Abstract
Background and Objectives
Low buffy coat mitochondrial DNA copy number (mtDNA-CN) is associated with incident risk of stroke and poststroke mortality; however, its prognostic utility has not been extensively explored. Our goal was to investigate whether low buffy coat mtDNA-CN is a marker and causal determinant of poststroke outcomes using epidemiologic and genetic studies.
Methods
First, we performed association testing between baseline buffy coat mtDNA-CN measurements and 1-month poststroke outcomes in 3,498 cases of acute, first stroke from 25 countries from the international, multicenter case-control study Importance of Conventional and Emerging Risk Factors of Stroke in Different Regions and Ethnic Groups of the World (INTERSTROKE). Then, we performed 2-sample mendelian randomization analyses to evaluate potential causative effects of low mtDNA-CN on 3-month modified Rankin Scale (mRS) score. Genetic variants associated with mtDNA-CN levels were derived from the UK Biobank study (N = 383,476), and corresponding effects on 3-month mRS score were ascertained from the Genetics of Ischemic Stroke Functional Outcome (GISCOME; N = 6,021) study.
Results
A 1-SD lower mtDNA-CN at baseline was associated with stroke severity (baseline mRS score: odds ratio [OR] 1.27, 95% confidence interval [CI] 1.19–1.36; p = 4.7 × 10−12). Independently of baseline stroke severity, lower mtDNA-CN was associated with increased odds of greater 1-month disability (ordinal mRS score: OR 1.16, 95% CI 1.08–1.24; p = 4.4 × 10−5), poor functional outcome status (mRS score 3–6 vs 0–2: OR 1.21, 95% CI 1.08–1.34; p = 6.9 × 10−4), and mortality (OR 1.35, 95% CI 1.14–1.59; p = 3.9 × 10−4). Subgroup analyses demonstrated consistent effects across stroke type, sex, age, country income level, and education level. In addition, mtDNA-CN significantly improved reclassification of poor functional outcome status (net reclassification index [NRI] score 0.16, 95% CI 0.08–0.23; p = 3.6 × 10−5) and mortality (NRI score 0.31, 95% CI 0.19–0.43; p = 1.7 × 10−7) beyond known prognosticators. With the use of independent datasets, mendelian randomization revealed that a 1-SD decrease in genetically determined mtDNA-CN was associated with increased odds of greater 3-month disability quantified by ordinal mRS score (OR 2.35, 95% CI 1.13–4.90; p = 0.02) and poor functional outcome status (OR 2.68, 95% CI 1.05–6.86; p = 0.04).
Discussion
Buffy coat mtDNA-CN is a novel and robust marker of poststroke prognosis that may also be a causal determinant of poststroke outcomes.
Classification of Evidence
This study provides Class II evidence that low buffy coat mtDNA-CN (>1 SD) was associated with worse baseline severity and 1-month outcomes in patients with ischemic or hemorrhagic stroke.
Patients with stroke from low- and middle-income countries bear a disproportionate burden of poststroke complications.1-3 Therefore, identifying cost-effective and highly predictive biomarkers that mediate poststroke recovery will allow better risk stratification and novel targets for acute stroke treatment.4
Mitochondrial health has an important role in both stroke pathogenesis and recovery,5,6 and mitochondrial DNA copy number (mtDNA-CN) is an emerging biomarker that has recently garnered interest due to its inexpensiveness, accessibility, and association with chronic diseases, including stroke.7 mtDNA-CN reflects the ratio of mitochondrial to nuclear DNA copies and acts as a rough surrogate for the number of mitochondria.8,9 Rare genetic disorders characterized by severe loss of mtDNA-CN, formally referred to as mtDNA depletion syndromes, can cause migraine, leukoencephalopathy, and stroke-like episodes.10,11 Conversely, higher mtDNA-CN levels have been reported in patients with primary mitochondrial disorders and some cancers.12,13 In the broader population, perturbations of leukocyte mtDNA-CN have been proposed to reflect general mitochondrial dysfunction, oxidative stress, impaired oxidative phosphorylation, and inflammation,8,9 which are key pathophysiologic determinants of stroke injury and prognosis. Indeed, low leukocyte mtDNA-CN is associated with increased risks of secondary hospitalization and mortality in patients with atherosclerotic and chronic kidney disease.14-16 To the best of our knowledge, only 1 study has investigated the association between mtDNA-CN and poststroke outcomes, wherein a prospective cohort study of 1,484 Chinese patients with stroke reported an association between mtDNA-CN and mortality.17 While these findings suggest a potential role for mtDNA-CN as a risk factor for poststroke outcomes, several important questions remain to be addressed regarding (1) the robustness of associations across stroke type and other clinically relevant subgroups, (2) whether associations are independent of baseline stroke severity, (3) whether mtDNA-CN is associated with the degree of functional disability among stroke survivors, and (4) whether mtDNA-CN is a causal determinant of poststroke outcomes.
To answer these questions, we investigated the relationships between both measured and genetically predicted mtDNA-CN levels and poststroke outcomes using large-scale datasets. First, we evaluated the association between buffy coat mtDNA-CN levels measured within 1 week of stroke symptom onset and 1-month outcomes in 3,498 patients with stroke from the Importance of Conventional and Emerging Risk Factors of Stroke in Different Regions and Ethnic Groups of the World (INTERSTROKE) study.18 Second, whereas epidemiologic analyses may suggest that low mtDNA-CN is a marker of stroke injury (reverse causation), there is also a possibility that low mtDNA-CN captures an underlying susceptibility to brain ischemia, engendering worse stroke outcome (causation). Therefore, to assess whether lower mtDNA-CN levels may be a causal risk factor for poor outcomes at 3 months after stroke, we conducted 2-sample mendelian randomization (MR) analyses using genetic effects derived from the UK Biobank (N = 383,476)19 and the Genetics of Ischemic Stroke Functional Outcome (GISCOME; N = 6,165).20 Overall, to address the primary research question of defining the extent to which low buffy coat mtDNA-CN is associated with poststroke outcomes, we evaluated whether low mtDNA-CN represents a marker and causal driver of poor poststroke outcomes.
Methods
INTERSTROKE Study
INTERSTROKE is a large international case-control study encompassing 32 countries across Asia, North America, South America, Europe, Australia, and Africa.18 The study design has been described in detail previously.21 In brief, participants were enrolled between January 11, 2007, and August 8, 2015. Cases consisted of patients with acute first stroke (ischemic or hemorrhagic) presenting within 5 days of symptom onset and 72 hours of hospital admission. Strokes were defined according to the World Health Organization definition, and subtypes were confirmed by neuroimaging (CT or MRI). Demographic characteristics, medical history, and risk factor data were collected through standardized questionnaires and physical examination. For patients who could not communicate, a proxy respondent was used (spouse or first-degree relative living in the same household aware of the patient's medical history and current treatments). The modified Rankin Scale (mRS)22 score was used as a marker of stroke severity and was measured at baseline and the 1-month follow-up. The presence of hemorrhagic transformation after ischemic stroke was assessed through neuroimaging (either CT or MRI) and adjudicated locally by a site investigator. The present analyses were performed on a subset of 3,498 INTERSTROKE cases with quantitative PCR (qPCR) mtDNA-CN measurements.
mtDNA-CN Measurement and Quality Control
At each recruitment center, nonfasting peripheral blood samples were collected in EDTA tubes from patients with stroke within 1 week of symptom onset (and within 72 hours of hospital admission). Blood samples were shipped from all regions (except South Asia and China due to sample transport restrictions) to the Clinical Research Laboratory and Biobank (Hamilton, Ontario, Canada). DNA was extracted from the buffy coat layer of centrifuged samples with the Qiagen (Venlo, the Netherlands) QIAsymphony DNA Midi (96.7%), DNA Mini (2.7%), or DSP DNA Midi (0.6%) kit. mtDNA-CN was assayed by the Genetic and Molecular Epidemiology Laboratory (Hamilton, Ontario, Canada) using a plasmid-normalized qPCR method developed previously described.23 Samples were run in duplicate, and those with a high coefficient of variation (>5%) were removed. On visual inspection of the distribution of mtDNA-CN values, a single sample with an extreme outlying value was removed. Additional outliers beyond 3 SDs of the mean were winsorized to the 99.7th percentile. mtDNA-CN values were normalized for known confounders by taking the residuals from a linear regression model for mtDNA-CN (dependent variable) vs age, sex, ethnicity, and qPCR batch (independent variables). The resulting numerical representation of mtDNA-CN was standardized to a mean of 0 and SD of 1 for subsequent analyses.
Statistical Analysis
All statistical analyses were performed with the statistical programming language R (version 3.6.0; R Foundation for Statistical Computing, Vienna, Austria). Plots were generated with a combination of the ggplot2, viridis, dplyr, grid, and gridExtra R packages. In INTERSTROKE, association testing was conducted to assess the relationship between low mtDNA-CN at baseline (continuous variable or discretized into quartiles) and stroke markers at 2 time points: markers collected at the time of the stroke event (hereafter referred to as baseline severity markers) and markers collected 1 month after the stroke event. The primary marker of baseline stroke severity was ordinal mRS score. Secondary markers included level of consciousness and hemorrhagic transformation after ischemic stroke. The primary stroke outcome at the 1-month follow-up was ordinal mRS score. Secondary outcomes at the 1-month follow-up included other formulations of mRS score, specifically poor functional outcome status (dichotomized mRS score 3–6 vs 0–2) and mortality status. Ordinal regression was used for analysis between ordinal mRS score and consciousness (polr R package). The proportional odds assumption was evaluated with the Brant test (Brant R package). Logistic regression analysis was conducted for dichotomous variables, including hemorrhagic transformation at baseline and 1-month poststroke outcomes (poor functional outcome and mortality statuses). All regression models were adjusted for age, sex, region, education level (none or primary school vs high school, trade school, college, or university), 2018 World Bank country income stratum (high, upper-middle, and lower-middle, or low income), household income (adjusted for country), primary stroke type (ischemic vs hemorrhagic stroke) and ischemic stroke Oxfordshire Community Stroke Project classification, prestroke dependency (prestroke mRS score 3–5 vs 0–2), Charlson comorbidity index, and stroke risk factors (hypertension, diabetes, hypercholesterolemia, atrial fibrillation or flutter, current smoker status, and waist-to-hip ratio) as defined previously.18 In addition to these covariates, baseline stroke severity (baseline mRS score) was included in models for 1-month poststroke outcomes. For analysis of dichotomous outcomes, additional subgroup analyses were performed stratifying by primary stroke type, baseline stroke severity, sex, age, country income level, and education level. The net reclassification index (NRI) was used to assess model reclassification improvement of 1-month poststroke outcomes on addition of mtDNA-CN to a baseline model including the same set of covariates as mentioned (Hmisc R package). Statistical analyses were adjusted for multiple hypotheses testing of 6 outcomes (3 markers of baseline stroke severity and three 1-month mRS score formulations), corresponding to a Bonferroni-corrected p value threshold of 0.008 (p < 0.05/6 = 0.008).
Mendelian Randomization
MR is a statistical genetics framework that leverages the random assortment of genetic alleles (the Mendel second law of independent assortment) to perform causal inference between an exposure and an outcome.24-26 The use of randomized genetic alleles as instrumental variables for an exposure endows several advantages, including robustness to traditional confounding factors and reverse causation. Indeed, evidence from animal models suggests that stroke induces changes in mtDNA-CN levels; therefore, reverse causality is a relevant concern that is addressed by MR.27,28 To evaluate the potential causal relationship between low mtDNA-CN (exposure) and stroke prognosis (outcome), we performed 2-sample MR analyses incorporating summary-level genome-wide association study (GWAS) data from 2 independent studies. Genetic variants associated with mtDNA-CN levels were identified from a previous GWAS we conducted in 383,476 White participants from the UK Biobank study.29 UK Biobank is a prospective cohort study including UK residents (age 40–69 years) recruited from 2006 to 2010.30 Eligibility criteria included White participants with suitable genetic microarray data who had nonoutlying blood cell count and array intensity values.29 UK Biobank mtDNA-CN estimates were derived with AutoMitoC, a computational pipeline that leverages array-based data to estimate mtDNA-CN.29 Corresponding genetic effects on 3-month mRS score were obtained from the GISCOME GWAS. GISCOME included 6,021 White patients with ischemic stroke from 12 studies across Europe, the United States, and Australia.20 Two formulations of 3-month mRS scores were tested in the present study: ordinal mRS score and poor functional outcome status (mRS score 3–6 vs 0–2). In GISCOME, 2,280 (63%) participants had poor functional outcome. There is no sample overlap between the UK Biobank and GISCOME datasets.
As previously described,29 an independent set of 26 genome-wide significant variants associated with mtDNA-CN located nearby or within genes expressed in the mitochondria were selected as instruments to genetically approximate mtDNA-CN levels (eAppendix and eTables 1 and 2, links.lww.com/WNL/B699). Collectively, these variants had an F statistic of 100, which is sufficient (F > 10) for the purposes of identifying a causal effect.
Two-sample MR analyses were executed with the TwoSampleMR (version 0.5.5) and MR-PRESSO (version 1.0) R packages.25,31 Three MR methods were used, including the inverse variance–weighted, weighted median, and MR-Egger methods. MR-PRESSO was used to detect global heterogeneity with p values derived from 1,000 simulations. The Egger intercept test was used to assess directional pleiotropy. Causal effects were expressed as odds of a higher mRS score category (or of poor functional outcome) per 1-SD decrease in genetically determined mtDNA-CN. MR analyses tested a similar set of hypotheses as epidemiologic analyses and were therefore viewed as confirmatory. Accordingly, a nominal p value threshold of 0.05 was considered statistically significant. To assess the potential for bidirectional effects (i.e., susceptibility to worse stroke outcome influencing mtDNA-CN), reverse MR analyses were also performed with suggestive loci (p < 5 × 10−6) from GISCOME used as genetic instruments.
Beyond commonly used tests, we also performed additional sensitivity analyses to address sources of confounding specific to mtDNA-CN and the prognostic nature of analyses. First, phenotypic mtDNA-CN measurements may reflect differences in immune cell proportions,32,33 so we examined the relationship between genetically determined blood cell traits and 3-month stroke outcomes. Blood cell traits entailed neutrophil, lymphocyte, white blood cell, and platelet counts, as well as the neutrophil-to-lymphocyte ratio. Genetic variants associated with blood cell counts were ascertained from a large European GWAS by the Blood Cell Consortium X (2021) comprising more than a half-million individuals.34 Genetic variants associated with neutrophil-to-lymphocyte ratio were derived from a UK Biobank GWAS we conducted in 340,002 British participants (unpublished data; eAppendix, links.lww.com/WNL/B699).35 Causal effect estimates were expressed per 1-SD increase in genetically determined blood cell traits. Second, because mtDNA-CN instruments were derived from a generally healthy population, the transferability of genetic effects to patients with stroke is unknown. Accordingly, we consolidated the mtDNA-CN instruments derived from the UK Biobank to calculate weighted polygenic scores for INTERSTROKE cases with genotyping data to test the association between genetically predicted and qPCR-measured mtDNA-CN levels in patients with stroke (eAppendix). Third, a potential challenge of evaluating prognostic factors among disease patients is vulnerability to index event bias, a form of selection bias that occurs during the study of risk factors for subsequent events among disease cases that can induce dependence among originally independent risk factors and lead to spurious associations.36 Accordingly, we repeated MR analyses using stroke outcome genetic effects corrected for this bias using a previously described correction method (eAppendix).20,37,38
Standard Protocol Approvals, Registrations, and Patient Consents
Research was approved by the Hamilton Integrated Research Ethics Board under project 06–331. All INTERSTROKE participants (or their proxies) provided written informed consent. INTERSTROKE analyses were reported following Strengthening the Reporting of Genetic Association Studies guidelines, and MR analyses were reported according to Strengthening the Reporting of Observational Studies in Epidemiology–MR guidelines.
Data Availability
The primary datasets in this study include INTERSTROKE, UK Biobank, and GISCOME. Anonymized INTERSTROKE data may be made available by request from any qualified investigator on approval of collaboration with the study principal investigator (M.O.). UK Biobank individual-level data can be acquired on application.30 UK Biobank mtDNA-CN GWAS summary statistics will be posted on the GWAS catalog.39 GISCOME summary statistics are freely available to download from the Cerebrovascular Disease Knowledge Portal.40
Results
Baseline Characteristics of INTERSTROKE Cases
A subset of 3,498 patients with stroke consented to genetic analysis, had peripheral blood specimen collected within 1 week of symptom onset, and had DNA samples that were successfully assayed for buffy coat mtDNA-CN (Figure 1). The patients with stroke analyzed in this study spanned 25 countries and 98 enrollment sites across Western Europe (26.6%), Eastern/Central Europe (11.8%), South America (28.1%), Africa (13.3%), Southeast Asia (6.8%), Western Asia (6.8%), and North America and Australia (6.5%) (Table 1). The average age of patients with stroke was 64.6 years (SD 14.4 years), and 1,482 (42.4%) individuals were female. The sample comprised 677 (19.4%), 1,259 (36.0%), and 1,562 (44.6%) individuals from lower-middle/low–income, upper-middle–income, and high-income countries, respectively. Primary stroke types consisted of 592 (16.9%) hemorrhagic, 2,889 (82.6%) ischemic, and 17 (0.5%) undefined cases. Among the 2,889 patients with ischemic stroke, 54 (1.9%) had hemorrhagic transformation of their infarct. At baseline, 2010 (57.5%) participants were functionally dependent on others to perform basic activities of daily living (mRS score 3–5). The level of consciousness was reduced (drowsy or unconscious) in 1,129 (29.9%) patients.
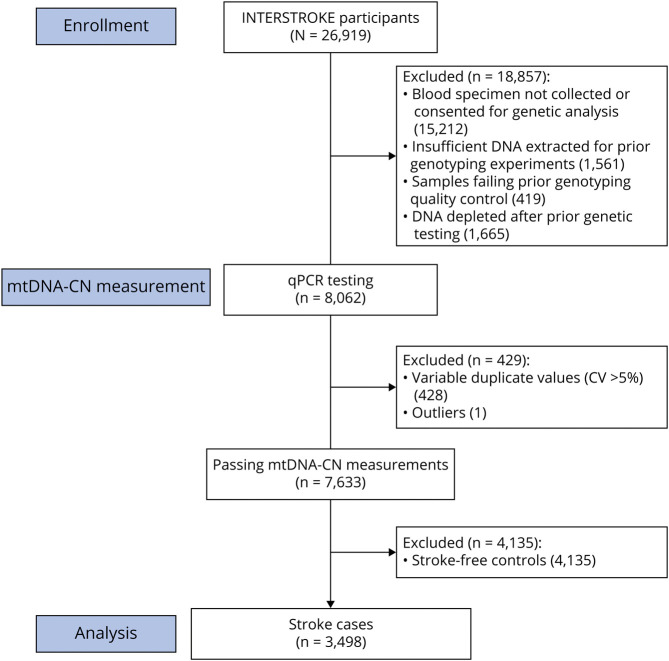
Figure 1. Participant Flowchart for the INTERSTROKE mtDNA-CN Substudy.
Of 26,919 research participants enrolled in the Importance of Conventional and Emerging Risk Factors of Stroke in Different Regions and Ethnic Groups of the World (INTERSTROKE) study, 11,707 (43%) consented to genetic analysis and had blood specimen collected at baseline. Previous DNA extraction yielded 10,146 (87%) samples with sufficient DNA for genotyping experiments (not included in the present investigation), of which 9,727 (96%) successfully genotyped samples passed quality control. Due to repeated genotyping and sequencing experiments performed in the same set of 9,727 extracted DNA samples, sufficient DNA remained for only 8,062 (83%) to be run on the mitochondrial DNA copy number (mtDNA-CN) quantitative PCR (qPCR) assay. Quality control of mtDNA-CN measurements led to 7,633 (95%) samples with suitable mtDNA-CN measurements. In the present analyses, we focus on a final subset of 3,498 participants with acute stroke.
Table 1.
Demographic Characteristics, Comorbid Conditions, and Stroke Characteristics for 3,498 INTERSTROKE Cases Included in This Study (N = 3,498)
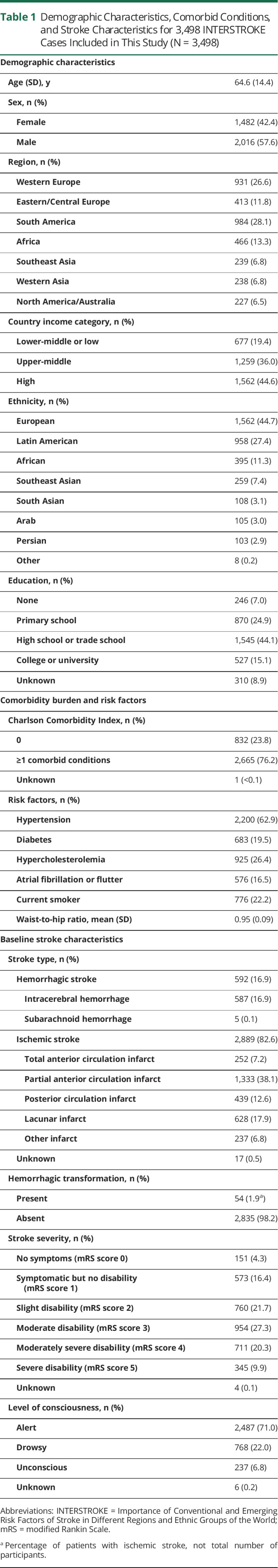
Notably, the characteristics of this INTERSTROKE subsample in this study differed from those of the whole sample because blood specimens from South Asia and China were precluded from genetic analyses (eTable 3, links.lww.com/WNL/B699). For example, cases included in mtDNA-CN analyses were more likely to live in a high-income country, to have at least a high school education, and o have certain risk factors (hypercholesterolemia and atrial fibrillation), whereas those not included in the present analyses were more likely to be current smokers and to have hemorrhagic stroke.
Lower mtDNA-CN Is Associated With Greater Stroke Severity at Baseline
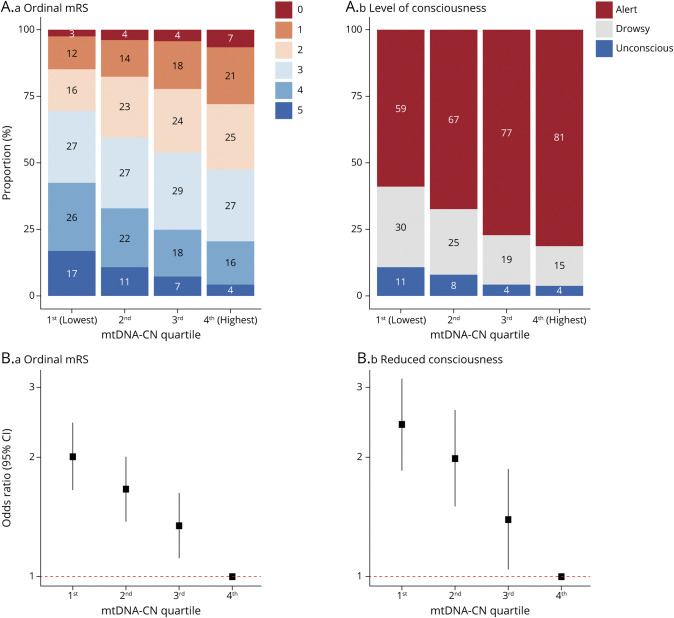
At baseline, a 1-SD lower mtDNA-CN was significantly associated with increased odds of having a more severe stroke (ordinal mRS score: odds ratio [OR] 1.27, 95% confidence interval [CI], 1.19–1.36; p = 4.7 × 10−12) and reduced consciousness (OR 1.34, 95% CI 1.21–1.48; p = 1.8 × 10−8) (eFigure 1, links.lww.com/WNL/B699). Among patients with ischemic stroke, the association with hemorrhagic transformation was nonsignificant (OR 1.33, 95% CI 0.92–1.93; p = 0.13). When patients with stroke were stratified by mtDNA-CN quartile, there was a stepwise increase in the proportion of individuals with higher stroke severity as mtDNA-CN decreased (eTable 4 and Figure 2A). Patients with stroke in the lowest mtDNA-CN quartile were at greatest risk of having a more severe stroke (OR 2.00, 95% CI 1.65–2.44; p = 2.9 × 10−12) and reduced consciousness (OR 2.42, 95% CI 1.84–3.17; p = 1.6 × 10−10) compared to those in the highest mtDNA-CN quartile (eTable 4 and Figure 2B). These associations were step-wise and graded, and there was no significant evidence suggesting that the proportional odds assumption had been violated in any ordinal analysis (Brant p > 0.05; eTable 4). Time from symptom onset to blood draw was not significantly associated with mtDNA-CN levels (p = 0.11).
Figure 2. Association Between mtDNA-CN and Baseline Stroke Severity.
(A) Stacked bar plots illustrate the proportion of each (a) ordinal modified Rankin Scale (mRS) score and (b) consciousness level category per mitochondrial DNA copy number (mtDNA-CN) quartile. (B) Forest plots illustrate the association between mtDNA-CN quartile and risk of having (a) more severe strokes as indicated by ordinal mRS score and (b) reduced consciousness. The highest (fourth) mtDNA-CN quartile was used as the reference group. CI = confidence interval.
Lower mtDNA-CN Is Associated With Poor Stroke Prognosis at 1 Month
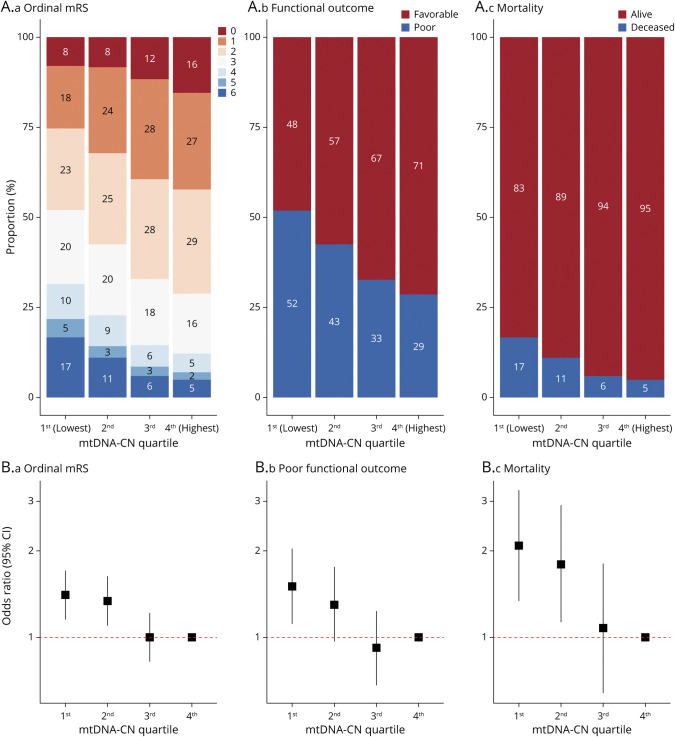
Of the 3,498 stroke patients, mRS score was recorded at follow-up for 3,470 (99.2%) individuals. At the 1-month follow-up, 1,354 (39.0%) patients had poor functional outcome (mRS score 3–6), including 337 (9.7%) patients who died. After adjustment for baseline stroke severity in addition to previous covariates, a 1-SD lower mtDNA-CN was significantly associated with higher 1-month mRS score (OR 1.16, 95% CI 1.08–1.24; p = 4.4 × 10−5), poor functional outcome (OR 1.21, 95% CI 1.08–1.34; p = 6.9 × 10−4), and mortality (OR 1.35, 95% CI 1.14–1.59; p = 3.9 × 10−4) (eFigure 2 and eTable 5, links.lww.com/WNL/B699). The magnitude of effect for mtDNA-CN on mortality risk was comparable to that of age, an established predictor of stroke outcomes (eFigure 3). Conversely, the effect of mtDNA-CN on poststroke disability (mRS score category and poor functional outcome status) was weaker than age (eFigure 3). There was no significant evidence suggesting that the proportional odds assumption had been violated in any ordinal analysis (Brant p > 0.05; eTable 5). Stratification by mtDNA-CN quartile revealed a consistent relationship between lower mtDNA-CN quartile and higher risk of adverse stroke outcomes (Figure 3). Patients with stroke in the lowest quartile had greater odds of being classified in a higher mRS score stratum (OR 1.40, 95% CI 1.15–1.71; p = 0.001) and having poor functional outcome (OR 1.51, 95% CI 1.11–2.04; p = 0.01) and mortality (OR 2.09, 95% CI 1.34–3.25; p = 0.001) compared to patients with stroke in the highest quartile (eTable 6 and Figure 3).
Figure 3. Association Between mtDNA-CN and 1-Month Poststroke Prognosis.
(A) Stacked bar plots illustrate the proportion of individuals belonging to (a) ordinal modified Rankin Scale (mRS) score, (b) functional outcome status, and (c) mortality categories per mitochondrial DNA copy number (mtDNA-CN) quartile. (B) Forest plots convey the association between mtDNA-CN quartile and poststroke outcomes with the fourth quartile as the reference for comparison. CI = confidence interval.
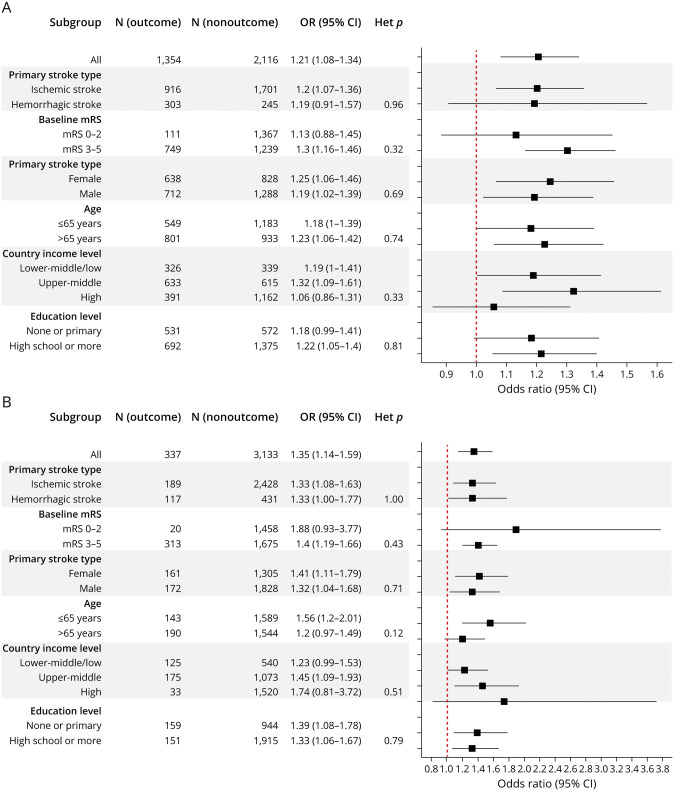
To further assess the robustness of mtDNA-CN-outcome associations, we performed subgroup analyses stratifying by primary stroke type, baseline stroke severity, sex, age, country income level, and education level. Directionally consistent associations were observed across all subgroups for both poor functional outcome and mortality status with no significant heterogeneity between subgroups detected (Cochran Q heterogeneity p > 0.10; Figure 4 and eTable 7, links.lww.com/WNL/B699).
Figure 4. Association Between mtDNA-CN and 1-Month Poststroke Prognosis Stratified by Subgroup.
Forest plots show the association between low mitochondrial DNA copy number (mtDNA-CN) (per 1-SD decrease) on (A) poor functional outcome (modified Rankin Scale [mRS] score 3–6) and (B) mortality status across various strata. Except for the subgroup variable used to stratify, regression models were adjusted for age, sex, region, education level, country income level, household income level, primary stroke type and Oxfordshire Community Stroke Project classification, Charlson comorbidity index, cardiovascular risk factors, prestroke disability, and baseline mRS score. CI = confidence interval; Het = heterogeneity; OR = odds ratio.
Last, we assessed whether incorporation of mtDNA-CN improved prediction of poststroke outcomes beyond known prognosticators, risk factors, and demographic characteristics. Addition of mtDNA-CN led to significant improvements in reclassification of functional outcome status (NRIoverall 0.16, 95% CI 0.08–0.23; p = 3.6 × 10−5) and mortality status (NRIoverall 0.31, 95% CI 0.19–0.43; p = 1.7 × 10−7). For both outcomes, NRI improvement was attributable to better reclassification of events (NRIPoor Outcome 0.20, 95% CI 0.15–0.26; p = 4.3 × 10−12; NRIDeath 0.33, 95% CI 0.22–0.44; p = 3.4 × 10−9) as opposed to nonevents (NRIFavorable Outcome −0.05; 95% CI −0.09 to −0.001; p = 0.045; NRIAlive −0.02; 95% CI −0.06 to 0.02; p = 0.30) (eTable 8, links.lww.com/WNL/B699).
Low mtDNA-CN Is a Putative Causal Risk Factor for 3-Month Stroke Outcomes
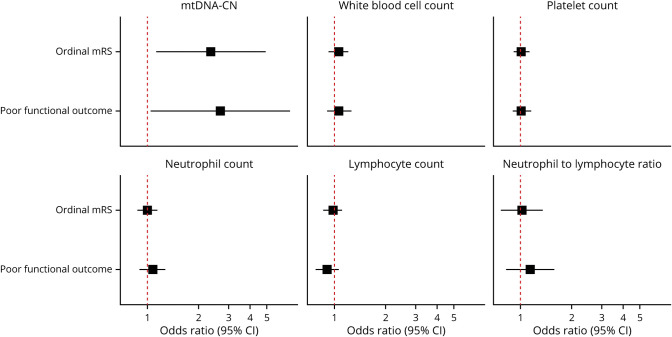
Using the UK Biobank and GISCOME studies (independently of INTERSTROKE), we found that genetically low mtDNA-CN was significantly associated with worse 3-month outcomes after stroke quantified by the ordinal mRS score (OR 2.35 per 1-SD decrease in genetically predicted mtDNA-CN, 95% CI 1.13–4.90; p = 0.02) and poor functional outcome (OR 2.68, 95% CI 1.05–6.86; p = 0.04) (Figure 5 and eTables 9–11, links.lww.com/WNL/B699). For all analyses, there was no significant evidence of directional pleiotropy (MR-Egger intercept p > 0.05) or global heterogeneity (Cochran Q and MR-PRESSO global test p > 0.05). Results were also directionally consistent when other MR methods (weighted median and MR-Egger) were used and when stroke outcome effects adjusted for index event bias were used (eTable 9). Because buffy coat mtDNA-CN is known to be correlated with immune cell counts, we also performed MR analyses for blood cell traits. Despite sufficient instrument strength for neutrophil (F = 100), platelet (F = 154), lymphocyte (F = 108), and total white blood cell (F = 106) counts and the neutrophil-to-lymphocyte ratio (F = 61), none were significantly associated with 3-month outcomes (Figure 5 and eTable 10). Reverse MR analyses did not suggest that susceptibility to worse stroke outcome affected mtDNA-CN levels (p > 0.10; eTable 11). Last, because genetic variants used to approximate genetically determined mtDNA-CN levels were originally derived from a healthy population, we verified that such genetic effects persist in INTERSTROKE stroke cases (β = 0.08-SD increase in mtDNA-CN per 1-SD increase in genetically predicted levels, 95% CI 0.02–0.05; p = 4.1 × 10−6; eTable 12).
Figure 5. MR Analyses Assessing the Effects of Low mtDNA-CN (and Blood Cell Traits) on 3-Month Post–Ischemic Stroke Prognosis.
Genetic predisposition to low mitochondrial DNA copy number (mtDNA-CN), but not blood cell counts, is associated with higher risk of 3-month outcomes after stroke. Effect estimates for mtDNA-CN are expressed per 1-SD decrease in genetically predicted mtDNA-CN, whereas those for blood cell traits were expressed per 1-SD increase in genetically predicted blood cell counts (or neutrophil-to-lymphocyte ratio). Causal effect estimates obtained by the inverse variance–weighted method are displayed because no significant heterogeneity or directional pleiotropy was detected for any analysis (eTables 9 and 10, links.lww.com/WNL/B699). CI = confidence interval; mRS = modified Rankin Scale.
This study provides Class II evidence that low buffy coat mtDNA-CN (>1 SD) was associated with worse baseline severity and 1-month outcomes in patients with ischemic or hemorrhagic stroke.
Discussion
Our study represents the first international multicenter exploration of buffy coat mtDNA-CN as a potential prognosticator of poststroke outcomes. First, lower buffy coat mtDNA-CN measured within 1 week of symptom onset correlated with functional and clinically relevant stroke severity indicators. Second, lower buffy coat mtDNA-CN was associated with greater risk of poor functional outcome and death at the 1-month follow-up consistently across primary stroke type, sex, age, country income, and education strata, as well as independent of baseline severity. Third, in addition to being a strong predictor of mortality with a magnitude of effect comparable to age, the inclusion of buffy coat mtDNA-CN improved the prediction of functional outcome and death. Fourth, MR analysis provided support for low buffy coat mtDNA-CN as a causal mediator of 3-month mRS score and poor functional outcome status. Together, our findings confirm the hypothesis that low buffy coat mtDNA-CN is a biomarker and mediator of worse stroke prognosis.
The main clinical implication of our study is that buffy coat mtDNA-CN may represent a useful prognostic marker of poststroke outcomes. First, buffy coat mtDNA-CN is a blood biomarker of poststroke outcomes that does not suffer from interrater variability and is not influenced by a patient's communication deficit. Second, the mtDNA-CN-outcome associations are consistent across stroke type, sex, age, country income level, education level, and baseline severity, which positions mtDNA-CN to have widespread utility for patients with stroke globally. We provide evidence suggesting that low mtDNA-CN may have consistent effects in both patients with ischemic stroke and those with hemorrhage stroke. This is particularly relevant for health systems in low-income settings, which bear a disproportionate global burden of hemorrhagic stroke,1-3 although further analyses in larger samples of patients with hemorrhagic stroke are warranted to confirm. Third, low mtDNA-CN represents a strong risk marker with effects comparable to established prognosticators, including older age. Moreover, the observed effect for mtDNA-CN on mortality is also comparable to that of carrying an APOE ε2 allele, which confers a 1.5-fold increased risk of 3-month mortality in patients with intracerebral hemorrhage and is present in ≈15% of the population.41 For comparison, we found that patients with stroke in the bottom 15% of mtDNA-CN levels had a 1.6-fold increased risk of 1-month mortality (OR 1.57, 95% CI 1.13–2.17; p = 0.007) relative to the remaining 85% of participants with higher levels. Fourth, mtDNA-CN is an easily accessible biomarker because it can be measured from peripheral blood after stroke, the assay necessitates only basic molecular laboratory techniques (qPCR), and the cost per sample is low (<US $5). Logistic and operational convenience, combined with evidence for robust, objective, and strong prognostic utility, raises the prospect of implementing mtDNA-CN clinically; however, replication of such findings in a prospective analysis and formal economic analyses is warranted.
Findings from MR analyses suggest that proper mtDNA regulation may be imperative for stroke protection and recovery, which aligns with animal model experiments demonstrating an important role for mtDNA-CN regulators in mediating protection against ischemia-reperfusion injury. For example, reoxygenation of rodents with acute kidney injury induces the formation of excessive mitochondrial reactive oxygen species, accompanied by a sharp decline in mtDNA-CN levels.28 In addition, genetic upregulation of the mtDNA replication initiation factor TFAM is sufficient to rescue this acute drop in mtDNA-CN, thereby attenuating ischemia-reperfusion injury. In the context of stroke models, mice with transient middle cerebral artery occlusion exhibit excessive cleavage of OPA1, another important mtDNA regulator, and treatment with either a cleavage-resistant form of OPA1 or mild overexpression of OPA1 markedly reduces infarct volume and neuronal apoptosis.27,42 In conjunction with prior mechanistic studies, our epidemiologic and genetic findings contribute to the mounting evidence that maintaining adequate mtDNA-CN may mediate cellular resilience to ischemic insults. Consistent epidemiologic associations in patients with hemorrhagic stroke suggest that mtDNA-CN may protect against stroke injury through general mechanisms pertinent to both etiologies such as the maintenance of blood-brain barrier integrity or anti-inflammatory effects.43 Indeed, circulating endothelial progenitor cells transfer their mitochondria to damaged endothelium, which restores the integrity of the blood-brain barrier, and this mitochondrial transfer may be mtDNA dependent, as has been shown for cancer cells.44,45 In addition, others have proposed that low blood mtDNA-CN may indicate a shift from anti-inflammatory to proinflammatory macrophage subtypes.9 Beyond potential mechanisms involving circulating leukocytes, it is plausible that genetic perturbation of blood mtDNA-CN reflects differences in mtDNA-CN levels in other cell and tissue types. Future experiments are required to decipher the underlying mechanisms and contributing cell types mediating this association, and additional MR analyses are necessary to assess potential causative effects in the posthemorrhage setting.
Our study had several limitations. First, because INTERSTROKE was an international multicenter study, measures of baseline severity (NIH Stroke Scale score) and outcome (3-month mRS score) that are common in smaller studies were substituted with b baseline mRS score and 1-month mRS score for feasibility, respectively, as was done previously.1 The interchangeability of such measures has been validated in previous studies showing high correlation between baseline NIH Stroke Scale and mRS scores (r = 0.69) and between 1-month and 3-month mRS scores (r = 0.87; weighted κ agreement 0.86).46,47 Granted, future studies are warranted to assess whether mtDNA-CN provides added utility to established measures of clinical and neuroanatomic severity. Second, complete blood cell counts were not measured in INTERSTROKE; thus, we cannot directly evaluate to what extent blood cell counts influence observational associations with poststroke outcomes. This is particularly important given that poststroke infection increases neutrophils that have low mtDNA-CN.48 However, most (>95%) blood samples were collected within 4 days of symptom onset, thereby mitigating confounding from infections occurring after this period. Moreover, our genetic analyses suggest that mtDNA-CN may have a direct role in stroke prognosis independently of changes in blood cell counts because mtDNA-CN GWAS effects had already been adjusted for major cell count determinants of mtDNA-CN levels (neutrophil, white blood cell, and platelet counts) and because no significant association was observed for genetically determined immune cell counts per se. Nonetheless, the genetic determinants of poststroke immune cell changes may differ from those influencing variation in cell counts within the general population, as shown previously.49 Third, although associations were corrected for a crude surrogate of infarct volume (Oxfordshire Community Stroke Project classification), direct measurements of infarct and hematoma volumes were not available. Fourth, survivorship bias may have led to conservative effect estimates because INTERSTROKE cases included patients surviving to hospital admission, and consequently, patients with severe, early fatal strokes were not represented. Last, MR analyses were limited by the following considerations: (1) causal effect estimates were imprecise albeit consistent in direction of effect with epidemiologic associations; (2) although sensitivity analyses did not show significant evidence of heterogeneity, directional pleiotropy, or outlying effects, it is impossible to completely exclude bias due to pleiotropy or index event bias; (3) mortality and hemorrhagic stroke outcomes could not be evaluated directly for lack of available GWAS summary statistics; and (4) analyses were based solely on Europeans, and while we found that genetically predicted mtDNA-CN was significantly associated with measured mtDNA-CN in an ethnically diverse sample of patients with stroke, future stroke outcome GWAS in non-Europeans are necessary to enable analyses addressing whether this causative relationship extends to other populations.
Low buffy coat mtDNA-CN measured within 1 week of symptom onset represents an accessible and robust biomarker of both stroke severity and prognosis. MR findings suggest that low mtDNA-CN may mediate poststroke outcomes. Additional investigations are warranted to replicate such findings in additional populations, to establish the temporal profile of poststroke mtDNA-CN changes in more detail, and to assess whether compounds that maintain mtDNA-CN levels after cerebral insult hold promise as a novel therapeutic strategy.
Acknowledgment
The authors acknowledge the investigators and participants of the INTERSTROKE, UK Biobank, and GISCOME studies for their important contributions to this research.
Glossary
- CI
confidence interval
- GISCOME
Genetics of Ischemic Stroke Functional Outcome
- GWAS
genome-wide association study
- INTERSTROKE
Importance of Conventional and Emerging Risk Factors of Stroke in Different Regions and Ethnic Groups of the World
- MR
mendelian randomization
- mRS
modified Rankin Scale
- mtDNA-CN
mitochondrial DNA copy number
- NRI
net reclassification index
- OR
odds ratio
- qPCR
quantitative PCR
Appendix. Authors

Footnotes
Class of Evidence: NPub.org/coe
Study Funding
Funding provided by the Canadian Institutes of Health Research (399497), Canadian Stroke Network, Heart and Stroke Foundation Canada (G-18-0022359).
Disclosure
M.R. Chong, S. Narula, R. Morton, C. Judge, L. Akhabir, N. Cawte, N. Pathan, R. Lali, P. Mohammadi-Shemirani, A. Shoamanesh, M. O'Donnell, S. Yusuf, and P. Langhorne report no disclosures relevant to the manuscript; G. Pare received support for genetic analyses for this work by the Canadian Institutes of Health Research (G-18-0022359) and Heart and Stroke Foundation of Canada (application 399497). Go to Neurology.org/N for full disclosures.
References
- 1.Langhorne P, O'Donnell MJ, Chin SL, et al. . Practice patterns and outcomes after stroke across countries at different economic levels (INTERSTROKE): an international observational study. Lancet. 2018;391(10134):2019-2027. [DOI] [PubMed] [Google Scholar]
- 2.Yusuf S, Rangarajan S, Teo K, et al. . Cardiovascular risk and events in 17 low-, middle-, and high-income countries. N Engl J Med. 2014;371(9):818-827. [DOI] [PubMed] [Google Scholar]
- 3.Dagenais GR, Leong DP, Rangarajan S, et al. . Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): a prospective cohort study. Lancet. 2020;395(10226):785-794. [DOI] [PubMed] [Google Scholar]
- 4.Montaner J, Ramiro L, Simats A, et al. . Multilevel omics for the discovery of biomarkers and therapeutic targets for stroke. Nat Rev Neurol. 2020;16(5):247-264. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, Lu J, Manaenko A, Tang J, Hu Q. Mitochondria in ischemic stroke: new insight and implications. Aging Dis. 2018;9(5):924-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Gao S. Mitochondrial dysfunction in ischemic stroke. In: Lapchak P, Yang GY, eds. Translational Research in Stroke. Springer; 2017:201-221. [Google Scholar]
- 7.Ashar FN, Zhang Y, Longchamps RJ, et al. . Association of mitochondrial DNA copy number with cardiovascular disease. JAMA Cardiol. 2017;2(11):1247-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13(5):481-492. [DOI] [PubMed] [Google Scholar]
- 9.Castellani CA, Longchamps RJ, Sun J, Guallar E, Arking DE. Mitochondrion thinking outside the nucleus: mitochondrial DNA copy number in health and disease. Mitochondrion. 2020;53:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paramasivam A, Venkatapathi C, Sandeep G, et al. . Homozygous R627W mutations in POLG cause mitochondrial DNA depletion leading to encephalopathy, seizures and stroke-like episodes. Mitochondrion. 2019;48:78-83. [DOI] [PubMed] [Google Scholar]
- 11.Bonora E, Chakrabarty S, Kellaris G, et al. . Biallelic variants in LIG3 cause a novel mitochondrial neurogastrointestinal encephalomyopathy. Brain. 2021;144(5):1451-1466. [DOI] [PubMed] [Google Scholar]
- 12.Brinckmann A, Weiss C, Wilbert F, et al. . Regionalized pathology correlates with augmentation of mtDNA copy numbers in a patient with myoclonic epilepsy with ragged-red fibers (MERRF-syndrome). PLoS One. 2010;5(10):e13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C, Bassig BA, Seow WJ, et al. . Mitochondrial DNA copy number and chronic lymphocytic leukemia/small lymphocytic lymphoma risk in two prospective studies. Cancer Epidemiol Biomarkers Prev. 2015;24(1):148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koller A, Fazzini F, Lamina C, et al. . Mitochondrial DNA copy number is associated with all-cause mortality and cardiovascular events in patients with peripheral arterial disease. J Intern Med. 2020;287(5):569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Guallar E, Ashar FN, et al. . Association between mitochondrial DNA copy number and sudden cardiac death: findings from the Atherosclerosis Risk in Communities study (ARIC). Eur Heart J. 2017;38(46):3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazzini F, Lamina C, Fendt L, et al. . Mitochondrial DNA copy number is associated with mortality and infections in a large cohort of patients with chronic kidney disease. Kidney Int. 2019;8(2):480-488. [DOI] [PubMed] [Google Scholar]
- 17.Song L, Liu T, Song Y, et al. . mtDNA copy number contributes to all-cause mortality of lacunar infarct in a Chinese prospective stroke population. J Cardiovasc Transl Res. 2020;13(5):783-789. [DOI] [PubMed] [Google Scholar]
- 18.O ’donnell MJ, Chin SL, Rangarajan S, et al. . Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet. 2016;388(10046):761-775. [DOI] [PubMed] [Google Scholar]
- 19.Bycroft C, Freeman C, Petkova D, et al. . The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soderholm M, Pedersen A, Lorentzen E, et al. . Genome-wide association meta-analysis of functional outcome after ischemic stroke. Neurology. 2019;92(12):e1271-e1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Donnell MJ, Xavier D, Liu L, et al. . Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet. 2010;376(9735):112-123. [DOI] [PubMed] [Google Scholar]
- 22.Uyttenboogaart M, Stewart RE, Vroomen PCAJ. Optimizing cutoff scores for the Barthel Index and the modified Rankin Scale for defining outcome in acute stroke trials. Stroke. 2005;36(9):1984-1987. [DOI] [PubMed] [Google Scholar]
- 23.Fazzini F, Schöpf B, Blatzer M, et al. . Plasmid-normalized quantification of relative mitochondrial DNA copy number. Sci Rep. 2018;8(1):15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith GD, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89-R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemani G, Zheng J, Elsworth B, et al. . The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandres-Ciga S, Noyce AJ, Traynor BJ. Mendelian randomization: a journey from obscurity to center stage with a few potholes along the way. JAMA Neurol. 2020;77(1):7-8. [DOI] [PubMed] [Google Scholar]
- 27.Lai Y, Lin P, Chen M, et al. . Restoration of L-OPA1 alleviates acute ischemic stroke injury in rats via inhibiting neuronal apoptosis and preserving mitochondrial function. Redox Biol. 2020;34:101503. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Zhao M, Wang Y, Li L, et al. . Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics. 2021;11(4):1845-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong M, Mohammadi-Shemirani P, Perrot N, et al. . GWAS and ExWAS of blood mitochondrial DNA copy number identifies 73 loci and highlights a potential causal role in dementia. medRxiv. Preprint posted online April 14, 2021. 10.1101/2021.04.08.21255031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudlow C, Gallacher J, Allen N, et al. . UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore AZ, Ding J, Tuke MA, et al. . Influence of cell distribution and diabetes status on the association between mitochondrial DNA copy number and aging phenotypes in the InCHIANTI study. Aging Cell. 2018;17(1):e12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurtado-Roca Y, Ledesma M, Gonzalez-Lazaro M, et al. . Adjusting mtDNA quantification in whole blood for peripheral blood platelet and leukocyte counts. PLoS One. 2016;11(10):e0163770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vuckovic D, Bao EL, Akbari P, et al. . The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182(5):1214-1231.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mbatchou J, Barnard L, Backman J, et al. . Computationally efficient whole genome regression for quantitative and binary traits. Nat Genet. 2021;53(7):1097-1103. [DOI] [PubMed] [Google Scholar]
- 36.Dahabreh I, Kent DM. Index event bias as an explanation for the paradoxes of recurrence risk research. JAMA. 2011;305(8):822-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dudbridge F, Allen RJ, Sheehan NA, et al. . Adjustment for index event bias in genome-wide association studies of subsequent events. Nat Commun. 2019;10(1):1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik R, Chauhan G, Traylor M, et al. . Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buniello A, Macarthur JAL, Cerezo M, et al. . The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005-D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawford KM, Gallego-Fabrega C, Kourkoulis C, et al. . Cerebrovascular disease knowledge portal: an open-access data resource to accelerate genomic discoveries in stroke. Stroke. 2018;49(2):470-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biffi A, Anderson CD, Jagiella JM, et al. . APOE genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: a genetic association study. Lancet Neurol. 2011;10(8):702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varanita T, Soriano ME, Sandri M, et al. . The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 2015;21(6):834-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W, Guo C, Feng H, Chen Y. Mitochondria: novel mechanisms and therapeutic targets for secondary brain injury after intracerebral hemorrhage. Front Aging Neurosci. 2021;12:615451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayakawa K, Chan SJ, Mandeville ET, et al. . Protective effects of endothelial progenitor cell-derived extracellular mitochondria in brain endothelium. Stem Cells. 2019;36(9):1404-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong LF, Kovarova J, Bajzikova M, et al. . Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. Elife. 2017;6:e22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruno A, Close B, Switzer JA, et al. . Simplified modified Rankin Scale questionnaire correlates with stroke severity. Clin Rehabil. 2013;27(8):724-727. [DOI] [PubMed] [Google Scholar]
- 47.Ovbiagele B, Lyden PD, Saver JL. Disability status at 1 month is a reliable proxy for final ischemic stroke outcome. Neurology. 2010;44364(8):688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Learoyd AE, Woodhouse L, Shaw L, et al. . Infections up to 76 days after stroke increase disability and death. Transl Stroke Res. 2017;8(6):541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres-Aguila NP, Carrera C, Giese AK, et al. . Genome-wide association study of white blood cell counts in patients with ischemic stroke. Stroke. 2019;50(12):3618-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The primary datasets in this study include INTERSTROKE, UK Biobank, and GISCOME. Anonymized INTERSTROKE data may be made available by request from any qualified investigator on approval of collaboration with the study principal investigator (M.O.). UK Biobank individual-level data can be acquired on application.30 UK Biobank mtDNA-CN GWAS summary statistics will be posted on the GWAS catalog.39 GISCOME summary statistics are freely available to download from the Cerebrovascular Disease Knowledge Portal.40