9020-03 | 87 | 0.099 | 0.370 | + |
| 9020-04 | 94 | 0.351 | 1.438 | + |
| 9020-05 | 97 | 1.595 | 1.260 | + |
| 9021-01 | 39 | 0.483 | 0.237 | − |
| 9021-02 | 43 | 0.793 | 0.271 | +* |
| 9021-03 | 50 | 0.138 | 3.068 | + |
| 9021-04 | 54 | 0.716 | 6.153 | + |
| 9021-05 | 57 | 2.345 | 28.644 | + |
Boldface numbers represent the first positive result by serological test.
*, the first positive result by AMPLINAT MPX assay.
TABLE 8.
HIV seroconversion window closure by AMPLINAT MPX assay
| Seroconversion panel ID | Days of closure
|
|
|---|---|---|
| Antibody test | Antigen test | |
| BBI PRB 923 | 12 | 2 |
| BBI PRB 929 | 25 | 18 |
| BBI PRB 936 | 14 | 7 |
| BBI PRB 937 | 14 | 7 |
| BBI PRB 943 | 9 | 7 |
| BBI PRB 949 | 14 | 12 |
| BCP 6247 | 16 | 9 |
| BCP 9010 | 20 | 16 |
| BCP 6240 | 7 | 2 |
| BCP 9020 | 14 | 11 |
| BCP 9021 | 14 | 7 |
| Avg | 14 | 9 |
Clinical performance.
Since October 1999, the Japanese Red Cross (JRC) has been conducting an evaluation of the AMPLINAT MPX assay (11, 19). Prescreened seronegative specimens were pooled by an automated pooling system (ALOKA, Tokyo, Japan) which prepared either 50- or 500-member plasma pools. Sample barcode identification, centrifugation, capping, and decapping of tubes were all performed automatically. For the subsequent PCR testing, the validity of each run was determined by evaluating the results for three NTCs and one positive control for each target. A run was considered valid when all NTCs were negative, all NTC-ICs were positive, and all positive controls were positive. Pools of 500 or 50 members that were positive by the AMPLINAT MPX assay were resolved to the single unit responsible for the positive pool result. Resolution was accomplished by an in-house nested RT-PCR virus-specific assay. The results of the clinical assay performance at the JRC are summarized in Table 9.
TABLE 9.
Summary of AMPLINAT MPX NAT at Japanese Red Cross testing centers
| Parameter | Value for group (dates tested)
|
||
|---|---|---|---|
| Pools of 500 (October 1999–February 2000) | Pools of 50 (February 2000–June 2000) | Total (October 1999–June 2000) | |
| No. of screened specimens | ≈2.0 million | ≈2.5 million | ≈4.5 million |
| No. of pools tested | 8,657 | 46,880 | 55,537 |
| Total runs | 515 | 1,118 | 1,633 |
| Valid runs | 499 | 1,080 | 1,579 |
| Valid runs (%) | 96.9% | 96.6% | 96.8% |
| False positive (no.) | 26 | 165 | 191 |
| False positive (%) | 0.30% | 0.35% | 0.34% |
| Invalid IC | 27 | 104 | 131 |
| IC failure (%) | 0.31% | 0.22% | 0.24% |
| No. HBV positivea | 17 | 33 | 50 |
| No. HCV positivea | 7 | 9 | 16 |
| No. HIV-1 positivea | 0 | 2 | 2 |
All of the results were confirmed by the JRC's nested PCR tests.
In the 500-member pool study, about 2.0 million single donations were tested by the AMPLINAT MPX assay during a 4-month period (October 1999 to February 2000). Twenty-four seronegative individual donors were found to be NAT positive (1:83,000). The majority of these donors were HBV positive (70.8%), and the remaining were HCV positive (29.2%). None were found to be HIV-1 positive. In the 50-member pool study, about 2.5 million single donations were tested by the AMPLINAT MPX assay during a 4-month period (February 2000 to June 2000). Forty-four seronegative individual donors were identified as NAT positive (1:76,000); 33 were HBV positive (75%), nine were HCV positive (20.4%), and two were HIV-1 positive (4.5%). While the overall rate of positivity was similar for both pool sizes, the HBV positivity rate was substantially higher for the 50-member pool. During the evaluation, the AMPLINAT MPX assay demonstrated valid runs 96.8% of the time, a false-positive rate of about 0.34%, and a low IC failure rate (0.24%).
DISCUSSION
A high-throughput and high-sensitivity automated multiplex assay is needed to meet the testing requirements of large blood testing centers. In this report, we describe a fully automated, high-throughput, multiplex viral detection system, the AMPLINAT MPX assay system, which simultaneously detects HBV DNA, HCV RNA, and HIV-1 RNA by using a fully automated sample preparation station (GT-X) and a target amplification and detection station (PRISM 7700 Analyzer). The time required for sample extraction is 1 h and 15 min. Amplification and detection require 2.5 h. The total time required to process 96 samples is about 4 h. Since this assay is designed to incorporate three negative controls and individual external positive controls for each virus, 90 test samples or pools can be tested every 4 h using one GT-X with one PRISM 7700 instrument combination. High-throughput testing (360 samples or pools per 8-h shift) can be achieved by using a combination of one GT-X with two PRISM 7700 Analyzers. With a pool size of 50 or 500 units, 18,000 or 180,000 units of blood, respectively, can be screened in a single 8-h shift.
False-positive results can often be problematic for in vitro nucleic acid amplification assays (12, 13). This is especially true in blood screening where the prevalence of viral infection is low, as is the case in volunteer donor populations. The problem is exacerbated when pooled units are tested for each false-positive test result; the entire pool must be quarantined until the true test status for each unit is resolved. In order to reduce the rate of false-positive results, a number of approaches have been applied in the AMPLINAT MPX assay design. (i) UNG is incorporated in the master mix to prevent false-positive results due to amplicon carryover. UNG also is effective in reducing primer-dimer formation during the early stages of PCR amplification and detection. Reducing nonspecific amplification before the first thermocycle is a key step for improving amplification efficiency (18) and consequently reducing the false-positive rate. (ii) Disposable reaction cartridges for sample extraction have been designed with splash barriers to prevent carryover contamination. (iii) Amplification and detection are carried out in a closed-vessel system. The contamination control features of the system were evaluated by testing alternating negative and positive samples (107 copies of HCV transcripts/ml) on the GT-X. The results showed 100% concordance of the corresponding negative and positive test results (data not shown). Assay specificity was evaluated using multiple replicates of 222 HBV, HCV, and HIV-1 seronegative specimens (Interstate Blood Bank). A total of 490 of 494 tests were negative by the AMPLINAT multiplex assay. Two of the four positive seronegative specimens were confirmed to be HCV RNA positive by the discriminatory tests while the remaining two samples represented those which could not be confirmed as positive. Therefore, the assay specificity from this study was 99.6% (data not shown). These data demonstrate that the AMPLINAT MPX assay is an automated, contained system with high specificity, sensitivity, and robustness.
The preseroconversion window period remains a source of viral infection in blood transfusion. Although the sensitivity of serological tests has improved in recent years, NATs provide a means to maximize the detection of window phase units prior to seroconversion. The AMPLINAT MPX system exhibited detection limits of 30 copies, 77 IU, and 42 copies per ml of HBV, HCV, and HIV-1, respectively. This sensitivity has been sufficient to detect virally infected, seronegative units when testing a pool containing 50 or 500 units. For HCV, the detection rate was similar for both pool sizes (approximately 1:290,000). It has been reported that the titer of HCV RNA can be extremely high early in infection; the concentration of HCV can reach 106 to 107 copies/ml within a brief interval, and it remains high until HCV antibody is detectable (M. P. Busch, B. D. Rawal, E. W. Feiburg, et al., Transfusion, abstr. 725, 1998, and S. L. Stramer, R. A. Porter, J. P. Brodsky, et al., Transfusion, abstr. 705, 1998). This high titer of virus makes HCV an attractive candidate for minipool NAT testing (S. Kleinman, personal communication) and is consistent with our observation that HCV positive units are detected at similar rates for both 50- and 500-member pools.
The current estimate for an HIV-1 window period is 22 days for a third-generation HIV antibody assay; this infectious window period is estimated to decrease to 16 days by HIV-1 p24 antigen testing (3, 4). The units in our clinical study were negative by both antibody and p24 antigen assays. The observation that AMPLINAT MPX detected HIV-1 in two seronegative specimens suggests that this test, as performed on the pooled samples, can further reduce the HIV-1 window period. It has been reported that early during the preseroconversion window period, prior to p24 antigen positivity, HIV-1 RNA levels in plasma range from 200 to 105 copies/ml, with the viral doubling time estimated to be 1 day (M. P. Busch, G. A. Satten, S. A. Herman, et al., Transfusion, abstr. 415, 1996). Therefore, pooled HIV-1 NAT is likely to be less effective than single-unit testing in reducing the window period.
Chronic HBV infection is often diagnosed with a persistent presence of HBsAg in the serum, which can be maintained at high levels even if virus replication in the liver has virtually ceased (20). During the HBV seroconversion window period, HBsAg is the earliest detectable serologic marker. However, it is known that in the early stage of HBV infection, the level of HBsAg is often undetectable by serological tests. In the JRC's evaluation of the AMPLINAT MPX system, several HBV-positive samples that were negative for HBsAg were identified by a sensitive licensed EIA. Five of these samples had relatively low levels of the virus; four of these samples contained pre-core mutants of the virus, and one sample was positive for a wild-type virus and was also positive for anti-HBs and anti-HBc. The high HBV sensitivity of the AMPLINAT MPX assay is further demonstrated in the JRC NAT trial where low HBV DNA viral loads (2.2 × 102 to 5.3 × 103 copies/ml) were observed in the pools that yielded positive DNA results by AMPLINAT MPX assay (11). The observation that positive samples were more frequent for a pool size of 50 than a pool size of 500 suggests that pooled HBV NAT is less sensitive than single-donation HBV NAT in determining the window period of infections.
In conclusion, we have developed an automated multiplex assay system, the AMPLINAT MPX assay, with high throughput, high sensitivity, and high specificity. The performance from both clinical and nonclinical studies demonstrates that the AMPLINAT MPX assay meets the workflow requirement for large-scale NAT screening in blood testing centers. Currently, no other system that allows for the simultaneous detection of HBV, HCV, and HIV-1 nucleic acids in the blood donation centers has been described. It is expected that such systems providing high-throughput, automated, multiplexed, qualitative tests will be increasingly relied upon in the evolution of NAT from mini-pool testing to single-unit screening across blood testing centers worldwide.
ACKNOWLEDGMENTS
We thank Jim Gallarda, Judy Weiss, and Maurice Rosenstraus for comments and help in writing the manuscript. We also thank Jim Araujo, Helen J. Lee, Dan Bristol, Hiroko Ohhashi, Tomomi Murata, Mitsunobu Nishigaya, and Katsushi Iwata for their excellent technical assistance.
We are very grateful to the Japanese Red Cross NAT screening research group (Kusuya Nishioka, Susumu Inoue, Shinichi Hirakawa, Hisao Yugi, Takeshi Murozuka, Yasushi Doi, Masaki Miyamoto, Hiroyuki Emura, Hideko Mine, Seiji Nakahira, Nobuko Suma, Yasufumi Mori, Hiroyuki Murokawa, Kiyoshi Minegishi, and other members) for their collaboration in the clinical performance evaluation.
REFERENCES
- 1.Abravaya K, Esping C, Hoenle R, Gorzowski J, Perry R, Kroeger P, Robinson J, Flanders R. Performance of a multiplex qualitative PCR LCx assay for detection of human immunodeficiency virus type 1 (HIV-1) group M subtypes, group O, and HIV-2. J Clin Microbiol. 2000;38:716–723. doi: 10.1128/jcm.38.2.716-723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruisten S, Van Gemen B, Koppelman M, Rasch M, Van Strijp D, Schukkink R, Beyer R, Weigel H, Lens P, Huisman H. Detection of HIV-1 distribution in different blood fractions by two nucleic acid amplification assays. AIDs Res Hum Retrovir. 1993;9:259–265. doi: 10.1089/aid.1993.9.259. [DOI] [PubMed] [Google Scholar]
- 3.Busch M P. HIV and blood transfusions: focus on seroconversion. Vox Sang. 1994;67(S3):13–18. doi: 10.1111/j.1423-0410.1994.tb04538.x. [DOI] [PubMed] [Google Scholar]
- 4.Busch M P, Lee L L L, Satten G A, et al. Time course of detection of viral and serological markers preceding HIV-1 seroconversion; implications for blood and tissue donor screening. Transfusion. 1995;35:91–97. doi: 10.1046/j.1537-2995.1995.35295125745.x. [DOI] [PubMed] [Google Scholar]
- 5.Defoort J P, Martin M, Casano B, Prato S, Camilla C, Fert V. Simultaneous detection of multiplex-amplified human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA using a flow cytometer microsphere-based hybridization assay. J Clin Microbiol. 2000;38:1066–1071. doi: 10.1128/jcm.38.3.1066-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. COBAS AMPLICORTM: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 7.Heid C A, Stevens J, Livak J K, William P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 8.Heredia A, Sorino V, Weiss S H, Bravo R, Vallejo A, Denny T N, Epstein J S, Hewlett I K. Development of a multiplex PCR assay for the simultaneous detection and discrimination of HIV-1, HIV-2, HTLV-I and HTLV-II. Clin Diagn Virol. 1996;7:85–92. doi: 10.1016/s0928-0197(96)00255-3. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi R, Dollinger G, Walsh P S, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 11.Iizuka H Y, Yamanaka R, Miyamoto M, Satoh S, Nakahira S, et al. Nationwide nucleic acid amplification testing of hepatitis B virus, hepatitis C virus and human immunodeficiency virus type 1 for blood transfusion and follow up study of nucleic acid amplification positive donors. Jpn J Infect Dis. 2000;53:116–123. [PubMed] [Google Scholar]
- 12.Kitchin P A, Szotyori A, Fromholc C, Almond N. Avoidance of false positive. Nature. 1990;344:201. doi: 10.1038/344201a0. [DOI] [PubMed] [Google Scholar]
- 13.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee S C, Antony A, Lee N, Leibow J, Yang J Q, Soviero S, Gutekunst K, Rosenstraus M. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J Clin Microbiol. 2000;38:4174–4179. doi: 10.1128/jcm.38.11.4171-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercier B, Burlot L, Ferec C. Simultaneous screening for HBV DNA and HCV RNA genomes in blood donations using a novel TaqMan PCR assay. J Virol Methods. 1999;77:1–9. doi: 10.1016/s0166-0934(98)00075-5. [DOI] [PubMed] [Google Scholar]
- 16.Miyachi H, Masukawa A, Ohshima T, Hirose T, Impraim C, Ando Y. Automated specific capture of hepatitis C virus RNA with probes and paramagnetic particle separation. J Clin Microbiol. 2000;38:18–21. doi: 10.1128/jcm.38.1.18-21.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullis K B. The polymerase chain reaction in an anemic mode: how to avoid oligodeoxyribonuclear fusion. PCR Methods Appl. 1991;1:1–4. doi: 10.1101/gr.1.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Otake K, Nishioka K. Nucleic acid amplification testing of hepatitis B virus. Lancet. 2000;355:1460. doi: 10.1016/S0140-6736(05)74655-9. [DOI] [PubMed] [Google Scholar]
- 20.Seeger C, Mason W S. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vet J A M, Majithia A R, Marras S A E, Tyagi S, Dube S, Poiesz B J, Kramer F R. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci USA. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]


